Abstract
The homothallic filamentous ascomycete Sordaria macrospora possesses genes which are thought to encode two pheromone precursors and two seven-transmembrane pheromone receptors. The pheromone precursor genes are termed ppg1 and ppg2. The putative products derived from the gene sequence show structural similarity to the α-factor precursors and a-factor precursors of the yeast Saccharomyces cerevisiae. Likewise, sequence similarity has been found between the putative products of the pheromone receptor genes pre2 and pre1 and the S. cerevisiae Ste2p α-factor receptor and Ste3p a-factor receptor, respectively. To investigate whether the α-factor-like pheromone-receptor pair of S. macrospora is functional, a heterologous yeast assay was used. Our results show that the S. macrospora α-factor-like pheromone precursor PPG1 is processed into an active pheromone by yeast MATα cells. The S. macrospora PRE2 protein was demonstrated to be a peptide pheromone receptor. In yeast MATa cells lacking the endogenous Ste2p receptor, the S. macrospora PRE2 receptor facilitated all aspects of the pheromone response. Using a synthetic peptide, we can now predict the sequence of one active form of the S. macrospora peptide pheromone. We proved that S. macrospora wild-type strains secrete an active pheromone into the culture medium and that disruption of the ppg1 gene in S. macrospora prevents pheromone production. However, loss of the ppg1 gene does not affect vegetative growth or fertility. Finally, we established the yeast assay as an easy and useful system for analyzing pheromone production in developmental mutants of S. macrospora.
The life cycle of ascomycetes can be either homothallic or heterothallic. Homothallic species are self-fertile and are able to complete the sexual cycle without a mating partner. In heterothallic ascomycetes, mating occurs only between cells of opposite mating type, which attract each other by secreting pheromones (for reviews see reference 9, 46, and 47). Each mating type produces its own specific pheromone. These can be divided into two groups depending on the pathway of synthesis and secretion.
The pheromone response system of the heterothallic ascomycetous yeast Saccharomyces cerevisiae is a well-studied model of pheromone and pheromone receptor interaction. Binding of pheromones to their specific receptors triggers a G-protein-linked signal transduction pathway that induces the expression of several genes, facilitating the fusion of MATa and MATα cells to form diploid MATa/α cells (44). The peptide synthesized by MATa cells is derived from a precursor with a C-terminal CaaX (where C is cysteine, a is an aliphatic residue, and X is any amino acid residue) motif, a signal for carboxymethylation and farnesylation. The mature lipopeptide pheromone, called a-factor, is secreted via an ATP-binding cassette transporter (7). MATα cells produce α-factor, which is secreted by the classical yeast secretory pathway (9, 35, 73). The α-factor is a 13-amino-acid peptide derived from a precursor containing several copies of the 13-amino-acid sequence in tandem repeats. The first step of precursor processing is the removal of the signal sequence of the prepro-α-factor (79). Further processing includes the Kex2 endoprotease, which cleaves the pro-protein at characteristic KR dipeptides (36), the dipeptidyl-aminopeptidase Ste13p, which recognizes the dipeptide XA or XP (35), and the carboxypeptidase Kex1p, which removes C-terminal extensions (16).
The S. cerevisiae pheromone receptors for a-factor (Ste3p) and α-factor (Ste2p) are members of the large family of G-protein-coupled receptors, which contain seven-transmembrane domains. In contrast to ascomycetes, only lipopeptide pheromones of the S. cerevisiae a-factor group and Ste3p-like lipopeptide receptors have been found in basidiomycetes (4, 12, 20, 43, 51, 70, 78). Interestingly, putative pheromone precursor genes encoding two different mating pheromones have been identified not only in heterothallic filamentous ascomycetes, such as Neurospora crassa, Cryphonectria parasitica, Magnaporthe grisea, and Podospora anserina, but also in the homothallic filamentous ascomycete Sordaria macrospora (3, 58, 69, 81).
The first step in the sexual reproduction of mycelial ascomycetes is to bring together two compatible nuclei in the same cell. In heterothallic species, a specialized hypha termed trichogyne is sent out from a female prefruiting body to grow towards a fertilizing male cell of the opposite mating type. A functional male cell may be a uninucleate spermatium or microconidium or a multinucleate macroconidium. Recently, it was demonstrated that male and female fertility of heterothallic mycelial ascomycetes depends on the interactions of pheromones with their specific receptors (17, 39, 40, 77). In the homothallic ascomycete S. macrospora, conidiospores, spermatia, and trichogynes are absent, suggesting that pheromones are not needed either for sensing a mating partner or for initializing fertilization events.
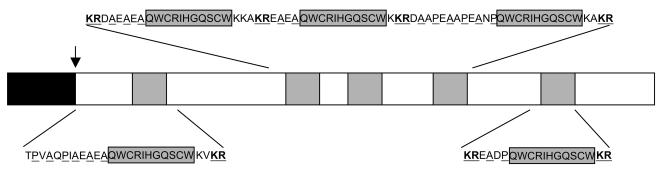
Nevertheless, two putative pheromone precursors genes, named ppg1 and ppg2, are transcriptionally expressed in S. macrospora. The ppg2 gene encodes a lipopeptide with a putative farnesylated and carboxymethylated C-terminal cysteine residue. The PPG1 precursor consists of a putative hydrophobic signal sequence and five repeats of the undecapeptide QWCRIHGQSCW. Each of the five repeated sequences is surrounded by maturation signals similar to those of the α-factor pheromone precursors of S. cerevisiae (58) (Fig. 1). The presence of these maturation motifs suggests that the S. macrospora PPG1 precursor is processed to an undecapeptide pheromone, hereafter referred to as pheromone PHE1. In addition to pheromone genes, two pheromone receptor genes, termed pre1 and pre2, have also been identified and shown to be transcriptionally expressed in S. macrospora (59). The predicted products of both genes are proteins with seven transmembrane domains. The pre1 gene product shows extensive amino acid similarity to the a-factor receptor Ste3p of S. cerevisiae and to lipopeptide pheromone receptors of basidiomycetes. The S. macrospora pheromone receptor PRE2 exhibits significant sequence similarity to the S. cerevisiae Ste2p α-factor receptor (59).
FIG. 1.
Schematic structure of the S. macrospora pheromone precursor PPG1. The predicted PPG1 pheromone precursor consists of a hydrophobic signal sequence (black box) and five repeats of the undecapeptide QWCRIHGQSCW (grey boxes). The putative cleavage site of the signal sequence is marked by a vertical arrow, KR dipeptides are indicated in bold letters and underlined, and XA/XP repeats, putative cleavage sites for a dipeptidyl-aminopeptidase, are underlined.
In filamentous ascomycetes, two functions of pheromones have been suggested. One is the regulation of initial recognition between trichogynes and spermatia of opposite mating types, which is essential for sexual reproduction in heterothallic filamentous ascomycetes. Second, pheromones are thought to play a role in postfertilization events (18). This may be equally important for heterothallic and homothallic fungi. In mycelial ascomycetes, an essential postfertilization event takes place during the development of dikaryotic hyphae, enabling karyogamy and meiosis. It has therefore been suggested that recognition between nuclei is mediated by the nucleus-limited expression of mating type-specific pheromones and receptors. In addition, these pheromones and receptors are thought to be limited to the plasma membrane region close to the individual nucleus. The spatial restriction of signaling components is proposed to facilitate recognition between two nuclei (21, 67, 72).
The fact that pheromone and pheromone receptor genes are transcribed in the homothallic ascomycete S. macrospora supports the idea that pheromones and receptors are functional in this fungus.
In order to examine the functionality of S. macrospora pheromone precursor PPG1 and pheromone receptor PRE2, the yeast S. cerevisiae was used as a heterologous assay system. A synthetic peptide was designed to analyze whether the undecapeptide predicted from the PPG1 sequence can act as an active peptide pheromone and trigger the pheromone response in the heterologous yeast system. Furthermore, we disrupted the pheromone precursor gene ppg1 of S. macrospora to investigate its involvement in fruiting body and ascospore development.
MATERIALS AND METHODS
Strains, culture conditions, and transformation.
Escherichia coli strain SURE was used as the host for plasmid amplification (30). Cloning and propagation of recombinant plasmids were done with standard protocols (65). All fungal strains used in this work are summarized in Table 1. Saccharomyces cerevisiae strain 7416-12-3 was used as a host for plasmids containing S. macrospora pheromone receptor gene pre2 or control plasmid pPGK in halo assays (42). Supernatants used in assays for pheromone activity were taken from cultures of strain W303-1B carrying either the S. macrospora ppg1 gene or the empty vector pPGK (22). To ensure that the synthetic yeast α-mating pheromone was functioning as intended in the halo assay set-up, strain 786-11-1 was used as a control (32). Strains 7416-12-3 and 786-11-1 are both derived from yeast strain 381G (31). As appropriate, yeast strains were grown in YEPD or SD minimal medium lacking uracil or uracil and tryptophan (71). Transformation of S. cerevisiae was done by electroporation according to Becker and Lundblad (2) in a Multiporator (Eppendorf) at 1.5 kV. For each transformation, 100 ng of plasmid DNA was used.
TABLE 1.
Strains used in this study
| Organism | Strain | Relevant genotype | Reference or source |
|---|---|---|---|
| Sordaria macro- spora | S48977 | Wild type, homothallic | This study |
| per44 (S48143) | Sterile, no ascospores | This study | |
| per46 (S43021) | Sterile, no ascospores | This study | |
| S52063 | ppg1::hygR, fertile | This study | |
| Saccharomyces cerevisiae | W303-1B | MATα | 22 |
| 7416-12-3 | MATacry1 ade2-1 his4-580 lys2 trp1 tyr1 SUP4-3 leu2 ura3 TYR1+sst2-1 ste2-10::LEU2 | 42 | |
| 786-11-1 | MATacry1 ade2-1 his4-580 lys2 trp1 tyr1 SUP4-3 ade3 leu2 ura3 TYR1+sst2-1 | 32 |
The Sordaria macrospora wild-type strain K (S48977) was available from our laboratory collection (Department of General and Molecular Botany, Bochum, Germany). The S. macrospora developmental mutant strains used for screening of halo formation are impaired in forming female gametangia (asc mutants), perithecia (pro mutants), or ascospores (per mutants) or produce unshaped, piled perithecia (pile mutants). They were generated by UV or ethyl methanesulfonate mutagenesis from the wild-type strain (49, 50, 54, 60). Transformation of S. macrospora was performed according to Nowrousian et al. (54). All S. macrospora strains were cultivated on corn meal medium (27). For the halo assays analyzing PPG1 pheromone secretion and for the Western analysis of enhanced green fluorescent protein (EGFP) secretion, S. macrospora wild-type and mutant strains and transformants were grown for 3 to 4 days in liquid CM medium (54).
Construction of plasmids.
In order to determine whether a peptide pheromone encoded by the ppg1 gene is secreted by S. macrospora, we fused the signal sequence (SS) of the ppg1 gene in-frame to the egfp gene of plasmid p82.9 (61). The SS region of the ppg1 gene was reconstituted with two annealed complementary oligonucleotides (ppg1S1 and ppg1S2). The sequences of all oligonucleotides used in this work are listed in Table 2. The signal sequence was inserted into the NcoI site of p82.9 in the sense (pSppg1-1) and in the antisense (pSppg1-2) orientations. As a control, the signal sequence of an aspartic proteinase gene (etp) was fused either in sense in-frame with the egfp gene (pSetp-1) or antisense (pSetp-2) (61). The 1.4-kb EcoRI hph cassette of pCB1003 was inserted into all constructs to provide a selectable marker (8).
TABLE 2.
Oligonucleotides used in this work
| Oligonucleotide | Sequence (5′-3′) |
|---|---|
| ppg1S1 | CATGAAGTTCACCCTCCCTCTTGTCATCTTCGCCGCCGTGGCCTCCGCCACCCCGGC |
| ppg1S2 | CATGGCCGGGGTGGCGGAGGCCACGGCGGCGAAGATGACAAGAGGGAGGGTGAACTT |
| ETP3 | CATGGTAGCCCTCACCAACCTCCTCCTCACTACCGTCCTCGCCTCTGCCGGCCTCGGTTCCGCCCTGCCAGC |
| ETP4 | CATGGCTGGCAGGCCGGAACCGAGGCCGGCAGAGGCGAGGACGGTAGTGAGGAGGAGGTTGGTGAGGGCTAC |
| pre2-1 | GGATCCATGGCTTCATGCGCTTCCCCAACA |
| pre2-2 | GGATCCCTACTCAAAAGACCTCGGCTTCTG |
| phe1-1 | GAATTCATGAAGTTCACCCTCCCTCTT |
| phe1-2 | GAATTCCTAGTTCTCATCAGAGTGAGCAG |
| pb41 | GATCTCAATAACGACACCATCAC |
| pb42 | GAATTCGAGATACTGAAAGGGTC |
| pb8 | GAGAATGTGGCTCCATCTCCA |
| tC1 | GATCCGCCTGGACGACTAAACC |
| h3 | ACTCGTCCGAGGGCAAAGGAATAG |
| pb6 | TGTCAAGGAAAATACCCAAGGCCG |
The yeast expression vector pPGK contains unique EcoRI, HindIII, and BamHI cloning sites between the phosphoglycerate kinase (PGK) promoter and the transcription terminator sequences (37). The open reading frames (ORFs) of the genes encoding PPG1 and PRE2 were inserted into pPGK to facilitate expression of the S. macrospora genes and to analyze the interaction of the two gene products. The genes were amplified from wild-type DNA by PCR with oligonucleotides pre2-1/pre2-2 (pre2) and phe1-1/phe1-2 (ppg1) as primers. The PCR products containing the ppg1 and pre2 coding sequence were inserted into the PCR cloning vector pDrive (Qiagen, Hilden, Germany). Sequence analysis of the PCR fragments (MWG-Biotech Customer Service, Ebersberg, Germany) confirmed the successful amplification and cloning of the desired fragments. Afterwards plasmids were digested with restriction enzyme BamHI or EcoRI. The resulting 1.7-kb pre2 BamHI and 835-bp ppg1 EcoRI fragments were cloned into the single BamHI and EcoRI restriction sites of the vector pPGK, respectively. An overview of all plasmids used in this work is given in Table 3.
TABLE 3.
Plasmids used in this work
| Plasmid | Vector | Insert | Reference |
|---|---|---|---|
| pSM1 | gpd promoter and egfp gene | 61 | |
| pSppg1-1 | p82.9 | ppg1 signal sequence fused sense to egfp gene | This study |
| pSppg1-2 | p82.9 | ppg1 signal sequence fused antisense to egfp gene | This study |
| pSetp-1 | p82.9 | etp signal sequence fused sense to egfp gene | 61 |
| pSetp-2 | p82.9 | etp signal sequence fused antisense to egfp gene | 61 |
| pPGK | PGK expression cassette | 37 | |
| pPre2 | pPGK | S. macrospora pre2 ORF | This study |
| pPpg1 | pPGK | S. macrospora ppg1 ORF | This study |
| pTCFL1 | FUS1-inducible lacZ reporter gene | 11 | |
| p204-20 | pBCKS(+) | 1,622-bp EcoRI fragment of cosmid clone H4 | This study |
| p210-1 | p204-20 | hph cassette from pCB1003 | This study |
Pheromones.
The synthetic S. macrospora PHE1 pheromone (Gln-Trp-Cys-Arg-Ile-His-Gly-Gln-Ser-Cys-Trp) was synthesized by Eurogentec (Seraing, Belgium). Synthetic S. cerevisiae α-factor (Trp-His-Trp-Leu-Gln-Leu-Lys-Pro-Gly-Gln-Pro-Met-Tyr) was obtained from Sigma-Aldrich (Munich, Germany; T6901). Pheromones were diluted in 2% dimethyl sulfoxide, and 1.5 nmol was used for halo and lacZ induction assays.
Determining the effects of S. macrospora pheromones on S. cerevisiae.
For halo assays, strain 7416-12-3 (MATa Δste2 sst2-1) containing plasmid pPre2 or control plasmid pPGK was grown in SD medium lacking uracil to an optical density at 600 nm (OD600) of 1.0 to 1.3. One milliliter of cells was embedded in 5 ml of soft agar. To obtain a lawn, the cell-containing soft agar was plated onto SD plates lacking uracil and containing 0.1 M citric acid (pH 4.5). To test the interaction between the PRE2 receptor produced by the cells in the lawn and certain pheromones, 20 μl of the following solutions was applied to filter disks (6 mm diameter; Schleicher & Schuell) and placed onto the lawn, either yeast culture supernatants (MATα strain W303-1B containing pPpg1 or control plasmid pPGK) or culture medium of S. macrospora strains grown for 3 to 4 days in CM medium or 1.5 nmol of synthetic pheromones. The plates with the yeast lawns were grown for 24 to 48 h and analyzed for pheromone-induced cell cycle arrest by the halo assay. Measuring the distance from the center of the filter disks to the edge of the zone of growth inhibition determined the size of the halo. Plates were scanned electronically and recorded images were edited with Adobe Photoshop 6.0.
Analysis of shmoo formation was done according to Sprague (74). Yeast MATa cells were grown to a density of 5 × 106 to 107 cells/ml and were treated either with pheromone at a concentration of 5.0 μM or with dimethyl sulfoxide. Samples were removed at 0 h, 2 h, and 4 h and examined by differential interference contrast light microscopy on a Zeiss Axiophot microscope. Images were recorded with an MC80DX camera, and the Adobe Photoshop 6.0 software was used for further processing.
Yeast FUS1 and lacZ induction assays.
Yeast cell cultures of MATα transformants carrying plasmid pPpg1 or plasmid pPGK as a control were grown in SD medium lacking uracil. Supernatants were obtained by centrifugation when growth had reached an OD600 of ≈1.0. Similarly, S. macrospora wild-type and mutant strains were grown in liquid CM medium, and cell-free supernatants were harvested after 3 to 4 days of growth.
Yeast MATa cells (7416-12-3) carrying pPre2 or the control vector pPGK, together with the pheromone-inducible reporter construct pTCFL1 (FUS1::lacZ) (11), were grown overnight to an OD600 of ≈1.0 in SD medium lacking uracil and tryptophan and harvested by centrifugation. Cell pellets were resuspended in 0.5 volume of fresh selective medium.
Equal volumes (10 ml) of receptor-containing MATa cells and supernatants of MATα S. cerevisiae or S. macrospora cultures containing the pheromones of interest were mixed and incubated in glass flasks at 30°C and 220 rpm for 2 h. In lacZ assays with synthetic pheromone, 1.5 nmol of synthetic PHE1 pheromone was added to 10 ml of MATa transformants expressing the S. macrospora pheromone receptor gene pre2. After the incubation period, the activity of β-galactosidase was measured. This was done by determining o-nitrophenyl-β-d-galactopyranoside cleavage by crude protein extracts (64). Crude protein extracts were obtained by destroying the yeast cells with glass beads (0.5-mm diameter) in the presence of the proteinase inhibitor phenylmethylsulfonyl fluoride. The method of Bradford (5) was used to determine the protein concentration. Enzyme activity was normalized to the protein concentration in the samples. Six to seven independent measurements of activity were taken from assays of two different transformants, and the mean activities were calculated.
Generation of ppg1-deficient S. macrospora mutants.
A 1,622-bp EcoRI fragment containing the ppg1 coding sequence and its flanking regions was subcloned from cosmid H4 (58) into pBCKS(+) (Stratagene) to obtain plasmid p204-20. The hygromycin phosphotransferase gene (hph) within pCB1003 (8) was isolated as a 1.4-kb SalI fragment and cloned into the SalI cleavage site of p204-20 (see Table 3). This caused the desired disruption of the ORF of the ppg1 gene. The resulting plasmid construct was referred to as p210-1.
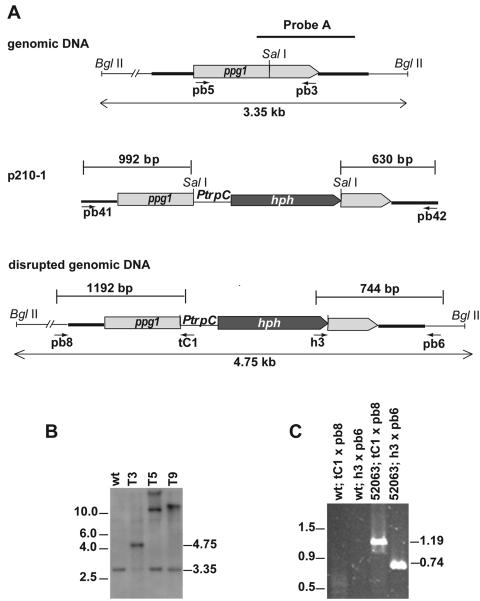
To insert the generated ppg1-hph construct into S. macrospora, a 3.4-kb fragment from p210-1 was amplified by PCR with primers pb41 and pb42 (Table 2). Transformation of S. macrospora was done according to Nowrousian et al. (54). ppg1 mutants were identified by Southern blot analysis. Genomic DNA was isolated as described previously (62). Successful homologous recombination was confirmed by PCR amplification with primers pb8 and tC1 for the 5′-flanking region and primers h3 and pb6 for the 3′-flanking region (see Table 2). Fungal transformants are often heterokaryotic, and mycelia carry transformed and nontransformed nuclei. A single spore isolate of a primary transformant carrying the disrupted ppg1 gene was therefore investigated further.
Immunodetection of secreted EGFP.
EGFP secretion in S. macrospora was put under the control of the signal sequences from S. macrospora genes ppg1 and etp. To achieve this, the signal sequences were fused in the sense and antisense orientations in frame with the egfp gene. Immunodetection of secreted EGFP was performed as previously described (61). As a control, a pSM1 transformant expressing the egfp gene at high levels was used. In the pSM1 transformant, the EGFP protein is not secreted and is therefore located in the cytoplasm. After S. macrospora transformants had grown for 3 days in liquid CM medium, 5 ml of the cell-free medium was removed from the culture, and proteins secreted into the medium were precipitated by adding 10% trichloroacetic acid. Subsequently, precipitated proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis according to standard protocols (45). The proteins were transferred to a polyvinylidene difluoride Western blotting membrane (Biometra), using a semidry blotting system (Biometra). A polyclonal anti-GFP living color peptide antibody (BD Bioscience) (diluted 1:500) was used for the detection of EGFP. The chemiluminescence Western blotting kit (Roche) was used for visualization as described by the manufacturer.
Confocal laser microscopy.
Confocal laser microscopy was carried out on a Zeiss LSM 510 META confocal system version 3.0 (excitation/emission 488 nm blue, 10% argon laser power, Ch2-1, BP505-550 filter, Ch2/72 μm pinhole, Zeiss Axiovert 100 M, Plan Apo 63/1.4 oil lens). Preparation of S. macrospora strains was done as described previously (61).
RESULTS
Functional expression of S. macrospora pheromones and pheromone receptors in S. cerevisiae.
In order to analyze the functionality of the S. macrospora peptide pheromone and the PRE2 pheromone receptor, we heterologously expressed the S. macrospora ppg1 and pre2 genes in S. cerevisiae and tested them for interaction. The coding sequences of the ppg1 and pre2 gene were amplified and put under the control of a constitutive promoter derived from the S. cerevisiae phosphoglycerate kinase (PGK) gene (37), resulting in plasmids pPpg1 and pPre2, respectively. To avoid competition between the S. cerevisiae α-factor receptor Ste2p and the S. macrospora PRE2 receptor, we used strain 7416-12-3 (MATa Δste2 sst2-1), lacking the S. cerevisiae STE2 gene, as a host for expression of the S. macrospora pre2 gene.
In an attempt to optimize the conditions for ppg1 expression and pheromone synthesis in an S. cerevisiae host, the MATα (W303-1B) strain was chosen because its MFα1p and MFα2p precursor proteins are structurally similar to the PPG1 prepro-protein of S. macrospora (58). To measure the activation of the heterologously expressed S. macrospora PRE2 receptor in MATa cells, an agar diffusion bioassay (halo assay) was employed. The assay is based on the fact that pheromone exposure triggers a cell division arrest in S. cerevisiae cells. In S. cerevisiae, binding of a pheromone to its receptor leads to the release of GDP from the G-protein α-subunit Gpa1p, followed by the binding of GTP and the liberation of the G-protein βγ-subunits (Ste4p/Ste18p). The βγ-dimer initiates the mitogen-activated protein kinase cascade, resulting in gene transcription, morphological changes, and a G1 cell cycle arrest (23, 25).
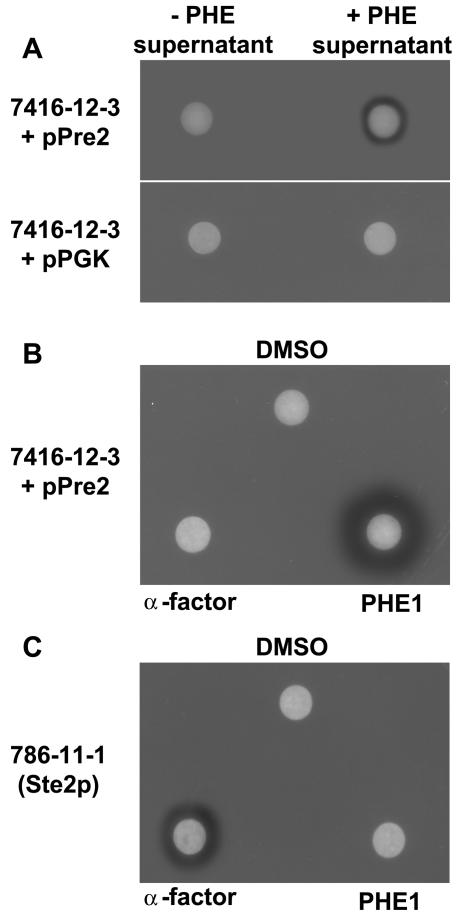
In addition to the ste2 mutation, the yeast MATa strain used in the halo assay also carried the sst2-1 mutation. The S. cerevisiae SST2 gene encodes a regulator of G-protein signaling (RGS) protein, which is crucial for recovery from pheromone-induced cell cycle arrest (22). Mutants lacking SST2 are approximately 100-fold more sensitive to pheromones (24). Supernatants of MATα transformants secreting an S. macrospora peptide pheromone were applied to filter disks and placed on a lawn of MATa cells expressing the S. macrospora pheromone receptor gene pre2 (Fig. 2A). As a control, the supernatant of ppg1-MATα transformants was applied to a MATa lawn containing only the mock vector pPGK. Alternatively, the supernatant of a MATα culture carrying pPGK was applied to a MATa pre2 lawn. Application of supernatants of S. cerevisiae cells secreting an S. macrospora peptide pheromone resulted in a bare zone immediately surrounding the filter disk. In the controls, no halo formation was observed (Fig. 2A). Hence, S. cerevisiae MATα cells expressing the S. macrospora ppg1 pheromone precursor gene must have been able to secrete an active pheromone which promotes cell cycle arrest in S. cerevisiae MATa cells expressing the S. macrospora pre2 gene.
FIG. 2.
Pheromone-induced growth arrest of S. cerevisiae transformants expressing the S. macrospora pre2 pheromone receptor gene. (A) Quantitative halo test of MATa lawns (7416-12-3; Δste2 sst2-1) either expressing the S. macrospora pre2 pheromone receptor gene (+PRE2) or carrying the mock vector pPGK (+pPGK). Sterile filter disks contained 20 μl of cell-free culture supernatant of MATα cells expressing (+PHE) or not expressing (−PHE) the S. macrospora ppg1 gene. (B) Response of MATa lawn (strain 7416-12-3) expressing the S. macrospora PRE2 receptor to a single dose of synthetic pheromone PHE1 (1.5 nmol/disk), α-factor (1.5 nmol/disk), and 2% dimethyl sulfoxide (DMSO; 10 μl/disk). (C) Functionality of α-mating factor was proved on a MATa sst2-1 lawn (strain 786-11-1) as described for B.
Although S. cerevisiae MATα cells seemed capable of processing the S. macrospora PPG1 precursor, the precise nature of the active pheromone remained unknown. So far, no pheromones have been purified from filamentous ascomycetes. However, since the overall structure of the PPG1 protein from S. macrospora resembles that of S. cerevisiae MFα1p and MFα2p with respect to signal sequence, cleavage sites for endopeptidases, dipeptidyl-aminopeptidases, and carboxypeptidases, we predicted that the active S. macrospora pheromone was an undecapeptide, QWCRIHGQSCW (Fig. 1).
To experimentally test the hypothesis that the undecapeptide PHE1 is one active form of an S. macrospora pheromone, the peptide QWCRIHGQSCW was synthesized in vitro; 1.5 nmol was added to an S. cerevisiae MATa Δste2 sst2-1 lawn producing the S. macrospora receptor PRE2 (Fig. 2B). Halo formation occurred in response to the synthetic undecapeptide pheromone. However, no halo formation was seen when synthetic S. cerevisiae α-factor or dimethyl sulfoxide was added in control experiments. As a positive control, we were able to induce halo formation when we applied the synthetic S. cerevisiae mating α-factor to strain 786-11-1 (MATa sst2-1), which produces the S. cerevisiae α-factor receptor Ste2p. Conversely, application of the synthetic PHE1 pheromone to MATa Ste2p cells resulted in no detectable halo (Fig. 2C). Thus, we were able to demonstrate that in S. cerevisiae cells expressing the S. macrospora pre2 gene, specific interaction occurs between the QWCRIHGQSCW undecapeptide PHE1 and the S. macrospora PRE2 receptor.
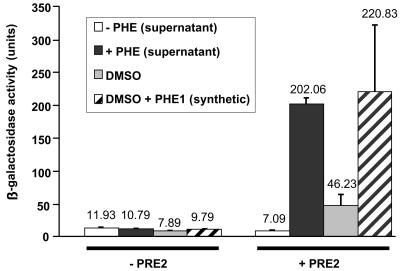
To quantify the extent of pheromone activation on gene expression, we used the pheromone-inducible FUS1-lacZ reporter gene (11, 76). S. cerevisiae cells expressing the S. macrospora receptor gene pre2 and the FUS1-lacZ reporter gene construct were treated with either culture supernatants of MATα cells containing pPpg1 or the control plasmid pPGK, synthetic pheromone, or 2% dimethyl sulfoxide (Fig. 3). The sixfold increase in β-galactosidase activity in the reporter strains after addition of 2% dimethyl sulfoxide might be due to a nonspecific activation of the PRE2 receptor by the solvent. However, a 20-fold increase in β-galactosidase activation in comparison to the negative controls was observed when the supernatant of MATα cells expressing the ppg1-encoded pheromone or the synthetic peptide QWCRIHGQSCW was added to the cells. This result suggests that the heterologous S. macrospora receptor PRE2 is capable of triggering the S. cerevisiae pheromone response pathway when activated by the synthetic peptide or the S. cerevisiae supernatant.
FIG. 3.
Activation of an inducible S. cerevisiae gene by the S. macrospora pheromone-receptor pair. Response to culture supernatants or synthetic pheromone was tested with a MATa Δste2 sst2-1 S. cerevisiae strain containing the pheromone-inducible FUS1-lacZ reporter plasmid and either pPre2 (+PRE2) or pPGK (−PRE2). Cell-free supernatants were collected from MATα transformants (W303-1B) secreting the S. macrospora peptide pheromone (black bar) or from MATα control transformants carrying pPGK (white bar). In addition, 1.5 nmol of synthetic PHE1 pheromone (striped bar) or an equal volume of the solvent dimethyl sulfoxide (DMSO) was used (grey bar). β-Galactosidase activity was assayed to assess pheromone response. Activities shown are averages from seven independent measurements taken from assays of two different transformants. Error bars are given as indicated.
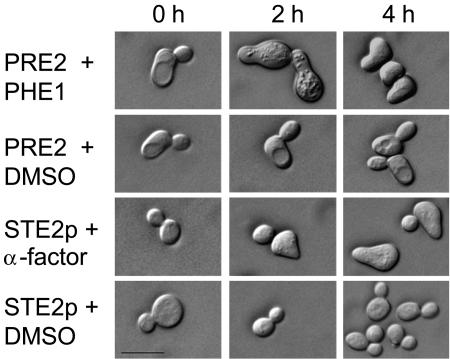
As a response to pheromone interaction, S. cerevisiae shows cell cycle arrest, pheromone-induced gene expression, and morphological changes. Cells exhibit polarized growth towards the mating partner, leading to pear-shaped forms (shmoos) of unconjugated haploid cells (19). Treatment of MATa Δste2 sst2-1 cells expressing the S. macrospora receptor PRE2 with synthetic pheromone for 2 h produced the characteristic morphological changes. The same extent of morphological changes was observed in the control experiments, where MATa sst2-1 cells (strain 786-11-1) carrying the endogenous Ste2p receptor were exposed to synthetic α-factor (Fig. 4). In summary, we have shown that the S. macrospora pre2 gene, when expressed in an S. cerevisiae MATa Δste2 mutant, can initiate the pheromone response pathway leading to cell cycle arrest, transcriptional activation of genes, and shmoo formation in the presence of S. macrospora peptide pheromone.
FIG. 4.
Shmooing of S. cerevisiae in response to the synthetic pheromone PHE1 and to S. cerevisiae α-factor. MATa Δste2 sst2-1 cells expressing the S. macrospora pre2 gene were treated for 0, 2, or 4 h with either synthetic pheromone PHE1 at 5 μM or dimethyl sulfoxide. As a control, a MATa sst2-1 strain was treated with synthetic α-factor or with dimethyl sulfoxide. Bar, 10 μm.
An α-factor-like pheromone is secreted by S. macrospora.
The secretion of diffusible peptide pheromones, which are recognized by pheromone receptors on the cell surface of a mating partner with an opposite mating type, initiates mating in heterothallic ascomycetes. In order to assess whether the homothallic fungus S. macrospora secretes a peptide pheromone into the extracellular medium and whether the ppg1-encoded pheromone was responsible for pheromone secretion, we disrupted the coding region of the ppg1 gene of S. macrospora by introducing a hygromycin resistance cassette (hph) by homologous recombination. The homologous recombination was confirmed by Southern blot and PCR analysis (Fig. 5). Phenotypic inspection of independent single-spore isolates revealed no differences from the wild type with respect to the timing of sexual development or the morphology and number of perithecia, asci, or ascospores.
FIG. 5.
Disruption of the S. macrospora ppg1 gene. (A) Structure of the ppg1 genomic region and construction of the ppg1::hph disruption strain. The positions of primers used to amplify the disruption construct from plasmid p210-1 and verify homologous recombination at the ppg1 locus are indicated. Ptrpc, Aspergillus nidulans trpC promoter. (B) Southern analysis. Genomic DNA from wild-type S. macrospora (wt) and three transformants (T3, T5, and T9) was digested with BglII, separated on a 1% agarose gel, blotted, and hybridized with the 1.2-kb 32P-labeled probe indicated in A. (C) PCR analysis for verification of homologous recombination from a single spore isolate (strain S52063) of transformant T3 and the wild type. The positions of primers are indicated in A.
Application of the cell-free culture supernatant of an S. macrospora wild-type strain onto the S. cerevisiae MATa lawn expressing PRE2 resulted in cell cycle arrest, proving that the culture medium of the wild-type S. macrospora was able to induce the pheromone response pathway in S. cerevisiae. However, when we applied the culture medium of the ppg1 disruption mutant onto the S. cerevisiae lawn expressing the pre2 gene, no halo was formed (Fig. 6A). Thus, the halo formation observed with the culture medium of the S. macrospora wild-type strain was due to the expression of the ppg1 gene and secretion of the encoded pheromone.
FIG. 6.
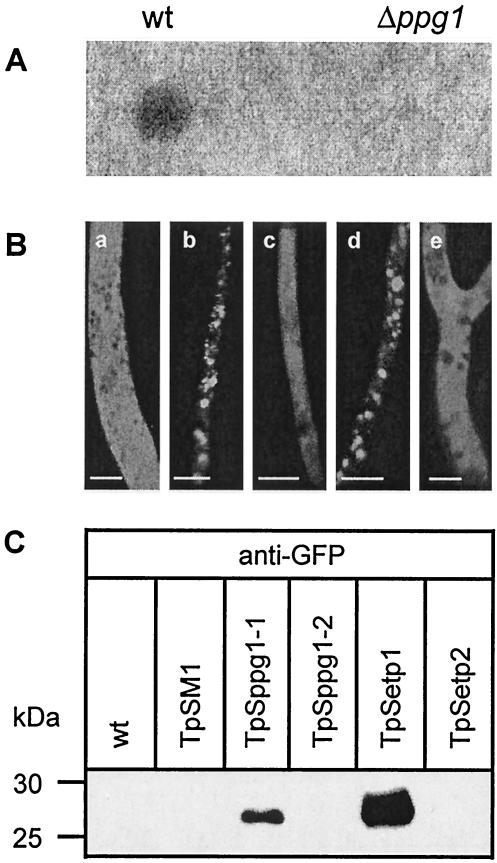
ppg1-encoded peptide pheromone of S. macrospora is secreted into the culture medium. (A) Halo assays with fungal culture supernatants. To measure the peptide pheromone which is secreted into the culture medium, 20 μl of culture medium of wild-type S. macrospora and an S. macrospora ppg1::hph disruption mutant (strain S52063) was spotted on an S. cerevisiae tester lawn (MATa Δste2 sst2-1) expressing the S. macrospora pheromone receptor PRE2. Analysis of halo formation was performed after 24 to 48 h of incubation of plates at 30°C. (B) Analysis of EGFP secretion mediated by N-terminal signal sequences of PPG1. Fluorescence microscopy. Hyphae of S. macrospora transformants carrying either pSM1 (a), pSppg1-1 (b), pSppg1-2 (c), pSetp1 (d), or pSetp2 (e) were analyzed by fluorescence microscopy. Scale bars represent 5 μm. (C) Western blot. Equal amounts of extracellular cell-free culture medium were analyzed from wild-type S. macrospora (wt) and transformants containing either pSM1 (TpSM1), pSppg1-1 (TpSppg1-1), pSppg1-2 (TpSppg1-2), pSetp1 (TpSetp1), or pSetp2 (TpSetp2). Culture medium was fractionated on a denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis and blotted on a polyvinylidene difluoride membrane. Western blot detection of EGFP protein was carried out with a polyclonal GFP antibody. Size markers were low-range polypeptides (Bio-Rad), and their sizes are indicated at the left.
The 16 N-terminal residues of the pheromone precursor PPG1 of S. macrospora constitute a hydrophobic region that may act as a signal sequence during translocation of the precursor across the membrane of the endoplasmic reticulum (58). This is the critical initial step of protein secretion, which is then followed by sorting into the Golgi network (15). To analyze whether the predicted signal sequence alone is able to mediate secretion, we fused the coding sequence of the PPG1 signal sequence in frame to an egfp reporter gene.
On the basis of plasmid p82.9, we constructed plasmids pSppg1-1 and pSppg1-2. Plasmid pSppg1-1 contains the signal sequence in the sense orientation. As a control, it was introduced in the antisense orientation into pSppg1-2. Both plasmids were transformed into wild-type S. macrospora, and transformants were analyzed for EGFP fluorescence (Fig. 6B). Microscopic investigation revealed a different localization of EGFP in transformants containing pSppg1-1 than in transformants containing pSppg1-2. In transformants expressing the PPG1 signal sequence in the sense orientation, the GFP fluorescence appeared to be distributed in distinct patches in the fungal hyphae, suggesting localization within cytoplasmic vesicles. Transformants carrying pSetp1 were analyzed as a control. The plasmid pSetp1 contains the signal sequence of the S. macrospora etp gene fused to the egfp gene. Previously it has been shown that the signal sequence of the etp gene can mediate the secretion of EGFP in S. macrospora (61). As can be seen from Fig. 6B, transformants containing pSetp1 show the same localization of EGFP within cytoplasmic vesicles as pSppg1-1 transformants. In contrast, EGFP was distributed throughout the cytoplasm in transformants containing the PPG1 signal sequence in the antisense orientation (TpSppg1-2). A similar pattern was observed for pSetp2 transformants, which carry an antisense ETP signal sequence fused to EGFP and for pSM1 transformants expressing the egfp gene.
Extracellular culture broth of S. macrospora transformants was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting with a polyclonal antibody against GFP in order to prove the presence of EGFP in the culture medium (Fig. 6C). The 27-kDa EGFP polypeptide was detected in the culture medium of pSppg1-1 and pSetp1 transformants. No signal was detected in culture supernatants of wild-type, pSM1, pSppg1-2, and pSetp2 transformants. Thus, the signal sequence derived from PPG1 was shown to be sufficient to mediate secretion of EGFP.
S. cerevisiae as a powerful screening system for the identification of pheromone mutants of S. macrospora.
In heterothallic species, mutations conferring male and/or female sterility can be detected directly because of their sterility effects in heterozygous crosses. However, recessive mutations that effect postfertilization fruiting-body development will remain undetected in heterothallic species until the mutant allele is available in both mating types, thus allowing homozygous crosses. In the homothallic S. macrospora, recessive mutations can be tested directly for defects in fruiting-body development. Therefore, S. macrospora was used to generate numerous mutants which are blocked at various stages of sexual development (28, 49, 50). These mutants were divided into four main groups: mutants displaying defects in ascogonium development, called asc mutants; mutants affected in early fruiting-body development, named pro (protoperithecia) mutants; mutants that form perithecia but no ascospores, termed per (perithecia) mutants; and mutants generating malformed and piled fruiting bodies, termed pile mutants (49). Using the halo assay, we have been able to demonstrate that the wild-type strain of S. macrospora does secrete a peptide pheromone.
Since pheromones are supposed to be involved in postfertilization events and fruiting-body development (18), we wanted to test if this assay may be suitable for analyzing pheromone production in the S. macrospora fruiting-body mutants. Three S. macrospora asc mutants, 37 pro mutants, 36 per mutants, and 5 pile mutants were screened by the halo assay. A lawn of S. cerevisiae strain 7416-12-3 (MATa Δste2 sst2-1) expressing the S. macrospora pre2 pheromone receptor gene was used. For each of the 81 S. macrospora mutants tested, 20 μl of culture medium was applied to an individual S. cerevisiae lawn. The culture medium of 54 mutants caused halo formation, which was approximately the same size as the halo seen with the culture medium of wild-type S. macrospora (Table 4). However, a change in production or secretion of the peptide pheromone was seen in several mutants. Compared to the halo formed with culture medium from wild-type S. macrospora, the culture medium of 12 mutants caused larger halos. In 15 cases, smaller halos were observed (Table 4).
TABLE 4.
Halo screening of developmental mutantsa
| Mutant type | No. screened | Halo size
|
||
|---|---|---|---|---|
| Wild type | > Wild type | < Wild type | ||
| asc | 3 | 2 | 1 | |
| pro | 37 | 25 | 5 | 7 |
| per | 36 | 22 | 7 | 7 |
| pile | 5 | 5 | ||
Halo sizes were larger than, smaller than, or comparable to the halo size caused by wild-type S. macrospora.
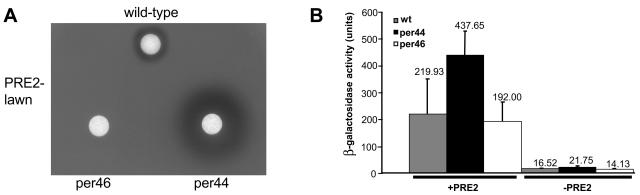
As an example, mutant per44, showing a broader halo, and mutant per46, showing no halo formation, were analyzed in more detail (Fig. 7A). The culture supernatants of both S. macrospora mutants were subsequently tested for their ability to activate the pheromone-inducible FUS1-lacZ reporter gene. In comparison to the wild-type culture medium, only a slight decrease in the level of reporter gene induction was seen with the supernatant of mutant per46. The application of supernatant of mutant per44 caused a twofold increase in FUS1-lacZ activation levels compared to the wild-type level. Activities were reduced to background level if the same supernatants were added to S. cerevisiae control strains lacking the S. macrospora PRE2 receptor (Fig. 7B). Thus, the heterologous S. cerevisiae system proved useful in identifying S. macrospora mutants affected in ppg1 gene expression.
FIG. 7.
Halo assays and FUS1-lacZ induction assays with cell-free culture medium of wild-type S. macrospora and developmental mutants per44 and per46. (A) Culture medium (20 μl) of wild-type S. macrospora and mutant strains per44 and per46 were applied on an S. cerevisiae tester lawn (MATa Δste2 sst2-1) expressing the S. macrospora pheromone receptor PRE2. (B) For S. cerevisiae FUS1-lacZ induction assays, culture medium of wild-type S. macrospora (grey bar) and mutant strains per44 (black bar) and per46 (white bar) was taken after growth of strains in liquid CM medium. Pheromone response to the culture medium was tested with a MATa Δste2 sst2-1 S. cerevisiae strain carrying the pheromone-inducible FUS1-lacZ reporter plasmid and either pPre2 (+PRE2), encoding the S. macrospora PRE2 receptor, or the mock vector pPGK (−PRE2); 10 ml of culture medium of S. macrospora strains was mixed with an equal volume of pre2-expressing S. cerevisiae cells. β-Galactosidase activity was assayed, and activities shown are averages of six independent measurements taken from assays of two different transformants. Error bars are given as indicated.
DISCUSSION
Interactions of pheromones and receptors of the homothallic filamentous ascomycete S. macrospora can be reconstituted in the yeast S. cerevisiae.
Despite being self-fertile, the filamentous ascomycete S. macrospora does carry and express two pheromone precursor genes and two pheromone receptor genes (58, 59). We expressed the α-factor-like pheromone precursor PPG1 and its cognate receptor PRE2 in the yeast S. cerevisiae to provide experimental evidence that the pheromones and receptors of S. macrospora are truly functional.
This study demonstrated that the S. macrospora pheromone precursor gene ppg1 and the pheromone receptor gene pre2 are functionally homologous to the S. cerevisiae MFα1 and MFα2 pheromone genes and the STE2 receptor gene, respectively. Despite the fact that the amino acid sequence of the S. macrospora PRE2 receptor shows limited similarity to the S. cerevisiae Ste2p α-factor receptor sequence (23.0% identity over 274 amino acids), PRE2 does act as a pheromone receptor in an ste2-deficient S. cerevisiae mutant (MATa Δste2 sst2-1). Activation of the S. cerevisiae pheromone response pathway through heterologously expressed receptors has so far been demonstrated for a number of mammalian G-protein-coupled receptors (26, 41, 63, 66, 75) and for a-factor-like pheromone receptors from basidiomycetes (29, 33, 57).
α-factor, which activates the Ste2p receptor in S. cerevisiae, is a 13-amino-acid peptide. It is processed from larger polypeptide precursors containing several copies of the active tridecapeptide. Similarly, an active S. macrospora peptide pheromone is thought to be an undecapeptide derived from the PPG1 precursor. This precursor contains five copies of the amino acid sequence QWCRIHGQSCW (58). In this work we demonstrated that a peptide pheromone secreted by S. cerevisiae MATα cells expressing the ppg1 gene interacts with the PRE2 receptor. So far, only a-factor-like pheromones of the basidiomycetes Coprinus cinereus and Schizophyllum commune have been heterologously expressed in S. cerevisiae (29, 57). Furthermore, we were able to show that the synthetic undecapeptide PHE1 (QWCRIHGQSCW) was biologically active when tested in the S. cerevisiae system, suggesting that the synthetic peptide might represent one active form of the S. macrospora peptide pheromone. The lowest concentration of synthetic PHE1 to cause visible halo formation was found to be 1.5 pmol.
The size of the halo formed when synthetic PHE1 was added to a MATa Δste2-PRE2 lawn was compared to the halo seen in a MATa lawn treated with synthetic α-factor. PHE1 seemed to have a more profound effect than the α-factor (Fig. 2B and C). One possible explanation for this is the presence of the extracellular protease Bar1p in S. cerevisiae cells. The enzyme encoded by the BAR1 gene cleaves S. cerevisiae α-factor between leucine 6 and lysine 7, rendering it nonfunctional. The enzymatic degradation of the α-factor enables the target cells to recover from arrest in the G1 phase of the cell cycle (13, 48). The PHE1 pheromone does not contain leucine or lysine residues. This may protect it from enzymatic cleavage, making it more stable than S. cerevisiae α-factor.
Activation of the heterologously expressed PRE2 receptor by synthetic pheromone PHE1 or an S. macrospora peptide pheromone secreted from MATα cells initiates the S. cerevisiae pheromone response pathway, resulting in characteristic changes. These include cell cycle arrest, gene activation, and the formation of one or more projections on the cell surface, commonly termed shmoo morphology (Fig. 2 to 4). It has previously been shown that pheromone receptors of basidiomycetes heterologously expressed in an S. cerevisiae host can initiate gene expression and cell cycle arrest in response to their cognate pheromone (29, 33, 57). However, formation of the shmoo morphology upon pheromone application has not been observed so far in experiments expressing basidiomycete receptors in S. cerevisiae (29). Dose-response curves have been determined for each of the three responses as a function of α-factor concentration in S. cerevisiae. The analysis revealed that approximately 100-fold-higher concentrations of pheromones are required for the induction of shmoo formation than for transcriptional activation or cell cycle arrest (52). In contrast to the S. commune receptor, the S. macrospora PRE2 receptor is able to mediate all pheromone responses when expressed in S. cerevisiae (29).
Since, in S. cerevisiae, PRE2 was activated by extracellular pheromones and was able to trigger the pheromone response, it seems likely that PRE2 has been incorporated into the plasma membrane and might interact with the Gpa1p protein of S. cerevisiae. This hypothesis is supported by the fact that the MATa Δste2 strain used in our analysis carried a mutation in the SST2 gene. Sst2p acts in desensitization to pheromone through negatively regulating the S. cerevisiae Gα-subunit Gpa1p. S. cerevisiae sst2-1 mutants are therefore at least 100-fold more sensitive to pheromones and fail to recover from pheromone-induced cell cycle arrest (10, 22, 24). Accordingly, increased sensitivity to pheromone was present in Sst2p-deficient MATa Δste2 cells expressing PRE2, but PHE1 never caused cell cycle arrest in wild-type SST2 strains (our unpublished results). The third intracellular loop of the S. cerevisiae Ste2p receptor is important for G-protein activation (14). However, no homologies in the amino acid sequences of Ste2p and PRE2 were found in the region predicted to form the third intracellular loop (59). Thus, properties other than sequence homology have to account for the ability of PRE2 and the S. cerevisiae G-protein to interact.
The importance of heterotrimeric G-proteins during fertilization and sexual development has been demonstrated for the heterothallic fungus Neurospora crassa, which is closely related to S. macrospora (1, 34, 38, 80). The N. crassa a-factor-like pheromone receptor PRE1 is thought to couple to the G-protein α-subunit GNA-1 of the heterotrimeric G-protein, thereby transferring the pheromone signal (39). A gene encoding a protein similar to the GNA-1 subunit of N. crassa has been identified in S. macrospora, and the PRE2 receptor may interact with this protein in a similar way as hypothesized for N. crassa. In addition to a gna-1 homologue, several genes encoding putative downstream components of the pheromone signal transduction cascade, including genes coding for subunits of G-proteins, RGS proteins, mitogen-activated protein kinase cascade components, and transcription factors have been isolated from S. macrospora (Table 5) (56). These findings suggest not only that pheromones and pheromone receptors are similar in structure and functionality, but also that the signal transduction pathways in S. macrospora, N. crassa, and S. cerevisiae are highly conserved.
TABLE 5.
Putative S. macrospora homologues of S. cerevisiae genes that encode components of the pheromone response pathway and processing of α-factora
| Functionb | S. cerevisiae gene; ORF name | Putative S. macrospora homologue (accession no. of partial gene sequence); E value |
|---|---|---|
| G-protein α subunit | GPA1; YHR005C | gna-1 (AJ879481); 5.2e-38 |
| G-protein β subunit | STE4; YOR212W | gnb-1 (AJ879478); 2.8e-33 |
| Serine threonine kinase | STE20; YHL007C | ste20 (AJ879480); 3.2e-26 |
| MAPKKK | STE11; YLR362W | nrc-1 (AJ879482); 1.3e-19 |
| MAPKK | STE7; YDL159W | ste7 (AJ879473); 4.1e-33 |
| MAPK | FUS3; YBL016W | fus3 (AJ879483); 3.4e-62 |
| Transcription factor | STE12; YHR08W | ste12 (AJ879472); 1.3e-43 |
| Regulator of G-protein signaling | SST2; YLR452C | rgs1 (AJ879475); 2.8e-17 |
| Protein tyrosine phosphatase; adaptation to pheromone | PTP3; YER075C | ptp3 (AJ879476); 2e-6 |
| Protein tyrosine phosphatase; adaptation to pheromone | MSG5; YNL053W | msg5 (AJ879477); 3.7e-15 |
| Carboxypeptidase; α-factor processing | KEX1; YGL203C | kex1 (AJ879474); 2.2e-59 |
| Subtilisin-like protease; α-factor processing | KEX2; YNL238W | kex2 (AJ879479); 9.3e-81 |
Partial gene sequences from S. macrospora genomic DNA were compared with S. cerevisiae ORF proteins (using BLASTX) available through the Saccharomyces genome database ( http://www.yeastgenome.org/). E (expected) values of S. macrospora partial ORFs are indicated with respect to the corresponding S. cerevisiae protein.
MAPK(KK), mitogen-activated protein kinase (kinase kinase).
S. macrospora secretes a peptide pheromone that activates the pheromone response pathway of MATa cells expressing the pre2 receptor gene.
Halo assays revealed that cell-free culture medium of S. macrospora wild-type strains contained components which can induce cell cycle arrest in MATa cells expressing the S. macrospora PRE2 receptor. It was previously demonstrated that the growth medium of the basidiomycete Schizophyllum commune applied to S. cerevisiae cells expressing a Schizophyllum commune receptor can activate expression of a FUS1-lacZ reporter gene (33). This suggests that filamentous fungi secrete pheromones into the culture medium.
The SignalP version 1.1 program (53) predicted a hydrophobic secretion signal in the N terminus of the S. macrospora PPG1 protein, which was expected to be cleaved between amino acids 16 and 17 (Fig. 1). The signal sequence of the S. cerevisiae α-factor precursor MFα1p has been used to manipulate secretion of heterologous proteins in S. cerevisiae for several years (6). The GFP fluorescence of transformants carrying the PPG1 signal sequence-egfp sense construct appeared as distinct patches distributed along the fungal hyphae, suggesting localization within cytoplasmic vesicles. Fluorescence signals in PPG1 signal sequence-egfp antisense transformants were distributed evenly throughout the cytoplasm (Fig. 6B). By means of Western blot analysis, we demonstrated that the PPG1 signal peptide carries sufficient information to enable EGFP to pass through the secretory pathway and to be released into the culture medium.
Using PCR, we identified the sequences of putative homologues of S. cerevisiae genes encoding the α-factor precursor processing enzymes Kex2p and Kex1p in the S. macrospora genome (Table 5). Reverse transcription-PCR and Northern blot analysis confirmed that these genes are transcriptionally expressed in S. macrospora (data not shown). Thus, processing of the PPG1 precursor in S. macrospora may follow the same pathway as described in S. cerevisiae. According to our findings, we postulate that the undecapeptide QWCRIHGQSCW most likely represents one active form of an α-factor-like pheromone in S. macrospora because it can activate the PRE2 receptor in the heterologous S. cerevisiae system. However, based on our data, we cannot exclude the possibility that longer or shorter forms of the peptide pheromone exert a pheromone function in S. macrospora or that the active peptide in the S. cerevisiae assay may have a different structure than the native, mature pheromone produced in S. macrospora.
Disruption of the S. macrospora ppg1 gene prevents secretion of the peptide pheromone but does not affect sexual development.
After disrupting the ppg1 gene, we were no longer able to induce halo formation in an S. cerevisiae MATa lawn expressing the pre2 gene when we added culture medium from the S. macrospora ppg1 mutant. Thus, we proved that disruption of the ppg1 gene resulted in a defect of peptide pheromone production and secretion. However, no other phenotypic changes have been observed in the S. macrospora ppg1 mutant. Despite lacking a functional ppg1 gene, the S. macrospora ppg1 mutant was able to develop fruiting bodies, asci, and ascospores, and its vegetative growth was not affected.
The homothallic ascomycete S. macrospora expresses both pheromone genes throughout its development, whereas heterothallic ascomycetes express pheromone genes predominantly in conidia, and expression occurs in a mating type-specific manner (3, 17, 58, 69, 81). Deletion of pheromone genes in the heterothallic ascomycetes Podospora anserina and Cryphonectria parasitica revealed that pheromones play an essential role in promoting fertilization. Strains carrying these mutations are male sterile because their conidia are unable to attract and mate with cells of the opposite mating type. In P. anserina, deletion of pheromone genes affects male fertility without impairing vegetative growth or postfertilization events (17). The same observation was made in C. parasitica, where the Mf1-1 gene encoding the α-factor-like pheromone was deleted by gene replacement (77). Our observations in S. macrospora were very similar, even though defects in male sterility could not be demonstrated because S. macrospora does not produce conidia. On the contrary, deletion of one of the two copies of the C. parasitica gene encoding the a-factor-like pheromone resulted in a pleiotropic phenotype. Mf2-2 mutants display reduced sexual reproduction. A cross between an Mf2-2 mutant (as the female) and a wild-type strain (as the male) produced only barren perithecia (82). It was therefore assumed that Mf2-2 of C. parasitica is required during a developmental phase after fertilization and that the CaaX-type pheromone acts in a dosage-specific manner in postfertilization events (77). Similarly, N. crassa mfa-1 mutants in which the critical cysteine residue of the open reading frame had been changed to the nonprenylatable residue tyrosine (YAAX mutants), as well as mutants with intact open reading frames but multiple mutations in the 3′ noncoding region (CAAX mutants), were shown to display delayed and reduced vegetative growth and aberrant sexual development. Surprisingly, this pleiotropic phenotype was observed in both mating types. It was therefore postulated that the lipopeptide pheromone of N. crassa may have an additional role in cementing hyphae together to stabilize the structure of the perithecium (40).
Northern blotting and reverse transcription-PCR analysis indicated that, in N. crassa, in contrast to pheromone precursor genes, expression of the receptor genes does not occur in a mating type-specific manner (39, 59). Deletion of the N. crassa pre-1 gene encoding the a-factor-like receptor does not affect vegetative growth or male fertility. On the other hand, Δpre1 matA mutants are sterile females, because their trichogynes are unable to recognize and fuse with mata conidia (39). Deletion of two putative pheromone receptor genes, gprA and gprB, in the homothallic filamentous ascomycete Aspergillus nidulans resulted in the production of fewer and smaller fruiting bodies carrying a reduced number of ascospores. Fruiting-body formation in homothallic conditions was absent in the ΔgprA ΔgprB double mutant (68). This result implies that a pheromone-receptor system is essential for postfertilization events during the sexual development of homothallic ascomycetes. It has to be expected that the second active pheromone-receptor pair (PPG2 and PRE1) of S. macrospora is able to functionally complement the disruption of the ppg1 gene in S. macrospora. This would explain why no defects in sexual development were detected in the S. macrospora ppg1 mutant.
The S. cerevisiae system provides a tool to measure pheromone secretion in developmental mutants of S. macrospora.
Expressing the functional S. macrospora receptor PRE2 in S. cerevisiae, we developed a powerful tool for studying pheromone secretion in developmental mutants of S. macrospora. Of 81 developmental mutants tested in this work, 27 secreted a different amount of peptide pheromone than the wild type (Table 4). The culture medium of these 27 strains led to the formation of either smaller or larger halos. The halo assay results for two mutants, per44 (larger halo) and per46 (no halo), were confirmed by testing their culture medium for activation of the pheromone-inducible FUS1-lacZ reporter (Fig. 7). Induction of the reporter gene was only slightly decreased with the supernatant of mutant per46. We detected a twofold increase in the induction of FUS1-lacZ with the supernatant of mutant per44. The FUS1-lacZ reporter assay was therefore more sensitive than the halo assay. Northern blot analysis showed that transcriptional expression of ppg1 was elevated in mutant per44, while in per46 the transcript level was only slightly decreased (our unpublished results). Currently we do not know which genes are defective in mutants per44 and per46.
However, four of the mutants screened (pro1, pro1, pro22, and per5) have previously been complemented by transformation with an indexed cosmid library. The pro1 gene encodes a transcription factor, and the pro22 gene codes for a putative membrane protein (50, 55). The PRO11 protein is a membrane-associated protein and thought to be a scaffolding protein in several signal transduction pathways (60). The defect in mutant per5 lies in the acl1 gene, encoding a subunit of an ATP-citrate lyase (54). The culture medium of per5 led to wild-type-like halo formation in the S. cerevisiae assay, whereas the culture medium of pro1, pro11, and pro22 caused increased halo sizes. In this context, it is of interest that interspecies microarray hybridization of N. crassa with targets of S. macrospora mutants pro1, pro11, and pro22 revealed that the pheromone precursor gene ppg1 is among the genes which are upregulated more than twofold in these mutants (55). These findings indicate that alterations in pheromone gene expression may be involved in the sexual development of homothallic ascomycetes.
The heterologous S. cerevisiae system offers an easy and valuable tool to identify mutants carrying a defect in ppg1 gene expression and pheromone secretion pathways. Further analysis of these mutants will help us to establish the way in which pheromones are processed in filamentous ascomycetes and how pheromones are involved in postfertilization events of filamentous ascomycetes.
Acknowledgments
We thank Silke Nimtz, Gisela Isowitz-Seidel, and Susanne Schlewinski for excellent technical assistance, Christian Würtz for help with some experiments, Ulrich Kück (Ruhr-University Bochum) for providing S. macrospora mutant strains and laboratory resources, and Carsten Theiss and Hans-Georg Mannherz (Ruhr-University Bochum Medical Faculty) for their support in confocal microscopy. The S. cerevisiae expression vectors and S. cerevisiae strains were kindly provided by Thomas Fowler (Vermont) and Duane Jenness (Worcester, Mass.). We thank Dörte and Martin Wren for help in manuscript preparation.
This work was funded by the Deutsche Forschungsgemeinschaft SFB480 (Bonn, Germany) and the Ruhr-University of Bochum.
REFERENCES
- 1.Baasiri, R. A., X. Lu, P. S. Rowley, G. E. Turner, and K. A. Borkovich. 1997. Overlapping functions for two G protein alpha subunits in Neurospora crassa. Genetics 147:137-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker, D. M., and V. Lundblad. 1994. Introduction of DNA into yeast cells, p. 13.7.1-13.7.10. In K. Struhl (ed.), Current protocols in molecular biology. Wiley, New York, N.Y. [DOI] [PubMed]
- 3.Bobrowicz, P., R. Pawlak, A. Correa, and D. J. Ebbole. 2002. The Neurospora crassa pheromone precursor genes are regulated by the mating type locus and the circadian clock. Mol. Microbiol. 45:759-804. [DOI] [PubMed] [Google Scholar]
- 4.Bölker, M., and R. Kahmann. 1993. Sexual pheromones and mating responses in fungi. Plant Cell. 5:1461-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Brake, A. J. 1990. Alpha-factor leader-directed secretion of heterologous proteins from yeast. Methods Enzymol. 185:408-421. [DOI] [PubMed] [Google Scholar]
- 7.Caldwell, G. A., F. Naider, and J. M. Becker. 1995. Fungal lipopeptide mating pheromones: a model system for the study of protein prenylation. Microbiol. Rev. 59:406-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carroll, A. M., Sweigard, J. A., and B. Valent. 1994. Improved vectors for selecting resistance to hygromycin. Fungal Genet. Newsl. 41:22. [Google Scholar]
- 9.Casselton, L. A. 2002. Mate recognition in fungi. Heredity 88:142-147. [DOI] [PubMed] [Google Scholar]
- 10.Chan, R. K., and C. A. Otte. 1982. Isolation and genetic analysis of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a-factor and α-factor pheromones. Mol. Cell. Biol. 2:11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, T., and J. Kurjan. 1997. Saccharomyces cerevisiae Mpt5p interacts with Sst2p and plays roles in pheromone sensitivity and recovery from pheromone arrest. Mol. Cell. Biol. 17:3429-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung, S., M. Karos, Y. C. Chang, J. Lukszo, B. L. Wickes, and K. J. Kwon-Chung. 2002. Molecular analysis of CPRalpha, a MATalpha-specific pheromone receptor gene of Cryptococcus neoformans. Eukaryot. Cell 1:432-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ciejek, E., and J. Thorner. 1979. Recovery of S. cerevisiae a cells from G1 arrest by alpha factor pheromone requires endopeptidase action. Cell 18:623-635. [DOI] [PubMed] [Google Scholar]
- 14.Clark, C. D., T. Palzkill, and D. Botstein. 1994. Systematic mutagenesis of the yeast mating pheromone receptor third intracellular loop. J. Biol. Chem. 269:8831-8841. [PubMed] [Google Scholar]
- 15.Conesa, A., P. J. Punt, N. van Luijk, and C. A. van den Hondel. 2001. The secretion pathway in filamentous fungi: a biotechnological view. Fungal Genet. Biol. 33:155-171. [DOI] [PubMed] [Google Scholar]
- 16.Cooper, A., and H. Bussey. 1989. Characterization of the yeast KEX1 gene product: a carboxypeptidase involved in processing secreted precursor proteins. Mol. Cell. Biol. 9:2706-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coppin, E., C. de Renty, and R. Debuchy. 2005. The function of the coding sequences for the putative pheromone precursors in Podospora anserina is restricted to fertilization. Eukaryot. Cell 4:407-420. [DOI] [PMC free article] [PubMed]
- 18.Coppin, E., R. Debuchy, S. Arnaise, and M. Picard. 1997. Mating types and sexual development in filamentous ascomycetes. Microbiol. Mol. Biol. Rev. 61:411-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cross, F., L. H. Hartwell, C. Jackson, and J. B. Konopka. 1988. Conjugation in Saccharomyces cerevisiae. Annu. Rev. Cell Biol. 4:429-457. [DOI] [PubMed] [Google Scholar]
- 20.Davidson, R. C., T. D. Moore, A. R. Odom, and J. Heitman. 2000. Characterization of the MFalpha pheromone of the human fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 38:1017-1026. [DOI] [PubMed] [Google Scholar]
- 21.Debuchy, R. 1999. Internuclear recognition: A possible connection between euascomycetes and homobasidiomycetes. Fungal Genet. Biol. 27:218-223. [DOI] [PubMed] [Google Scholar]
- 22.Dietzel, C., and J. Kurjan. 1987. Pheromonal regulation and sequence of the Saccharomyces cerevisiae SST2 gene: a model for desensitization to pheromone. Mol. Cell. Biol. 7:4169-4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dohlman, H. G. 2002. G proteins and pheromone signaling. Annu. Rev. Physiol. 64:129-152. [DOI] [PubMed] [Google Scholar]
- 24.Dohlman, H. G., J. Song, D. Ma, W. E. Courchesne, and J. Thorner. 1996. Sst2, a negative regulator of pheromone signaling in the yeast Saccharomyces cerevisiae: expression, localization, and genetic interaction and physical association with Gpa1 (the G-protein alpha subunit). Mol. Cell. Biol. 16:5194-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dohlmann, H. G., and J. W. Thorner. 2001. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu. Rev. Biochem. 70:703-754. [DOI] [PubMed] [Google Scholar]
- 26.Dowell, S. J., and A. J. Brown. 2002. Yeast assays for G-protein-coupled receptors. Receptors Channels 8:343-352. [PubMed] [Google Scholar]
- 27.Esser, K. 1982. Cryptogams—cyanobacteria, algae, fungi, lichens. Cambridge University Press, London, England.
- 28.Esser, K., and J. Straub. 1958. Genetische Untersuchungen an Sordaria macrospora Auersw.: Kompensation und Induktion bei genbedingten Entwicklungsdefekten. Z. Vererbungslehre. 89:729-746. [PubMed] [Google Scholar]
- 29.Fowler, T. J., S. M. DeSimone, M. F. Mitton, J. Kurjan, and C. A. Raper. 1999. Multiple sex pheromones and receptors of a mushroom-producing fungus elicit mating in yeast. Mol. Biol. Cell. 10:2559-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greener, A. 1990. New competent cells for highest transformation efficiencies. Strategies 3:5-6. [Google Scholar]
- 31.Hartwell, L. H. 1980. Mutants of Saccharomyces cerevisiae unresponsive to cell division control by polypeptide mating hormone. J. Cell Biol. 85:811-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hasson, M. S., D. Blinder, J. Thorner, and D. D. Jenness. 1994. Mutational activation of the STE5 gene product bypasses the requirement for G protein β and γ subunits in the yeast pheromone response pathway. Mol. Cell. Biol. 14:1054-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hegner, J., C. Siebert-Bartholmei, and E. Kothe. 1999. Ligand recognition in multiallelic pheromone receptors from the basidiomycete Schizophyllum commune studied in yeast. Fungal Genet. Biol. 26:190-197. [DOI] [PubMed] [Google Scholar]
- 34.Ivey, F. D., P. N. Hodge, G. E. Turner, and K. A. Borkovich. 1996. The G alpha i homologue gna-1 controls multiple differentiation pathways in Neurospora crassa. Mol. Biol. Cell 7:1283-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Julius, D., L. Blair, A. Brake, G. Sprague, and J. Thorner. 1983. Yeast alpha factor is processed from a larger precursor polypeptide: the essential role of a membrane-bound dipeptidyl aminopeptidase. Cell 32:839-852. [DOI] [PubMed] [Google Scholar]
- 36.Julius, D., A. Brake, L. Blair, R. Kunisawa, and J. Thorner. 1984. Isolation of the putative structural gene for the lysine-arginine-cleaving endopeptidase required the processing of yeast prepro-α-factor. Cell 36:309-318. [DOI] [PubMed] [Google Scholar]
- 37.Kang, Y. S., J. Kane, J. Kurjan, J. M. Stadel, and D. J. Tipper. 1990. Effects of expression of mammalian Gα and hybrid mammalian-yeast Gα proteins on the yeast pheromone response signal transduction pathway. Mol. Cell. Biol. 10:2582-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kays, A. M., and K. Borkovich. 2004. Severe impairment of growth and differentiation in a Neurospora crassa mutant lacking all heterotrimeric G-alpha proteins. Genetics 166:1229-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim, H., and K. A. Borkovich. 2004. A pheromone receptor gene, pre1, is essential for mating type-specific directional growth and fusion of trichgynes and female fertility in Neurospora crassa. Mol. Microbiol. 52:1781-1798. [DOI] [PubMed] [Google Scholar]
- 40.Kim, H., R. L. Metzenberg, and M. A. Nelson. 2002. Multiple functions of mfa-1, a putative pheromone precursor gene of Neurospora crassa. Eukaryot. Cell 1:987-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.King, K., H. G. Dohlman, J. Thorner, M. G. Caron, and R. J. Lefkowitz. 1990. Control of yeast mating signal transduction by a mammalian β2-adrenergic receptor and Gsα subunit. Science 250:121-123. [DOI] [PubMed] [Google Scholar]
- 42.Konopka, J. B., and D. D. Jenness. 1991. Genetic fine-structural analysis of the Saccharomyces cerevisiae α-pheromone receptor. Cell Regul. 2:439-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kothe, E. 1999. Mating types and pheromone recognition in the homobasidiomycete Schizophyllum commune. Fungal Genet. Biol. 27:146-152. [DOI] [PubMed] [Google Scholar]
- 44.Kurjan, J. 1993. The pheromone response pathway in Saccharomyces cerevisiae. Annu. Rev. Genet. 27:147-179. [DOI] [PubMed] [Google Scholar]
- 45.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 46.Leberer, E., D. Y. Thomas, and M. Whiteway. 1997. Pheromone signalling and polarized morphogenesis in yeast. Curr. Opin. Genet. Dev. 7:59-66. [DOI] [PubMed] [Google Scholar]
- 47.Lengeler, K. B., R. C. Davidson, C. D'Souza, T. Harashima, W. C. Shen, P. Wang, X. Pan, M. Waugh, and J. Heitman. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64:746-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.MacKay, V. L., S. K. Welch, M. Y. Insley, T. R. Manney, J. Holly, G. C. Saari, and M. L. Parker. 1988. The Saccharomyces cerevisiae BAR1 gene encodes an exported protein with homology to pepsin. Proc. Natl. Acad. Sci. USA 85:55-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masloff, S., and U. Kück. Unpublished data.
- 50.Masloff, S., S. Pöggeler, and U. Kück. 1999. The pro1+ gene from Sordaria macrospora encodes a C6 zinc finger transcription factor required for fruiting body development. Genetics 152:191-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McClelland, C. M., J. Fu, G. L. Woodlee, T. S. Seymour, and B. L. Wickes. 2002. Isolation and characterization of the Cryptococcus neoformans MATa pheromone gene. Genetics 160:935-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moore, S. A. 1983. Comparison of dose-response curves for alpha factor-induced cell division arrest, agglutination, and projection formation of yeast cells. J. Biol. Chem. 258:13849-13856. [PubMed] [Google Scholar]
- 53.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 54.Nowrousian, M., S. Masloff, S. Pöggeler, and U. Kück. 1999. Cell differentiation during sexual development of the fungus Sordaria macrospora requires ATP citrate lyase activity. Mol. Cell. Biol. 19:450-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nowrousian, M., C. Ringelberg, J. C. Dunlap, J. J. Loros, and U. Kück. Cross-species microarray hybridization to identify developmentally regulated genes in the filamentous fungus Sordaria macrospora. Mol. Genet. Genomics, in press. [DOI] [PubMed]
- 56.Nowrousian, M., C. Würtz, S. Pöggeler, and U. Kück. 2004. Comparative sequence analysis of Sordaria macrospora and Neurospora crassa as a means to improve genome annotation. Fungal Genet. Biol. 41:285-292. [DOI] [PubMed] [Google Scholar]
- 57.Olesnicky, N. S., A. J. Brown, S. J. Dowell, and L. Casselton. 1999. A constitutively active G-protein-coupled receptor causes mating self-compatibility in the mushroom Coprinus. EMBO J. 18:2756-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pöggeler, S. 2000. Two pheromone precursor genes are transcriptionally expressed in the homothallic ascomycete Sordaria macrospora. Curr. Genet. 37:403-411. [DOI] [PubMed] [Google Scholar]
- 59.Pöggeler, S., and U. Kück. 2001. Identification of transcriptionally expressed pheromone receptor genes in filamentous ascomycetes. Gene 280:9-17. [DOI] [PubMed] [Google Scholar]
- 60.Pöggeler, S., and U. Kück. 2004. A WD40 repeat protein regulates fungal cell differentiation and can functionally be replaced by the mammalian homologue striatin. Eukaryot. Cell 3:232-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pöggeler, S., S. Masloff, B. Hoff, S. Mayrhofer, and U. Kück. 2003. Versatile EGFP reporter plasmids for cellular localization of recombinant gene products in filamentous fungi. Curr. Genet. 43:54-61. [DOI] [PubMed] [Google Scholar]
- 62.Pöggeler, S., S. Risch, U. Kück, and H. D. Osiewacz. 1997. Mating-type genes from the homothallic fungus Sordaria macrospora are functionally expressed in a heterothallic ascomycete. Genetics 147:567-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Price, L. A., E. M. Kajkowski, J. R. Hadcock, and B. A. Ozenberger. 1995. Functional coupling of a mammalian somatostasin receptor to the yeast pheromone response pathway. Mol. Cell. Biol. 15:6188-6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rose, M., and D. Botstein. 1983. Construction and use of gene fusions to lacZ (β-galactosidase) that are expressed in yeast. Methods Enzymol. 101:167-180. [DOI] [PubMed] [Google Scholar]
- 65.Sambrook, J., E. F. Fritsch,, and T. Maniatis. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 66.Sander, P., S. Grunewald, M. Bach, W. Haase, H. Reilander, and H. Michel. 1994. Heterologous expression of the human D2S dopamine receptor in protease-deficient Saccharomyces cerevisiae strains. Eur. J. Biochem. 226:697-705. [DOI] [PubMed] [Google Scholar]
- 67.Schuurs, T. A., H. J. Dalstra, J. M. Scheer, and J. G. Wessels. 1998. Positioning of nuclei in the secondary mycelium of Schizophyllum commune in relation to differential gene expression. Fungal Genet. Biol. 23:150-161. [DOI] [PubMed] [Google Scholar]
- 68.Seo, J. A., K. H. Han, and J. H. Yu. 2004. The gprA and gprB genes encode putative G protein-coupled receptors required for self-fertilization in Aspergillus nidulans. Mol. Microbiol. 53:1611-1623. [DOI] [PubMed] [Google Scholar]
- 69.Shen, W. C., P. Bobrowicz, and D. J. Ebbole. 1999. Isolation of pheromone precursor genes of Magnaporthe grisea. Fungal Genet. Biol. 27:253-263. [DOI] [PubMed] [Google Scholar]
- 70.Shen, W. C., R. C. Davidson, G. M. Cox, and J. Heitman. 2002. Pheromones stimulate mating and differentiation via paracrine and autocrine signaling in Cryptococcus neoformans. Eukaryot. Cell 1:366-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sherman, F., G. R. Fink, and C. W. Lawrence. 1979. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 72.Shiu, P. K., and N. L. Glass. 2000. Cell and nuclear recognition mechanisms mediated by mating type in filamentous ascomycetes. Curr. Opin. Microbiol. 3:183-188. [DOI] [PubMed] [Google Scholar]
- 73.Singh, A., E. Y. Chen, J. M. Lugovoy, C. N. Chang, R. A. Hitzeman, and P. H. Seeburg. 1983. Saccharomyces cerevisiae contains two discrete genes coding for the alpha-factor pheromone. Nucleic Acids Res. 25:4049-4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sprague, J., G.F. 1991. Assays of yeast mating. Methods Enzymol. 194:77-93. [DOI] [PubMed] [Google Scholar]
- 75.Tate, C. G., and R. Grisshammer. 1996. Heterologous expression of G-protein-coupled receptors. Trends Biotechnol. 14:426-430. [DOI] [PubMed] [Google Scholar]
- 76.Trueheart, J., and G. R. Fink. 1989. The yeast cell fusion protein FUS1 is O-glycosylated and spans the plasma membrane. Proc. Natl. Acad. Sci. USA 86:9916-9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Turina, M., A. Prodi, and N. K. Alfen. 2003. Role of the Mf1-1 pheromone precursor gene of the filamentous ascomycete Cryphonectria parasitica. Fungal Genet. Biol. 40:242-251. [DOI] [PubMed] [Google Scholar]
- 78.Vaillancourt, L. J., M. Raudaskoski, C. A. Specht, and C. A. Raper. 1997. Multiple genes encoding pheromones and a pheromone receptor define the Bβ1 mating-type specificity in Schizophyllum commune. Genetics 146:541-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Waters, M. G., E. A. Evans, and G. Blobel. 1988. Prepro-α-factor has a cleavable signal sequence. J. Biol. Chem. 263:6209-6214. [PubMed] [Google Scholar]
- 80.Yang, Q., S. I. Poole, and K. A. Borkovich. 2002. A G-protein beta subunit required for sexual and vegetative development and maintenance of normal G alpha protein levels in Neurospora crassa. Eukaryot. Cell 1:378-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang, L., R. A. Baasiri, and N. K. van Alfen. 1998. Viral repression of the fungal pheromone precursor gene expression. Mol. Cell. Biol. 18:953-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang, L., A. C. Churchill, P. Kazmeirczak, D. H. Kim, and N. K. van Alfen. 1993. Hypovirulence-associated traits induced by a mycovirus of Cryphonectria parasitica are mimicked by targeted inactivation of a host gene. Mol. Cell. Biol. 13:7782-7792. [DOI] [PMC free article] [PubMed] [Google Scholar]