Abstract
Quorum sensing, also known as cell-density sensing in the unicellular eukaryote Dictyostelium discoideum, is required for efficient entry into the differentiation and development segment of its life cycle. Quorum sensing is accomplished by simultaneously secreting and sensing the glycoprotein Conditioned Medium Factor, or CMF. When the density of starving cells is high, CMF levels are high, which leads to aggregation followed by development. Here, we describe the role of pldB, a gene coding for a putative phospholipase D (PLD) homologue, in quorum sensing. We find that in submerged culture, adding butanol, an inhibitor of PLD-catalyzed phosphatidic acid production, allows cells to bypass the requirement for CMF mediated quorum sensing and aggregate at low cell density. Deletion of pldB mimics the presence of butanol, allowing cells to aggregate at low cell density. pldB− cells also initiate and finish aggregation rapidly. Analysis of early developmental gene expression in pldB− cells reveals that the cyclic AMP receptor cAR1 is expressed at higher levels earlier than in wild-type cells, which could explain the rapid aggregation phenotype. As would be predicted, cells overexpressing pldB are unable to aggregate even at high cell density. Adding CMF to these pldB− overexpressing cells does not rescue aggregation. Both of these phenotypes are cell autonomous, as mixing a small number of pldB− cells with wild-type cells does not cause the wild-type cells to behave like pldB− cells.
Development in multicellular organisms requires quorum-sensing mechanisms that allow monitoring of the density of different cell types. In general, these mechanisms use secreted signaling molecules produced by specific cell types. These secreted molecules turn on signaling cascades which change the developmental program of the cell. Examples of quorum sensing are present in bacteria (14), Dictyostelium discoideum (6), and mammals (46). The simple Dictyostelium developmental cycle and its amenity to biochemistry, cell biology, and molecular genetics provides an excellent system in which to study eukaryotic quorum sensing. Dictyostelium cells normally exist as individual amoebae that feed upon bacteria. However, when the cells deplete their food source and begin to starve, they stop cell division and aggregate into a multicellular organism of 2 × 104 to 105 cells by using pulses of cyclic AMP (cAMP) as a chemoattractant to recruit more starving cells. The aggregated cells then differentiate and develop into a fruiting body, consisting of a mass of spores situated upon a column of stalk cells (24, 28). Once food becomes available, the spores germinate into the next generation of amoebae. Without a quorum-sensing mechanism to measure the density of starving cells, small cohorts of cells that happen to starve at the same time would form small, ineffectual fruiting bodies.
When starved in the presence of a high concentration of other starving cells, a Dictyostelium cell responds in a typical manner upon receiving a pulse of cAMP; it releases a burst of cAMP to relay the signal to other starving cells, moves toward the source of cAMP, and expresses specific classes of genes required for the cell to undergo development (7-9, 11, 16, 28, 34, 36, 41). cAR1, a cell surface cAMP receptor, senses the cAMP (39). Binding of cAMP to cAR1 causes a transient influx of Ca2+ (27) and activates an associated heterotrimeric G protein. The Gβγ subunit, with the assistance of a cytosolic protein called cytosol regulator of adenylyl cyclase (CRAC), transiently activates adenylyl cyclase, while the Gα2 subunit activates guanylyl cyclase (20, 25, 40, 42).
The activation of this G protein signaling event and subsequent aggregation and development depends upon an 80-kDa secreted glycoprotein called conditioned medium factor (CMF) (15, 29). CMF is synthesized in actively dividing cells but not secreted until starvation. Since only starving cells can secrete and sense CMF, it is an ideal molecule for quorum sensing during development (22). When only a few cells are starving, CMF levels are low and preclude aggregation. Once a large number of cells are starving, the high levels of CMF released cause the initiation of aggregation. As can be expected, cells lacking CMF are unable to aggregate unless exogenous or recombinant CMF is added (42). Thus, CMF may coordinate the development of appropriate sized fruiting bodies by allowing aggregation only when most of the cells in a given area are starving, as determined by high levels of CMF.
CMF exerts control over development by regulating cAMP signaling through cAR1 (45). When at high cell density, and thus in the presence of high levels of CMF, cAMP binds to cAR1, activating its associated G protein. Gα2 binds GTP and releases Gβγ. These subunits then activate guanylyl and adenylyl cyclase, respectively. In the absence of CMF, cAMP can still bind to cAR1, and the activation of cAR1 can still cause Gα2 to release GDP and bind GTP. However, activation of adenylyl and guanylyl cyclase is greatly inhibited. This inhibition of adenylyl and guanylyl cyclase can be removed by a 10-s exposure of CMF, showing that lack of CMF is the direct cause of inhibition. The presence or absence of CMF has no effect on the levels of cAR1- and cAMP-induced binding of GTP to membranes, arguing that the lack of CMF has no effect on the interaction between cAR1 and its G protein in vitro. Thus, CMF controls aggregation by regulating cAMP signaling at a point after G protein activation but before the activations of adenylyl and guanylyl cyclases. CMF accomplishes this by controlling the GTPase rate of Gα2. We found that in lysates, approximately 250 molecules of GTP bind to a cell's membrane in response to a pulse of cAMP, regardless of whether CMF is present or absent (4, 45). After cAMP stimulation, GTP is hydrolyzed to GDP at a rate of approximately 240 molecules in 3 min in the absence of CMF. However, in the presence of CMF, the rate of GTP hydrolysis is drastically reduced to approximately 51 molecules in 3 min. Since lysates from cells lacking Gα2 have no cAMP-stimulated GTP binding or hydrolysis (4), these results argue that quorum sensing through CMF is accomplished by controlling the cAMP-stimulated GTPase activity of Gα2.
We have previously shown that CMF exerts its effect on cAR1 signaling by activating its own G protein signaling pathway, as opposed to its G protein-independent pathway (5). We demonstrated that CMF uses Gα1 and Gβ to control the activity of phospholipase C, which in turn regulates the cAMP-stimulated GTPase activity of Gα2. In many organisms, the GTPase activity of Gα proteins is regulated by RGS (regulator of G protein signaling) proteins. In turn, RGS proteins can be controlled by the levels of phosphatidic acid, a by-product of phospholipase D (30). This leads to the interesting possibility that phospholipase D may be involved in quorum sensing. In this report, we describe the role of a phospholipase D (PLD), PldB, in Dictyostelium development and present evidence that PldB is a negative regulator of quorum sensing.
MATERIALS AND METHODS
Cell culture.
Ax2 wild-type cells were grown in shaking culture in HL5 medium as described by Gomer et al. (15). pldB− cells were grown in submerged culture in the above medium containing 10 μg of blasticidin/ml or on bacterial plates. For development, cells at mid-log phase (2 × 106 cells/ml) were washed in buffer and plated on filter pads following the procedure of Jain et al. (22). Conditioned medium was prepared by starving Ax2 cells at 5 × 106 cells/ml in PBM (20 mM KH2PO4, 10 μM CaCl2, 1 mM MgCl2 [pH 6.1] with KOH) in shaking culture for 20 h and then clarifying the conditioned medium as described by Gomer et al. (15). To examine the effect of conditioned medium on cell aggregation, cells were starved in submerged monolayer culture as described by Gomer et al. (15) at surface densities of 224 × 103, 112 × 103, 56 × 103, 28 × 103, 14 × 103, and 7 × 103 cells/cm2 in PBM, PBM supplemented with 0.1% butanol, or PBM supplemented with 10% conditioned medium.
Generation of pldB deletion mutant and overexpression mutant.
To generate a deletion mutant of pldB, we replaced 719 nucleotides in the middle of the pldB gene with the blasticidin resistance marker under control of the actin 15 promoter. A partial cDNA clone of pldB obtained from the Dictyostelium genome project was digested with NdeI to remove part of the pldB coding region, and the ends were blunted. The plasmid carrying the blasticidin cassette (pBsr519) (35) was digested with BamHI to isolate the cassette. The ends of the blasticidin cassette were blunted; it was then ligated into the modified cDNA clone of pldB. The resulting plasmid contained 0.5 kbp of pldB, the blasticidin cassette replacing 719 nucleotides of pldB sequence, and 0.6 kbp of pldB sequence. For transformation, 10 μg of the plasmid was linearized with PvuI and gel purified with Geneclean (Bio 101, Vista, Calif.). The fragment was then used to transform Ax2 cells by following the procedure of Shaulsky et al. (37). Multiple clones with the same phenotypes were isolated, and the ones showing the strongest phenotypes were used for subsequent experiments. To generate an overexpression mutant, the pldB gene, isolated by PCR from genomic DNA, was placed under the control of the constitutively active actin 15 promoter by using the Gateway System (Invitrogen, Carlsbad, Calif.). The plasmid was used to transform Ax2 and pldB− cells by following the procedure of Shaulsky et al. (37).
Antibody production and Western blots.
The synthetic peptide GCFLIVYKKKKHDEDKPS from amino acids 30 to 47 of PldB was used to immunize a rabbit at Biosynthesis, Inc. (Lewisville, Tex.). Serum was collected 8 weeks after the second injection and purified by using the above peptide attached to agarose beads. Cells (106) were collected from different stages of development and boiled for 3 min in sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer in a final volume of 500 μl. A 20-μl aliquot was loaded onto a 10% polyacrylamide gel electrophoresis gel, and Western blots were done according to the method of Jain and Gomer (21).
Northern blotting and reverse transcription-PCR (RT-PCR).
RNA was isolated from cells starved on filter pads at the times indicated with the Rneasy mini kit (QIAGEN, Inc., Valencia, Calif.) or Trizol reagent (Invitrogen). RNA was then run on an agarose gel, visualized by ethidium bromide staining, and transferred to a Hybond-N+ membrane (Amersham, Piscataway, N.J.). Northern blot analysis was performed following the procedure of Bishop et al. (3) with the following partial gene fragments as DNA probes: pldB (250 bp), cAR1 (710 bp), CRAC (616 bp), ACA (705 bp), and IG7 (368 bp). Blots with the same probe were incubated together in the same hybridization buffer and exposed to film for the same length of time in the same film cassette. Densitometry was performed by using a Molecular Dynamics personal densitometer SI.
RT-PCR was performed with the GeneAmp RNA PCR kit (Applied Biosystems, Foster City, Calif.) as per the manufacturer's directions. The upstream primer for pldB was 5′-CACCATTAGCTTCCCATTGTC-3′, and the downstream primer was 5′-GGTGATCTTGCATTAGCATCCTC-3′. This set of primers provides a 170-bp product. RT-PCR was also performed on the IG7 gene as a control for the total amount of RNA used. The upstream primer was 5′-TTACATTTATTAGACCCGAAACCAAGCG-3′, and the downstream primer was 5′TTCCCTTTAGACCTATGGACCTTAGCG-3′. This set of primers yields a 370-bp product. The PCR products were separated on a 2% agarose gel and visualized with ethidium bromide. Duplicate reactions were performed without reverse transcriptase to control for DNA contamination in the RNA preparation.
RESULTS
Dictyostelium has at least two phosphatidylcholine-specific PLD genes.
To identify genes that encode PLD proteins in D. discoideum, we performed a BLAST search against the database of the Japanese Dictyostelium cDNA project by using the human phosphatidylcholine-specific PLD1 amino acid sequence as a query (30). Two different partial cDNAs were identified. We then performed a BLAST search against the database of the Dictyostelium Genome project by using the partial cDNAs as queries (13). Contigs containing these sequences were identified, yielding large putative open reading frames for both genes. These genes were named pldA and pldB. pldA was expressed consistently in both vegetative and developing cells (data not shown), while pldB was differentially regulated. pldB was chosen for further study due to its expression pattern.
The pldB gene resides on chromosome 3, is 2,747 nucleotides long with one intron, and codes for a hypothetical protein of 867 amino acids. The translated protein contains the conserved regions found in all PLD1's including a pleckstrin homology (PH) domain, catalytic region I (CRI), CRII, a loop, CRIII, CRIV, and the C-terminal tail (12, 26). Overall, the protein shares 32% similarity and 21% identity with PLD1 from humans. Excluding the N terminus and loop domains, which vary widely from species to species, PldB has 51% similarity and 34% identity to PLD1 from humans. A comparison of catalytic regions of PldB with other PLDs is shown in Fig. 1.
FIG. 1.
Comparison of the predicted amino acid sequences of the catalytic regions of PldB (D. disc), human Pld1 (H. sap1), human Pld2 (H. sap2), and Drosophila melanogaster Pld (D. mela). Amino acid identities are in black boldface type, while amino acid similarities are in gray.
Expression pattern of pldB.
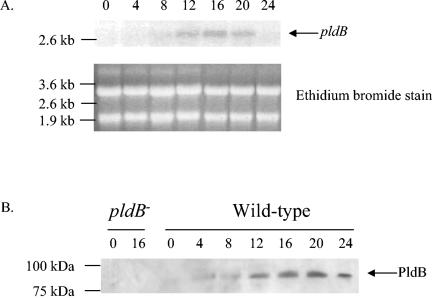
The developmental expression of pldB was examined by probing a Northern blot of total RNA from developing cells with a DNA probe generated from a fragment of pldB. The 2.8-kb pldB mRNA exhibited little or no expression in vegetative cells (Fig. 2A). Expression was apparent by 8 h and peaked by 16 h. To examine production of the PldB protein, we raised antibodies against a peptide of PldB (see Materials and Methods). These antibodies were then affinity purified with the same peptide and used to probe Western blots. To ensure that the antibodies were specific for PldB, they were used on a Western blot of lysates from both wild-type Ax2 cells and pldB− cells. A band at approximately 100 kDa appears in the wild-type sample but is absent from the pldB− sample (Fig. 2B), suggesting that the antibodies specifically recognize PldB. To examine developmental production of PldB, we stained a Western blot of cell lysates from developing cells. As seen with the Northern blot, PldB is not produced in vegetative cells. PldB protein could be detected by 4 h and peaked at 20 h. This pattern roughly paralleled that of the pldB mRNA expression. Thus, while the PldB transcript and protein appear to be absent in vegetative cells, it is present during aggregation and throughout development.
FIG. 2.
The expression of pldB is developmentally regulated. (A) A Northern blot of total cellular RNA isolated from Ax2 cells at 4-h intervals after starvation on filter pads was probed with a radiolabeled fragment of the pldB gene. (B) A Western blot of lysates from pldB− cells and Ax2 cells taken at 4-h intervals after starvation on filter pads was probed with affinity-purified anti-PldB peptide antibodies.
PLD activity regulates quorum sensing.
It has previously been observed that extracellular CMF is required for wild-type cells to aggregate when starved at low cell density (21). To examine whether PLD activity plays a role in quorum sensing, we measured the ability of cells to aggregate at different densities after treating them with butanol, which inhibits PLD function by blocking its ability to produce phosphatidic acid (38). In agreement with previously published data (5), wild-type cells starved in the absence of conditioned medium are only able to form aggregates down to 28 × 103 cells/cm2 (Table 1). Adding conditioned media allows aggregation at 7 × 103 cells/cm2. Butanol (0.1%) in the absence of conditioned medium also allows aggregation to occur as low as 7 × 103 cells/cm2, thus mimicking conditioned medium. To ensure that this effect is due specifically to butanol and not the presence of alcohol, the same experiment was performed with 0.1% t-butanol, an isomer of butanol, which has no effect on PLD activity (34); these cells behaved exactly like the untreated cells, aggregating only down to 28 × 103 cells/cm2. Therefore, when plated at low cell density, cells treated with butanol and presumably lacking PLD activity respond to starvation as if they are in the presence of CMF. This suggests that PLD activity may activate quorum sensing and may allow cells to aggregate at much lower cell densities than normal.
TABLE 1.
Effect of butanol on low-cell-density aggregationa
| Cell type and treatment | Presence of aggregates at cell density (103 cells/cm2) of:
|
|||||
|---|---|---|---|---|---|---|
| 224 | 112 | 56 | 28 | 14 | 7 | |
| Ax2 | + | + | + | ± | − | − |
| Ax2 + CMF | + | + | + | + | + | ± |
| Ax2 + butanol | + | + | + | + | + | + |
| Ax2 + t-butanol | + | + | + | ± | − | − |
Cells were starved at various cell densities in submerged monolayer culture in the absence or presence of CMF, butanol, or t-butanol (as a negative control). The field of cells was then examined with an inverted microscope at 18 h. The presence of aggregates is represented by a plus sign, while the absence of aggregates is represented by a minus sign. Densities allowing only partial or substandard aggregation are shown as ±. These are the representative results of five separate assays.
pldB is a negative regulator of quorum sensing.
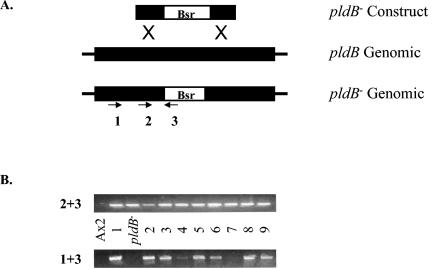
Since butanol allowed cells to aggregate at low cell density, we reasoned that disrupting the gene for pldB would also allow cells to aggregate at low cell density. We therefore inserted a blasticidin resistance cassette in the pldB locus by homologous recombination (Fig. 3A). Genomic integration and disruption of the pldB gene was confirmed by PCR (Fig. 3B). Strains exhibiting proper integration of the disruption construct into the pldB gene were examined for their ability to aggregate at low cell density (Table 2). pldB− cells, in the absence of conditioned medium, were able to aggregate at 7 × 103 cells/cm2, as seen with the addition of butanol to wild-type cells. Thus, disruption of phosphatidic acid production by butanol and deletion of pldB both allow aggregation at low cell density. The addition of exogenous CMF had no apparent effect on aggregation, suggesting that pldB is required for cells to properly respond to CMF. Therefore, when starved, cells lacking pldB behave as though they are at high cell density and are nonresponsive to conditioned medium. This suggests that pldB normally blocks quorum sensing through the CMF pathway and that loss of pldB mimics the presence of CMF.
FIG. 3.
Construction of the pldB deletion mutant. (A) After homologous pairing between the linearized targeting vector, pldB− construct, and the genomic pldB locus, homologous recombination in the regions labeled X replaces the endogenous pldB sequence with an exogenous sequence containing the blasticidin resistance cassette (Bsr), resulting in a deletion of the pldB gene. The cell lines produced by this event are resistant to blasticidin. Black box, gene; white box, blasticidin resistance cassette. (B) PCR was performed on DNA prepared from wild-type Ax2 cells, the gene disruption construct (pldB−), and 9 blasticidin-resistant clones, and the resulting products were separated on an agarose gel. Primer pair 2-3 tests for the presence of the disruption construct in the genome, while primer pair 1-3 tests for proper insertion of the disruption construct in the pldB gene. Strains 1, 2, 3, 5, 6, 8, and 9 show proper integration of the disruption construct.
TABLE 2.
Ability of mixed populations of wild-type and pldB− cells to aggregate at low cell densitya
| Cell type(s) (ratio) | Presence of aggregates at cell density (103 cells/cm2) of:
|
|||||
|---|---|---|---|---|---|---|
| 224 | 112 | 56 | 28 | 14 | 7 | |
| Ax2 | + | + | + | ± | − | − |
| pldB− | + | + | + | + | + | ± |
| Ax2:pldB− (9:1) | + | + | + | ± | − | − |
| Ax2:pldB− (1:1) | + | + | + | + | ± | − |
| Ax2:pldB− (1:9) | + | + | + | + | + | ± |
Wild-type Ax2 and pldB− cells were mixed in different ratios and were starved at various cell densities in submerged monolayer culture. The field of cells was then examined with an inverted microscope at 18 h. The presence of aggregates is represented by a plus sign, while the absence of aggregates is represented by a minus sign. Densities allowing only partial or substandard aggregation are shown as ±. These are the representative results of three separate assays.
It is possible that pldB− cells improperly secrete a diffusible factor, such as CMF, that allows them to aggregate at low cell density. To examine this possibility, we mixed wild-type cells and pldB− cells together at ratios of 1:9, 1:1, and 9:1 and examined their ability to aggregate at low cell density (Table 2). We found that the addition of 10% mutant cells to wild-type cells did not induce aggregation at low cell density. Conversely, the addition of 10% wild-type cells to mutant cells did not prevent aggregation at low cell density. Therefore, the ability of pldB− cells to aggregate at low cell density is not due to a change in cell secretion and is thus cell autonomous.
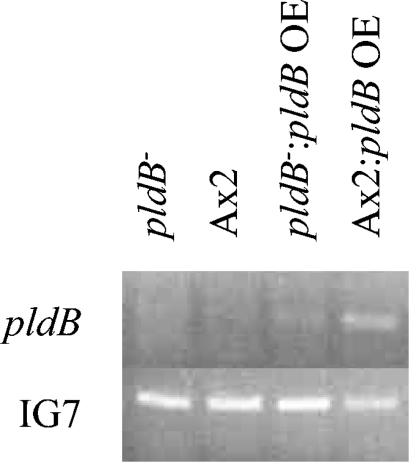
Since pldB gene activity normally blocks quorum sensing, we reasoned that cells overexpressing pldB should be unable to aggregate at high cell density. To test this, we created strains that overexpress pldB by introducing a plasmid-borne copy of the pldB gene constitutively expressed from the actin 15 promoter into both wild-type and pldB− cells. RT-PCR was performed on RNA isolated from vegetative cells to confirm overexpression of pldB (Fig. 4). As expected, very little if any product is seen in the Ax2 wild-type or pldB− cells (pldB is not expressed in vegetative cells). However, both Ax2 wild-type and pldB− cells containing the overexpression plasmid show the presence of pldB mRNA. The same RNA samples were used to perform RT-PCR to monitor the expression of IG7 (a transcript used as an internal control) and demonstrate that the differences seen in expression of pldB were not due to differences in starting RNA amounts. In addition, Northern blots of vegetative Ax2 wild-type and pldB− cells containing the overexpression plasmid identified a 2.8-kb band, corresponding to the full-length pldB transcript (data not shown). Thus, we were able to confirm overexpression of pldB. The effect of overexpressing pldB is shown in Table 3. In both wild-type and pldB− cells overexpressing pldB, aggregation was inhibited even at 224 × 103 cells/cm2, the highest cell density examined. Addition of 0.1% butanol is able to partially reverse this inhibition and allow aggregation at 112 × 103 cells/cm2, suggesting that the inability to aggregate may be ameliorated by inhibition of PLD activity. In addition, exogenous CMF was unable to induce aggregation at a lower cell density, corroborating our previous results suggesting that pldB is involved in the cellular response to CMF. If pldB is a negative regulator of aggregation, then enhancing its activity would preclude aggregation at higher and higher cell densities; our data show that this is the case. Likewise, removing its activity would allow cells to aggregate at any density, including extremely low densities, which our data also show to occur. Taken together, these data suggest that pldB is a negative regulator of quorum sensing and lies in the CMF pathway.
FIG. 4.
Overexpression of pldB in wild-type and pldB− cells. RNA from vegetative wild-type and pldB− cells, with or without the pldB overexpression plasmid, were collected, and RT-PCR with pldB-specific primers was performed. Simultaneous reactions were performed with primers for the IG7 gene to ensure that equal amounts of total RNA were used in each of the samples.
TABLE 3.
Effect of pldB expression on low-cell-density aggregationa
| Cell type and treatment | Presence of aggregates at cell density (103 cells/cm2) of:
|
|||||
|---|---|---|---|---|---|---|
| 224 | 112 | 56 | 28 | 14 | 7 | |
| Ax2 | + | + | + | ± | − | − |
| Ax2 + CMF | + | + | + | + | + | ± |
| pldB− | + | + | + | + | + | + |
| pldB OE | − | − | − | − | − | − |
| pldB− OE | − | − | − | − | − | − |
| pldB OE + CMF | − | − | − | − | − | − |
| pldB OE + butanol | + | + | − | − | − | − |
Wild-type Ax2 and pldB− cells containing or lacking a pldB overexpressing plasmid (OE) were starved at various cell densities in submerged monolayer culture in the absence or presence of CMF. The field of cells was then examined with an inverted microscope at 18 h. The presence of aggregates is represented by a plus sign, while the absence of aggregates is represented by a minus sign. Densities allowing only partial or substandard aggregation are shown as ±. These are the representative results of four separate assays.
pldB− cells develop rapidly.
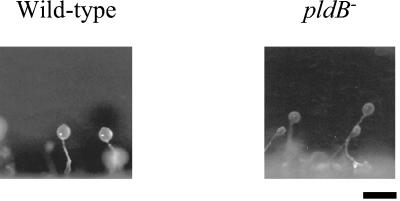
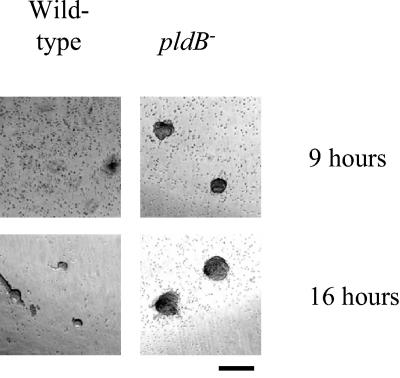
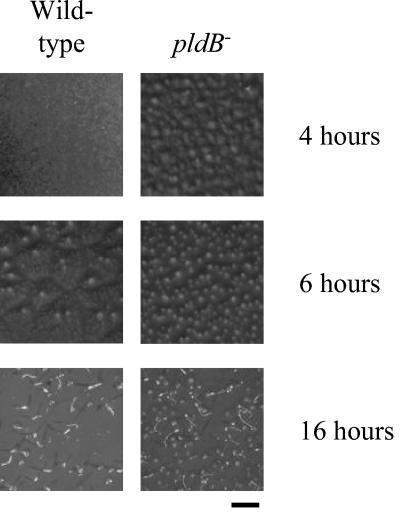
To determine whether pldB is required for proper development of the fruiting body, pldB− cells were starved on agar and allowed to develop. The resulting fruiting bodies were relatively normal, with the possibility that pldB− fruiting bodies may have slightly longer stalks than wild-type fruiting bodies (Fig. 5). To determine whether pldB is involved in the timing of aggregation, we examined the time it takes for pldB− cells to aggregate in submerged culture. Normally in this assay, wild-type cell aggregates appear at 16 h. pldB− cells form aggregates by 9 h, demonstrating that they aggregate more rapidly than wild-type cells (Fig. 6). We next examined whether this is a phenomenon associated with submerged cultures by determining whether pldB−cells can develop as rapidly on filter pads. On filter pads, pldB− cells show an accelerated rate of development compared to wild-type cells. pldB− cells form aggregates by 4 h, while wild-type cells show no signs of development (Fig. 7). By 6 h, pldB− cells form tight aggregates, whereas wild-type cells only start streaming, the initial stage of aggregation. pldB− cells develop consistently more rapidly, forming fruiting bodies by 16 h, 6 h ahead of the wild-type cells. Thus, loss of pldB expression allows an earlier initiation of the developmental cycle, leading to early aggregation and culminating in early fruiting body formation.
FIG. 5.
pldB− cells form fruiting bodies with slightly elongated stalks. Wild-type Ax2 and pldB− cells were plated with bacteria on agar. As the bacteria were consumed, development was triggered, and after 24 h, fruiting bodies formed. A side view of these fruiting bodies is shown. Bar, 1 mm.
FIG. 6.
pldB− cells aggregate rapidly in submerged culture. Cells were starved at 224 × 103 cells/cm2 in PBM, and photos were taken 9 and 16 h later. Bar, 1 mm.
FIG. 7.
pldB− cells develop rapidly on filter pads. Cells were deposited on filters and allowed to develop. Photos were taken at the indicated times. Bar, 3 mm.
pldB− cells have altered cAR1 expression.
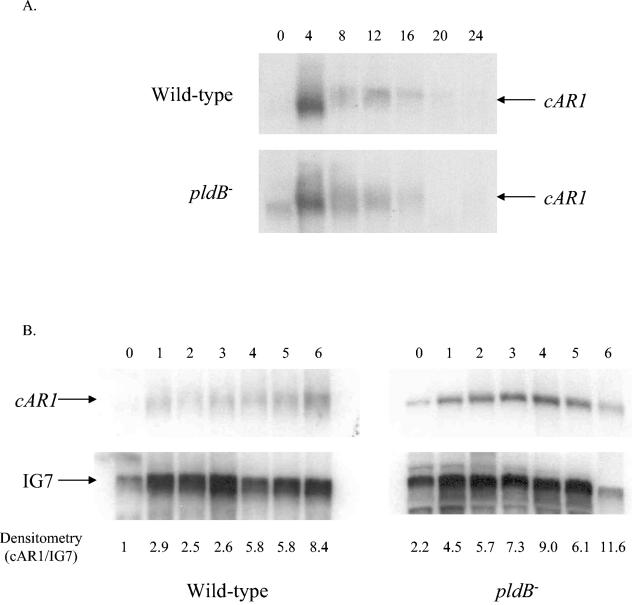
To determine whether the accelerated development seen in pldB− cells is due to earlier expression of genes required for development, we did a series of Northern blots to examine the expression of three genes required early in development, cAR1 (cAMP receptor on cell surface), CRAC, and ACA (adenylyl cyclase A), in wild-type and pldB− cells. For CRAC and ACA mRNA expression patterns, we found no apparent difference between wild-type cells and pldB− cells (data not shown). However, we found that cAR1 is more highly expressed in pldB− cells under vegetative conditions than in wild-type cells (Fig. 8A).
FIG. 8.
car1 is expressed strongly and early in pldB− cells. Northern blots of total cellular RNA isolated from wild-type Ax2 cells and pldB− cells at 4-h intervals (A) or 1-h intervals (B) after starvation were probed with a radiolabeled fragment of the car1 gene. The same blots were stripped and reprobed with a radiolabeled fragment of the IG7 gene to control for RNA loading. The densitometry measurements are a ratio of cAR1 to IG7, normalized to the wild-type Ax2 vegetative sample.
To take a closer look at the expression pattern of cAR1 at the earlier stages of development, we isolated total RNA from vegetative cells and cells developed on filter pads at 1, 2, 3, 4, 5, and 6 h. We confirmed that cAR1 mRNA is higher in pldB− cells under vegetative conditions than in wild-type cells (Fig. 8B). In addition, cAR1 expression in pldB− cells remains consistently higher than in wild-type cells throughout the first 6 h of development. Thus, cAR1 is expressed earlier and at higher levels in pldB− cells.
DISCUSSION
We previously described how the extracellular signaling protein CMF regulates the initiation of aggregation by controlling the GTPase activity of Gα2, the G protein associated with the cAMP receptor cAR1 (5). This regulation is mediated by a signal transduction pathway involving Gα1, Gβ, and phospholipase C. Here we describe the identification of a gene with sequence similarity to phospholipase D, PldB, and present evidence that it is also involved in the quorum-sensing pathway.
Cells lacking pldB are able to aggregate at low cell density, thus bypassing the requirement for CMF. Cells overexpressing pldB are unable to aggregate at high cell density. In fact, while adding CMF to wild-type cells allows aggregation at low cell density, adding CMF to pldB-overexpressing cells has no effect, demonstrating that they are insensitive to CMF. Taken together, these two observations suggest that pldB is a negative regulator of quorum sensing in the CMF responsive pathway. Thus, we hypothesize that in wild-type cells, the presence of CMF triggers a decrease in PldB activity, which then allows cells to aggregate.
In addition to allowing cells to aggregate in suboptimal concentrations of CMF, loss of pldB also accelerates the developmental program. In both submerged monolayer conditions and on filter pads, pldB− cells aggregate well before wild-type cells. Interestingly, this developmental phenotype is not seen in other mutants, such as Gα1− cells, or drug treatments, such as addition of protein kinase C activators, which cause aggregation at low cell density (5; unpublished data). This argues that pldB may also be playing a role in aggregation or development that is outside of the previously described CMF pathway.
To address the rapid development phenotype observed in pldB− cells, we examined the expression patterns of the early development genes cAR1, CRAC, and ACA to uncover the cause of the accelerated development phenotype of pldB− cells. These genes perform indispensable roles in the cAMP signaling pathway responsible for aggregation. While there is no apparent difference in expression of CRAC and ACA between wild-type and pldB− cells, cAR1 is expressed earlier and at higher levels in pldB− cells than in wild-type cells. pldB− cells may respond to extracellular cAMP earlier and thus initiate development earlier. These data indicate that altered cAR1 expression is likely one of the factors that cause the rapid development of pldB− cells, suggesting that PldB plays a role in the timing of development.
Sequence analysis suggests that PldB belongs to the phospholipase D family of enzymes. Specifically, it contains all of the PLD conserved regions including a PH domain, CRI to CRIV, a loop, and a C-terminal domain, which suggests that PldB belongs to the PLD1 subfamily (12, 26). One of the more interesting aspects of the structure of PldB is the PH domain. PH domains are best known as phospholipid binding domains, but they have also been shown to be involved in binding to the βγ-subunits of heterotrimeric G proteins and protein kinase C (27, 44). In Dictyostelium cells, the PH domains of CRAC and protein kinase B are believed to be responsible for translocating these proteins to the plasma membrane during chemotaxis (17, 18, 32). It has been suggested that the selective localization of such PH-containing proteins to the leading edge of the cell is what allows a directional response to a chemoattractant (10, 33). Specifically, it is proposed that the localized activities of phosphatidylinositol 3-kinase and PTEN cause the accumulation of phosphatidylinositol-(3,4,5)-triphosphate at the leading edge, forming binding sites for PH-domain-containing proteins (17, 19). It is therefore feasible that the PH domain of PldB also allows it to be localized to the leading edge of a chemotaxing cell. This would in turn cause PldB activity, in the form of phosphatidic acid (PA) production, to be concentrated at the leading edge. It has been shown that PA is a regulator of RGS activity. Specifically, mammalian RGS4 is inhibited by phosphatidic acid (PA) (31). RGS proteins act as GTPase-activating proteins for heterotrimeric G proteins. We showed previously that CMF controls the GTPase activity of Gα2 (4). The effect of CMF on GTPase activity is indirect and involves a G protein-coupled signal transduction pathway (5). An RGS protein is most likely directly responsible for controlling GTPase. This RGS protein could be regulated by PA produced by PldB. Therefore, PldB, through localization provided by its PH domain, could be involved in the localized regulation of Gα2, the G protein mediating cAMP chemotaxis. This is especially intriguing given that G proteins in chemotaxing cells are evenly distributed along the membrane (23). Localized control of G protein activity could help to influence the polarization of a chemotaxing cell. In such a scenario, CMF would decrease PldB association with the membrane, leading to decreased PA production. This would consequently decrease RGS activity, allowing increased signaling through Gα2.
In mammalian cells, PLD1 has been implicated in a number of cellular processes including Golgi function (2), clathrin coat assembly in endosomes and lysosomes (1), vesicle-dependent secretion (43), and the production of phosphatidic acid as a second messenger. It is reasonable to believe that PldB may also function in these processes in Dictyostelium. Interestingly, pldB disruptants exhibit phenotypes that suggest the disruption of one of these cellular pathways. Given the genetic pliability of D. discoideum, we are now in a position to uncover many other roles of PLD that are not easily identifiable in mammalian systems.
Acknowledgments
We thank Peter Thomason for the Gateway overexpression vector.
This work was supported by NIGMS grant S06-GM606564 from the National Institutes of Health and grant 0346975 from the National Science Foundation. The infrastructure and instrumentation of the Biological Sciences Department at Hunter are supported by Research Centers in Minority Institutions Award RR-03037 from the National Center for Research Resources of the National Institutes of Health.
REFERENCES
- 1.Arneson, L. S., J. Kunz, R. A. Anderson, and L. M. Traub. 1999. Coupled inositide phosphorylation and phospholipase D activation initiates clathrin-coat assembly on lysosomes. J. Biol. Chem. 274:17794-17805. [DOI] [PubMed] [Google Scholar]
- 2.Bi, K., M. G. Roth, and N. T. Ktistakis. 1997. Phosphatidic acid formation by phospholipase D is required for transport from the endoplasmic reticulum to the Golgi complex. Curr. Biol. 7:301-307. [DOI] [PubMed] [Google Scholar]
- 3.Bishop, J. D., B. C. Moon, F. Harrow, D. Ratner, R. H. Gomer, J. P. Dottin, and D. T. Brazill. 2002. A second UDP-glucose pyrophosphorylase is required for differentiation and development in Dictyostelium discoideum. J. Biol. Chem. 277:32430-32437. [DOI] [PubMed] [Google Scholar]
- 4.Brazill, D. T., R. Gundersen, and R. H. Gomer. 1997. A cell-density sensing factor regulates the lifetime of a chemoattractant-induced Galpha-GTP conformation. FEBS Lett. 404:100-104. [DOI] [PubMed] [Google Scholar]
- 5.Brazill, D. T., D. F. Lindsey, J. D. Bishop, and R. H. Gomer. 1998. Cell density sensing mediated by a G protein-coupled receptor activating phospholipase C. J. Biol. Chem. 273:8161-8168. [DOI] [PubMed] [Google Scholar]
- 6.Clarke, M., and R. H. Gomer. 1995. PSF and CMF, autocrine factors that regulate gene expression during growth and early development of Dictyostelium. Experientia 51:1124-1134. [DOI] [PubMed] [Google Scholar]
- 7.Devreotes, P. 1989. Dictyostelium discoideum: a model system for cell-cell interactions in development. Science 245:1054-1058. [DOI] [PubMed] [Google Scholar]
- 8.Dottin, R. P., S. R. Bodduluri, J. F. Doody, and B. Haribabu. 1991. Signal transduction and gene expression in Dictyostelium discoideum. Dev. Genet. 12:2-5. [DOI] [PubMed] [Google Scholar]
- 9.Firtel, R. A. 1995. Integration of signaling information in controlling cell-fate decisions in Dictyostelium. Genes Dev. 9:1427-1444. [DOI] [PubMed] [Google Scholar]
- 10.Firtel, R. A., and C. Y. Chung. 2000. The molecular genetics of chemotaxis: sensing and responding to chemoattractant gradients. Bioessays 22:603-615. [DOI] [PubMed] [Google Scholar]
- 11.Firtel, R. A., P. J. M. van Haastert, A. R. Kimmel, and P. N. Devreotes. 1989. G protein linked signal transduction pathways in development: Dictyostelium as an experimental system. Cell 58:235-239. [DOI] [PubMed] [Google Scholar]
- 12.Frohman, M., T. Sung, and A. Morris. 1999. Mammalian phospholipase D structure and regulation. Biochim. Biophys. Acta 1439:175-186. [DOI] [PubMed] [Google Scholar]
- 13.Glockner, G., L. Eichinger, K. Szafranski, J. A. Pachebat, A. T. Bankier, P. H. Dear, D. Lehmann, C. Baumgart, G. Parra, J. F. Abril, R. Guigo, K. Kumpf, B. Tunggal, E. Cox, M. A. Quail, M. Platzer, A. Rosenthal, and A. A. Noegel. 2002. Sequence and analysis of chromosome 2 of Dictyostelium discoideum. Nature 418:79-85. [DOI] [PubMed] [Google Scholar]
- 14.Gomer, R. H. 1994. Knowing that you're among friends. Curr. Biol. 4:734-735. [DOI] [PubMed] [Google Scholar]
- 15.Gomer, R. H., I. S. Yuen, and R. A. Firtel. 1991. A secreted 80x10(3)Mr protein mediates sensing of cell density and the onset of development in Dictyostelium. Development 112:269-278. [DOI] [PubMed] [Google Scholar]
- 16.Gross, J. D. 1994. Developmental decisions in Dictyostelium discoideum. Microbiol. Rev. 58:330-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, Y. E., M. Iijima, C. A. Parent, S. Funamoto, R. A. Firtel, and P. N. Devreotes. 2003. Receptor mediated regulation of PI3Ks confines PI(3,4,5) to the leading edge of chemotaxing cells. Mol. Biol. Cell 14:1313-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iijima, M., and P. Devreotes. 2002. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell 109:599-610. [DOI] [PubMed] [Google Scholar]
- 19.Iijima, M., Y. E. Huang, and P. Devreotes. 2002. Temporal and spatial regulation of chemotaxis. Dev. Cell 3:469-478. [DOI] [PubMed] [Google Scholar]
- 20.Insall, R., A. Kuspa, P. J. Lilly, G. Shaulsky, L. R. Levin, W. F. Loomis, and P. Devreotes. 1994. CRAC, a cytosolic protein containing a pleckstrin homology domain, is required for receptor and G protein-mediated activation of adenylyl cyclase in Dictyostelium. J. Cell Biol. 126:1537-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain, R., and R. H. Gomer. 1994. A developmentally regulated cell surface receptor for a density-sensing factor in Dictyostelium. J. Biol. Chem. 269:9128-9136. [PubMed] [Google Scholar]
- 22.Jain, R., I. S. Yuen, C. R. Taphouse, and R. H. Gomer. 1992. A density-sensing factor controls development in Dictyostelium. Genes Dev. 6:390-400. [DOI] [PubMed] [Google Scholar]
- 23.Jin, T., N. Zhang, Y. Long, C. A. Parent, and P. N. Devreotes. 2000. Localization of the G protein betagamma complex in living cells during chemotaxis. Science 287:1034-1036. [DOI] [PubMed] [Google Scholar]
- 24.Kessin, R. H. 2001. Dictyostelium—evolution, cell biology, and the development of multicellularity. Cambridge University Press, Cambridge, United Kingdom.
- 25.Lilly, P. J., and P. N. Devreotes. 1994. Identification of CRAC, a cytosolic regulator required for guanine nucleotide stimulation of adenylyl cyclase in Dictyostelium. J. Biol. Chem. 269:14123-14129. [PubMed] [Google Scholar]
- 26.Lisovitch, M., M. Czarny, G. Fiucci, and X. Tang. 2000. Phospholipase D: molecular and cell biology of a novel gene family. Biochem. J. 345:401-415. [PMC free article] [PubMed] [Google Scholar]
- 27.Lodowski, D. T., J. A. Pitcher, W. D. Capel, R. J. Lefkowitz, and J. J. Tesmer. 2003. Keeping G proteins at bay: A complex between G protein-coupled receptor kinase 2 and Gbg. Science 300:1256-1262. [DOI] [PubMed] [Google Scholar]
- 28.Loomis, W. F. 1975. Dictyostelium discoideum. A developmental system. Academic Press, New York, N.Y.
- 29.Mehdy, M. C., and R. A. Firtel. 1985. A secreted factor and cyclic AMP jointly regulate cell-type-specific gene expression in Dictyostelium discoideum. Mol. Cell. Biol. 5:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morio, T., H. Urushihara, T. Saito, Y. Ugawa, H. Mizuno, M. Yoshida, R. Yoshino, B. N. Mitra, M. Pi, T. Sato, K. Takemoto, H. Yasukawa, J. Williams, M. Maeda, I. Takeuchi, H. Ochiai, and Y. Tanaka. 1998. The Dictyostelium developmental cDNA project: generation and analysis of expressed sequence tags from the first-finger stage of development. DNA Res. 5:335-340. [DOI] [PubMed] [Google Scholar]
- 31.Ouyang, Y. S., Y. Tu, S. A. Barker, and F. Yang. 2003. Regulators of G-protein signaling (RGS) 4, insertion into model membranes and inhibition of activity by phosphatidic acid. J. Biol. Chem. 278:11115-11122. [DOI] [PubMed] [Google Scholar]
- 32.Parent, C. A., B. J. Blacklock, W. M. Froehlich, D. B. Murphy, and P. N. Devreotes. 1998. G protein signaling events are activated at the leading edge of chemotactic cells. Cell 95:81-91. [DOI] [PubMed] [Google Scholar]
- 33.Parent, C. A., and P. N. Devreotes. 1999. A cell's sense of direction. Science 284:765-770. [DOI] [PubMed] [Google Scholar]
- 34.Peters, D. J. M., M. Cammans, S. Smit, W. Spek, M. M. van Lookeren Campagne, and P. Schaap. 1991. Control of cAMP-induced gene expression by divergent signal transduction pathways. Dev. Genet. 12:25-34. [DOI] [PubMed] [Google Scholar]
- 35.Puta, F., and C. Zeng. 1998. Blasticidin resistance cassette in symmetrical polylinkers for insertional inactivation of genes in Dictyostelium. Folia Biol. (Prague) 44:185-188. [PubMed] [Google Scholar]
- 36.Schaap, P. 1991. Intercellular interactions during Dictyostelium development, p. 147-178. In M. Dworkin (ed.), Microbial cell-cell interactions. American Society for Microbiology, Washington, D.C.
- 37.Shaulsky, G., R. Escalante, and W. F. Loomis. 1996. Developmental signal transduction pathways uncovered by genetic suppressors. Proc. Natl. Acad. Sci. USA 93:15260-15265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen, Y., L. Xu, and D. A. Foster. 2001. Role for phospholipase D in receptor-mediated endocytosis. Mol. Cell. Biol. 21:595-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Theibert, A., and P. Devreotes. 1986. Surface receptor-mediated activation of adenylate cyclase in Dictyostelium. Regulation by guanine nucleotides in wild-type cells and aggregation deficient mutants. J. Biol. Chem. 261:15121-15125. [PubMed] [Google Scholar]
- 40.van Haastert, P. J. M., M. J. de Vries, L. C. Penning, F. Roovers, J. van der Kaay, C. Erneux, and M. M. van Lookeren Campagne. 1989. Chemoattractant and guanosine 5′-[g-thio]triphosphate induce the accumulation of inositol 1,4,5,-trisphophate in Dictyostelium cells that are labelled with [3H]-inositol by electroporation. Biochem. J. 258:577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Haastert, P. J. M., P. M. W. Janssens, and C. Erneux. 1991. Sensory transduction in eukaryotes—a comparison between Dictyostelium and vertebrate cells. Eur. J. Biochem. 195:289-303. [DOI] [PubMed] [Google Scholar]
- 42.van Haastert, P. J. M., B. E. Snaar-Jagalska, and P. M. W. Janssens. 1987. The regulation of adenylate cyclase by guanine nucleotides in Dictyostelium discoideum membranes. Eur. J. Biochem. 162:251-258. [DOI] [PubMed] [Google Scholar]
- 43.Way, G., N. O'Luanaigh, and S. Cockcroft. 2000. Activation of exocytosis by cross-linking of the IgE receptor is dependent on ADP-ribosylation factor 1-regulated phospholipase D in RBL-2H3 mast cells: evidence that the mechanism of activation is via regulation of phosphatidylinositol 4,5-bisphosphate synthesis. Biochem. J. 346:63-70. [PMC free article] [PubMed] [Google Scholar]
- 44.Yang, X. L., Y. L. Zhang, Z. S. Lai, F. Y. Xing, and Y. H. Liu. 2003. Pleckstrin homology domain of G protein coupled receptor kinase-2 binds to PKC and affects activity of PKC kinase. World J. Gastroenterol. 9:800-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuen, I. S., R. Jain, J. D. Bishop, D. F. Lindsey, W. J. Deery, P. J. M. van Haastert, and R. H. Gomer. 1995. A density-sensing factor regulates signal transduction in Dictyostelium. J. Cell Biol. 129:1251-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, Y. P., Ricardo. 1994. Positional cloning of the mouse obese gene and its human homologue. Nature 372:425-432. [DOI] [PubMed] [Google Scholar]