ABSTRACT
Background: Stressful events increase the risk for treatment-resistant depression (TRD), and trauma-focused psychotherapy can be useful for TRD patients exposed to early life stress (ELS). Epigenetic processes are known to be related to depression and ELS, but there is no evidence of the effects of trauma-focused psychotherapy on methylation alterations.
Objective: We performed the first epigenome-wide association study to investigate methylation changes related to trauma-focused psychotherapies effects in TRD patients.
Method: Thirty TRD patients assessed for ELS underwent trauma-focused psychotherapy, of those, 12 received trauma-focused cognitive behavioural therapy, and 18 Eye Movement Desensitization and Reprocessing (EMDR). DNA methylation was profiled with Illumina Infinium EPIC array at T0 (baseline), after 8 weeks (T8, end of psychotherapy) and after 12 weeks (T12 – follow-up). We examined differentially methylated CpG sites and regions, as well as pathways analysis in association with the treatment.
Results: Main results obtained have shown 110 differentially methylated regions (DMRs) with a significant adjusted p-value area associated with the effects of trauma-focused psychotherapies in the entire cohort. Several annotated genes are related to inflammatory processes and psychiatric disorders, such as LTA, GFI1, ARID5B, TNFSF13, and LST1. Gene enrichment analyses revealed statistically significant processes related to tumour necrosis factor (TNF) receptor and TNF signalling pathway. Stratified analyses by type of trauma-focused psychotherapy showed statistically significant adjusted p-value area in 141 DMRs only for the group of patients receiving EMDR, with annotated genes related to inflammation and psychiatric disorders, including LTA, GFI1, and S100A8. Gene set enrichment analyses in the EMDR group indicated biological processes related to inflammatory response, particularly the TNF signalling pathway.
Conclusion: We provide preliminary valuable insights into global DNA methylation changes associated with trauma-focused psychotherapies effects, in particular with EMDR treatment.
KEYWORDS: Treatment-resistant depression; methylomic; epigenome-wide association study; trauma-focused psychotherapy; EMDR; TRD
HIGHLIGHTS
Stressful events increase treatment-resistant depression, and trauma-focused psychotherapy can be useful for these patients.
Epigenome-wide data shows changes associated with trauma-focused psychotherapies, especially eye movement desensitization and reprocessing therapy, in treatment-resistant depression patients.
Genes and biological pathways related to inflammatory and immune systems are among the most statistically significant results.
Abstract
Antecedentes: Los eventos estresantes aumentan el riesgo de depresión resistente al tratamiento (DRT), y la psicoterapia centrada en trauma puede ser útil para los pacientes con DRT expuestos a estrés en su vida temprana (EVT). Se sabe que los procesos epigenéticos están relacionados con la depresión y EVT, pero no hay evidencia de los efectos de la psicoterapia centrada en trauma sobre las alteraciones de la metilación.
Objetivo: Realizamos el primer estudio de asociación de amplio-epigenoma para investigar los cambios de metilación relacionados con los efectos de las psicoterapias centradas en trauma en pacientes con DRT.
Método: Treinta pacientes con DRT evaluados para EVT se sometieron a psicoterapia centrada en trauma, de ellos, 12 recibieron terapia cognitivo-conductual centrada en trauma y 18, desensibilización y reprocesamiento por movimientos oculares (EMDR). La metilación del ADN se perfiló con la matriz Illumina Infinium EPIC en T0 (valor inicial), después de 8 semanas (T8, final de la psicoterapia) y después de 12 semanas (T12 – seguimiento). Examinamos sitios y regiones CpG metilados diferencialmente, así como análisis de vías en asociación con el tratamiento.
Resultados: Los principales resultados obtenidos mostraron 110 regiones diferencialmente metiladas (RDMs) con un área de valor p ajustado significativa asociada con los efectos de las psicoterapias centradas en el trauma en toda la cohorte. Varios genes anotados están relacionados con procesos inflamatorios y trastornos psiquiátricos, como LTA, GFI1, ARID5B, TNFSF13 y LST1. Los análisis de enriquecimiento genético revelaron procesos estadísticamente significativos relacionados con el receptor del factor de necrosis tumoral (FNT) y la vía de señalización del FNT. Los análisis estratificados por tipo de psicoterapia centrada en trauma mostraron un área de valor p ajustado estadísticamente significativo en 141 RDMs solo para el grupo de pacientes que recibieron EMDR, con genes anotados relacionados con la inflamación y los trastornos psiquiátricos, incluidos LTA, GFI1 y S100A8. Los análisis de enriquecimiento de conjuntos de genes en el grupo EMDR indicaron procesos biológicos relacionados con la respuesta inflamatoria, particularmente la vía de señalización del FNT.
Conclusión: Proporcionamos información preliminar valiosa sobre los cambios globales en la metilación del ADN asociados con los efectos de las psicoterapias centradas en trauma, en particular con el tratamiento EMDR.
PALABRAS CLAVE: Depresión resistente al tratamiento, metilómico, estudio de asociación de amplio-epigenoma, psicoterapia centrada en trauma, EMDR, DRT
1. Introduction
Major depressive disorder (MDD) is the most prevalent psychiatric disorder worldwide, represents the third leading cause of years lived with disability (GBD, 2016 Disease and Injury Incidence and Prevalence Collaborators, 2016), and associates very frequently with impaired quality of life and functioning (Hasin et al., 2018). In spite of advances in pharmacological treatment, this approach is often ineffective, especially when used as a single strategy. Indeed, approximately 68% of individuals do not achieve remission after a first course of antidepressant medication and up to 30% of patients with depressive episodes do not adequately respond to two different trials of antidepressants (Dodd et al., 2021; Jaffe et al., 2019). This condition is defined as treatment-resistant depression (TRD).
One of the many prognostic variables used to predict treatment outcomes in MDD is exposure to stressful events, especially when experienced during childhood (so defined early life stress – ELS), which associates with unfavourable outcomes and increased probability to develop TRD (Kautzky et al., 2017; Kraus et al., 2019). Evidence-based trauma-focused psychotherapies, including trauma-focused cognitive behavioural therapy (TF-CBT) and eye movement desensitization and reprocessing (EMDR), have been proposed as beneficial approaches for MDD management and TRD patients being exposed to ELS, showing evidence of positive therapeutic responses (Minelli et al., 2019; Yan et al., 2021).
Epigenetic mechanisms, such as DNA methylation, are involved in many neurobiological processes, being altered in stress-related psychiatric disorders, including MDD, and available methylation studies point to epigenetic signatures associated with trauma exposure. A systematic review on methylation studies in relation to maltreatment performed in clinical and non-clinical samples of children and adults revealed some promising markers identified with epigenome-wide approaches, with significant genes involved in neural cell development (BDNF, KITLG, and POU3F1), cell signalling and apoptosis (LINGO3 and 2NPFF2), neural influences on motor system (ALS2) and nervous system inflammation (ITGB1) (Parade et al., 2021). A systematic review and meta-analysis on epigenome-wide association studies (EWAS) and candidate gene studies in correlation with childhood maltreatment found 44 significant cytosine-phosphate-guanine dinucleotide (CpG) sites, 97.7% hypomethylated, with two CpG sites situated near the RPTOR gene, involved in cell growth regulation. Network analysis identified genes associated with the PI3K-AKT and AMPK signalling pathways and gene ontology analysis showed enrichment of biological mechanisms associated with nervous system development and regulation of multicellular organismal processes (Neves et al., 2021).
Studies performed in clinical samples revealed some differentially methylated regions in relation to childhood maltreatment. An EWAS performed by Labonté and colleagues studied hippocampal samples of suicide victims, with and without a history of ELS, and found 362 differentially methylated promoters in maltreated individuals (Labonté et al., 2012). In a genome-wide gene expression and DNA methylation study in patients with post-traumatic stress disorders (PTSD), differential methylation profile was seen in 69.3% of transcripts in individuals exposed to child abuse, while 33.6% of probes were differentially methylated in patients without this exposure (Mehta et al., 2013). An EWAS on patients with borderline personality disorders (BPD), with and without ELS history, and controls, revealed that differential methylation patterns were stronger when comparing patients with trauma and controls than when comparing patients without trauma and controls, suggesting that trauma associates with epigenomic alterations that modulate or increase disease symptoms (Arranz et al., 2021). Finally, an EWAS showed that development of combat-related PTSD was associated with diverse methylation profiles in different genomic positions, including genes involved in the immune system through the human leucocyte antigen region and HEXDC, and MAD1L1, known to be associated with immune processes and PTSD (Snijders et al., 2020).
In non-clinical samples, different epigenetic and epigenome-wide data also point to differential methylomic profiles in relation to maltreatment history. A DNA methylome variation study performed in a prospective controlled trial on a psychosocial intervention found that childhood abuse and neglect were associated with 14% of the inter-individual variation in DNA methylation across the human genome in adulthood (O’Donnell et al., 2018). Analysis of genome-wide DNA methylation profiles in elderly people suffering from ELS and controls showed 71 differentially methylated CpG sites, annotated to genes involved in neuronal projections and neurodevelopment, including SKAP2, DLGAP2, and MTOR, all associated with traumatic stress (Marinova et al., 2017). Studying maltreated individuals in methylomic variation analyses, Cecil and collaborators found 118 differentially methylated probes for physical neglect, 34 for physical abuse and 7 for sexual abuse, including probes annotated in genes previously implicated in stress-related disorders (GABBR1, GRIN2D, CACNA2D4, PSEN2) (Cecil et al., 2016).
Few studies performed on clinical samples assessed methylation variations in response to different kinds of psychotherapy. One was performed in PTSD patients undergoing trauma-focused psychotherapy and trauma-exposed controls who did not receive any trauma-focused treatment, and showed 12 differentially methylated genomic regions specific for PTSD improvement, but not related to depressive symptoms changes, which included hypermethylation of ZFP57, involved in stress vulnerability (Vinkers et al., 2021). Instead, the EWAS performed by Yang and colleagues detected methylation changes in PTSD patients following prolonged-exposure psychotherapy, with CREB-BDNF signalling pathway predicting symptoms changes and severity, and resilience markers, including FKBP5, NR3C1, SDK1, and MAD1L1, associated with disease recovery (Yang et al., 2021). In a candidate gene study (Perroud et al., 2013), patients with BPD presented significantly higher brain-derived neurotrophic factor (BDNF) methylation status in comparison to controls, with increased methylation paralleling a higher number of childhood trauma. After undergoing intensive dialectical behaviour therapy, non-responders presented higher BDNF methylation status, while responders showed a decreased methylation over time (Perroud et al., 2013). Also, in another candidate genes study, methylation status of two genes known to be dysregulated in BPD, APBA3 and MCF2, was studied in BPD patients undergoing dialectical behavioural therapy and controls, with results showing genes’ hypermethylation at the beginning of therapy in responders, in comparison to non-responders, underlining possible epigenetic biomarkers predictive of therapy outcomes (Knoblich et al., 2018). Patients with panic disorder subjected to CBT presented differential candidate gene MAOA methylation patterns according to response status, with responders showing increased methylation up to the level of healthy controls and non-responders revealing decreased methylation along the therapy course (Ziegler et al., 2016).
Based on the information presented, the available literature on epigenetic and on epigenome-wide approaches primarily focuses on epigenetic biomarkers of childhood maltreatment in both non-clinical and clinical samples, particularly on PTSD and BPD patients. Only two studies, directed to the study of PTSD patients, evaluated DNA methylation changes in response to trauma-focused psychotherapies (Vinkers et al., 2021; Yang et al., 2021).
Currently, no studies have longitudinally assessed the genome-wide methylation effects of trauma-focused psychotherapies in patients with TRD who have experienced childhood trauma. The goal was to identify possible relations between global methylation alterations and the effects of trauma-focused psychotherapies on symptom changes, disease response, and relapse, in order to find putative epigenome-wide biomarkers associated with psychotherapy effects.
2. Materials and methods
2.1. Study participants and clinical assessment
Thirty TRD patients were voluntarily enrolled in the study. The diagnostic criterion for inclusion was a diagnosis of MDD according to the Diagnostic and Statistical Manual of Mental Disorders-IV (DSM-IV) classification system. The exclusion criteria were the following: (a) cognitive impairment or mental retardation; (b) history of bipolar disorder, schizophrenia or schizoaffective disorder; (c) primary diagnosis of substance abuse, alcohol abuse or dependency, obsessive-compulsive disorder, personality disorder or PTSD; and (d) comorbidity with eating disorders; (e) comorbidity with alcohol and substance dependence; (f) neurological disorders (i.e. Parkinson's disease, multiple sclerosis, Alzheimer's and other dementias, epilepsy, stroke, brain tumours, traumatic conditions of the nervous system); (g) comorbidity with other severe medical illness and severe autoimmune diseases (i.e. cancers, Crohn's Disease, Rheumatoid Arthritis (RA), Scleroderma, Psoriasis, Myasthenia gravis, Sjögren syndrome, Systemic lupus erythematosus); (h) pregnancy. Patients were referred to the Psychiatric Hospital ‘Villa Santa Chiara’ in Verona, Italy. The study was approved by the Ethics Committee for Clinical Trials of province of Verona and Rovigo (N: 234777/11.05.16). Participants received full explanation about study procedures and gave written informed consents to participate.
TRD definition was the failure to respond to at least two trials with two or more classes of antidepressant drugs and to a trial with a tricyclic drug (TCA), corresponding to stage III or above, in accordance to Thase and Rush staging system (Thase & Rush, 1997).
Assessment of ELS was performed with the Italian version of the Childhood Experience of Care and Abuse Questionnaire (CECA.Q) (Bifulco et al., 2005).
All patients received trauma-focused psychotherapies and were assigned blinded to TF-CBT or EMDR. Each patient received three individual sessions per week, lasting 60 min each, over a period of 8 weeks, in addition to drug treatment as usual (TAU), for a total of 24 sessions of TF-CBT or EMDR carried out by experienced psychotherapists.
The symptomatological assessment was carried out at 4 timepoints: baseline (T0), after four weeks of treatment, after eight weeks of treatment/end of psychotherapy (T8), and four weeks after the end of the treatment when the patients came back to the hospital for the follow-up visit (T12). Clinical depressive symptoms evaluations were made using Montgomery-Åsberg Depression Rating Scale (MADRS) (Montgomery & Asberg, 1979). The variation of the score between visits (Delta MADRS) was used in the statistical analysis in order to evaluate the influence of symptoms changes. Response to trauma-focused psychotherapies was defined as a reduction greater than 50% in MADRS score at the T12 assessment. Finally, after 6 months from the beginning of the treatment (T26), a clinical evaluation of relapse was carried out by phone. The clinician evaluated the condition of relapse if the patient reported at least one of the following conditions: a significant worsening of one's condition concerning mental health; a score of ≤ 5 on a Likert scale, in which the patient evaluated their own mood from 0 to 10 (where 10 is very good and 0 is completely negative); the pharmacological treatment was significantly increased in dosage or new drugs were added.
2.2. DNA extraction and methylation analysis
Fasting blood samples were collected at T0, T8 and T12 using EDTA Tube and the DNA was extracted from whole blood samples using the Gentra Puregene Blood kit (Qiagen), according to the manufacturer’s instructions. DNA quantification and quality evaluation were performed through spectro-photometric analysis (NanoDrop 2000, Thermo Scientific). DNAs were pipetted on 96-well processing plates: same-subject T0, T8 and T12 DNAs on the same plate; between-subjects randomized based on sex and age on separate plates.
Methylation was profiled at T0, T8 and T12 with Illumina Infinium Methylation EPIC BeadChip array (850k) using HiScan array scanning systems (chips and scanner from Illumina, San Diego, CA). Methylation data were available for thirty patients at T0 and T8, and for 27 patients at T12, since methylation data at T12 was not available for 3 patients undergoing EMDR.
Methylation levels were quantified after quality control and normalization using ChAMP R package. Probes with detection p-value cut off below .01 and with a beadcount less than 3 in at least 5% of patients were removed. We also removed probes containing single nucleotide polymorphisms (SNPs) with minor allele frequency above 0.01 within 10 base pair (bp) of the single base extension position based on the list from Pidsley and collaborators (Pidsley et al., 2016). Probes linked to X- and Y-chromosomes were removed. None of the samples have more that 10% of not available (NA) and all were retained for analysis. Normalization was performed on beta-values using Beta-Mixture Quantile (BMIQ) Normalisation as implemented in ChAMP R package. After normalization beta values were transformed to M-values to perform association (Du et al., 2010).
2.3. Statistical analysis
To evaluate clinical efficacy, MADRS scores were analysed using repeated measures ANOVA, including the outcome score at different times as the dependent variable, time as the within-subject factor, and the psychotherapy treatment group as the between-subjects factor.
A mixed linear model approach from the Limma R package was used to perform association analysis using patient as blocking factor. To adjust for confounding factors, we included white blood cell fractions estimated for granulocytes, monocytes, B cells, NK cells, CD4 + T cells, and CD8 + T cells, and two principal components from control probes.
We estimated white blood cellular composition using the estimateCellCounts function from the R minfi package (Fortin et al., 2017). Principal component analysis (PCA) was performed on EPIC chip control probes to correct for technical artefacts. Optimal number of principal components to use were determined using findPC R package (Zhuang et al., 2022).
Differentially methylated probes (DMPs) were reported at the suggestive threshold of p ≤ 10−5. Cytosine guanine dinucleotide (CpG) site annotation was performed using IlluminaHumanMethylationEPICanno.ilm10b2.hg19 R package (hg19 genome reference).
Differentially methylation region (DMR) analysis was performed using bumphunter R package. Single probe statistic was calculated using univariate model with patient as blocking factor and the phenotype/feature of interest as explanatory variable. Probes were aggregated in clusters/regions with at least 7 probes, with maximum distance of 300 bp within each probe. Region p-values were computed by permutation procedure over 250 permutations. Differentially methylated regions (DMRs) were considered those with adjusted p-value ≤ .05, but also those with adjusted p-value ≤ .1 are reported in supplementary tables. DMRs were considered as relevant those with adjusted p-value area ≤ .05.
Enrichment analysis was performed with enricher and enrichGO functions from clusterProfiler R package. Gene sets were obtained from graphite (Sales et al., 2019), msigdbr and has-ord-db R packages. Significant enriched gene sets were considered as those with an adjusted p-value ≤ .1. P-values were adjusted using the Benjamini & Hochberg method unless otherwise stated.
3. Results
The socio-demographic and clinical characteristics of the 30 TRD patients who experienced ELS treated with trauma-focused psychotherapies, including the TF-CBT (N = 12) and EMDR (N = 18) groups, are shown in Table 1. The mean age of the enrolled participants was 51.9 years (standard deviation of 8.9 years) and 76.7% were females. Twelve patients underwent TF-CBT and eighteen patients followed EMDR programme. As reported in the original paper on the clinical trial (Minelli et al., 2019) performed on 22 patients, even in this larger group where we obtained EWAS data at three timepoints (T0, T8 and T12), trauma-focused psychotherapies reduced depression symptomatology (F2,56 = 60.03; p = 3.16 × 10−12), whereas the treatment group by timepoint interaction did not show any significant difference. However, the treatment variable resulted significant (p < .05), as well as MADRS post hoc comparisons showing, as in the previous paper, a significant difference in scores between the two treatment groups at T12 (follow-up visit), with a lower score in the EMDR group than in the TF-CBT group (8.89 ± 10.90 vs. 16.92 ± 12.10; p < .05). Although our study included only patients with MDD as primary diagnosis, a portion of them had PTSD in comorbidity (secondary diagnosis). Since these patients could benefit more from trauma-focused psychotherapy, leading to possible confounding effects, we conducted the same analyses by excluding patients with PTSD in comorbidity, and the same results were confirmed (results not shown).
Table 1.
Demographic and clinical characteristics of TRD patients treated with TF-CBT and EMDR.
| Characteristics | Whole cohort (n = 30) | TF-CBT group (n = 12) | EMDR group (n = 18) |
|---|---|---|---|
| Age in years, mean (SD) | 51.9 (±8.9) | 52.5 (±5.9) | 51.5 (±10.3) |
| Gender, n (%F) | 23 (76.7) | 7 (58.4) | 16 (88.9) |
| Education in years, mean (SD) | 12.4 (±3.2) | 12.8 (±3.2) | 12.1 (±3.1) |
| Smokers, n (%) | 12 (40.0) | 6 (50.0) | 6 (33.4) |
| Body Mass Index (BMI), mean (SD) | 26.0 (±4.3) | 25.2 (±2.9) | 26.6 (±5.0) |
| Age of onset in years, mean (SD) | 32.2 (±13.5) | 33.3 (±13.8) | 31.3 (±13.2) |
| MADRS total score at the baseline, mean (SD) | 28.4 (±7.0) | 27.0 (±6.3) | 30.5 (±7.4) |
| Recurrent MDD, n (%) | 30 (100) | 12 (100) | 18 (100) |
| Psychotic symptoms, n (%) | 5 (16.6) | 3 (25.0) | 2 (11.1) |
| Comorbidity with personality disorders, n (%) | 23 (76.7) | 9 (75.0) | 14 (77.8) |
| Comorbidity with anxiety disorders, n (%) | 25 (83.4) | 11 (91.7) | 14 (77.8) |
| Comorbidity with PTSD, n (%) | 9 (30.0) | 3 (25.0) | 6 (33.0) |
| Comorbidity with alcohol and/or substance abuse, n (%) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Comorbidity with psychiatric disorders among first-degree relatives, n (%) | 27 (90.0) | 12 (100%) | 15 (83.4) |
| Trauma (CECA-Q): Mother antipathy, n (%) | 22 (73.3) | 11 (91.6) | 11 (61.1) |
| Trauma (CECA-Q): Father antipathy, n (%) | 4 (13.3) | 1 (8.3) | 3 (16.6) |
| Trauma (CECA-Q): Mother neglect, n (%) | 16 (53.3) | 8 (66.6) | 8 (44.4) |
| Trauma (CECA-Q): Father neglect, n (%) | 22 (73.3) | 8 (66.6) | 14 (77.7) |
| Trauma (CECA-Q): Physical abuse mother, n (%) | 10 (33.3) | 6 (50.0%) | 4 (22.2) |
| Trauma (CECA-Q): Physical abuse father, n (%) | 5 (16.6) | 2 (16.6) | 3 (16.6) |
| Trauma (CECA-Q): Sexual abuse, n (%) | 14 (46.6) | 5 (41.6) | 9 (50.0) |
Note: CECA-Q: Childhood Experience of Care and Abuse Questionnaire; EMDR: Eye Movement Desensitization and Reprocessing; F: female, PTSD: Post Traumatic Stress Disorder; SD: standard deviation; TF-CBT: trauma-focused cognitive behavioural therapy; TRD: treatment resistant depression
3.1. Single CpG site analysis
In order to evaluate possible methylation changes related to the trauma-focused psychotherapies treatment, we performed longitudinal analyses to evaluate DMPs between T0 and T12 in the whole cohort. We identified 9 DMPs with nominal p-values ≤ 10−5 but, after FDR correction, none of these probes remained significant (Supplementary Table 1Sa). Additionally, we conducted T0-T12 analyses adding covariates into the model and we found 11 DMPs when including type of trauma-focused psychotherapy, 27 DMPs for clinical symptoms variations as measured by MADRS, 19 for response at T12, and 10 for relapse at T26, all with nominal p-values ≤ 10−5 (Supplementary Table 1Sb, 1Sc, 1Sd, and 1Se). However, after FDR correction only the analysis including clinical symptoms variations as covariate presented significant adjusted p-values (q ≤ 0.05) with 5 DMPs, three of which annotated in the genes SRC1N1, RGS20 and INPPL1 (Supplementary Table 1Sc).
We also performed additional longitudinal analyses taking into account all three timepoints (T0, T8, and T12) to investigate whether the results obtained might differ across different time frames. The results revealed the presence of 9 DMPs with nominal p-values ≤ 10−5, but none remained significant after FDR correction (Supplementary Table 2Sa). When adding the covariates, we identified 9 DMPs for type of trauma-focused psychotherapies, 40 DMPs for clinical symptoms variations, 16 for response and 11 for relapse, all with nominal p-values ≤ 10−5 (Supplementary Table 2Sb, 2Sc, 2Sd, and 2Se). However, after FDR correction, only the analysis including clinical symptoms variations as covariate presented significant adjusted p-values (q ≤ 0.05), with 5 DMPs, 4 of which annotated in the genes INPPL1, HDC, SRCIN1 and CTPS1 (Supplementary Table 2Sc).
3.2. Differentially methylated region analysis
To identify DMRs associated with the effects of trauma-focused psychotherapies, regional analyses were conducted in the entire cohort. The DMR analysis between T0 and T12 resulted in 160 DMRs with nominal p-value area ≤ .05, of which 110 DMRs remained significant after FDR adjustment. 110 of these regions were annotated in genes, with the most significant ones being LTA, GFI1, ARID5B, CD52, LDLRAD4, C11orf21, and GAS7 (Supplementary Table 3Sa). When considering the covariates into the model, we found several DMRs with nominal p-value area ≤ .05: 3 DMRs when considering type of trauma-focused psychotherapy, nine for clinical symptoms variation, 22 for response, and 12 for relapse. After FDR correction, none of these DMRs remained significant considering the adjusted p-value area (Supplementary Table 3Sb, 3Sc, 3Sd and 3Se).
We did not perform regional analyses considering all three timepoints because we did not find significant data in the corresponding DMPs analyses to allow us to carry out further analysis
3.3. Gene and pathway enrichment analysis
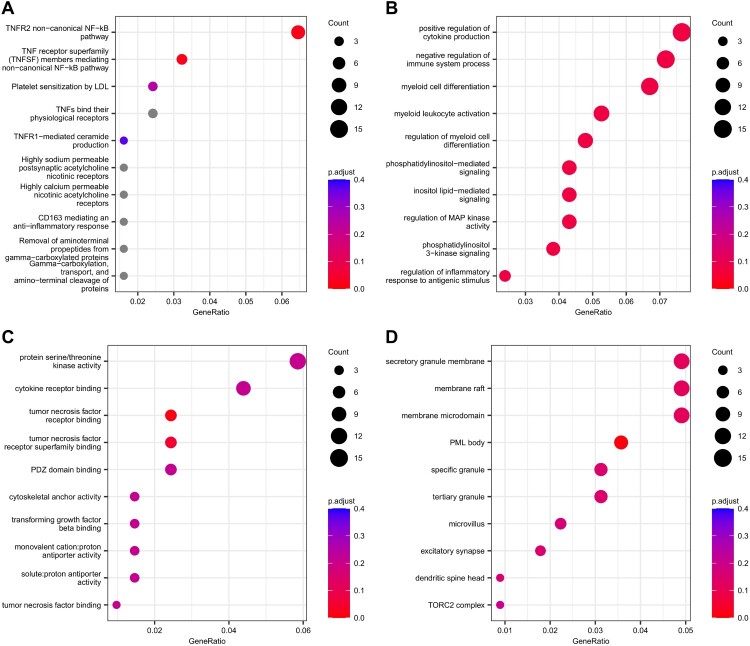
Gene enrichment analyses were conducted on the 251 genes laying on the DMRs found in the T0-T12 analyses performed on the entire group of patients. The ‘TNFR2 non-canonical NF-kB pathway’ and ‘TNF receptor superfamily (TNFSF) members mediating non-canonical NF-kB pathway’ Reactome pathways were found to be significantly enriched (adjusted p-value ≤.1, Figure 1(A) and Supplementary Table 4Sa). Additionally, 33 gene ontology (GO) biological processes (BPs; Figure 1(B) and Supplementary Table 4Sb), 2 GO molecular functions (MF; Figure 1(C) and Supplementary Table 4Sc) related to ‘tumor necrosis factor receptor binding’ and ‘tumor necrosis factor receptor superfamily binding’, and 1 GO cellular components (CC; Figure 1(D) and Supplementary Table 4Sd) were found to be significant (adjusted p-value ≤0.1).
Figure 1.
Gene set enrichment analyses on the genes annotated to significant differentially methylated regions in the longitudinal analysis performed between T0 and T12. (A) Top 10 enriched Reactome pathways. Top 10 enriched gene sets for (B) GO BP, (C) GO MF and (D) GO CC. Dots are coloured according to adjusted p-value (in grey pathways with adjusted p-value >.4). Dot size represents the number of genes that belong to the gene set/pathway.
3.4. Stratified analyses by type of trauma-focused psychotherapy
In the prior clinical study by our research group (Minelli et al., 2019), we observed a decrease in depressive symptomatology in patients with TRD treated with trauma-focused psychotherapy, with a greater efficacy of EMDR. Therefore, we also performed longitudinal analysis (T0-T12) separately for the EMDR and TF-CBT patients’ subgroups.
We found 1 differentially methylated CpG site in EMDR subgroup (Supplementary Table 5Sa), and six differentially methylated CpG sites in TF-CBT subgroup (Supplementary Table 5Sb), all with nominal p-values ≤ 10−5. However, none of these CpG sites remained significant after FDR correction.
Regional analyses revealed 259 DMRs with nominal p-value area ≤ .05, of which 141 DMRs also presented significant adjusted p-value area. Among these regions, 141 were annotated in genes, with the most significant ones being LTA, GFI1, MIR4526, TRIM39-RPP21, and S100A8 (Supplementary Table 6Sa). Regional analysis on the methylation profiles of patients undergoing TF-CBT revealed 9 DMRs with nominal p-value area ≤ .05, but none remained significant after FDR correction (Supplementary Table 6Sb).
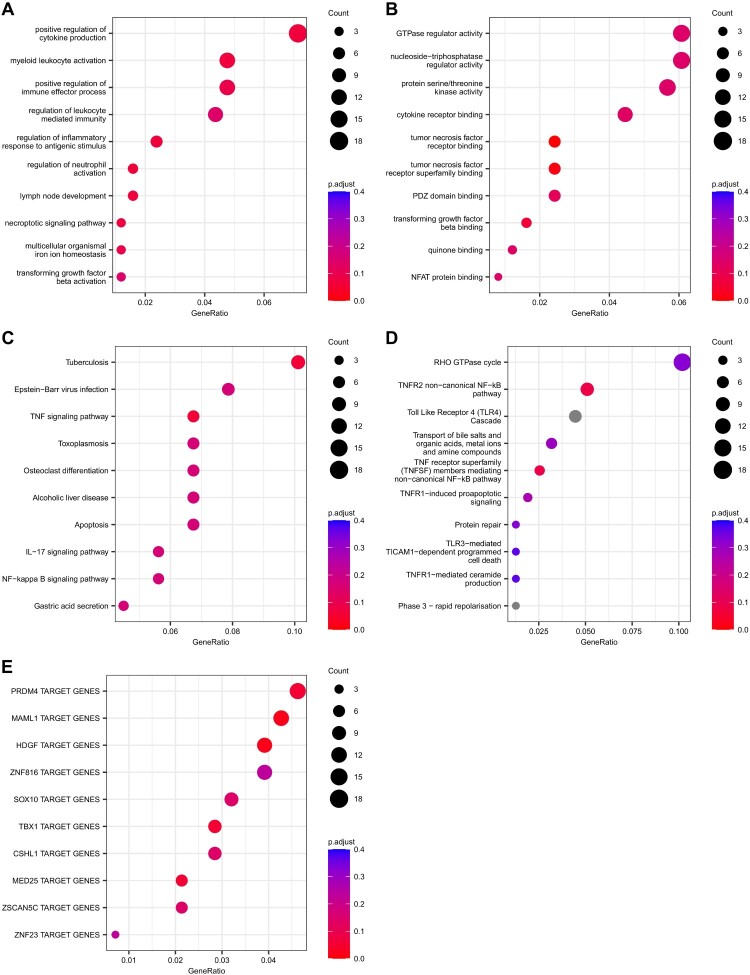
Gene set enrichment analyses were performed on the genes with significant DMRs (adjusted p-value ≤ .1) in the EMDR subgroup. We found 8 significant (adjusted p-value ≤ .1) GO BPs mainly related to inflammatory response (Figure 2(A); Supplementary Table 7Sa) and 3 GO MFs (Figure 2(B); Supplementary Table 7Sb). Using biological pathways, we found 2 significantly enriched KEGG pathways (Figure 2(C); Supplementary Table 7Sc) and 2 Reactome pathways (Figure 2(D); Supplementary Table 7Sd). Both pathway sources pointed out the TNF signalling pathway as enriched in the DMRs associated genes. Finally, we searched for enrichment in transcription factor target using gene set defined in the Molecular Signature Database. We found enrichment for targets of the transcription factors MAML1, HDGF, TBX1, MED25, and PRDM4 (adjusted p-value ≤ .1, Figure 2(E); Supplementary Table 7Se).
Figure 2.
Gene set enrichment analyses on the genes annotated to significant differentially methylated regions in the longitudinal analysis performed between T0 and T12, in patients who underwent EMDR. Top 10 enriched gene set for (A) GO BP and (B) GO MF. Top 10 enriched pathways for (C) KEGG and (D) Reactome. Top 10 enriched Transcription Factor Target gene set defined by the MSigDB (collection C3: regulatory target gene sets, GTRD subset)
In Table 2, a synthesis of the main results from the different analyses performed is reported.
Table 2.
Summary of some the most significant results in the subgroup of patients who underwent EMDR trauma-focused psychotherapy.
| (A) Regional analyses | |
|---|---|
| Genes in significant DMRs | LTA |
| GFI1 | |
| MIR4526 | |
| TRIM39-RPP21 | |
| S100A8 | |
| DMRs in genes related to inflammatory processes and psychiatric disorders | LTA |
| GFI1 | |
| S100A8 | |
| (B) Gene set and biological pathways enrichment analyses | |
| Gene ontology biological processes | Positive regulation of cytokine production |
| Myeloid leukocyte activation | |
| Positive regulation of immune effector process | |
| Regulation of inflammatory response to antigenic stimulus | |
| Regulation of neutrophil activation | |
| Lymph node development | |
| Necroptotic signalling pathway | |
| Multicellular organismal iron ion homeostasis | |
| Gene ontology molecular functions | TNF receptor binding |
| TNF receptor subfamily binding | |
| TGF beta binding | |
| KEGG biological pathways | Tuberculosis |
| TFN signalling pathway | |
| Reactome biological pathways | TNFR2 non-canonical NF-kB pathway |
| TNF receptor superfamily members mediating non-canonical NF-kB pathway | |
| Transcription factor target genes | MAML1 |
| HDGF | |
| TBX1 | |
| MED25 | |
| PRDM4 | |
Notes: (A) Regional analyses in correlational analyses performed in T0-T12, highlighting some significant genes related to inflammatory/immune system and psychiatric disorders. The identified genes presented an adjusted p-value area ≤ .05. (B) Gene set and biological pathways enrichment analyses conducted on the genes with significant differentially methylated regions in the longitudinal analysis performed between T0 and T12. Enriched gene sets were considered those with adjusted p-value ≤ 0.1. DMRs: differentially methylated regions; EMDR: Eye Movement Desensitization and Reprocessing; GFI1: Growth Factor Independent 1 Transcriptional Repressor; HDGF: Hepatoma-derived growth factor; LTA: Lymphotoxin-alpha; MAML1: Mastermind Like Transcriptional Coactivator 1; MED25: Mediator Complex Subunit 25; MIR4526: MicroRNA 4526; NF-κB: Nuclear factor-κB; PRDM4: PR/SET Domain 4; S100A8: S100 Calcium Binding Protein A8; TBX1: T-Box Transcription Factor 1; TGF: Transforming Growth Factor; TNF: Tumour Necrosis Factor; TNFR2: Tumor Necrosis Factor Receptor 2; TRIM39-RPP21: Tripartite motif-containing 39 and Ribonuclease P/MRP 21 kDa subunit.
4. Discussion
This is the first EWAS to investigate how changes in global DNA methylation in TRD patients experiencing ELS were related to the outcomes of trauma-focused psychotherapies (TF-CBT and EMDR). Considering the available literature, our study contributes to filling a gap in the current scientific knowledge, identifying potential epigenetic biomarkers that could predict therapy response and disease outcomes.
We found several DMPs and DMRs related to therapy outcomes in T0-T12 analyses, particularly when considering specific covariates, such as clinical response and trauma-focused psychotherapy, into the analyses. Also, gene and pathways enrichment analyses revealed significant biological processes, in particular related to inflammatory and immune pathways. The most significant results were obtained for patients undergoing EMDR.
When analysing differentially methylated CpG sites, our findings revealed several DMPs between T0 and T12, with significant adjusted p-values when accounting for changes in clinical symptoms. Among these significant probes, we identified annotated genes such as SRCIN1, RGS20, and INPPL. The gene SRCIN1 (SRC kinase signalling inhibitor 1) is a protein-coding gene that acts as a tumour suppressor and plays a role in inhibiting SRC kinase-mediated signalling pathways, involved in some oncogenic and cellular processes (Wang et al., 2016; Zhang et al., 2017). RGS20 encodes for the Regulator of G protein Signalling 20 protein, which is involved in regulating the activity of G protein-coupled receptors (GPCRs) and plays a role in various physiological processes such as neurotransmission, hormone signalling, and immune responses (Huang et al., 2018). The gene INPPL (Inositol Polyphosphate Phosphatase-Like) encodes the protein INPPL1, regulates insulin function and plays a role in the regulation of epidermal growth factor receptor turnover and actin remodelling. It has also been associated with cancer progression in various types of cancer (Pedicone et al., 2021). To date, there is no evidence from any studies indicating a connection between these genes and psychiatric disorders or stressful life events.
Looking at regional analysis, we have identified several significant DMRs annotated in genes related to inflammatory and immune processes, such as LTA, GFI1, ARID5B, CD52, TNFSF13, SLFN13, LST1, TNFRSF1A, CCRL2, and IL32. Moreover, some significant genes are known to be related to schizophrenia risk, such as LTA (Arab & Elhawary, 2015; Pandey et al., 2018), ARID5B (Drago et al., 2014), TNFSF13 (Catts & Weickert, 2012), TNFRSF1A (Gandal et al., 2018), IL32 (Keshavarz et al., 2022), LDLRAD4 (Kikuchi et al., 2003), and GAS7 (Z. Zhang et al., 2016), attention-deficit hyperactivity disorder, like GFI1 (Miyake et al., 2021), bipolar disorder, including TNFRSF1A (Gandal et al., 2018), and autism spectrum disorder, like TNFRSF1A (Gandal et al., 2018) and C11orf21 (Dall’Aglio et al., 2018). Notably, another significant gene is LST1 (Leukocyte Specific Transcript 1), which plays a role in immune system regulation. It is primarily expressed in immune cells, particularly macrophages and lymphocytes, and is known to be associated with MDD in genome-wide association studies of depression (Wu et al., 2021).
Gene set and pathways enrichment analyses revealed significant pathways and gene ontology processes related to the TNF receptor and TNF signalling pathway. This finding, along with the numerous genes involved in inflammatory and immune responses reported above, is consistent with the inflammatory/immune hypothesis of MDD pathophysiology, and also with the known association between inflammatory pathways and traumatic life events, particularly ELS (Müller et al., 2019; Silva et al., 2021). Also, patients with MDD present increased blood expression of pro-inflammatory cytokines, as well as elevated levels of various genes and proteins associated with innate immunity, including TNF, which has been observed in the brains of MDD patients (Maffioletti et al., 2020).
We also performed stratified subgroup analyses among patients who underwent TF-CBT or EMDR and the results highlighted several significant DMRs in those who were treated with EMDR. These regions were annotated in genes related to inflammatory and immune processes, such as LTA, GFI1, S100A8, ARID5B, CD52, PTX4, CD300A and CDKL1. Moreover, some significant genes are known to be related to schizophrenia risk, such as LTA (Arab & Elhawary, 2015; Pandey et al., 2018), S100A8 (Lanz et al., 2019), bipolar disorder, including S100A8 (Lanz et al., 2019), and attention-deficit hyperactivity disorder, such as GFI1 (Miyake et al., 2021). Furthermore, gene set enrichment analyses performed on the genes with significant DMRs in the EMDR subgroup revealed biological processes primarily associated with inflammatory responses, particularly the TNF signalling pathway. Additionally, we identified enrichment in transcription factor targets and we found significant values for targets of the transcription factors MAML1, HDGF, TBX1, MED25, and PRDM4. To date, there is no direct evidence linking these particular transcription factors to psychiatric disorders or stress-related responses. However, in a study performed by our group, a gene of the MED family, MED22, related to inflammatory responses, was found to be downregulated in MDD patients experiencing emotional neglect in childhood, suggesting that emotional neglect in infancy might involve the activation of genes targeting MED22, with consequent expression dysregulation (Minelli et al., 2018).
Analysing the findings of our study in the light of the current scientific literature, two studies reported DNA methylation changes in patients with PTSD following psychotherapy. One was performed by Vinkers and collaborators (Vinkers et al., 2021). This study investigated longitudinal changes of peripheral DNA methylation levels in relation to trauma-focused psychotherapies, EMDR including TF-CBT techniques, for PTSD in soldiers who obtained remission, non-remitted PTSD patients, and trauma-exposed military controls, also investigating whether these DMRs were relevant for the development of deployment-related PTSD in an independent prospective cohort. The authors found that recovery from PTSD following trauma-focused psychotherapies was related to specific DNA methylation alterations. Significant changes in DNA methylation were found at 12 DMRs in the genes APOB, MUC4, EDN2, ZFP57, GPX6, CFAP45, AFF3, TP73, UBCLP1, RPL13P and two intergenic regions, with consistent prospective evidence for ZFP57 methylation alterations related to changing PTSD symptoms. Similar to our study, the authors studied longitudinal DNA methylation changes following trauma-focused psychotherapies. However, differently from our study, the authors did not employ an epigenome-wide approach. Instead, they examined changes in DNA methylation in candidate-genes before and after trauma-focused psychotherapy in responding and non-responding PTSD patients and controls. Moreover, unlike our study, they did not consider MDD psychopathology or depressive symptoms as primary outcomes. Also, differently from our study, their patients received EMDR including TF-CBT techniques or TF-CBT without EMDR, and the authors did not consider stratified separate analyses by type of trauma-focused psychotherapy. The second study was conducted by Yang and colleagues, who performed an EWAS to identify epigenetic markers in PTSD patients undergoing prolonged-exposure psychotherapy with and without a hydrocortisone augmentation prior to each session (Yang et al., 2021). The analyses conducted in this study have identified the CREB–BDNF signalling pathway, linked to startle reaction and fear learning and memory processes, as a common marker that predicted symptom change and severity. Additionally, several resilience markers previously reported (FKBP5, NR3C1, SDK1, and MAD1L1) were found to be linked with the recovery from PTSD. Particularly, the levels of methylation in the gene body region of FKBP5 decreased significantly as the clinician-administered PTSD scale score decreased in individuals who responded to treatment, while there were no changes observed in non-responders. Similar to our study, the authors employed longitudinal epigenome-wide analyses to identify methylation changes in clinical samples. However, they focused on patients with PTSD, without considering depressive symptoms, and employed a different psychotherapeutic approach.
Our study has several strengths that highlight the significance of the findings and also some limitations that should be acknowledged when interpreting the results. It is the first to specifically analyse the longitudinal effects of trauma-focused psychotherapies in TRD patients characterized for ELS and is one of the few pieces of evidence supporting the biological effects of different types of trauma-focused psychotherapy in a longitudinal epigenomic approach. Other strengths of our study include the use of standardized clinical assessments conducted before and after trauma-focused psychotherapeutic treatment, ensuring that the measurements are reliable and comparable, enabling an accurate evaluation of symptoms changes. Additionally, we adopted an unbiased epigenome-wide approach and conducted a comprehensive biological longitudinal characterization of DMPs, DMRs, and different genes and pathways enrichment analyses. One limitation that may be addressed is the relatively small sample size. Although this may limit the generalizability of the findings, it should be regarded in the context of the originality of the study design, as it is the first epigenome-wide longitudinal approach to specifically depict the effects of trauma-focused psychotherapy in a well-characterized cohort of TRD patients. Moreover, the difficulties in conducting a longitudinal study with TRD patients, who have previously suffered from different types of ELS, should also be considered and explain the small sample size. Indeed, to date, four manuscripts available in the literature performed EWAS analyses in longitudinal studies on similar sample sizes with a similar duration (12 weeks) or shorter time (4, 5 or 8 weeks). In details, (1) Moschny et al. (2020), in which 17 MDD/TRD patients were treated with electroconvulsive therapy (ECT) for 4 weeks; (2) Yang et al. (2021), in which 42 PTSD patients were randomized to be treated with prolonged-exposure therapy vs placebo and were followed for 3 months/12 weeks; (3) Sirignano et al. (2021), in which 34 MDD/TRD patients were treated with ECT for about 4/5 weeks; (4) Van Assche et al. (2023), in which 84 MDD patients were randomized to be treated with personalized cognitive intervention compared to standard cognitive treatment for 8 weeks. Moreover, clinical intervention studies use more homogenous patient groups, reducing the need for very large samples. These studies also test specific interventions, suggesting that distinct epigenetic changes related to the intervention may occur. Another limitation is the relevance of peripheral blood methylation to the brain and the biological correspondence between the tissues. It is important to note that methylation differences can vary significantly across different tissues, even though consistent effects of various methylation quantitative trait loci have been observed across tissues (Liew et al., 2006). Regarding this point, it is important to take into account the difficulties of directly evaluating brain tissue, as well as the advantages of analysing peripheral blood samples, since epigenetic and transcriptional alterations in peripheral blood in some part reflect the molecular and cellular changes occurring in the brain. A further limitation is the absence of evaluations at the different timepoints for PTSD symptoms. While our primary outcome measure was depression, a proportion of participants in this study had PTSD as comorbid diagnosis. Evaluating PTSD symptom changes during treatment could add valuable information on possible underlying mechanisms of trauma-focused treatment in TRD, as it is possible that positive changes in depression symptoms during trauma-focused treatment were mediated by a decrease in PTSD symptoms. As many MDD and, in particular, TRD patients have a PTSD comorbidity (Flory & Yehuda, 2015), the treatment implications, as well as the risk and biological correlates of this frequent comorbidity should represent a great challenge. Additionally, according to a recent review (Zheng et al., 2022), Bumphunter and other supervised methods for regional analysis were shown to have high rates of false positives even using EPIC arrays. Aware of this limit, we made our analysis more stringent by analysing only those regions with at least 8 probes clustered with a maximum distance between probes of 300 bp. Finally, we did not account for potential confounding factors such as gender-related effects, and our analyses were not structured in a control group design. These points should be addressed in future studies.
In conclusion, our study offers valuable insights into the DNA methylation changes associated with trauma-focused psychotherapy. It highlights potential differentially methylated CpG sites, DMRs and biological pathways that play a role in trauma-focused psychotherapy outcomes. The findings point to numerous genes and biological pathways involved in the inflammatory and immune system, which is consistent with the inflammatory/immune hypothesis of MDD pathophysiology. Furthermore, our stratified analyses revealed that the most significant findings were related to the group of patients undergoing EMDR treatment. Numerous significant DMRs and biological pathways, mainly related to the inflammatory and immune system, were found in this subgroup. In summary, our study makes a valuable contribution in addressing a significant gap in the literature. These findings may enhance our understanding of the interplay between psychiatric and biological systems, ultimately leading to improved and personalized treatment outcomes.
Supplementary Material
Acknowledgements
We would like to express our sincere gratitude to all the volunteers who participated in the study. We thank all the staff of the Psychiatric Hospital ‘Villa Santa Chiara’.
Funding Statement
This work was supported by the Italian Ministry of Health under Grant [Ricerca Corrente 2022]. The post-doc position of Dr. Rosana Carvalho Silva was partly funded by the Psychiatric Hospital ‘Villa Santa Chiara’, Verona, Italy. The assistant research position of Valentina Menesello is funded by ERA-PerMed PROMPT project [IT-MoH ERP-2020-23671059].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Available upon request.
References
- Arab, A. H., & Elhawary, N. A. (2015). Association between ANKK1 (rs1800497) and LTA (rs909253) Genetic Variants and Risk of Schizophrenia. BioMed Research International, 1. 10.1155/2015/821827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arranz, M. J., Gallego-Fabrega, C., Martín-Blanco, A., Soler, J., Elices, M., Dominguez-Clavé, E., Salazar, J., Vega, D., Briones-Buixassa, L., & Pascual, J. C. (2021). A genome-wide methylation study reveals X chromosome and childhood trauma methylation alterations associated with borderline personality disorder. Translational Psychiatry, 11(1), 5. 10.1038/s41398-020-01139-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bifulco, A., Bernazzani, O., Moran, P. M., & Jacobs, C. (2005). The childhood experience of care and abuse questionnaire (CECA.Q): Validation in a community series. British Journal of Clinical Psychology, 44(4), 563–581. 10.1348/014466505X35344 [DOI] [PubMed] [Google Scholar]
- Catts, V. S., & Weickert, C. S. (2012). Gene expression analysis implicates a death receptor pathway in schizophrenia pathology. PLoS One, 7(4), e35511. 10.1371/journal.pone.0035511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecil, C. A. M., Smith, R. G., Walton, E., Mill, J., McCrory, E. J., & Viding, E. (2016). Epigenetic signatures of childhood abuse and neglect: Implications for psychiatric vulnerability. Journal of Psychiatric Research, 83, 184–194. 10.1016/j.jpsychires.2016.09.010 [DOI] [PubMed] [Google Scholar]
- Dall’Aglio, L., Muka, T., Cecil, C. A. M., Bramer, W. M., Verbiest, M. M. P. J., Nano, J., Hidalgo, A. C., Franco, O. H., & Tiemeier, H. (2018). The role of epigenetic modifications in neurodevelopmental disorders: A systematic review. Neuroscience & Biobehavioral Reviews, 94, 17–30. 10.1016/j.neubiorev.2018.07.011 [DOI] [PubMed] [Google Scholar]
- Dodd, S., Bauer, M., Carvalho, A. F., Eyre, H., Fava, M., Kasper, S., Kennedy, S. H., Khoo, J.-P., Lopez Jaramillo, C., Malhi, G. S., McIntyre, R. S., Mitchell, P. B., Castro, A. M. P., Ratheesh, A., Severus, E., Suppes, T., Trivedi, M. H., Thase, M. E., Yatham, L. N., … Berk, M. (2021). A clinical approach to treatment resistance in depressed patients: What to do when the usual treatments don’t work well enough? The World Journal of Biological Psychiatry, 22(7), 483–494. 10.1080/15622975.2020.1851052 [DOI] [PubMed] [Google Scholar]
- Drago, A., Giegling, I., Schäfer, M., Hartmann, A. M., Konte, B., Friedl, M., Serretti, A., & Rujescu, D. (2014). Genome-wide association study supports the role of the immunological system and of the neurodevelopmental processes in response to haloperidol treatment. Pharmacogenetics and Genomics, 24(6), 314–319. 10.1097/FPC.0000000000000052 [DOI] [PubMed] [Google Scholar]
- Du, P., Zhang, X., Huang, C.-C., Jafari, N., Kibbe, W. A., Hou, L., & Lin, S. M. (2010). Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics, 11(1), 587. 10.1186/1471-2105-11-587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory, J. D., & Yehuda, R. (2015). Comorbidity between post-traumatic stress disorder and major depressive disorder: alternative explanations and treatment considerations. Dialogues in Clinical Neuroscience, 17(2), 141–150. 10.31887/DCNS.2015.17.2/jflory [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin, J.-P., Triche, T. J., & Hansen, K. D. (2017). Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics (Oxford, England), 33(4), 558–560. 10.1093/bioinformatics/btw691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandal, M. J., Zhang, P., Hadjimichael, E., Walker, R. L., Chen, C., Liu, S., Won, H., van Bakel, H., Varghese, M., Wang, Y., Shieh, A. W., Haney, J., Parhami, S., Belmont, J., Kim, M., Moran Losada, P., Khan, Z., Mleczko, J., Xia, Y., … Geschwind, D. H. (2018). Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science, 362(6420), 10.1126/science.aat8127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GBD 2016. Disease and injury incidence and prevalence collaborators . (2016). Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. The Lancet, 388(10053), 1545–1602. 10.1016/S0140-6736(16)31678-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin, D. S., Sarvet, A. L., Meyers, J. L., Saha, T. D., Ruan, W. J., Stohl, M., & Grant, B. F. (2018). Epidemiology of Adult DSM-5 Major Depressive Disorder and Its Specifiers in the United States. JAMA Psychiatry, 75(4), 336–346. 10.1001/jamapsychiatry.2017.4602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, G., He, X., & Wei, X.-L. (2018). lncRNA NEAT1 promotes cell proliferation and invasion by regulating miR–365/RGS20 in oral squamous cell carcinoma. Oncology Reports, 39(4), 1948–1956. 10.3892/or.2018.6283 [DOI] [PubMed] [Google Scholar]
- Jaffe, D. H., Rive, B., & Denee, T. R. (2019). The humanistic and economic burden of treatment-resistant depression in Europe: a cross-sectional study. BMC Psychiatry, 19(1), 247. 10.1186/s12888-019-2222-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kautzky, A., Baldinger-Melich, P., Kranz, G. S., Vanicek, T., Souery, D., Montgomery, S., Mendlewicz, J., Zohar, J., Serretti, A., Lanzenberger, R., & Kasper, S. (2017). A New prediction model for evaluating treatment-resistant depression. The Journal of Clinical Psychiatry, 78(2), 215–222. 10.4088/JCP.15m10381 [DOI] [PubMed] [Google Scholar]
- Keshavarz, F., Soltani, M., Mokhtarian, K., Beshkar, P., Majidi, J., Azadegan-Dehkordi, F., Anjomshoa, M., & Bagheri, N. (2022). Autoantibodies against central nervous system antigens and the serum levels of IL-32 in patients with schizophrenia. Neuroimmunomodulation, 29(4), 493–499. 10.1159/000526425 [DOI] [PubMed] [Google Scholar]
- Kikuchi, M., Yamada, K., Toyota, T., Itokawa, M., Hattori, E., Yoshitsugu, K., Shimizu, H., & Yoshikawa, T. (2003). Two-step association analyses of the chromosome 18p11.2 region in schizophrenia detect a locus encompassing C18orf1. Molecular Psychiatry, 8(5), 467–469. 10.1038/sj.mp.4001280 [DOI] [PubMed] [Google Scholar]
- Knoblich, N., Gundel, F., Brückmann, C., Becker-Sadzio, J., Frischholz, C., & Nieratschker, V. (2018). DNA methylation of APBA3 and MCF2 in borderline personality disorder: Potential biomarkers for response to psychotherapy. European Neuropsychopharmacology, 28(2), 252–263. 10.1016/j.euroneuro.2017.12.010 [DOI] [PubMed] [Google Scholar]
- Kraus, C., Kadriu, B., Lanzenberger, R., Zarate, C. A., & Kasper, S. (2019). Prognosis and improved outcomes in major depression: a review. Translational Psychiatry, 9(1), 127. 10.1038/s41398-019-0460-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonté, B., Suderman, M., Maussion, G., Navaro, L., Yerko, V., Mahar, I., Bureau, A., Mechawar, N., Szyf, M., Meaney, M. J., & Turecki, G. (2012). Genome-wide epigenetic regulation by early-life trauma. Archives of General Psychiatry, 69(7), 722–731. 10.1001/archgenpsychiatry.2011.2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz, T. A., Reinhart, V., Sheehan, M. J., Rizzo, S. J. S., Bove, S. E., James, L. C., Volfson, D., Lewis, D. A., & Kleiman, R. J. (2019). Postmortem transcriptional profiling reveals widespread increase in inflammation in schizophrenia: A comparison of prefrontal cortex, striatum, and hippocampus among matched tetrads of controls with subjects diagnosed with schizophrenia, bipolar or major. Translational Psychiatry, 9(1), 151. 10.1038/s41398-019-0492-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew, C.-C., Ma, J., Tang, H.-C., Zheng, R., & Dempsey, A. A. (2006). The peripheral blood transcriptome dynamically reflects system wide biology: A potential diagnostic tool. Journal of Laboratory and Clinical Medicine, 147(3), 126–132. 10.1016/j.lab.2005.10.005 [DOI] [PubMed] [Google Scholar]
- Maffioletti, E., Minelli, A., Tardito, D., & Gennarelli, M. (2020). Blues in the brain and beyond: Molecular bases of major depressive disorder and relative pharmacological and Non-pharmacological treatments. Genes, 11(9), 10.3390/genes11091089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinova, Z., Maercker, A., Küffer, A., Robinson, M. D., Wojdacz, T. K., Walitza, S., Grünblatt, E., & Burri, A. (2017). DNA methylation profiles of elderly individuals subjected to indentured childhood labor and trauma. BMC Medical Genetics, 18(1), 21. 10.1186/s12881-017-0370-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta, D., Klengel, T., Conneely, K. N., Smith, A. K., Altmann, A., Pace, T. W., Rex-Haffner, M., Loeschner, A., Gonik, M., Mercer, K. B., Bradley, B., Müller-Myhsok, B., Ressler, K. J., & Binder, E. B. (2013). Childhood maltreatment is associated with distinct genomic and epigenetic profiles in posttraumatic stress disorder. Proceedings of the National Academy of Sciences, 110(20), 8302–8307. 10.1073/pnas.1217750110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minelli, A., Magri, C., Giacopuzzi, E., & Gennarelli, M. (2018). The effect of childhood trauma on blood transcriptome expression in major depressive disorder. Journal of Psychiatric Research, 104, 50–54. 10.1016/j.jpsychires.2018.06.014 [DOI] [PubMed] [Google Scholar]
- Minelli, A., Zampieri, E., Sacco, C., Bazzanella, R., Mezzetti, N., Tessari, E., Barlati, S., & Bortolomasi, M. (2019). Clinical efficacy of trauma-focused psychotherapies in treatment-resistant depression (TRD) in-patients: A randomized, controlled pilot-study. Psychiatry Research, 273, 567–574. 10.1016/j.psychres.2019.01.070 [DOI] [PubMed] [Google Scholar]
- Miyake, K., Miyashita, C., Ikeda-Araki, A., Miura, R., Itoh, S., Yamazaki, K., Kobayashi, S., Masuda, H., Ooka, T., Yamagata, Z., & Kishi, R. (2021). DNA methylation of GFI1 as a mediator of the association between prenatal smoking exposure and ADHD symptoms at 6 years: The Hokkaido Study on Environment and Children’s Health. Clinical Epigenetics, 13(1), 74. 10.1186/s13148-021-01063-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery, S. A., & Asberg, M. (1979). A new depression scale designed to be sensitive to change. British Journal of Psychiatry, 134(4), 382–389. 10.1192/bjp.134.4.382 [DOI] [PubMed] [Google Scholar]
- Moschny, N., Zindler, T., Jahn, K., Dorda, M., Davenport, C. F., Wiehlmann, L., Maier, H. B., Eberle, F., Bleich, S., Neyazi, A., & Frieling, H. (2020). Novel candidate genes for ECT response prediction-a pilot study analyzing the DNA methylome of depressed patients receiving electroconvulsive therapy. Clinical Epigenetics, 12(1), 114. 10.1186/s13148-020-00891-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, N., Krause, D., Barth, R., Myint, A.-M., Weidinger, E., Stettinger, W., Zill, P., Drexhage, H., & Schwarz, M. J. (2019). Childhood adversity and current stress are related to Pro- and anti-inflammatory cytokines in major depression. Journal of Affective Disorders, 253, 270–276. 10.1016/j.jad.2019.04.088 [DOI] [PubMed] [Google Scholar]
- Neves, I., Dinis-Oliveira, R. J., & Magalhães, T. (2021). Epigenomic mediation after adverse childhood experiences: A systematic review and meta-analysis. Forensic Sciences Research, 6(2), 103–114. 10.1080/20961790.2019.1641954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell, K. J., Chen, L., MacIsaac, J. L., McEwen, L. M., Nguyen, T., Beckmann, K., Zhu, Y., Chen, L. M., Brooks-Gunn, J., Goldman, D., Grigorenko, E. L., Leckman, J. F., Diorio, J., Karnani, N., Olds, D. L., Holbrook, J. D., Kobor, M. S., & Meaney, M. J. (2018). DNA methylome variation in a perinatal nurse-visitation program that reduces child maltreatment: A 27-year follow-up. Translational Psychiatry, 8(1), 15. 10.1038/s41398-017-0063-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, G. N., Rizavi, H. S., Zhang, H., & Ren, X. (2018). Abnormal gene and protein expression of inflammatory cytokines in the postmortem brain of schizophrenia patients. Schizophrenia Research, 192, 247–254. 10.1016/j.schres.2017.04.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parade, S. H., Huffhines, L., Daniels, T. E., Stroud, L. R., Nugent, N. R., & Tyrka, A. R. (2021). A systematic review of childhood maltreatment and DNA methylation: Candidate gene and epigenome-wide approaches. Translational Psychiatry, 11(1), 134. 10.1038/s41398-021-01207-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedicone, C., Meyer, S. T., Chisholm, J. D., & Kerr, W. G. (2021). Targeting SHIP1 and SHIP2 in Cancer. Cancers, 13(4)), 10.3390/cancers13040890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perroud, N., Salzmann, A., Prada, P., Nicastro, R., Hoeppli, M. E., Furrer, S., Ardu, S., Krejci, I., Karege, F., & Malafosse, A. (2013). Response to psychotherapy in borderline personality disorder and methylation status of the BDNF gene. Translational Psychiatry, 3(1), e207. 10.1038/tp.2012.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidsley, R., Zotenko, E., Peters, T. J., Lawrence, M. G., Risbridger, G. P., Molloy, P., Van Djik, S., Muhlhausler, B., Stirzaker, C., & Clark, S. J. (2016). Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biology, 17(1), 208. 10.1186/s13059-016-1066-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sales, G., Calura, E., & Romualdi, C. (2019). metaGraphite-a new layer of pathway annotation to get metabolite networks. Bioinformatics (Oxford, England), 35(7), 1258–1260. 10.1093/bioinformatics/bty719 [DOI] [PubMed] [Google Scholar]
- Silva, R. C., Maffioletti, E., Gennarelli, M., Baune, B. T., & Minelli, A. (2021). Biological correlates of early life stressful events in major depressive disorder. Psychoneuroendocrinology, 125, 105103. 10.1016/j.psyneuen.2020.105103 [DOI] [PubMed] [Google Scholar]
- Sirignano, L., Frank, J., Kranaster, L., Witt, S. H., Streit, F., Zillich, L., Sartorius, A., Rietschel, M., & Foo, J. C. (2021). Methylome-wide change associated with response to electroconvulsive therapy in depressed patients. Translational Psychiatry, 11(1), 347. 10.1038/s41398-021-01474-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders, C., Maihofer, A. X., Ratanatharathorn, A., Baker, D. G., Boks, M. P., Geuze, E., Jain, S., Kessler, R. C., Pishva, E., Risbrough, V. B., Stein, M. B., Ursano, R. J., Vermetten, E., Vinkers, C. H., Smith, A. K., Uddin, M., Rutten, B. P. F., PGC PTSD EWAS Consortium, & Nievergelt, C. M. (2020). Longitudinal epigenome-wide association studies of three male military cohorts reveal multiple CpG sites associated with post-traumatic stress disorder. Clinical Epigenetics, 12(1), 11. 10.1186/s13148-019-0798-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thase, M. E., & Rush, A. J. (1997). When at first you don’t succeed: Sequential strategies for antidepressant nonresponders. The Journal of Clinical Psychiatry, 58(Suppl 1), 16–21. 10.4088/JCP.v58n0103 [DOI] [PubMed] [Google Scholar]
- Van Assche, E., Hohoff, C., Zang, J., Knight, M. J., & Baune, B. T. (2023). Epigenetic modification related to cognitive changes during a cognitive training intervention in depression. Progress in Neuro-Psychopharmacology and Biological Psychiatry, 127, 110835. 10.1016/j.pnpbp.2023.110835 [DOI] [PubMed] [Google Scholar]
- Vinkers, C. H., Geuze, E., van Rooij, S. J. H., Kennis, M., Schür, R. R., Nispeling, D. M., Smith, A. K., Nievergelt, C. M., Uddin, M., Rutten, B. P. F., Vermetten, E., & Boks, M. P. (2021). Successful treatment of post-traumatic stress disorder reverses DNA methylation marks. Molecular Psychiatry, 26(4), 1264–1271. 10.1038/s41380-019-0549-3 [DOI] [PubMed] [Google Scholar]
- Wang, P., Wang, H., Li, X., Liu, Y., Zhao, C., & Zhu, D. (2016). Srcin1 suppressed osteosarcoma cell proliferation and invasion. PLoS One, 11(8), e0155518. 10.1371/journal.pone.0155518 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wu, W., Howard, D., Sibille, E., & French, L. (2021). Differential and spatial expression meta-analysis of genes identified in genome-wide association studies of depression. Translational Psychiatry, 11(1), 8. 10.1038/s41398-020-01127-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, S., Shan, Y., Zhong, S., Miao, H., Luo, Y., Ran, H., & Jia, Y. (2021). The effectiveness of Eye movement desensitization and reprocessing toward adults with major depressive disorder: A meta-analysis of randomized controlled trials. Frontiers in Psychiatry, 12, 700458. 10.3389/fpsyt.2021.700458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, R., Xu, C., Bierer, L. M., Flory, J. D., Gautam, A., Bader, H. N., Lehrner, A., Makotkine, I., Desarnaud, F., Miller, S. A., Jett, M., Hammamieh, R., & Yehuda, R. (2021). Longitudinal genome-wide methylation study of PTSD treatment using prolonged exposure and hydrocortisone. Translational Psychiatry, 11(1), 398. 10.1038/s41398-021-01513-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M., Ma, F., Xie, R., Wu, Y., Wu, M., Zhang, P., Peng, Y., Zhao, J., Xiong, J., Li, A., Kequan, C., Zhang, Y., Liu, S., Wang, J., & Chen, X. (2017). Overexpression of Srcin1 contributes to the growth and metastasis of colorectal cancer. International Journal of Oncology, 50(5), 1555–1566. 10.3892/ijo.2017.3952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Zheng, F., You, Y., Ma, Y., Lu, T., Yue, W., & Zhang, D. (2016). Growth arrest specific gene 7 is associated with schizophrenia and regulates neuronal migration and morphogenesis. Molecular Brain, 9(1), 54. 10.1186/s13041-016-0238-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Y., Lunetta, K. L., Liu, C., Katrinli, S., Smith, A. K., Miller, M. W., & Logue, M. W. (2022). An evaluation of the genome-wide false positive rates of common methods for identifying differentially methylated regions using illumina methylation arrays. Epigenetics, 17(13), 2241–2258. 10.1080/15592294.2022.2115600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang, H., Wang, H., & Ji, Z. (2022). findPC: An R package to automatically select the number of principal components in single-cell analysis. Bioinformatics (Oxford, England), 38(10), 2949–2951. 10.1093/bioinformatics/btac235 [DOI] [PubMed] [Google Scholar]
- Ziegler, C., Richter, J., Mahr, M., Gajewska, A., Schiele, M. A., Gehrmann, A., Schmidt, B., Lesch, K.-P., Lang, T., Helbig-Lang, S., Pauli, P., Kircher, T., Reif, A., Rief, W., Vossbeck-Elsebusch, A. N., Arolt, V., Wittchen, H.-U., Hamm, A. O., Deckert, J., & Domschke, K. (2016). MAOA gene hypomethylation in panic disorder-reversibility of an epigenetic risk pattern by psychotherapy. Translational Psychiatry, 6(4), e773. 10.1038/tp.2016.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Available upon request.