Abstract
The phosphorylation of transcription factors by mitogen-activated protein kinases (MAP) is a pivotal event in the cellular response to the activation of MAP kinase signal transduction pathways. Mitogenic and stress stimuli activate different pathways and lead to the activation of distinct groups of target proteins. Elk-1 is targeted by three distinct MAP kinase pathways. In this study, we demonstrate that the MAP kinase ERK2 is targeted to Elk-1 by a domain which is distinct from, and located N-terminally to, its phosphoacceptor motifs. Targeting via this domain is essential for the efficient and rapid phosphorylation of Elk-1 in vitro and full and rapid activation in vivo. Specific residues involved in ERK targeting have been identified. Our data indicate that the targeting of different classes of MAP kinases to their nuclear substrates may be a common mechanism to increase the specificity and efficiency of this signal transduction pathway.
Extracellular signals are transduced to the nucleus by a series of pathways. Several transcription factors are subsequently phosphorylated by protein kinases which lie at the end of these pathways to elicit a specific program of gene expression (reviewed in reference 31). The mitogen-activated protein (MAP) kinase cascades represent some of the best-studied signal transduction pathways which directly target nuclear proteins (reviewed in reference 56). In humans, at least three parallel pathways exist. The ERK pathway primarily transmits mitogenic and differentiation stimuli, whereas the JNK and p38 pathways primarily transduce stress stimuli to the nucleus (reviewed in references 56 and 60). These pathways are conserved in a diverse range of organisms including yeast, Drosophila, and Caenorhabditis elegans (reviewed in reference 56).
Several nuclear targets for MAP kinase pathways have been identified. For example, c-Jun is phosphorylated by the JNK MAP kinases (10, 33). ATF-2 (19, 36, 57) and ATFa (1, 19) also represent JNK targets, whereas CHOP (58) and MEF2C (20) are targets for the p38 pathway. Members of the ternary complex factor (TCF) subfamily of ETS-domain transcription factors are also targets of MAP kinase pathways (reviewed in references 56 and 60). The TCF Elk-1 is a target for all three pathways (27, 43, 61; reviewed in reference 56). However, SAP-1 appears to be targeted efficiently only by the ERK and p38 pathways (43, 54, 59, 61), although it can also act as a JNK substrate (28). The TCFs share three regions of sequence similarity with known functions: the N-terminal ETS DNA-binding domain, the B-box SRF-binding domain, and the C-terminal MAP kinase-regulated transcriptional activation domain (C domain) (reviewed in reference 55). Multiple MAP kinase sites are located in the C-terminal domain which are phosphorylated by ERK, JNK, and p38 MAP kinases in vitro and in vivo (43; reviewed in references 55 and 56). An additional short domain of sequence similarity, the D domain, is located N-terminally from the C domain (37, 42). However, to date, no function has been ascribed to this domain.
MAP kinases preferentially phosphorylate sites containing the consensus sequence Pro-Xaa-Ser/Thr-Pro, although in most cases the minimal consensus sequence Ser/Thr-Pro is sufficient for phosphorylation (8). This is apparent in Elk-1, where the most critical phosphorylation site (Ser383) conforms to this minimal consensus sequence (24, 32, 40). Due to this limited consensus site, specific targeting of MAP kinases to transcription factors has been proposed as a mechanism to ensure phosphorylation of the correct proteins by individual MAP kinases (9, 31). Indeed, targeting and binding of the JNK MAP kinases to c-Jun has been shown to be a prerequisite for efficient phosphorylation (6, 10, 30, 52). A short region of c-Jun, the delta domain, appears to be sufficient for this interaction (6, 10, 30, 52). Similar interactions occur between JNK MAP kinases and ATF-2 (18, 19, 36) via a short motif which is distinct from the phosphoacceptor sites (19, 36). JNK MAP kinases also bind to ATFa (1) and Elk-1 (15), although in these cases it is unclear whether binding to a distinct domain occurs. Interactions between ERK MAP kinases and their substrates have been demonstrated for p90rsk (22), c-Myc (17), Spi-B (39), and Elk-1 (3, 44), although to date, their site of interaction has not been identified. In the last of these, the significance of MAP kinase binding for activation of the Elk-1 transcription factor has not been addressed.
In this study, we have investigated the targeting of ERK2 MAP kinase to Elk-1. Targeting of ERK2 to Elk-1 is essential for efficient phosphorylation in vitro and correlates with the formation of stable complexes. The targeting domain was mapped to the D-domain homology region of Elk-1 that is distinct from the phosphoacceptor motifs in its C-terminal transcriptional activation domain and is sufficient for kinase binding. Activation of Elk-1 in vitro and in vivo by ERK2 is dependent upon this MAP kinase targeting domain. Our results demonstrate that ERK MAP kinases are targeted to the Elk-1 transcription factor. A general mechanism therefore appears to be emerging whereby the substrate specificity of distinct MAP kinases is determined at least in part by recognition and binding to short domains within transcription factors that are distinct from the phosphorylated residues.
MATERIALS AND METHODS
Plasmid constructs.
The following plasmids were constructed for expressing GST fusion proteins in Escherichia coli. pAS407 (encoding GST-Elk205; Elk-1 amino acids 205 to 428), pAS545 (encoding GST-Elk310; Elk-1 amino acids 310 to 428), pAS406 (encoding GST-Elk330; Elk-1 amino acids 330 to 428), and pAS405 (encoding GST-Elk349; Elk-1 amino acids 349 to 428) were generated by inserting BamHI-EcoRI-cleaved PCR-derived fragments into the same sites of pGEX-3X. pAS547, encoding Elk-1 amino acids 310 to 348 fused to c-Jun amino acids 55 to 223, was constructed by ligating an EcoRI-cleaved PCR fragment (encoding c-Jun amino acids 55 to 223) into the NaeI-EcoRI sites of pAS545. pAS569, encoding glutathione S-transferase (GST) fused to Elk-1 amino acids 310 to 348 and c-Jun amino acids 197 to 223 (GST-ElkD), was constructed by cleavage of pAS547 with NaeI (to remove c-Jun amino acids 55 to 197) followed by religation of the vector. pAS548, pAS549, pAS550, and pAS564 (encoding GST-Elk205 mutants) are derivatives of pAS407 with the site-directed mutations R314A/K315A (GST-Elk205M1), R317A/L319A (GST-Elk205M2), L323A/S324A (GST-Elk205M3), and L327A/L328A (GST-Elk205M4), respectively. pAS565 (GST-Elk307M2) and pAS566 (GST-Elk307M3) were constructed by cleavage of pAS549 and pAS550, respectively, with BamHI-BglII (to remove DNA encoding Elk-1 amino acids 205 to 306) followed by religation of the vector. pAS567 (GST-Elk310S383A/S389A) was constructed by PCR-mediated site-directed mutagenesis with pAS545 as a template, and pAS568 (encoding GST-Elk310M2/S383A/S389A) was constructed with pAS567 as a template.
Mutations were introduced by a two-step PCR protocol with a mutagenic primer and two flanking primers as described previously (50).
pAS278 and pAS380 were constructed for expressing Elk-1 derivatives as hexahistidine/FLAG-tagged proteins in E. coli. pAS278 was constructed by ligating the NcoI-BglII fragment from pQE6/16Elk (14) and the BglII-XhoI fragment from pAS276 (35) into the NcoI and XhoI sites of pET-Hnef-PFH (62). pAS379 was constructed by inserting a BamHI-XhoI PCR fragment of Elk-1, encoding amino acids 322 to 428, into the same sites in pBS-SK+. pAS380 (encoding full-length His/FLAG-tagged Elk-1 with an internal deletion of amino acids 312 to 321 (Elk-1ΔD), was constructed by ligating a NcoI-BglII fragment from pQE6/16Elk and a BamHI-XhoI fragment from pAS379 into the NcoI and XhoI sites of pET-Hnef-PFH.
The following plasmids were constructed for use in mammalian cell transfections. pG5E1b contains five GAL4 DNA-binding sites cloned upstream of a minimal promoter element and the firefly luciferase gene (46). The vector pSG5 (Stratagene), which expresses wild-type mouse MAP kinase phosphatase 1 (MKP-1), was provided by N. Tonks (53). The expression vectors for HA-tagged ERK2 (pCMV5-HA-ERK2) and constitutively active MEK1(ΔN S218E-S222D) (pCMV-MEK-DN) (38) were provided by M. Weber and N. Ahn, respectively. pSG424 encodes the GAL4 DNA-binding domain (45). pAS551 (GAL4-Elk205), pAS552 (GAL4-Elk349), pAS553 (GAL4-Elk205M1), pAS554 (GAL4-Elk205M2), pAS555 (GAL4-Elk205M3), and pAS570 (GAL4-Elk205M4) were constructed by ligating the BamHI-XbaI fragments from pAS407, pAS405, pAS548, pAS549, pAS550, and pAS564, respectively, into the same sites of pSG424. pAS571 (pCMV-GAL4), pAS572 (pCMV-GAL4-Elk205), pAS574 (pCMV-GAL4-Elk205M1), pAS575 (pCMV-GAL4-Elk205M2), pAS576 (pCMV-GAL4-Elk205M3), and pAS577 (pCMV-GAL4-Elk205M4) were constructed by ligating the HindIII-XbaI fragments from pSG424, pAS551, pAS553, pAS554, pAS555, and pAS570, espectively, into the same sites of pCMV5. pRSETB–Elk-1 was generated by inserting the NcoI-HindIII fragment from pAS278 into the same sites of pRSETB (Invitrogen). pAS383 (encoding full-length Elk-1 with a C-terminal FLAG tag) was constructed by ligating a KpnI-HindIII fragment from pRSETB–Elk-1 into the same sites of pCMV5. pAS387, encoding Elk-1ΔD, was constructed by ligating the XbaI-BamHI fragment of pAS380 into the same sites of pCMV5.
The following constructs were made for expressing proteins in yeast. pAS591 (encoding a TRP1 selectable marker and a fusion of the GAL4 DNA-binding domain and ERK2 [GAL-ERK2]) was constructed by insertion of an XmaI-PstI-cleaved PCR fragment into the same sites of pAS2-1 (Clontech). pAS467 (encoding a LEU2 selectable marker and a fusion of the GAL4 activation domain and full-length Elk-1 [Elk-AD]) was constructed by insertion of an EcoRI-cleaved PCR fragment encompassing the first 90 nucleotides of Elk-1 (and introducing an NcoI site at the N terminus) and an EcoRI-BamHI fragment from pAS278 into pGAD424 (Clontech). pAS504 (encoding a fusion of the GAL4 activation domain and Elk-1ΔD [ElkΔD-AD]) was constructed by insertion of an NcoI-XhoI fragment from pAS380 into the same sites of pAS467. Constitutively activated MEK-1 (CA-MEK1) was PCR amplified from pCMV–MEK-1 (38), cleaved with EcoRV, and ligated into the same site in pHAM8 (kindly provided by H. Mountain) to give pAS592 (containing a MET3 promoter-driven MEK-1 gene). pAS593 was constructed by insertion of a BamHI-SacI-cleaved PCR fragment amplified from pAS592 (encompassing the MET3 promoter and MEK-1 gene) into the same sites of the yeast expression vector pRS313 (51), which carries the HIS3 selectable marker gene.
All plasmid constructs encoding Elk-1-derived proteins made by PCR were verified by automated dideoxy sequencing.
Protein expression and purification.
GST fusion proteins were expressed in E. coli JM101 and purified as described previously (49). Full-length hexahistidine-tagged polypeptides were expressed in E. coli BL21(DE3) pLysS with the pET vector system. Following nickel affinity purification (Novagen), the proteins were further purified by gel filtration and fast protein liquid chromatography (Superose 12 column; Pharmacia). Fractions containing recombinant Elk-1 proteins were identified, and the integrity of the proteins was verified by Western blot analysis. The concentrations of full-length GST- or His-tagged proteins were estimated after sodium dodecyl sulfate-polyacrylamide gel electrophoresis by Coomassie blue staining with bovine serum albumin as a standard.
Tissue culture, cell transfection, and reporter gene assays.
COS-1 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS) (Gibco BRL). Transfection experiments were carried out by the Lipofectamine method (Gibco BRL) or with Superfect transfection reagent (Qiagen).
For reporter gene assays, GAL4-driven promoters were cotransfected with various vectors encoding GAL4–Elk-1 fusion proteins. Following transfection by the Lipofectamine method, cells were left for 24 h in serum-free medium prior to stimulation. To stimulate the ERK2 pathway, COS-1 cells were treated with 50 nM epidermal growth factor (EGF) (Sigma) and left for 12 h. Transfections by the Superfect method were essentially the same, except that the cells were left in 10% FBS for 12 h and then transferred to serum-free medium for a further 12 h prior to stimulation. The cells were solubilized with glycylglycine lysis buffer (1% Triton, 25 mM glycylglycine [pH 7.8], 15 mM MgSO4, 4 mM EGTA, 1 mM dithiothreitol [DTT]), and the extracts were centrifuged at 14,000 × g for 15 min at 4°C to remove cellular debris. The activities of the GAL4 DNA-binding domain (amino acids 1 to 147), and GAL4–Elk-1 deletion fusion proteins (250 ng of plasmid DNA) were measured in cotransfection assays in COS-1 cells with 250 ng of reporter plasmid pG5E1bLuc (containing five GAL4 DNA-binding sites cloned upstream of a minimal promoter element) and the firefly luciferase gene. GAL-Elk fusions containing point mutations were analyzed in a similar way, except that 50 ng of expression vector and 1 μg of reporter construct were used. Luciferase assays were carried out in 350 μl of luciferase assay buffer (25 mM glycylglycine [pH 7.8], 15 mM MgSO4, 15 mM K3PO4, 4 mM EGTA, 2 mM ATP, 1 mM DTT) with 100 μl of luciferin buffer (1 mM d-luciferin, 25 mM glycylglycine [pH 7.8], 10 mM DTT) and the appropriate volume of total-cell extracts. The light emission was measured for 10 s with a Turner TD-20/20 luminometer. Transfection efficiencies were normalized by measuring the activity of a cotransfected plasmid (1 μg) which expresses β-galactosidase (pCH110; Pharmacia KB Biotechnology Inc.). β-Galactosidase activity was determined with a chemiluminescent substrate in the Galacto-light Plus kit (Tropix).
Yeast transformation, extract preparation, and β-galactosidase filter assays.
Yeast strain Y187 (MATα ura3-52 his3 ade2-101 trp1-901 leu2-3,112 met- gal4Δ gal80Δ URA3::GAL1-lacZ) was used throughout. Cells were grown in YPD or the appropriate selective minimal medium. Cotransformation of yeast with the GAL4 DNA-binding domain, GAL4 activation domain fusion plasmids, and the MEK-1 expression plasmid was carried out by the standard lithium acetate method with 2 μg of each plasmid (12). Transformants containing fusion proteins were plated onto synthetic dextrose (SD) medium lacking the appropriate amino acids (tryptophan for pAS2-1-, leucine for pGAD424-, and histidine for pRS313-derived constructs respectively) and incubated at 30°C for 3 days. β-Galactosidase filter assays were carried out in the presence of isopropyl-β-d-thiogalactopyranoside (IPTG) as described in the Clontech MATCHMAKER two-hybrid system manual and performed on at least five independent colonies from each cotransformed strain. Yeast whole-cell extracts were prepared for Western blot analysis as described previously (41).
Expression of the GAL-ERK2 and Elk-AD fusion proteins was verified by Western blotting.
Protein kinase assays.
To prepare recombinant ERK2 MAP kinase, COS-1 cells were transfected with constructs encoding HA-epitope-tagged ERK2. Cells were maintained in 10% FBS for 12 h, starved overnight in serum-free medium, and then either stimulated with EGF (to produce activated ERK) or harvested immediately (to produce inactive ERK). EGF-stimulated cells were harvested 5 min after stimulation. The cells were solubilized with Triton lysis buffer (20 mM Tris [pH 7.4], 1% Triton X-100, 10% glycerol, 137 mM NaCl, 2 mM EDTA, 25 mM β-glycerophosphate, 1 mM sodium orthovanadate, 2 mM pyrophosphate, 1 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin per ml, 0.5 mM DTT). The extracts were centrifuged at 14,000 × g for 15 min at 4°C. Epitope-tagged ERK2 (HA) was immunoprecipitated (3 h at 4°C) from extracts with the appropriate monoclonal antibodies immobilized on protein G-Sepharose (Pharmacia). Immunoprecipitates were washed three times with Triton lysis buffer and once with kinase buffer (25 mM HEPES [pH 7.4], 25 mM β-glycerophosphate, 25 mM MgCl2, 0.5 mM DTT, 0.1 mM sodium orthovanadate). Purified kinases were then eluted from beads by competition with 0.1 mg of HA peptide per ml. Recombinant active ERK2 was also obtained from New England Biolabs (NEB). The kinase assays were initiated by the addition of substrate protein (as specified), 50 μM ATP, and 50 μM [γ-32P]ATP (10 Ci/mmol) in kinase buffer in 20 μl (final volume). The reactions were carried out at 30°C and terminated at the times specified by the addition of 4 μl of 5× Laemmli sample buffer. The phosphorylation of substrate proteins was examined after SDS-PAGE by autoradiography and quantified by phosphorimaging (Fuji BAS1500 phosphorimager; Tina 2.08e software).
Binding and phosphorylation assays.
Binding assays were performed by incubating 100 pmol of GST fusion proteins or His-FLAG-tagged full-length proteins in 350 μl of kinase-binding buffer with immunopurified ERK2 MAP kinase or transfected whole-cell extracts. The complexes were collected with either 10 μl of glutathione-agarose or protein G-M2 FLAG antibody-coupled beads and washed three times with kinase-binding buffer (40 mM HEPES [pH 7.5], 150 mM NaCl, 5 mM MgCl2, 0.2 mM EDTA, 0.1% Triton X-100, 40 mM β-glycerophosphate, 0.5 mM DTT, 0.1 mM sodium orthovanadate) and twice with kinase buffer. Protein kinase assays were carried out on the remaining complexes as described above. When required, myelin basic protein (5 μg; Sigma) was added to the kinase reactions. Alternatively, phosphorylation-induced mobility shifts on substrate proteins were detected by Western blotting analysis with antibodies against the FLAG-epitope tag.
Western blot analysis.
HA-tagged ERK2 was detected by immunoblot analysis with a mouse monoclonal anti-HA antibody (clone 12CA5; Boehringer Mannheim). FLAG-tagged Elk-1 and Elk-1ΔD in extracts from COS-1 or yeast cells were detected by immunoblot analysis with a mouse monoclonal anti-M2 FLAG antibody (Kodak). GAL4-ERK fusion proteins were detected with an anti-ERK monoclonal antibody (Santa Cruz). Immune complexes were detected by using horseradish peroxidase-conjugated secondary antibody followed by enhanced chemiluminescence (Amersham). GAL4 fusion proteins were detected with the anti-GAL4 antibody directed against the amino-terminal DNA-binding domain (Santa Cruz).
Gel retardation assays.
Gel retardation assays were performed with a 32P-labelled 134-bp c-fos promoter fragment containing the serum response element (SRE) (SRE*) as described previously (59). Phosphorylation of 0.2 pmol of His-tagged Elk-1 and Elk-1ΔD was performed at 30°C for the specified times with activated ERK2 (NEB) in 20 μl of kinase buffer supplemented with 50 μM ATP. Phosphorylated Elk-1 or Elk-1ΔD (0.02 pmol) was used in DNA-binding reactions. Antibody supershift experiments were carried out by the addition of 1 μl of anti-Phosphoplus Elk-1(Ser383) antibody (NEB). Binding-reaction mixtures for reactions on the SRE contained purified bacterially expressed coreSRF (48). Protein-DNA complexes were analyzed on nondenaturing 5% polyacrylamide gels cast in 0.5× or 1× Tris-borate-EDTA and visualized by autoradiography and phosphorimaging. Data were quantified by phosphorimaging, and the data are presented graphically after curve fitting with the appropriate equation using BIOSOFT Fig.P or Microsoft Excel software.
Figure generation. All figures were generated electronically from scanned images of autoradiographic images by using Picture Publisher (Micrografix) and Powerpoint 7.0 (Microsoft) software. Results are representative of the original autoradiographic images.
RESULTS
Elk-1 contains a MAP kinase targeting motif.
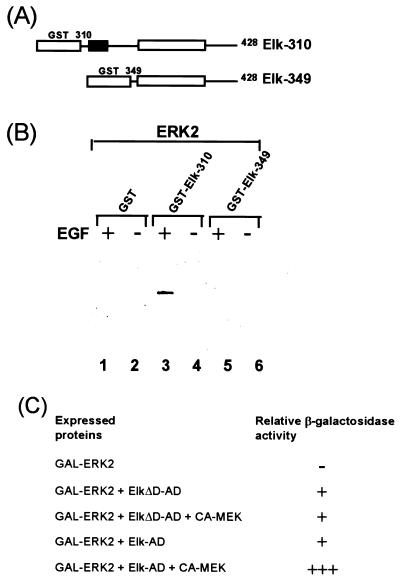
Elk-1 is phosphorylated by the ERK2 MAP kinase within its C-terminal domain (reviewed in reference 56), and truncated Elk-1 fusion proteins associate with ERK MAP kinases in vitro (3, 44). To map the ERK2-binding site on Elk-1, a series of truncated Elk-1 proteins fused to GST were constructed. Fusion proteins containing Elk-1 amino acids 310 to 428 are efficient substrates and bind to ERK2 (data not shown; see Fig. 7B and C). However, N-terminal deletions beyond amino acid 330 resulted in a reduction in the efficiency of phosphorylation by ERK2 and abolished the binding of the kinase (data not shown). The N-terminal end of the ERK2-binding motif therefore appears to map between amino acids 310 and 330, which encompasses the conserved D domain (Fig. 1A).
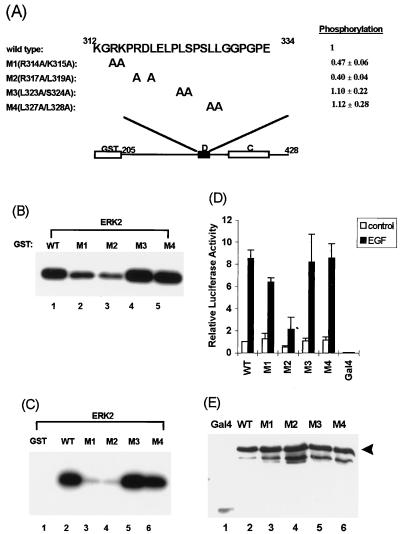
FIG. 7.
Mapping the D domain residues required for targeting of ERK2. (A) Amino acid sequences of the WT and D-domain mutants R314A/K315A (M1), R317A/L319A (M2), L323A/S324A (M3), and L327A/L328A (M4). Numbers above the sequence represent the N- and C-terminal residues in the D domain. The degree of phosphorylation of each protein (relative to WT Elk-205) is indicated. Standard deviations of the data from four independent experiments are indicated. (B) Immune-complex kinase assays of mutant GST–Elk-205 fusions by ERK2. The phosphorylation of GST–Elk-205 fusion proteins by ERK2 MAP kinase was examined by the immune-complex protein kinase assay. Kinase assays were performed for 15 min at 30°C with 2 U of activated ERK2 (NEB) and equal molar quantities (5 pmol) of GST–Elk-1 fusion proteins as substrates. (C) Binding and phosphorylation assays of wild-type and mutant GST-Elk proteins. Equal molar quantities of GST–Elk-205 fusion proteins were bound to ERK2 (50 U; NEB) and washed, and the remaining bound kinases were assayed by incubating with [γ-32P]ATP in kinase buffer for 2 h at 30°C. (D) EGF-stimulated transcriptional activation by wild-type and mutant GAL4–Elk-1 fusion proteins. COS-1 cells were transfected with cytomegalovirus promoter-driven constructs encoding GAL4 fusions to either WT or D-domain mutant Elk-1 derivatives and a GAL4-driven luciferase reporter plasmid. The cells were either unstimulated or stimulated with EGF. Transfection efficiency was monitored by using the β-galactosidase expression vector pCH110. The luciferase activities relative to GAL4–Elk-205-mediated reporter activation in unstimulated cells (means ± standard deviations; n = 3) are presented. (E) Expression levels of the GAL4 fusion proteins in COS-1 cells were examined with total-cell extracts for Western blot analysis with an anti-GAL4 antibody. Bands representing full-length GAL4–Elk-205 are indicated by the arrowhead.
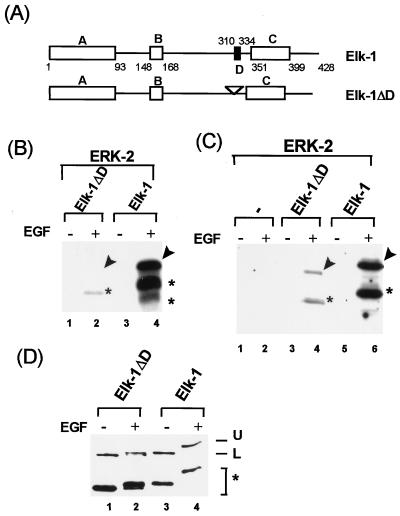
FIG. 1.
ERK2 MAP kinase is targeted to Elk-1 via the D domain. (A) Diagram of full-length Elk-1 and Elk-1 with a 9-amino-acid deletion in the D-domain (Elk-1ΔD). Bacterially expressed FLAG epitope-tagged full-length Elk-1 and Elk-1ΔD proteins were purified, and equimolar concentrations were used in all assays. (B) Immune complex kinase assays of full-length Elk-1ΔD (lanes 1 and 2) and Elk-1 (lanes 3 and 4) phosphorylated by ERK2 with 3.75 pmol of full-length Elk-1 or Elk-1ΔD for 30-min reactions. (C) Binding and phosphorylation assays. Equal molar quantities (37.5 pmol) of Elk-1 and Elk-1ΔD were immobilized onto protein G-FLAG antibody-coupled beads and incubated with immunopurified ERK2 for 18 h at 4°C. After extensive washing, the beads were incubated with [γ-32P]ATP in kinase buffer for 2 h at 30°C. The activation of ERK2 by prior cell stimulation with EGF is indicated above each lane (+, activation; −, no activation). Arrows indicate the positions of full-length proteins, and asterisks indicate N-terminal degradation products. (D) Western blot analysis (M2 anti-FLAG antibody) of Elk-1ΔD (lanes 1 and 2) and Elk-1 (lanes 3 and 4) after phosphorylation for 2 h at 30°C with immunopurified ERK2 MAP kinase. The upper band (U) corresponds to the phosphorylated form, and the lower band (L) corresponds to the unphosphorylated form of the full-length proteins. The asterisks represent N-terminal degradation products.
To assess the importance of the D domain as an ERK2-binding site and to investigate the interaction with full-length proteins, a 9-amino-acid deletion was made in the Elk-1 D domain (Elk-1ΔD) and its proficiency as an ERK2 substrate was compared to that of the wild-type (WT) full-length protein. ERK2 was immunopurified from transfected COS1 cells and was incubated with WT and mutated Elk-1 proteins. Activation of ERK2 was achieved by EGF stimulation of the cells prior to purification. In comparison to WT Elk-1, Elk-1ΔD was inefficiently phosphorylated by ERK2 (Fig. 1B, compare lanes 2 and 4). The binding of ERK2 to WT Elk-1 and Elk-1ΔD was investigated by incubation of the immunopurified kinase with each Elk-1 protein. Specifically bound proteins were obtained by coimmunoprecipitation with antibodies directed to the FLAG epitope tag on the Elk-1 proteins followed by removal of nonspecifically bound proteins by extensive washing. Bound kinases were subsequently detected by incubation of the final immunoprecipitates with [γ-32P]ATP to detect Elk-1 phosphorylation. Elk-1ΔD bound ERK2 much less efficiently than did WT Elk-1 (Fig. 1C, lanes 4 and 6). Western blotting demonstrated that equal molar quantities of WT Elk-1 and Elk-1ΔD were used in the kinase assays (Fig. 1D). The D domain is therefore involved in targeting of ERK2 to Elk-1 in the context of the full-length protein.
ERK2 targeting to Elk-1 is essential for efficient phosphorylation and activation in vitro.
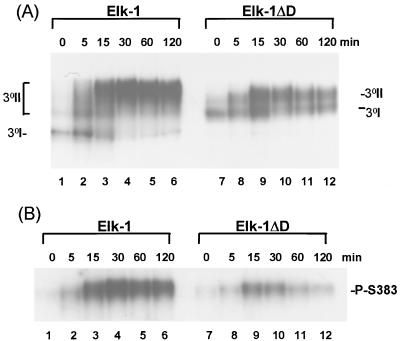
Targeting of ERK2 to the Elk-1 D domain is essential for maximal phosphorylation by ERK2 (Fig. 1). The effect of ERK2 targeting on the kinetics of Elk-1 phosphorylation were analyzed in vitro. Phosphorylation was monitored first by a reduction in the mobility of Elk-1 on SDS-PAGE (Fig. 2A) and second by the incorporation of [γ-32P]ATP (Fig. 2B).
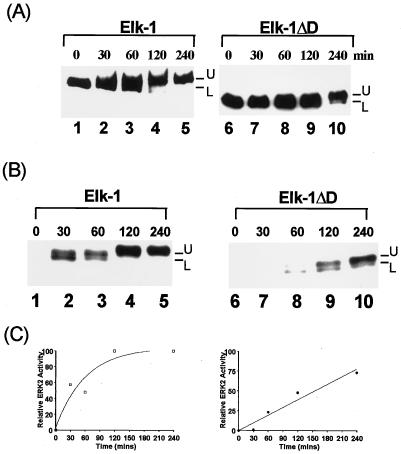
FIG. 2.
Deletion of the D domain alters the kinetics of Elk-1 phosphorylation by ERK2 MAP kinase (A and B). Elk-1 (lanes 1 to 5) and Elk-1ΔD (lanes 6 to 10) were phosphorylated by ERK2 for the times indicated above each lane, and samples were analyzed by Western blot analysis with the anti-FLAG antibody (A) and autoradiography of kinase reaction products in the presence of [γ-32P]ATP (B). The upper band (U) corresponds to the phosphorylated form of the proteins, and the lower band (L) indicates the unphosphorylated form of the proteins. (C) Graphic representation of the data from panel B. Data are presented relative to phosphorylation of WT Elk-1 after 240 min (taken as 100).
The reduction in mobility of WT Elk-1 occurred much more rapidly than for Elk-1ΔD (Fig. 2B and C). Indeed, the maximal shift to the slower-migrating form occurred between 60 and 120 min for WT Elk-1 but was not achieved until after 240 min for Elk-1ΔD. Similarly, maximal phosphorylation of full-length WT-Elk-1 was obtained after 120 min (Fig. 2B, lanes 1 to 5, and Fig. 2C) whereas in Elk-1ΔD, the kinetics of phosphorylation were severely delayed and maximal phosphorylation was still not reached after 240 min (Fig. 2B, lanes 6 to 10, and Fig. 2C).
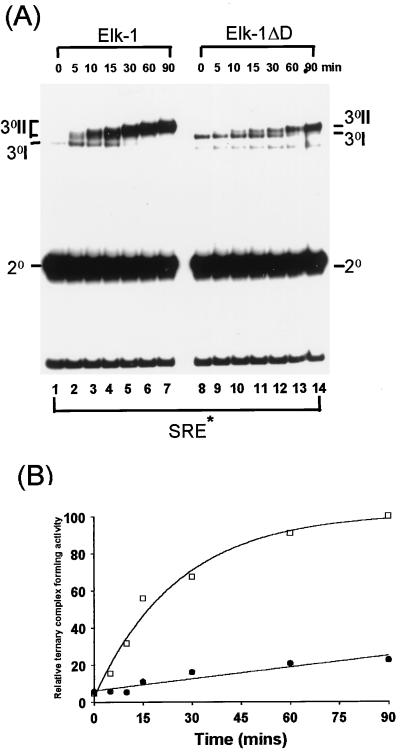
Phosphorylation of Elk-1 by ERK2 stimulates its autonomous DNA-binding capacity to “high-affinity” ets motifs such as the E74 site (47), recruitment into ternary complexes with serum response factor (SRF) and the c-fos SRE (13, 14) and the formation of a lower-mobility complex with long SRE fragments (SRE*) (40). The formation of ternary DNA-bound complexes by Elk-1 was monitored after phosphorylation by ERK2 over a 90-min period. Stimulation of a lower-mobility ternary complex containing WT Elk-1 on the SRE* site was observed after 5 min, with maximal stimulation achieved after 60 min (Fig. 3A, lanes 1 to 7, and Fig. 3B). In contrast, stimulation of ternary-complex formation with Elk-1ΔD to the SRE* site was observed with delayed kinetics (15 min; Fig. 3A, lanes 8 to 14, and Fig. 3B) and was maximally increased after 2 h (data not shown). Stimulation of autonomous DNA binding of WT Elk-1 to the E74 site also occurred with similar kinetics, whereas the activation of DNA binding by Elk-1ΔD was severely delayed (data not shown).
FIG. 3.
Deletion of the Elk-1 D domain perturbs the kinetics of ERK2 phosphorylation-induced ternary-complex formation. Elk-1 and Elk-1ΔD were phosphorylated in vitro by activated-ERK2 MAP kinase for the indicated times. (A) Kinetic study of ternary-complex formation of Elk-1 or Elk-1ΔD with SRF and a 134-bp fragment of the c-fos promoter containing the SRE (SRE*). The locations of the binary SRF-SRE complex (2°), unphosphorylated ternary complex (3°I), and multiple phosphorylated forms of ternary complex (3°II) are indicated. A band resulting from C-terminally truncated Elk-1ΔD runs below these ternary complexes. (B) The graph represents the quantification of the data shown in panel A. Data are presented relative to WT Elk-1 binding after 90 min (taken as 100). Open squares indicate wild-type Elk-1; solid circles represent Elk-1ΔD.
Taken together, these data are consistent with the notion that the Elk-1 D domain is required for targeting of ERK2 to Elk-1 and its subsequent rapid activation in vitro.
Activation of ERK2 is required for binding to the Elk-1 D domain.
To assess whether ERK2 activation is required for binding to the Elk-1 D domain, the interaction of ERK2 with Elk-1 and truncated derivatives was analyzed. GST–Elk-1 fusion proteins were incubated with extracts from COS-1 cells which had been transfected with HA-tagged ERK2 and either stimulated with EGF or left unstimulated. Following the removal of nonspecifically bound proteins by extensive washing, the presence of ERK2 was detected by Western blotting with an antibody against the ERK2 HA-epitope tag. No binding of ERK2 to GST or GST-Elk349 was detected (Fig. 4B, lanes 1 and 5). In contrast, ERK2 binding to GST–Elk-310 was detected. This interaction was dependent upon prior activation of ERK2 by EGF stimulation (Fig. 4B, lanes 3 and 4). Prior activation of ERK2 in mammalian cells is therefore required for binding to Elk-1 via the D domain.
FIG. 4.
Activation of ERK2 MAP kinase is required for interaction with Elk-1. (A) Diagrammatic representation of GST–Elk-1 fusions. The black box represents the D domain of Elk-1. (B) Whole-cell extracts from COS-1 cells transfected with HA-epitope-tagged ERK-2 were bound to equal molar quantities (200 pmol) of GST or GST–Elk fusion proteins. Following extensive washing, the remaining ERK2 protein was detected by Western blot analysis with the anti-HA antibody (+, cells activated by EGF; −, no activation). (C) Yeast two-hybrid analysis of ERK2 interaction with Elk-1 and Elk-1ΔD. Plasmids encoding the indicated proteins were introduced into a yeast strain harboring a β-galactosidase reporter gene driven by a GAL4-responsive promoter. Relative activities given are scored according to the intensity of the color on a plate assay. −, white colonies; +, barely detectable blue color; +++, strong blue color.
The interaction between ERK2 and Elk-1 was also investigated by the yeast two-hybrid assay (Fig. 4C). ERK2 was fused to the GAL4 DNA-binding domain (GAL-ERK2) and coexpressed in yeast with fusions of the GAL4 activation domain with either Elk-1 (Elk-AD) or Elk-1ΔD (ElkΔD-AD). Interactions were detected by the ability of the two proteins to reconstitute a functional transcription factor and activate a GAL4-driven β-galactosidase reporter gene. Expression of GAL-ERK2 alone gave no detectable reporter gene activity. However, coexpression of Elk-AD or ElkΔD-AD caused activation of the reporter gene, indicating an interaction with GAL-ERK2. The low level of activation and apparent lack of requirement for the Elk-1 D domain suggested that this represented a weak basal level of interaction. Constitutively active MEK1 protein (CA-MEK), the kinase directly upstream from ERK2, was therefore introduced to activate ERK2. Coexpression of CA-MEK further stimulated reporter gene activation in the presence of Elk-AD but not in the presence of ElkΔD-AD (which lacks the D domain). Activation of ERK2 is therefore required to allow efficient interaction of ERK2 with the Elk-1 D domain in intact cells.
Taken together, these data demonstrate that efficient binding of ERK2 to Elk-1 requires prior activation of the kinase and the presence of the D domain in Elk-1.
The D domain is essential for efficient ERK2 targeting and Elk-1 activation in vivo.
Disruption of the Elk-1 D domain correlates with delayed phosphorylation kinetics in vitro. One possible consequence of targeting ERK2 to Elk-1 in vivo would be to ensure rapid and efficient phosphorylation of Elk-1 in response to mitogenic stimulation. To investigate such a possibility, we determined the response of WT Elk-1 and Elk-1ΔD in vivo to cellular stimulation by EGF. Activation of Elk-1 was investigated by gel retardation analysis with cell extracts from transfected COS-1 cells in the presence of SRF and the long c-fos SRE as a binding site. Western blotting indicated that Elk-1 and Elk-1ΔD were expressed to similar levels (data not shown). Levels of transfected Elk-1 and Elk-1ΔD were in vast excess over those of endogenous TCFs, and these do not contribute significantly to the complexes shown (data not shown). The formation of a slower-migrating ternary complex by WT Elk-1 was rapidly stimulated after 5 min (Fig. 5A, lane 2), reached maximal levels of binding within 30 min (lanes 1 to 6), and was maintained for 120 min (lane 6). In contrast, the induction of a slower-migrating ternary complex containing Elk-1ΔD was reduced with only partial conversion into the slower-migrating form (lanes 7 to 12), and the mobility of the complex began to return to basal levels after 120 min (lane 12). The phosphorylation status of Elk-1 and Elk-1ΔD was directly monitored by supershifting the Elk-1-containing ternary complexes with antibodies directed against phospho-Ser383. Phosphorylation of Ser383 closely mirrored the stimulation of a lower-mobility complex. Phosphorylation of Ser383 in WT Elk-1 occurred maximally 30 min after induction and was maintained for 2 h (Fig. 5B lanes 1 to 6), whereas in Elk-1ΔD, the degree and persistence of Ser383 phosphorylation were severely reduced (lanes 7 to 12).
FIG. 5.
Deletion of the ERK-binding motif alters the kinetics of EGF-stimulated Elk-1 binding to the SRE. (A) COS-1 cells were transfected with 5 μg of cytomegalovirus promoter-driven expression vectors encoding either WT Elk-1 or Elk-1ΔD. Total-cell extracts were taken at the indicated times after EGF stimulation and bound to coreSRF and the c-fos SRE (SRE*). Ternary complexes containing unphosphorylated Elk-1 (3°I) and phosphorylated forms (3°II) are shown. (B) A parallel experiment was carried out with the same extracts in the presence of the anti-Phosphoplus Elk-1(Ser383) antibody. Supershifted bands representing Ser383-phosphorylated Elk-1 derivatives are indicated.
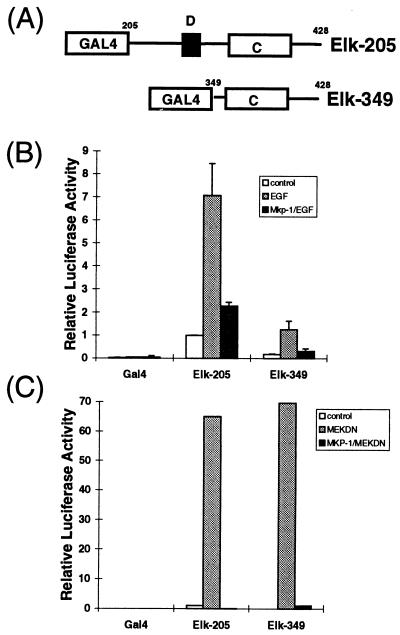
The Elk-1 C-terminal domain acts as an autonomous transcriptional activation domain in vivo whose activity is stimulated by phosphorylation with ERK MAP kinases (reviewed in reference 55). GAL4 fusion proteins containing the intact Elk-1 C-terminal domain preceded by progressively shorter N-terminal sequences (Fig. 6A) were tested for their ability to activate a GAL4-driven luciferase reporter gene in transient-transfection assays. COS-1 cells were transfected with the GAL4–Elk-1 fusions and treated with either EGF alone (to activate endogenous ERK2) or EGF plus MAP kinase phosphatase 1 (to deactivate ERK2). This treatment regime should ensure a normal, transient level of flux through the ERK pathway. In the absence of stimulation, all fusion proteins activated the reporter gene to low levels (Fig. 6B). Following EGF stimulation, both GAL4–Elk-205 (Fig. 6B) and GAL4–Elk-310 (data not shown) showed greatly enhanced transcriptional activation. In contrast, further truncation of the Elk-1 moiety which removes the D domain caused a drop in the level of activation (GAL4–Elk-349) (Fig. 6B). Cotransfection of MKP1 caused the EGF-stimulated transcriptional activation by all the GAL4 fusion proteins to be reduced to near basal levels (Fig. 6B). The reduced activation of GAL4–Elk-349 by EGF may result from defects in ERK signalling to GAL4–Elk-349 or from defects in transcriptional activation by GAL4–Elk-349. To distinguish between these possibilities, we performed control experiments to examine the effect of supraphysiological activation of ERK MAP kinase by a constitutively activated allele of MEK1, a kinase that activates ERK2. Both GAL4–Elk-205 and GAL4–Elk-349 (which lacks the D domain) were efficiently activated by constitutive MEK1. This activation was blocked by cotransfection of the MAP kinase phosphatase MKP-1 (Fig. 6C). Thus, all the GAL4–Elk-1 fusion proteins are fully activatable by supraphysiological (Fig. 6C) but not by physiological (Fig. 6B) activation of the ERK MAP kinase pathway.
FIG. 6.
The Elk-1 D domain is essential for efficient ERK2-inducible transcriptional activation in vivo. (A) Diagrammatic representation of a series of truncated Elk-1 proteins fused to the DNA-binding domain of GAL4. (B) COS-1 cells were transfected with GAL4–Elk-1 fusions and a GAL4-driven luciferase reporter plasmid. The cells were either unstimulated or stimulated with EGF for 30 min. MKP-1 was cotransfected where indicated to block the ERK2 MAP kinase pathway. (C) COS-1 cells were transfected with GAL4–Elk-1 fusions, a constitutively active form of MEK-1, and a GAL4-driven luciferase reporter plasmid. MKP-1 was cotransfected where indicated to block the ERK2 MAP kinase pathway. Transfection efficiency was monitored by using the β-galactosidase expression vector pCH110. The luciferase activities relative to GAL4–Elk-205-mediated reporter activation in unstimulated cells (means ± standard deviations; n = 5) are presented.
These data indicate that the D domain is required for efficient stimulation of Elk-1-mediated transcriptional activation by ERK MAP kinases in vivo. This is consistent with the role for this domain in ERK2 targeting to Elk-1 in vitro and subsequent activation of DNA binding in vitro and in vivo. The Elk-1 D domain is therefore required for targeting and the subsequent rapid phosphorylation by ERK2 both in vitro and in vivo.
Mapping Elk-1 residues that are critical for targeting by ERK2.
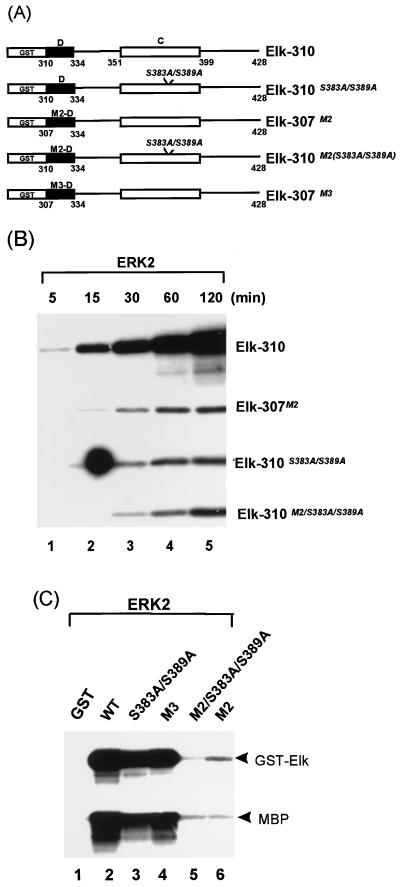
Site-directed mutagenesis was used to identify residues which play critical roles in ERK2 targeting to Elk-1. Pairs of conserved amino acids in the Elk-1 D domain were mutated to alanine residues (Fig. 7A). Such mutations will remove side chains that are available for intermolecular interactions but should cause minimal structural disruption in comparison to large amino acid deletions. WT and mutant GST fusion proteins containing Elk-1 amino acids 205 to 428 (Elk-205) (Fig. 7A) were tested as ERK2 substrates (Fig. 7B) and for ERK2 binding (Fig. 7C). Mutations M1 and M2 caused a reduction in the efficiency of phosphorylation which correlates with reduced ERK2 binding (Fig. 7B, lanes 2 and 3; Fig. 7C, lanes 3 and 4). In contrast, mutations M3 and M4 had minimal effects on Elk-1 binding and phosphorylation by ERK2 (Fig. 7B, lanes 4 and 5; Fig. 7C, lanes 5 and 6). GAL4 fusion proteins were also constructed with each of the mutant Elk-1 derivatives to investigate their activation by ERK MAP kinases in vivo. In comparison to WT GAL4–Elk-205, the M1 and M2 mutant GAL4–Elk-1 fusion proteins exhibit reduced activation of transcription in response to EGF stimulation in vivo (Fig. 7D). Of these two proteins, the M2 mutant exhibits the largest reduction in transactivation whereas the M3 and M4 mutants show minimal alterations in their transcriptional activation activity (Fig. 7D). Western blotting indicates that all the mutant proteins were expressed to equivalent levels (Fig. 7E). A correlation therefore exists between reduced binding and phosphorylation of Elk-1 by ERK2 in vitro and stimulation of Elk-1-mediated transcriptional activation by the ERK pathway in vivo. The effect of the M1 mutation on Elk-1 activity in vivo is not as great as that observed for the M2 mutation, although both apparently cause similar reductions in ERK2 phosphorylation in vitro. A likely explanation for this difference is that the threshold of the assays for detecting ERK2 binding is reached in the M1 mutant in vitro but not in vivo, allowing further differentiation between the effects of each mutant. This is consistent with the observation that the LXL motif altered in the M2 mutant is the most critical in vivo determinant and is conserved among various MAP kinase targets (see Discussion).
Collectively, these data demonstrate that residues within the Elk-1 D domain play key roles in the targeting of ERK2 and subsequent phosphorylation and activation of the Elk-1 transcription factor.
Role of the phosphoacceptor residues Ser383/Ser389 in ERK2 binding.
Ser383 and Ser389 have previously been shown to be the major ERK2 phosphorylated residues in Elk-1 (14, 32, 40). In addition, these two residues are the major determinants of ERK2-stimulated DNA binding and transcriptional activation by Elk-1 (14, 24, 40). To investigate a potential role for these phosphoacceptor sites in ERK2 binding, GST–Elk-1 fusion proteins containing mutations of these residues were compared to the WT protein and mutants containing substitutions in the D domain (Fig. 8A). Mutation of either the phosphoacceptor sites (Ser383/Ser389) or the D domain (mutant M2 [Fig. 7A]) or both caused a reduction in the rate and extent of phosphorylation of each of these mutants by ERK2 (Fig. 8B). To investigate whether this reduction in ERK2-mediated phosphorylation correlated with a reduction in the ability of the mutant proteins to bind to ERK2, GST–Elk-1 fusion proteins were incubated with activated ERK2, unbound proteins were removed by extensive washing, and the remaining bound proteins were detected by phosphorylation of the fusion protein. The ERK2 substrate myelin basic protein was also added to detect bound kinase on mutants that lack some of the ERK2 phosphoacceptor motifs. Mutations within the D domain have differential effects on ERK2 binding (Fig. 7C and Fig. 8C, lanes 4 and 6). Binding to the D-domain mutant, M2, is virtually abolished (as judged from phosphorylation of GSTElk-310M2 and MBP [Fig. 8C, lane 6]). Similarly, GST–Elk-310M2(Ser383A/Ser389A), which includes additional mutations in the phosphoacceptor motifs, exhibits negligible kinase-binding activity (Fig. 8C, lane 5). However, the GST–Elk-310(Ser383A/Ser389A) protein, which contains mutations in just the phosphoacceptor motifs, efficiently binds ERK2 (lane 3).
FIG. 8.
The phosphoacceptor sites of Elk-1, Ser383 and Ser389, are not involved in binding ERK2. (A) Diagram illustrating a series of GST–Elk-310/GST–Elk-307 mutant proteins. The solid box represents the D-domain of Elk-1 (amino acids 310 to 334) with the indicated mutations (M2-D, R317A/L319A; M3-D, L323A/S324A). Additional mutations of the phosphoacceptor residues (S383A/S389A) are indicated within the C domain (open box) in italics. The numbers of the N- and C-terminal amino acids of each domain in the Elk-1 moiety are indicated. (B) Mutation of the Elk-1 D domain and the phosphoacceptor sites (S383A/S389A) alters the kinetics of phosphorylation by ERK2. A 5-pmol portion of each GST–Elk-1 fusion protein was phosphorylated by activated ERK2 in the presence of [γ-32P]ATP at the times indicated above each lane, and samples were analyzed by autoradiography. (C) Binding and phosphorylation of GST–Elk-1 fusion proteins by ERK2. Activated ERK2 (50 U) was bound to equal molar quantities (100 pmol) of WT and mutant GST–Elk-1 fusion proteins and subsequently incubated for 2 h at 30°C with [γ-32P]ATP in kinase buffer containing 5 μg of MBP (Sigma). Bands representing phosphorylated GST–Elk-1 fusion proteins and MBP are indicated.
Taken together, these data demonstrate that in contrast to the D-domain, the phosphoacceptor motifs which play major roles in Elk-1 function (Ser383/Ser389) are not essential for ERK2 binding.
The Elk-1 D domain is sufficient for binding ERK2.
The Elk-1 D domain is required in its natural context for efficient phosphorylation by ERK2 on phosphoacceptor motifs in the adjacent transcriptional activation domain. However, to investigate whether this domain is sufficient for binding ERK2, the Elk-1 D domain was fused to GST (Fig. 9A) and its ability to bind ERK2 was compared to that of a GST fusion containing the entire Elk-1 C terminus (GST–Elk-205). GST and GST–Elk-1 fusion proteins were incubated with activated ERK2, unbound proteins were removed by extensive washing, and the remaining kinases were detected by including the ERK2 substrate MBP in a phosphorylation reaction. ERK2 was detected bound to both GST–Elk-205 and GST–ElkD, whereas little kinase activity was bound by GST alone (Fig. 9B).
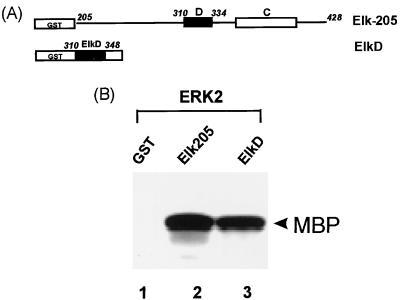
FIG. 9.
The D domain of Elk-1 is sufficient to bind ERK2. (A) Diagram illustrating a GST fusion to Elk-1. The solid box represents the D domain of Elk-1 (amino acids 310 to 334). (B) Equal molar quantities (100 pmol) of GST, GST–Elk-205, and GST-ElkD were incubated with activated ERK2 (50 U). After extensive washing, 5 μg of MBP was added, and the mixture was incubated with [γ-32P]ATP in kinase buffer for 2 h at 30°C. Phosphorylated MBP (arrowhead) was detected by autoradiography and represents the phosphorylation of MBP by the remaining bound ERK2.
These results therefore demonstrate that Elk-1 amino acids 310 to 348 (encompassing the D domain) are sufficient for binding ERK2.
DISCUSSION
The recognition and binding of c-Jun by JNK MAP kinases has been established as a paradigm for how MAP kinases achieve substrate specificity (reviewed in references 9, 31, and 60). However, to date, the binding of ERK MAP kinases to their substrates has not been investigated in detail. In this study, we demonstrate that ERK2 also recognizes and binds short motifs in transcription factors. The ETS-domain transcription factor Elk-1 is activated by three distinct MAP kinase pathways including the ERKs (reviewed in references 56 and 60). We demonstrate that ERK2 binds to Elk-1 via the conserved D domain. Binding is a prerequisite for efficient phosphorylation and subsequent activation of Elk-1 in vitro and in vivo. The targeting of MAPKs to Elk-1 is therefore a pivotal event in transducing extracellular mitogenic signals into a nuclear response. A model emerges in which ERK MAP kinases are targeted to Elk-1 via the D domain, which allows subsequent phosphorylation of phosphoacceptor motifs in the adjacent C-terminal transcriptional activation domain (Fig. 10).
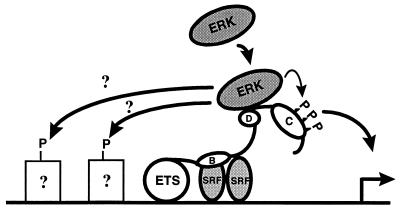
FIG. 10.
Model of a mechanism of ERK2 targeting and phosphorylation of Elk-1. ERK2 is targeted to Elk-1 by binding the D domain. Once bound, the kinases can phosphorylate the C terminus of Elk-1 and potentially other neighboring transcription factors. Once phosphorylated, Elk-1 can activate transcription at the promoter (indicated by a hinged arrow). The ETS domain, B box, MAP kinase targeting (D) and transcriptional activation domain (C) of Elk-1 are indicated by open ellipses. Dimeric SRF proteins are indicated as shaded ellipses, whereas other putative (indicated by question marks) transcription factor targets are shown as open squares.
MAP kinase targeting domains.
The D domain represents a region of sequence similarity conserved among all known TCFs. Due to its basic N-terminal region, it has been proposed to represent a nuclear localization signal (37). We demonstrate, however, that one function of this domain is for targeting of ERK2 and that it is essential for efficient Elk-1 phosphorylation and the subsequent activation of its DNA-binding and transcriptional activation properties.
The N-terminal 108 amino acids of the ETS-domain transcription factor Spi-B are sufficient for binding the ERK and JNK MAP kinases (39). A short motif within this region shows significant sequence homology to the TCF D domain (Fig. 11), indicating that this may represent a conserved binding motif for ERK MAP kinases. Indeed, mutation of Leu319 in Elk-1, which is conserved with Spi-B, severely impairs the targeting and binding of ERK2 in vitro and in vivo (Fig. 7).
FIG. 11.
Similarity among MAP kinase-binding motifs. The sequences of the ERK-binding domain of Elk-1, the homologous domain in SAP-1 and SAP-2, the JNK-binding domain of c-Jun and JIP-1, and the putative ERK/JNK-binding motif from Spi-B are shown. Amino acid numbers of the N- and C-terminal residues are given based on their location in either human Elk-1, SAP-1, SAP-2, Spi-B, c-Jun, or mouse JIP-1 proteins. Identical or highly conserved (Arg/Lys, Ser/Thr, and Leu/Ile) amino acids are highlighted. Brackets indicate the putative central MAP kinase-binding motif.
c-Jun contains a JNK binding motif known as the delta domain (6, 10, 30). This motif is a similar size (30 amino acids) but shows only limited homology to the Elk-1 D-domain (Fig. 11). The Elk-1 amino acid Leu319 is conserved in the c-Jun delta domain. The common role of this conserved amino acid in ERK and JNK targeting to Elk-1 and c-Jun, respectively, suggests that the central LXL motif (containing Leu319 in Elk-1) contained in these targeting domains may act as a scaffold around which specificity-determining residues can be added. This hypothesis is further supported by the observation that the JNK-binding domain of the JNK inhibitor protein JIP-1 also contains a similar motif (Fig. 11) (11). It therefore appears that short conserved domains which allow specific targeting of MAP kinases to transcription factors have evolved. Conversely, MAP kinases themselves also contain domains which are responsible for generating their target specificity (2). In the case of JNK1 and JNK2, a short motif in c-Jun has been shown to direct the specificity and affinity of kinase binding (18, 30). In addition, the local context of the phosphoacceptor sites plays a role in kinase targeting to c-Jun. A bipartite mechanism therefore exists for directing first the interaction of the JNKs with c-Jun and second the phosphorylation of c-Jun once bound (18, 29). The context of the phosphoacceptor motifs may also play a role in the specificity of phosphorylation of Elk-1 by ERK2. Indeed, it has previously been suggested that the local context of the phosphoacceptor motifs themselves may determine ERK2 targeting to Elk-1 (3). However, the critical phosphoacceptor motifs in Elk-1 (Ser383 and Ser389) are not required for ERK2 binding (Fig. 8). Moreover, a deletion mutant which contains the D domain but lacks the C terminus of Elk-1 (including Ser383, Ser389, and several other minor ERK2 phosphorylation sites) can still efficiently bind ERK2 (Fig. 9). The Elk-1 D domain is therefore sufficient for ERK2 binding. Further work is required to determine the role of the context of the phosphoacceptor motifs in ERK2-mediated phosphorylation of Elk-1.
Complementarity between the recognition interfaces of MAP kinases and their nuclear targets therefore appears to be an important specificity determinant in directing signal transduction pathways to the correct transcription factors and hence activation of the correct program of gene expression.
Role of ERK MAP kinase targeting to TCFs.
MAP kinases phosphorylate sites based on the optimum consensus sequence Pro-Xaa-Ser/Thr-Pro but often phosphorylate sites conforming to the relaxed consensus Ser/Thr-Pro (reviewed in reference 8). In the case of the TCFs, few of the multiple MAP kinase sites conform to the consensus sequence and none of the consensus sites are strictly conserved amongst all family members. Based on this low consensus sequence, it is unclear how MAP kinases recognize and phosphorylate TCFs rather than other potential nuclear substrates containing Ser/Thr-Pro motifs. Moreover, phosphorylation of the TCFs is very rapid, occurring within 5 minutes of cell stimulation with phorbol myristate acetate (21). This event is preceded by translocation of the ERK MAP kinases into the nucleus (5, 16, 34, 46), whereupon the kinases must rapidly locate their targets. Similarly, UV causes a rapid translocation of JNK MAP kinases to the nucleus (4). The presence of a targeting domain on Elk-1 would greatly increase the speed and specificity of this recognition process. This is consistent with the observation that only the activated ERK2 protein binds to Elk-1 via the D domain (Fig. 4).
In the case of ERK binding to Elk-1 and JNK binding to c-Jun, the interaction appears to be stable. A stable interaction of JNK MAP kinases with Elk-1 has also been reported (15), although under our experimental conditions, we were unable to see such stable interactions (reference 18 and data not shown). Therefore, it appears that ERK and JNK do not interact with Elk-1 in an identical manner. This may reflect a change in the binding kinetics and/or the interaction sites of the different MAP kinases. For other kinases, binding to their transcription factor substrates may also occur with different kinetic parameters. For example, interactions with rapid off-rates would not be detected by the assays used in this study, although targeting may still occur. Indeed, such transitory interactions may represent a common mode of interaction of kinases with their substrates.
A second potential role for MAP kinase binding could be the recruitment of the kinases to phosphorylate other substrates which are bound in the vicinity of Elk-1 (Fig. 10). Such a scenario has been documented for the JNK MAP kinases bound to c-Jun, which can phosphorylate heterodimer partners that lack a JNK-binding site (29). Potential targets for ERKs bound to Elk-1 are members of the STAT complex which bind to the c-sis inducible element (SIE) immediately upstream from the TCFs in the c-fos promoter. Indeed, STAT-1 has been shown to be phosphorylated by ERKs (7). Alternatively, the coactivator CREB-binding protein (CBP) could be a potential target, since this protein binds to both Elk-1 and SAP-1a and appears to be an in vitro substrate for ERK2 (25, 26).
Once bound to the D domain, ERK2 must phosphorylate the multiple Ser/Thr-Pro sites in the adjacent C-terminal domain. In Elk-1, the D-domain is separated by a linker peptide which is rich in glycine residues and thus is likely to be highly flexible. An attractive model is that this linker may act as a hinge to allow each Ser/Thr-Pro site within the C domain to be sequentially brought into the kinase-active site, although it is currently unknown whether each site is phosphorylated in a specific order. Such a role for the D domain in directing ERK2 action toward specific residues is supported by the observation that mutations in this domain alter the phosphorylation pattern of Elk-1 (data not shown). Indeed, stimulation of DNA binding occurs at submaximal phosphorylation levels (Fig. 2 and 3), which may reflect that a subset of critical sites are initially phosphorylated which is sufficient for activation of this process. Alternatively, a subset of the proteins may become fully phosphorylated and active in DNA binding. Further experiments are required to differentiate between these two possibilities.
In conclusion, our data contribute to our understanding of how ERK MAP kinases recognize and phosphorylate their nuclear targets. Elk-1 contains an ERK2 MAP kinase-binding motif which allows efficient kinase targeting and subsequent transcription factor activation. It is likely that in the future, more specific links between different MAP kinases and short recognition motifs in their transcription factor targets will be identified.
ACKNOWLEDGMENTS
We thank Margaret Bell and Catherine Pyle for excellent technical and secretarial assistance and Bob Liddell for DNA sequencing and oligonucleotide synthesis. We are grateful to Steve Yeaman and Adam West for comments on the manuscript and to members of our laboratories for stimulating discussions. We are also grateful to Stefan Roberts for providing reagents and to Brian Morgan for providing advice and reagents.
This work was supported by the North of England Cancer Research Campaign, the Wellcome Trust, and the U.S. National Cancer Institute. R.J.D. is an investigator of the Howard Hughes Medical Institute. A.D.S. is supported by the Lister Institute of Preventative Medicine.
REFERENCES
- 1.Bocco J L, Bahr A, Goetz J, Hauss C, Kallunki T, Kedinger C, Chatton B. In vivo association of ATFa with JNK/SAP kinases activities. Oncogene. 1996;12:1971–1980. [PubMed] [Google Scholar]
- 2.Brunet A, Pouysségur J. Identification of MAP kinase domains by redirecting stress signals into growth factor responses. Science. 1996;272:1652–1655. doi: 10.1126/science.272.5268.1652. [DOI] [PubMed] [Google Scholar]
- 3.Cano E, Hazzalin C A, Kardalinou E, Buckle R S, Mahadevan L C. Neither ERK nor JNK/SAPK MAP kinases subtypes are essential for histone H3/HMG-14 phosphorylation or c-fos and c-jun induction. J Cell Sci. 1995;108:3599–3609. doi: 10.1242/jcs.108.11.3599. [DOI] [PubMed] [Google Scholar]
- 4.Cavigelli M, Dolfi F, Claret F X, Karin M. Induction of c-fos expression through JNK-mediated TCF/Elk-1 phosphorylation. EMBO J. 1995;14:5957–5964. doi: 10.1002/j.1460-2075.1995.tb00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen R-H, Sarnecki C, Blenis J. Nuclear localisation and regulation of erk- and rsk-encoded protein kinases. Mol Cell Biol. 1992;12:915–927. doi: 10.1128/mcb.12.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai T, Rubie E, Franklin C C, Kraft A, Gillespie D A F, Avruch J, Kyriakis J M, Woodgett J R. Stress-activated protein kinases bind directly to the δ domain of c-Jun in resting cells: implications for repression of c-jun function. Oncogene. 1995;10:849–855. [PubMed] [Google Scholar]
- 7.David M, Petricoin III E, Benjamin C, Pine R, Weber M J, Larner A C. Requirement for MAP kinase (ERK2) activity in interferon α- and interferon β-stimulated gene expression through STAT proteins. Science. 1995;269:1721–1723. doi: 10.1126/science.7569900. [DOI] [PubMed] [Google Scholar]
- 8.Davis R J. The mitogen-activated protein kinase signal transduction pathway. J Biol Chem. 1993;268:14553–14556. [PubMed] [Google Scholar]
- 9.Davis R J. MAPKs: new JNK expands the group. Trends Biochem Sci. 1994;19:470–473. doi: 10.1016/0968-0004(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 10.Dérijard B, Hibi M, Wu I-H, Barrett T, Su B, Deng T, Karin M, Davis R J. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 11.Dickens M, Rogers J, Cavanagh J, Raitano A, Xia Z, Halpern J R, Greenberg M E, Sawyers C, Davis R J. A cytoplasmic inhibitor of the JNK signal transduction pathway. Science. 1997;277:693–696. doi: 10.1126/science.277.5326.693. [DOI] [PubMed] [Google Scholar]
- 12.Gietz R D, Woods R A. High efficiency transformation with lithium acetate. In: Johnston J R, editor. Molecular genetics of yeast: a practical approach. Oxford, United Kingdom: IRL Press; 1994. pp. 121–134. [Google Scholar]
- 13.Gille H, Sharrocks A D, Shaw P E. Phosphorylation of transcription factor p62TCF by MAP kinase stimulates ternary complex formation at c-fos promoter. Nature. 1992;358:415–417. doi: 10.1038/358414a0. [DOI] [PubMed] [Google Scholar]
- 14.Gille H, Kortenjann M, Thomae O, Moomaw C, Slaughter C, Cobb M H, Shaw P E. ERK phosphorylation potentiates Elk-1-mediated ternary complex formation and transactivation. EMBO J. 1995;14:951–962. doi: 10.1002/j.1460-2075.1995.tb07076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gille H, Strahl T, Shaw P E. Activation of ternary complex factor Elk-1 by stress-activated protein kinases. Curr Biol. 1995;5:1191–1200. doi: 10.1016/s0960-9822(95)00235-1. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez F A, Seth A, Raden D L, Bowman D S, Fay F S, Davis R J. Serum-induced translocation of mitogen-activated protein kinase to the cell surface ruffling membrane and the nucleus. J Cell Biol. 1993;122:1089–1101. doi: 10.1083/jcb.122.5.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta S, Davis R J. MAP kinase binds to the NH2-terminal activation domain of c-Myc. FEBS Lett. 1994;353:281–285. doi: 10.1016/0014-5793(94)01052-8. [DOI] [PubMed] [Google Scholar]
- 18.Gupta S, Barrett T, Whitmarsh A J, Cavanagh J, Sluss H K, Dérijard B, Davis R J. Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J. 1996;15:2760–2770. [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta S, Campbell D, Dérijard B, Davis R J. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 20.Han J, Jiang Y, Li Z, Kravchenko V V, Ulevitch R J. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature. 1997;386:296–299. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- 21.Hipskind R A, Büscher D, Nordheim A, Baccarini M. Ras/MAP kinase-dependent and -independent signalling pathways target distinct ternary complex factors. Genes Dev. 1994;8:1803–1816. doi: 10.1101/gad.8.15.1803. [DOI] [PubMed] [Google Scholar]
- 22.Hsiao K M, Chou S Y, Shih S J, Ferrell J E. Evidence that inactive p42 mitogen-activated protein kinase and inactive Rsk exist as a heterodimer in vivo. Proc Natl Acad Sci USA. 1994;91:5480–5484. doi: 10.1073/pnas.91.12.5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ihle J N. STATs and MAPKs: obligate or opportunistic partners in signaling. Bioessays. 1996;18:95–98. doi: 10.1002/bies.950180204. [DOI] [PubMed] [Google Scholar]
- 24.Janknecht R, Ernst W H, Pingoud V, Nordheim A. Activation of ternary complex factor Elk-1 by MAP kinases. EMBO J. 1993;12:5097–5104. doi: 10.1002/j.1460-2075.1993.tb06204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janknecht R, Nordheim A. Regulation of the c-fos promoter by the ternary complex factor Sap-1a and its coactivator CBP. Oncogene. 1996;12:1961–1969. [PubMed] [Google Scholar]
- 26.Janknecht R, Nordheim A. MAP kinase-dependent transcriptional coactivation by Elk-1 and its cofactor CBP. Biochem Biophys Res Commun. 1996;228:831–837. doi: 10.1006/bbrc.1996.1740. [DOI] [PubMed] [Google Scholar]
- 27.Janknecht R, Hunter T. Convergence of MAP kinase pathways on the ternary complex factor SAP-1a. EMBO J. 1997;16:1620–1627. doi: 10.1093/emboj/16.7.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janknecht R, Hunter T. Activation of the SAP-1a transcription factor by the c-Jun N-terminal kinase (JNK) mitogen-activated protein kinase. J Biol Chem. 1997;272:4219–4224. doi: 10.1074/jbc.272.7.4219. [DOI] [PubMed] [Google Scholar]
- 29.Kallunki T, Deng T, Hibi M, Karin M. c-Jun can recruit JNK to phosphorylate dimerization partners via specific docking interactions. Cell. 1996;87:929–939. doi: 10.1016/s0092-8674(00)81999-6. [DOI] [PubMed] [Google Scholar]
- 30.Kallunki T, Su B, Tsigelny I, Sluss H K, Dérijard B, Moore G, Davis R J, Karin M. JNK2 contains a specificity-determining region responsible for efficient c-jun binding and phosphorylation. Genes Dev. 1994;8:2996–3007. doi: 10.1101/gad.8.24.2996. [DOI] [PubMed] [Google Scholar]
- 31.Karin M. Signal transduction from the cell surface to the nucleus through the phosphorylation of transcription factors. Curr Opin Cell Biol. 1994;6:415–424. doi: 10.1016/0955-0674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 32.Kortenjann M, Thomae O, Shaw P E. Inhibition of v-raf-dependent c-fos expression and transformation by a kinase-defective mutant of the mitogen activated protein kinase Erk2. Mol Cell Biol. 1994;14:4815–4824. doi: 10.1128/mcb.14.7.4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kyriakis J M, Banerjee P, Nikolakaki E, Dai T, Rubie E A, Ahmad M F, Avruch J, Woodgett J R. The stress-activated protein kinase subfamily of c-Jun kinases. Nature. 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 34.Lenormand P, Sardet C, Pagès G, L’Allemain G, Brunet A, Pouysségur J. Growth factors induce nuclear translocation of MAP kinases (p42 (mapK) and p44 (mapK)) but not their activator MAP kinase kinase (p45 (mapKK)) in fibroblasts. J Cell Biol. 1993;122:1079–1088. doi: 10.1083/jcb.122.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ling Y, Lakey J H, Roberts C E, Sharrocks A D. Molecular characterisation of the B-box protein-protein interaction motif of the ETS-domain transcription factor Elk-1. EMBO J. 1997;16:2431–2440. doi: 10.1093/emboj/16.9.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Livingstone C, Patel G, Jones N. ATF-2 contains a phosphorylation-dependent transcriptional activation domain. EMBO J. 1995;14:1785–1797. doi: 10.1002/j.1460-2075.1995.tb07167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez M, Oettgen P, Akbarali Y, Fendorfer U, Liberman T A. ERP, a new member of the ets transcription factor/oncoprotein family: cloning, characterization, and differential expression during B-lymphocyte development. Mol Cell Biol. 1994;14:3292–3309. doi: 10.1128/mcb.14.5.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mansour S J, Matten W T, Hermann A S, Candia J M, Rong S, Fukasawa K, Vande-Woude G F, Ahn N G. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- 39.Mao C, Ray-Gallet D, Tavitian A, Moreau-Gachelin F. Differential phosphorylations of Spi-B and Spi-1 transcription factors. Oncogene. 1996;12:863–873. [PubMed] [Google Scholar]
- 40.Marais R, Wynne J, Treisman R. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell. 1993;73:381–393. doi: 10.1016/0092-8674(93)90237-k. [DOI] [PubMed] [Google Scholar]
- 41.Peterson C L, Dingwall A, Scott M P. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc Natl Acad Sci USA. 1994;91:2905–2908. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price M A, Rogers A E, Treisman R. Comparative analysis of the ternary complex factors Elk-1, SAP-1a and SAP-2 (ERP/NET) EMBO J. 1995;14:2589–2601. doi: 10.1002/j.1460-2075.1995.tb07257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price M A, Cruzalegui F H, Treisman R. The p38 and ERK MAP kinase pathways co-operate to activate ternary complex factors and c-fos transcription in response to UV light. EMBO J. 1996;15:6552–6563. [PMC free article] [PubMed] [Google Scholar]
- 44.Rao V N, Reddy E S P. Elk-1 proteins interact with MAP kinases. Oncogene. 1994;9:1855–1860. [PubMed] [Google Scholar]
- 45.Sadowski I, Ptashne M. A vector for expressing GAL4(1–147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seth A, Gonzalez F A, Gupta S, Raden D L, Davis R J. Signal transduction within the nucleus by mitogen-activated protein kinase. J Biol Chem. 1992;267:24796–24804. [PubMed] [Google Scholar]
- 47.Sharrocks A D. ERK2/p42 MAP kinase stimulates both autonomous and SRF-dependent DNA binding by Elk-1. FEBS Lett. 1995;368:77–80. doi: 10.1016/0014-5793(95)00604-8. [DOI] [PubMed] [Google Scholar]
- 48.Shore P, Sharrocks A D. The transcription factors Elk-1 and serum response factor interact by direct protein-protein contacts mediated by a short region of Elk-1. Mol Cell Biol. 1994;14:3283–3291. doi: 10.1128/mcb.14.5.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shore P, Bisset L, Lakey J, Waltho J P, Virden R, Sharrocks A D. Characterization of the Elk-1 ETS DNA-binding domain. J Biol Chem. 1995;270:5805–5811. doi: 10.1074/jbc.270.11.5805. [DOI] [PubMed] [Google Scholar]
- 50.Shore P, Whitmarsh A J, Bhaskaran R, Waltho J, Davis R J, Sharrocks A D. Determinants of DNA binding specificity of ETS-domain transcription factors. Mol Cell Biol. 1996;16:3338–3349. doi: 10.1128/mcb.16.7.3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sikorsi R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sluss H K, Barrett T, Derijard B, Davis R J. Signal transduction by tumor necrosis factor mediated by JNK protein kinases. Mol Cell Biol. 1994;14:8376–8384. doi: 10.1128/mcb.14.12.8376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun H, Charles C H, Lau L F, Tonks N K. MKP-1 (3CH134), an immediate early gene product, is a dual specificity phosphatase that dephosphorylates MAP kinase in vivo. Cell. 1993;75:487–493. doi: 10.1016/0092-8674(93)90383-2. [DOI] [PubMed] [Google Scholar]
- 54.Strahl T, Gille H, Shaw P E. Selective response of ternary complex factor Sap-1a to different mitogen-activated protein kinase sub-groups. Proc Natl Acad Sci USA, 1996;93:11563–11568. doi: 10.1073/pnas.93.21.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Treisman R. Ternary complex factors: growth regulated transcriptional activators. Curr Opin Genet Dev. 1994;4:96–101. doi: 10.1016/0959-437x(94)90097-3. [DOI] [PubMed] [Google Scholar]
- 56.Treisman R. Regulation of transcription by MAP kinase cascades. Curr Opin Cell Biol. 1996;8:205–215. doi: 10.1016/s0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- 57.Van Dam H, Wilhelm D, Herr I, Steffen A, Herrlich P, Angel P. ATF-2 is preferentially activated by stress-activated protein kinases to mediate c-jun induction in response to genotoxic agents. EMBO J. 1995;14:1798–1811. doi: 10.1002/j.1460-2075.1995.tb07168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang X, Ron D. Stress-induced phosphorylation and activation of the transcription factor CHOP (GADD153) by p38 MAP kinase. Science. 1996;272:1347–1349. doi: 10.1126/science.272.5266.1347. [DOI] [PubMed] [Google Scholar]
- 59.Whitmarsh A J, Shore P, Sharrocks A D, Davis R J. Integration of MAP kinase signal transduction pathways at the serum response element. Science. 1995;269:403–407. doi: 10.1126/science.7618106. [DOI] [PubMed] [Google Scholar]
- 60.Whitmarsh A J, Davis R J. Transcription factor AP-1 regulation by mitogen-activated protein kinase signal transduction pathways. J Mol Med. 1996;74:589–607. doi: 10.1007/s001090050063. [DOI] [PubMed] [Google Scholar]
- 61.Whitmarsh A J, Yang S-H, Su MS-S, Sharrocks A D, Davis R J. Role of p38 and JNK MAP kinases in the activation of ternary complex factors. Mol Cell Biol. 1997;17:2360–2371. doi: 10.1128/mcb.17.5.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao L-J, Narayan O. A gene expression vector useful for protein purification and studies of protein: protein interactions. Gene. 1993;137:345–346. doi: 10.1016/0378-1119(93)90032-x. [DOI] [PubMed] [Google Scholar]