Abstract
The von Hippel-Lindau tumor suppressor protein (pVHL) binds to elongins B and C and posttranscriptionally regulates the accumulation of hypoxia-inducible mRNAs under normoxic (21% O2) conditions. Here we report that pVHL binds, via elongin C, to the human homolog of the Caenorhabditis elegans Cul2 protein. Coimmunoprecipitation and chromatographic copurification data suggest that pVHL-Cul2 complexes exist in native cells. pVHL mutants that were unable to bind to complexes containing elongin C and Cul2 were likewise unable to inhibit the accumulation of hypoxia-inducible mRNAs. A model for the regulation of hypoxia-inducible mRNAs by pVHL is presented based on the apparent similarity of elongin C and Cul2 to Skp1 and Cdc53, respectively. These latter proteins form complexes that target specific proteins for ubiquitin-dependent proteolysis.
von Hippel-Lindau (VHL) disease, a hereditary cancer syndrome characterized by the development of renal carcinomas, pheochromocytomas, and vascular tumors of the central nervous system and retina, is caused by germ line mutations in the VHL tumor suppressor gene (9, 17, 27, 35). Tumor development in this setting is due to somatic loss or mutation of the remaining wild-type VHL allele (9, 17).
Functional inactivation of both VHL alleles has also occurred in the majority of sporadic renal cell carcinomas and cerebellar hemangioblastomas examined to date (7, 10, 14, 21, 40, 45). Loss of heterozygosity at the VHL gene locus has been described for premalignant lesions of the kidney, such as atypical cysts (32, 49), suggesting that inactivation of the VHL gene is an early event in renal carcinogenesis. Furthermore, reintroduction of a wild-type but not mutant VHL cDNA into VHL (−/−) renal carcinoma cells suppresses their ability to form tumors in nude mouse xenograft assays (11, 18).
The VHL gene product, pVHL, binds to elongins B and C (6, 22, 24, 44). These two proteins, when bound to elongin A, generate a transcriptional elongation complex called elongin or SIII (1). pVHL competes with elongin A, at least in vitro, for binding to elongins B and C and thereby inhibits elongin activity (6). The majority of inherited pVHL mutants are known (or can be predicted) to be defective for binding to elongins B and C (22, 24, 48). Thus, inhibition of elongin (SIII) activity may contribute to tumor suppression by pVHL.
VHL-associated neoplasms are typically hypervascular and overproduce angiogenic peptides, such as vascular endothelial growth factor (VEGF) (3, 39, 43, 47). Several groups have shown that pVHL is a negative regulator of hypoxia-inducible mRNAs, such as the VEGF and Glut1 glucose transporter mRNAs, under normoxic conditions (11, 16, 37, 41). Surprisingly, this effect of pVHL appears to be largely at the level of mRNA stability rather than at the level of transcript elongation (11, 16). Thus, there is currently no direct evidence that pVHL regulates transcriptional elongation in vivo. Furthermore, pVHL appears to reside primarily but not exclusively in the cytoplasm, again raising the possibility that pVHL performs a function(s) unrelated to transcriptional elongation (5, 18, 28, 31).
Cullins are a recently identified family of proteins that exhibit significant sequence similarity to the yeast Cdc53 protein and hence are suspected of functioning to target certain proteins for ubiquitin-dependent proteolysis (15, 19, 23). Here we show that a cullin, Cul2 (23), binds specifically to pVHL-elongin complexes and that the formation of these complexes is linked to the regulation of hypoxia-inducible mRNAs by pVHL. A model for the regulation of hypoxia-inducible mRNAs by pVHL is proposed based on the apparent similarity of elongins C, B, and A to Skp1, ubiquitin, and F-box proteins, respectively (2, 8).
MATERIALS AND METHODS
Cell culture.
786-O and A498 renal carcinoma cells, obtained from the American Type Culture Collection (Rockville, Md.), were grown in Dulbecco’s modified Eagle’s medium containing 10% defined and supplemented bovine calf serum (Hyclone) at 37°C in a humidified, 10% CO2-containing atmosphere. 786-O subclones stably transfected with pRc/CMV, pRc/CMV-VHL(wt), and pRc/CMV(1-115) were described previously and were maintained in the presence of G418 (500 μg/ml) (18). The 786-O and A498 subclones described in this article were generated and propagated in the same manner. Radioisotopic labelling was performed by methionine starvation for 1 h, followed by growth in methionine-free Dulbecco’s modified Eagle’s medium supplemented with [35S]methionine (0.5 mCi/ml of medium; EXPRE35S35S protein labelling mix [New England Nuclear Corp.]) and 2% dialyzed fetal bovine serum for 4 h. Cell were lysed in 50 mM Tris (pH 8.0)–120 mM NaCl–0.5% Nonidet P-40 (NP-40) unless otherwise indicated.
Primer sets.
The primer sets used for plasmid construction were as follows: A, 5′-CAGAGGATCCGCCACCATGGATGGGCTTCTGGTTAAC-3′ and 5′-AGCCGAATTCAGTCTTTCTGCACATTTGG-3′; B, 5′-CAGAGGATCCGCCACCATGGATGGGCTTCTGGTTAAC-3′ and 5′-CGTTGAATTCAAATGCGATCCTGTGTCAG-3′; C, 5′-AGGTGGATCCTGGCTCTTCAGAGATGCCA-3′ and 5′-CTGTCTAGAATTCAATCTTCGTAGAGCGACCTGACG-3′; D, 5′-AGGTGGATCCTGGCTCTTCAGAGATGCCA-3′ and 5′-CTGTCTAGAATTCACCGGACAACCTGGAGGCATCG-3′; E, 5′-GCGCGGATCCGCCACCATGTCCCAGGTCATCTTCTGC-3′ and 5′-CTGTGAATTCAAGGCTTGACTAGGCTCCG-3′; F, 5′-GCGCGGATCCGCCACCATGGAGCCGCAGCCCTACCCA-3′ and 5′-CTGTGAATTCAAGGCTTGACTAGGCTCCG-3′; G, 5′-GCGCGGATCCAACAATTTTATCAAAAACCAAGAC-3′ and 5′-CTACTTATGTCGATGCAGCGCACTTAAGCGCG-3′; H, 5′-GCGCAAGCTTGCC ACCATGGCTAGCATGACTGGTGGACAGCAAATGGGATCCATGGAT GGAGAGGAGAAAACC-3′ and 5′-GCGCAATTGGAGACCTTAACAATCTAAGAAGTTCGC-3′; and I, 5′-GCGCAAGCTTGGATCCGCCACCATGGACGTGTTTCTCATG-3′ and 5′-GCGCGAATTCACTGCACAGCTTGTTC-3′.
Plasmids.
pRc/CMV was obtained from Invitrogen. pcDNAI-HA-VHL, pRc/CMV-VHL(wt), and pRc/CMV-VHL(1-115), each of which introduces an N-terminal hemagglutinin (HA) epitope tag, were described previously (18). To make pRc/CMV-VHL(1-197) and pRc/CMV-VHL(1-206), a human VHL cDNA was PCR amplified with primer sets A and B, respectively. The PCR products were restricted with Bst1107 and EcoRI and used to replace the corresponding cDNA fragment in pcDNAI-HA-VHL. The resulting HA-VHL cDNAs were then excised as HindIII-XbaI fragments and cloned into pRc/CMV that had been linearized with these two enzymes.
To make pRc/CMV-VHL(1-187) and pRc/CMV-VHL(1-167), a human VHL cDNA was PCR amplified with primer sets C and D, respectively. The PCR products were restricted with HpaI and XbaI and used to replace the corresponding cDNA fragments in pRc/CMV-VHL.
To make pRc/CMV-VHL(1-155), pcDNAI-HA-VHL was digested with Bst1107 and EcoRI. The backbone plasmid was electrophoretically separated from the corresponding VHL cDNA fragment, blunt ended with Klenow DNA polymerase, and recircularized. The resulting HA-VHL cDNA was then excised as a HindIII-XbaI fragment and cloned into pRc/CMV that had been linearized with these two enzymes.
To make pRc/CMV-VHL(72-213) and PRc/CMV-VHL(94-213), a human VHL cDNA was PCR amplified with primer sets E and F, respectively. The PCR products were restricted with BamHI and BspEI and used to replace the corresponding cDNA fragments in pcDNAI-HA-VHL. The resulting HA-VHL cDNAs were then excised as HindIII-XbaI fragments and cloned into pRc/CMVI that had been linearized with these two enzymes.
To make pRc/CMV-VHL(126-206), the BamHI-EcoRI VHL cDNA insert from pGEX2TK-VHL(126-206) (22) was cloned into the pcDNAI-HA backbone that had been linearized with these two enzymes. The resulting HA-VHL cDNA was then excised as a HindIII-XbaI fragment and cloned into pRc/CMV that had been linearized with these two enzymes.
pRc/CMV-VHL(L158S), pRc/CMV-VHL(C162F), and pRc/CMV-VHL (157NAAIRS) were generated by site-directed mutagenesis with mutator oligonucleotides A (5′-CCAGTGTATACTTCGAAAGAGCGATGCCTC-3′), B (5′-GAAAGAGCGATTCCTCCAGGTTGTCCG-3′), and C (5′-ACACTGCCAG TGTATAATGCGGCAATACGATCGCTCCAGGTTGTCCGG-3′), respectively, by using a Transformer Site-Directed Mutagenesis kit (Version 2; Clontech) according to the manufacturer’s instructions and a selection primer designed to eliminate the XbaI site in the pRc/CMV polylinker (5′-TCGAGCATGCAGCTTAGCGGGCCCTATTCTATA-3′).
A partial human Cul2 cDNA cloned into pBSK (Stratagene) was kindly provided by Edward Kipreos. A BamHI-NcoI Cul2 cDNA fragment was used to screen a human embryonic kidney (293 cells) λZAP library (18). Positive clones were isolated, and plasmids were rescued by in vivo excision according to the manufacturer’s instructions. Multiple independent, overlapping cDNA clones were sequenced along both DNA strands and used to assemble a consensus Cul2 cDNA sequence. Clone 16-1 contained the entire open reading frame for human Cul2.
To make pGEX2TK-Cul2(491-745), a human Cul2 cDNA was PCR amplified with primer set G. The PCR product was restricted with BamHI and EcoRI and subcloned into pGEX2TK (Pharmacia PL) that had been linearized with these two enzymes.
To make pcDNAIII-T7-elongin C, a human elongin C cDNA was PCR amplified with primer set H. The PCR product, encoding a T7 epitope tag fused to the N terminus of elongin C, was restricted with HindIII and BsaI and subcloned into pcDNAIII (Invitrogen) that had been linearized with HindIII and EcoRI.
To make the pGEX2TK-elongin C plasmids, a human elongin C cDNA was PCR amplified with sense primers containing BamHI sites and antisense primers containing either EcoRI sites or, for fragments containing internal EcoRI sites, BsaAI sites that had been designed to generate overhangs that were compatible with EcoRI cohesive ends. The products were digested and ligated into pGEX2TK that had been linearized with BamHI and EcoRI.
To make pSP72-elongin B, a human elongin B cDNA was PCR amplified with primer set I. The product was digested with BamHI and EcoRI and ligated into pSP72 (Promega) that had been cut with these two enzymes.
All PCRs were performed with Pfu DNA polymerase (Stratagene), and the regions of the final cDNAs generated by PCR were confirmed by DNA sequence analysis. Site-directed VHL gene mutations were confirmed by DNA sequence analysis.
Immunoprecipitation and immunoblotting.
Immunoprecipitation and immunoblotting were performed as described previously (22). Immunoprecipitation was performed with approximately 1 μg of monoclonal antibody, and immunoprecipitates were washed five times with 20 mM Tris (pH 8.0)–900 mM NaCl–1 mM EDTA–0.5% NP-40 unless otherwise indicated prior to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. Anti-Cul2 immunoblotting was performed with crude anti-Cul2 rabbit sera at a 1:100 dilution. Protein concentrations of whole-cell extracts were determined by the Bradford method and were used to normalize protein loading in whole-cell immunoblot assays.
Antibodies.
Monoclonal anti-HA and anti-T7 antibodies were obtained from Boehringer Mannheim and Novagen, respectively. The polyclonal anti-Glut1 antibody GT-11A was obtained from Alpha Diagnostic. The monoclonal anti-VHL antibody IG32 and polyclonal anti-elongin B sera were described previously (8, 22). The anti-T-antigen monoclonal antibody pAB419 was used as a control (12). To generate polyclonal anti-Cul2 sera, GST-Cul2(491-745) was expressed in Escherichia coli, purified by preparative SDS-polyacrylamide gel electrophoresis, and injected into New Zealand White rabbits as previously described (18).
Peptide microsequence analysis.
Isolation of p70 by preparative SDS-polyacrylamide gel electrophoresis, digestion with trypsin, and sequencing of tryptic peptides were performed as described previously (22).
In vitro translation.
Coupled in vitro transcription and translation were performed with TNT reticulocyte lysate (Promega) according to the manufacturer’s instructions.
Partial proteolytic peptide mapping.
Partial proteolytic peptide mapping of radiolabelled Cul2 was performed precisely as described elsewhere (13).
Purification of pVHL complexes from rat liver.
A postlysosomal supernatant was prepared from the livers of 360 male Sprague-Dawley rats and fractionated by (NH4)2SO4 precipitation as described previously (4). The 0 to 38% (NH4)2SO4 fraction was resuspended in buffer A (20 mM HEPES-NaOH [pH 7.9], 0.5 mM EDTA, 1 mM dithiothreitol [DTT], 10% [vol/vol] glycerol) containing 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 10 mg of antipain per ml, and 10 mg of leupeptin per ml and was brought to a conductivity equivalent to that of buffer A containing 100 mM KCl by a combination of 3 h of dialysis against buffer A containing 0.5 mM PMSF and of dilution with the same buffer. Following centrifugation for 30 min at 28,000 × g, the resulting supernatant was mixed with 800 ml of phosphocellulose (P11; Whatman) preequilibrated in buffer A containing 100 mM KCl and 0.5 mM PMSF. After 45 min, the slurry was filtered at 500 ml/h in a 10.5-cm-diameter column. The phosphocellulose flowthrough fraction was mixed with 250 ml of Toyopearl DEAE-650M (TosoHaas) preequilibrated in buffer C (40 mM Tris-HCl [pH 7.9], 0.5 mM EDTA, 1 mM DTT, 10% [vol/vol] glycerol) containing 80 mM KCl. After 45 min, the slurry was filtered at 150 ml/h in a 5.0-cm-diameter column and then was washed at the same flow rate with buffer C containing 80 mM KCl. The column contents were eluted stepwise at 250 ml/h with buffer A containing 220 mM KCl, and 50-ml fractions were collected. Fractions containing pVHL were concentrated by 0.3 mg/ml (NH4)2SO4 precipitation, resuspended in 10 ml of buffer A containing 10 mg of antipain per ml and 10 mg of leupeptin per ml, and dialyzed against buffer A containing 300 mM KCl to a conductivity equivalent to that of buffer A containing 500 mM (NH4)2SO4. Following centrifugation for 15 min at 12,000 × g, the resulting supernatant was applied at 20 ml/h to a 500-ml, 2.6-cm-diameter Ultrogel AcA34 gel filtration column (IBF Biotechnics) preequilibrated with buffer A containing 400 mM KCl. The column contents were eluted at 20 ml/h, and 10-ml fractions were collected. Fractions containing pVHL, which eluted with an apparent native molecular mass of between 330 and 200 kDa, were diluted with an equal volume of buffer E (40 mM HEPES-NaOH [pH 7.9], 0.1 mM EDTA, 1 mM DTT, 10% [vol/vol] glycerol) containing 2.0 M (NH4)2SO4. Following centrifugation for 20 min at 60,000 × g, the resulting supernatant was applied to a Spherogel TSK phenyl 5-PW column (21.5 by 150 mm; Beckman Instruments) preequilibrated with buffer E containing 1.0 M (NH4)2SO4.
The column contents were eluted at 5 ml/min with a linear gradient of 250 ml of 1.0 M (NH4)2SO4 in buffer E to no (NH4)2SO4 in buffer E, and 5-ml fractions were collected. Fractions containing pVHL, which eluted at between 170 and 80 mM (NH4)2SO4, were pooled and dialyzed against buffer C to a conductivity equivalent to that of buffer C containing 60 mM KCl. Following centrifugation for 20 min at 60,000 × g, the resulting supernatant was applied to a Bio-Gel SEC DEAE 5-PW column (7.5 by 75 mm; Bio-Rad) preequilibrated with buffer C containing 60 mM KCl. The column contents were eluted at 0.8 ml/min with a 40-ml linear gradient of 60 to 250 mM KCl in buffer C, and 0.7-ml fractions were collected. Fractions containing pVHL, which eluted at between 120 and 140 mM KCl, were pooled and brought to 5 mM potassium phosphate. Following centrifugation for 20 min at 60,000 × g, the resulting supernatant was applied to a crystalline hydroxylapatite Bio-Scale CHT-I column (7 by 52 mm; Bio-Rad) preequilibrated with buffer P (5 mM potassium phosphate [pH 7.6], 0.1 mM EDTA, 1 mM DTT, 10% [vol/vol] glycerol). The column contents were eluted at 0.6 ml/min with a 24-ml linear gradient of 50 to 400 mM potassium phosphate (pH 7.6) in buffer P, and 0.3-ml fractions were collected. Fractions containing pVHL, which eluted at between 50 and 80 mM potassium phosphate, were pooled and diluted with buffer C containing 50 mM KCl to a conductivity equivalent to that of buffer C containing 80 mM KCl. Following centrifugation for 20 min at 60,000 × g, the resulting supernatant was applied to a Mono Q column (5 by 50 mm; Pharmacia) preequilibrated with buffer C containing 80 mM KCl. The column contents were eluted at 0.4 ml/min with a 12-ml linear gradient of 80 to 300 mM KCl in buffer C, and 0.2-ml fractions were collected. Fractions containing pVHL eluted at between 180 and 200 mM KCl.
GST pull-down assay.
Approximately 1 μg of glutathione S-transferase (GST)–elongin C fusion protein, as determined by Coomassie blue staining, and 5 μl of elongin B in vitro translate were incubated in 500 μl of NETN (20 mM Tris-HCl [pH 8.0], 120 mM NaCl, 1 mM EDTA, 0.5% NP-40) for 1 h at 4°C with rocking. Twenty-five microliters of glutathione-Sepharose (1:1 in NETN supplemented with 0.5% milk) was then added. After 30 min of incubation, the glutathione-Sepharose was washed five times with NETN. Bound proteins were eluted by boiling in SDS-containing sample buffer, resolved by electrophoresis in SDS–15% polyacrylamide gels, and detected by fluorography.
Northern blot analysis.
RNA was extracted from nearly confluent monolayer cultures in RNAzol B (Tel-Test, Inc.) according to the manufacturer’s protocol. Ten micrograms of total RNA was denatured in a solution containing 20 mM MOPS [3-(N-morpholino)propanesulfonic acid], 6.3% formaldehyde, and 48% formamide, resolved in a 1% agarose–2.2 mM formaldehyde gel in 20 mM MOPS–5 mM sodium acetate–1 mM EDTA (pH 8) (in the presence of ethidium bromide), and transferred to a Zeta-Probe GT nylon membrane (Bio-Rad) in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). To detect the Glut1 mRNA, the membrane was hybridized with a radiolabelled oligonucleotide complementary to a region encoding the intracellular C-terminal domain of the Glut1 protein (5′-GCTCCTCGGGTGTCTTATCACTTTG-3′) in a solution containing 0.655 M NaCl, 50 mM Na2HPO4 (pH 7.2), 7% SDS, 10 mg of bovine serum albumin per ml, and 25 mg of polyadenylic acid per ml. The membrane was washed in 2× SSC–1% SDS at 50°C twice for 30 min each time, followed by a wash with 0.5× SSC–0.1% SDS at 50°C for 2 min. To detect the VEGF mRNAs, the membrane was hybridized with a radiolabelled VEGF121 cDNA (a gift from Mark Goldberg) in a solution containing 50 mM Tris (pH 7.5), 1 M NaCl, 1% SDS, 10% dextran sulfate, 50% formamide, and 100 mg of herring sperm DNA per ml. The membrane was washed twice in 2× SSC–0.2% SDS at 25°C for 30 min each time, twice in 1× SSC–0.2% SDS at 25°C for 15 min each time, and finally in 0.5× SSC–0.2% SDS at 65°C for 15 min.
RESULTS
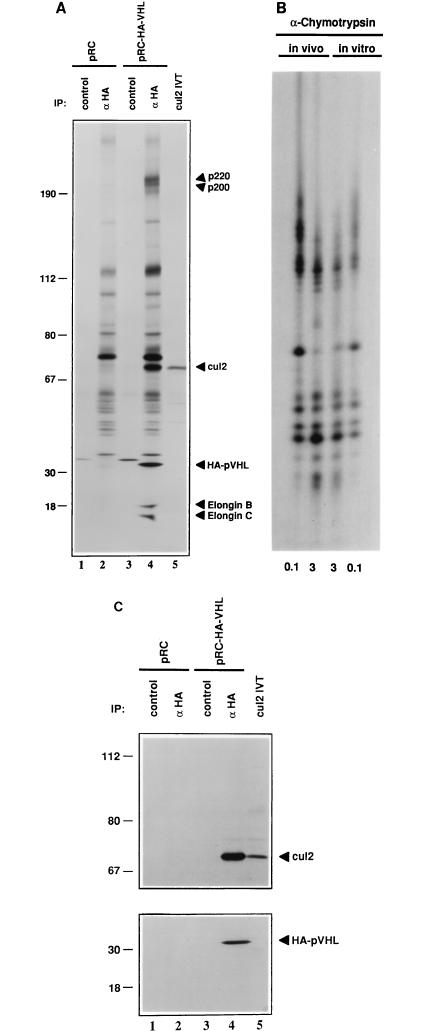
To look for pVHL-binding proteins, 786-O [VHL (−/−)], renal carcinoma cells that were stably transfected with a plasmid encoding HA-tagged pVHL (Fig. 1A, lanes 3 and 4) or the backbone expression plasmid (Fig. 1A, lanes 1 and 2) were metabolically labelled with [35S]methionine, lysed, and immunoprecipitated with an anti-HA antibody (Fig. 1A, lanes 2 and 4) or a control antibody (Fig. 1A, lanes 1 and 3) under stringent conditions. Bound proteins were detected by autoradiography (Fig. 1A) or immunoblot analysis (Fig. 1C). pVHL coimmunoprecipitated with cellular proteins with molecular masses of ∼200, 70, 18, and 15 kDa (compare Fig. 1A, lane 4, to lanes 2 and 3). These proteins were also detected in monoclonal and polyclonal anti-VHL immunoprecipitates but not in isotype-matched control immunoprecipitates (data not shown). The 18- and 15-kDa proteins were shown previously to be elongins B and C, respectively (6, 22, 24). The characterization of p200 will be described elsewhere.
FIG. 1.
Coimmunoprecipitation of Cul2 with ectopically produced pVHL. (A and C) VHL (−/−) renal carcinoma cells stably transfected with a plasmid encoding HA-tagged pVHL (lanes 3 and 4) or with the backbone expression plasmid (lanes 1 and 2) were metabolically labelled with 35S, lysed, and immunoprecipitated (IP) with control or anti-HA antibody (αHA) as indicated. Bound proteins (lanes 1 to 4) as well as 35S-labelled Cul2 in vitro translate (IVT) (lane 5) were resolved by SDS-polyacrylamide gel electrophoresis and detected by autoradiography (A), anti-Cul2 immunoblot analysis (C, upper panel), or anti-HA immunoblot analysis (C, lower panel). Numbers at left are in kilodaltons. (B) The 35S-labelled Cul2 bands in panel A, lanes 4 and 5 (in vivo and in vitro, respectively), were excised and digested with 0.1 and 3 μg of α-chymotrypsin as indicated. Proteolytic fragments were resolved in an SDS–20% polyacrylamide gel and detected by autoradiography.
p70 was purified by preparative anti-HA immunoelectrophoresis and subjected to microsequence analysis following tryptic digestion. Three peptide sequences that were present in the conceptual open reading frame of the human Cul2 gene were obtained (23). A full-length human Cul2 cDNA clone was isolated by screening a cDNA library with the available partial human Cul2 cDNA clone and was translated in vitro. The major Cul2 in vitro translation product comigrated with p70 (Fig. 1A, compare lanes 4 and 5), and the partial proteolytic peptide maps of these two proteins generated with α-chymotrypsin (Fig. 1B) and Staphylococcus aureus V8 protease (data not shown) were identical. In some experiments, translation of Cul2, whether in vitro or in vivo, also gave rise to a second band that migrated more slowly in SDS-polyacrylamide gels (see, for example, Fig. 1C and 4). The nature of this second band is currently under investigation. Finally, both p70 and the Cul2 in vitro translation product were specifically recognized by a polyclonal anti-Cul2 antibody in Western blot assays (Fig. 1C). Thus, pVHL, at least when overproduced, can bind to human Cul2. While these experiments were in progress, the same conclusion was reported by Pause and coworkers (38).
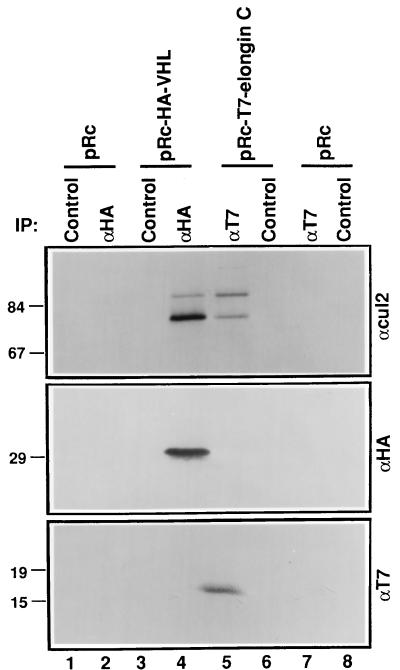
FIG. 4.
Cul2 binds to elongin C in the absence of pVHL. VHL (−/−) renal carcinoma cells stably transfected with a plasmid encoding HA-tagged pVHL (lanes 3 and 4), T7 epitope-tagged elongin C (lanes 5 and 6), or the backbone expression plasmid (lanes 1, 2, 7, and 8) were lysed and immunoprecipitated (IP) with anti-HA (αHA), anti-T7 (αT7), or control antibody as indicated. Bound proteins were resolved by SDS-polyacrylamide gel electrophoresis and immunoblotted with anti-Cul2 polyclonal (upper panel), anti-HA monoclonal (middle panel), or anti-T7 monoclonal (lower panel) antibodies. Numbers at left are in kilodaltons.
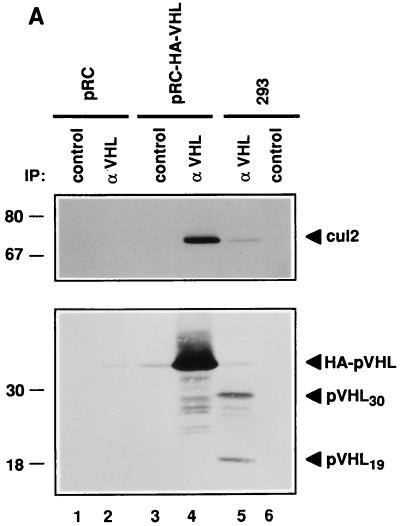
To ask whether pVHL and Cul2 might associate under physiological conditions, 293 human embryonic kidney cells containing endogenous, wild-type pVHL (18, 22) were immunoprecipitated with anti-VHL antibody or a control antibody (Fig. 2A). The stable transfectants described above were analyzed in parallel. As expected, anti-VHL but not control 293 cell immunoprecipitates contained wild-type pVHL, as determined by anti-VHL immunoblot analysis (Fig. 2A, compare lanes 5 and 6). Two species of pVHL (pVHL30 and pVHL19) were detected in these and other cells (data not shown). pVHL19 results from translation initiation at an internal methionine codon (17a). In addition, anti-Cul2 immunoblot analysis confirmed the presence of Cul2 specifically in the anti-VHL immunoprecipitate (compare Fig. 2A, lane 5, to lanes 4 and 6). The available anti-Cul2 sera do not immunoprecipitate Cul2 efficiently from mammalian cell extracts (data not shown). Thus, the reciprocal experiment, in which an anti-Cul2 immunoprecipitate would be analyzed for the presence of pVHL, cannot yet be performed.
FIG. 2.
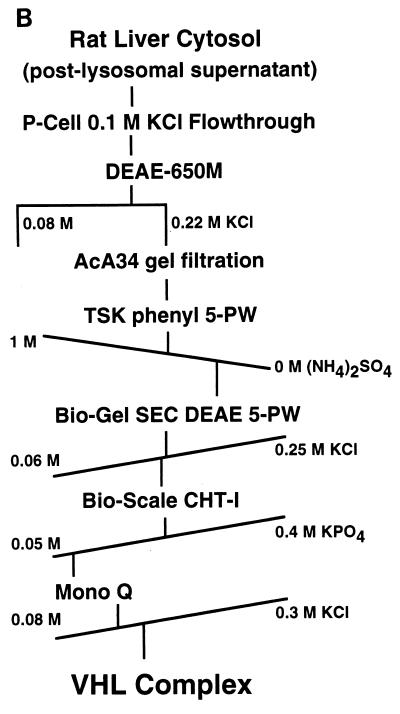
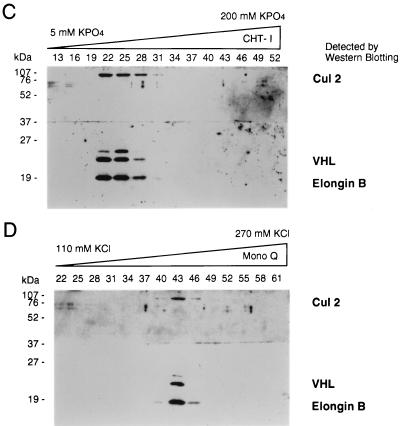
Copurification of Cul2 and endogenous pVHL. (A) 293 [VHL (+/+)] human embryonic kidney cells (lanes 5 and 6) and VHL (−/−) renal carcinoma cells stably transfected with a plasmid encoding HA-tagged pVHL (lanes 3 and 4) or with the backbone expression plasmid (lanes 1 and 2) were lysed and immunoprecipitated (IP) with anti-VHL (αVHL) or control antibody as indicated. Bound proteins were detected by anti-Cul2 (upper panel) or anti-VHL (lower panel) immunoblot analysis. Numbers at left are in kilodaltons. (B) Scheme for biochemical purification of pVHL complexes. P-cell, phosphocellulose P11. (C and D) Cochromatography of Cul2 with pVHL-elongin complexes. Aliquots of column fractions from the final two steps (hydroxylapatite Bio-Scale CHT-I high-pressure liquid chromatography and Mono Q chromatography [C and D, respectively]) were resolved by SDS–12% polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane. The top half of the membrane was immunoblotted with rabbit anti-Cul2 sera, and the bottom half was immunoblotted with a cocktail containing an anti-VHL monoclonal antibody and rabbit polyclonal anti-elongin B sera. Fraction numbers are shown above the lanes.
In addition, a stable multiprotein complex containing endogenous pVHL, Cul2, and elongins B and C could be extensively purified from rat liver cytosolic extracts according to the procedure outlined in Fig. 2B. Analysis of column fractions from the final two steps in the purification of this complex revealed close cochromatographic elution of pVHL with Cul2 and elongin B (Fig. 2C and D) as well as elongin C (data not shown). The coimmunoprecipitation and chromatographic data, taken together, suggest that pVHL-Cul2 complexes exist naturally in cells.
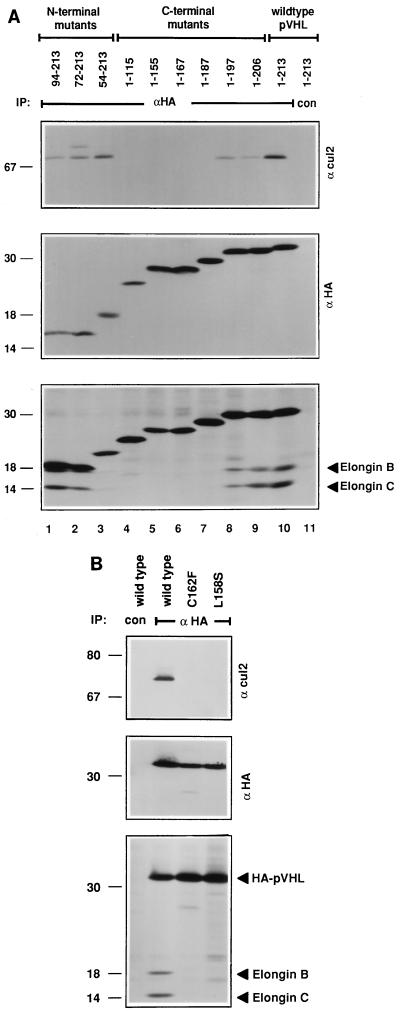
The minimal region of pVHL capable of binding to elongins B and C, at least in vitro, corresponds to residues 157 to 172 (22, 24). To map the region of pVHL required for binding to Cul2, 786-O subclones that stably produced N-terminal HA epitope-tagged pVHL mutants were metabolically labelled with [35S]methionine, lysed, and immunoprecipitated with an anti-HA antibody under stringent conditions (Fig. 3A). Comparable recovery of each of the pVHL mutants was confirmed by anti-HA immunoblot analysis (Fig. 3A, middle panel). Elongins B and C were detected by autoradiography (lower panel), and Cul2 was detected by anti-Cul2 immunoblot analysis (upper panel). In keeping with the earlier in vitro mapping studies, pVHL(1-197) and pVHL(94-213) retained the ability to bind to elongins B and C, whereas pVHL(1-167) did not. pVHL(1-187), surprisingly, did not stably associate with elongins B and C, suggesting that residues C terminal to pVHL residue 172 contribute to elongin B and C binding in vivo. The ability of pVHL to bind to Cul2 mirrored its ability to bind to elongins B and C in these assays (compare upper and lower panels in Fig. 3A).
FIG. 3.
Binding of Cul2 and elongins to pVHL mutants. (A) Deletion mutants; (B) missense mutants. VHL (−/−) renal carcinoma cells ectopically producing the indicated HA-tagged pVHL species were metabolically labelled with 35S, lysed, and immunoprecipitated (IP) with anti-HA (αHA) or control (con) antibody as indicated. Bound proteins were resolved by SDS-polyacrylamide gel electrophoresis and detected by anti-Cul2 immunoblot analysis (upper panel), anti-HA immunoblot analysis (middle panel), or autoradiography (lower panel). The apparent decrease in the binding of elongins B and C to pVHL(54-213) in panel A was not a reproducible finding. Numbers at left are in kilodaltons.
pVHL(157-172), a synthetic peptide, blocks the binding of pVHL to elongins B and C in vitro (22). Scanning mutagenesis of this peptide, in which single amino acid residues were converted to either alanine or a residue corresponding to a tumor-derived mutation, suggested that T157, L158, and C162 were likely to form critical contacts between pVHL and the elongins (4a). We therefore generated stable 786-O renal carcinoma subclones that ectopically produced HA-tagged versions of pVHL(L158S), pVHL(C162F), or pVHL(157-162NAAIRS). The former two correspond to naturally occuring VHL gene mutations (48). The latter replaces pVHL residues 157 to 162 with the sequence NAAIRS, which is believed to be highly flexible based on its appearance in both alpha-helical and beta-sheet structures (34, 46). These three mutants were, as expected, unable to bind to elongins B and C (Fig. 3B and data not shown). In addition, none of these mutants bound to Cul2.
pVHL binding to elongins B and C appeared, based on these studies, to be necessary and sufficient for binding to Cul2, raising the possibility that Cul2 can bind to elongins B and C in the absence of pVHL. To test this hypothesis, 786-O cells that stably produced a T7 epitope-tagged version of elongin C were immunoprecipitated with an anti-T7 antibody or a control antibody (Fig. 4). In parallel, 786-O cells that stably produced HA-pVHL were immunoprecipitated with an anti-HA antibody or a control antibody. Bound proteins were detected by anti-Cul2 (Fig. 4, upper panel), anti-HA (middle panel), or anti-T7 (lower panel) immunoblot analysis. Cul2 coimmunoprecipitated with elongin C (compare Fig. 4, lane 5, to lanes 4 and 6). As 786-O cells lack endogenous, wild-type pVHL, this result suggests that elongin C can, at least indirectly, bind to Cul2 in the absence of pVHL.
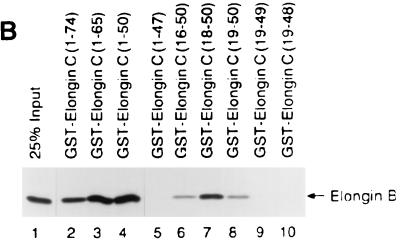
Cul2 and elongin C share significant sequence similarity with Saccharomyces cerevisiae proteins Cdc53 and Skp1, respectively (Fig. 5 and 6) (see Discussion) (2, 38). The potential significance of the elongin C similarity to Skp1 is underscored by the following observation. A peptide containing elongin C residues 18 to 50, produced as either a GST fusion protein (Fig. 6) or a synthetic peptide (data not shown), was sufficient to bind to elongin B. An earlier study had shown that elongin C residues 21 to 30 were necessary for binding to elongin B (42). Thus, the region of highest similarity between elongin C and Skp1, corresponding to elongin C residues 18 to 50, is a functional domain.
FIG. 5.
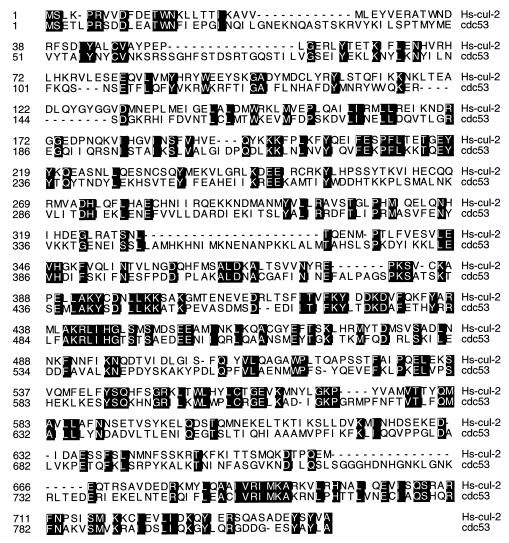
Similarity of human Cul2 to S. cerevisiae Cdc53 protein. Identical amino acid residues are shaded with black.
FIG. 6.
The region of elongin C that is most similar to Skp1 is an elongin B-binding domain. (A) Similarity of elongin C to Skp1. Identical amino acid residues are shaded with black. Elongin C residues 18 to 50 are underlined. (B) Binding of radiolabelled elongin B to the indicated GST-elongin C fusion proteins (lanes 2 to 10). Bound proteins were resolved by SDS-polyacrylamide gel electrophoresis and detected by fluorography. Lane 1 contained 25% of the elongin B present in each binding reaction mixture.
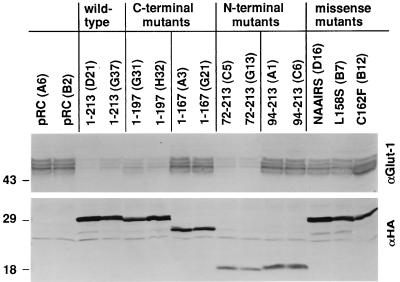
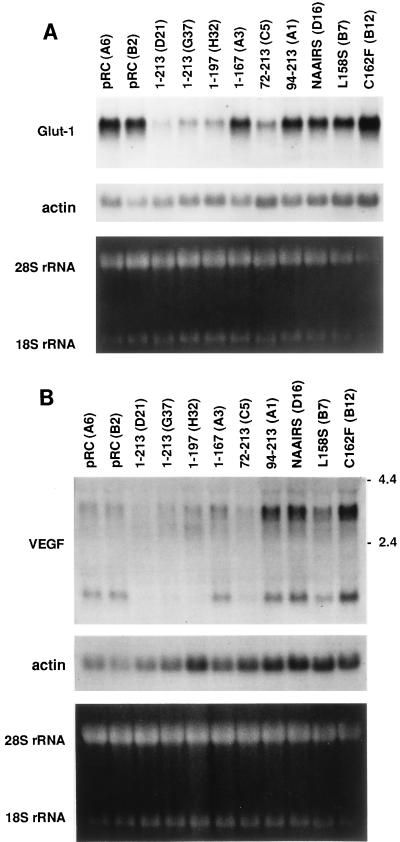
pVHL plays a critical role in the degradation of hypoxia-inducible mRNAs (11, 16). Consequently, cells lacking pVHL overproduce proteins such as VEGF and the Glut1 glucose transporter, which are both encoded by hypoxia-inducible mRNAs (11, 16, 37, 41). To determine whether this activity might be linked to the ability of pVHL to bind to elongins and Cul2, 786-O subclones producing the various pVHL mutants described above were tested for Glut1 protein production by steady-state anti-Glut1 immunoblot analysis (Fig. 7). Three or more independent clones were tested for each pVHL mutant. In parallel, total RNA was harvested from representative clones and analyzed for Glut1 and VEGF mRNA abundance by Northern blot analysis (Fig. 8). It was shown previously that the inhibition of VEGF mRNA accumulation by pVHL leads to a commensurate decrease in VEGF protein production (11, 16, 41).
FIG. 7.
Inhibition of Glut1 protein production by pVHL mutants. Whole-cell extracts (∼50 μg of protein per lane, as determined by the Bradford method) prepared from VHL (−/−) renal carcinoma cells ectopically producing the indicated HA-tagged pVHL species were resolved by SDS-polyacrylamide gel electrophoresis and immunoblotted with anti-Glut1 polyclonal (upper panel) or anti-HA monoclonal (lower panel) antibodies. Clone identifiers are indicated in parentheses.
FIG. 8.
Inhibition of hypoxia-inducible mRNA accumulation by pVHL mutants. Total RNA was isolated from VHL (−/−) renal carcinoma cells ectopically producing the indicated HA-tagged pVHL species and analyzed by Northern hybridization with radiolabelled probes specific for either the Glut1 mRNA (A) or the VEGF mRNA (B). Clone identifiers are indicated in parentheses. Comparable loading of RNA was confirmed by ethidium bromide staining of the rRNAs as well as by rehybridization of the filters to a radiolabelled actin probe. The presence of multiple VEGF mRNA isoforms was noted previously by others (26, 36, 43).
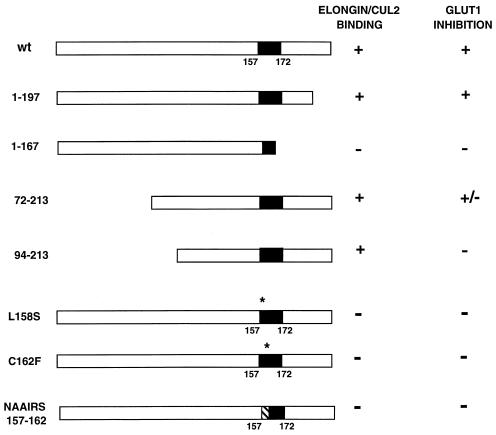
Wild-type pVHL, as expected, inhibited Glut1 and VEGF expression, whereas pVHL(1-167) did not (Fig. 7 and 8). Similar results were obtained following the reintroduction of wild-type pVHL and pVHL(1-167) into A498 VHL (−/−) renal carcinoma cells (data not shown). pVHL(1-197), which did not bind to p200 (data not shown) but which retained the ability to bind to elongins B and C and Cul2 (Fig. 3A), also inhibited Glut1 and VEGF expression (Fig. 7 and 8). These results suggested that the binding of pVHL to elongins and Cul2 might be linked to its ability to regulate Glut1 and VEGF. In keeping with this view, all of the missense mutants that were unable to bind to elongins and Cul2 were likewise unable to inhibit Glut1 and VEGF mRNA accumulation. Some, but not all, clones producing pVHL(72-213) produced levels of Glut1 comparable to those produced by cells producing wild-type pVHL. The reason for this clonal variation is unclear (shown in Fig. 7 and 8 are data for clones in which Glut1 production was inhibited). Finally, pVHL(94-213), although able to bind to elongins and Cul2, was unable to inhibit Glut1 and VEGF expression (Fig. 7 and 8). The simplest interpretation of these data, summarized in Fig. 9, is that binding to elongins and Cul2 is necessary but not sufficient for pVHL to regulate hypoxia-inducible genes.
FIG. 9.
Summary of elongin-Cul2 binding and Glut1 inhibition by pVHL mutants. For Glut1 inhibition, + indicates that all clones tested produced Glut1 at levels similar to those in cells producing wild-type (wt) pVHL, − indicates that all clones tested produced Glut1 at levels similar to those in parental VHL (−/−) cells, and +/− indicates that some clones produced Glut1 at levels similar to those in cells producing wild-type pVHL and that some clones produced Glut1 at levels similar to those in parental VHL (−/−) cells. The reason for the variation in Glut1 production among these clones is currently unknown. Shown in Fig. 7 and 8 are data for pVHL(72-213) clones that scored positive. Black boxes indicate minimal elongin B and C binding domains based on in vitro studies (22). Asterisks indicate sites of missense mutations. The hatched box indicates the site of an NAAIRS substitution mutation.
DISCUSSION
We found that pVHL, when overproduced in cells, forms complexes that contain elongin B, elongin C, and Cul2. While the manuscript was in preparation, similar conclusions were reached by Pause and coworkers (38). Our work confirms and extends these observations by providing evidence that these complexes form under native conditions and, more importantly, by linking the formation of these complexes to a biological activity, namely, the previously described ability of pVHL to regulate hypoxia-inducible mRNAs.
Furthermore, we provide evidence that Cul2 can bind, at least indirectly, to elongin C in the absence of pVHL. This observation can account for the earlier observation that Cul2 does not bind to pVHL in the absence of elongin C in vitro (38). Specifically, in the simplest model, Cul2 binds to elongin C which, in turn, binds directly to pVHL (6). Importantly, the pVHL elongin-binding domain is a hot spot for naturally occurring mutations in VHL kindreds, and ∼70% of the VHL-associated mutations characterized to date would be predicted to abrogate elongin binding (22, 24, 48). Thus, the available genetics suggest that binding to elongin-Cul2 complexes contributes to the ability of pVHL to suppress tumor growth in vivo. Conceivably, this property is due, at least in part, to the ability of pVHL, when bound to elongins and Cul2, to suppress hypoxia-inducible mRNAs.
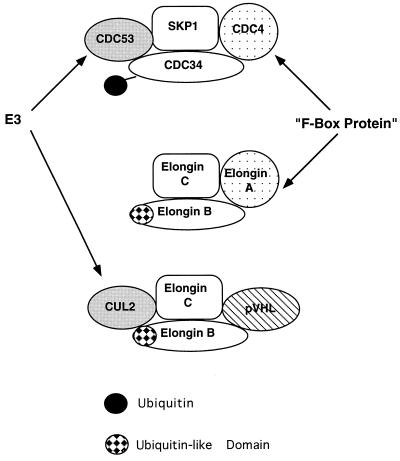
How might binding to elongins and Cul2 contribute to the regulation of hypoxia-inducible mRNA stability by pVHL? One clue may come from a careful inspection of the primary sequences of Cul2 and the elongins. Targeted proteolysis of proteins by ubiquitin requires the concerted action of an E1 ubiquitin-activating enzyme, an E2 ubiquitin-conjugating enzyme and, at least in some cases, an E3 ubiquitin ligase (15, 19). Cdc53, a putative E3 protein, forms complexes which contain Skp1, Cdc34 (an E2 protein), and an F-box protein (so called because of apparent homology to cyclin F), such as Grr1 or Cdc4 (Fig. 10). These complexes target proteins such as G1 cyclins (or Clns) as well as cyclin-dependent kinase inhibitors, such as Sic1, for ubiquitin-mediated proteolysis (15, 19).
FIG. 10.
Both Skp1 and elongin C participate in the formation of multiprotein complexes. Cdc4 and elongin A contain F-box motifs. Cdc53 and Cul2 are putative E3 ubiquitin ligases. Cdc34 is a putative E2 ubiquitin-conjugating enzyme, and elongin B contains a ubiquitin-like domain.
Cul2 appears to be a homolog of Cdc53 (Fig. 5) and is a member of a family of putative E3-like ubiquitin ligases (23, 38). Likewise, elongin C, as first reported by Bai and coworkers, contains a region that is homologous to Skp1 (Fig. 6), and elongin A is an F-box protein (2). Finally, elongin B, although not a recognizable E2 protein, contains a region which, as first reported by Garrett and coworkers (8), is similar to ubiquitin. The potential significance of these homologies is underscored by the observation that the minimal region of elongin C capable of binding to elongin B (elongin C residues 18 to 50) corresponds to the region of greatest similarity between elongin C and Skp1 (Fig. 6) (2).
Thus, one model, which remains to be tested, is that complexes containing Cul2 and elongins B and C target certain proteins for ubiquitin-dependent proteolysis. A more speculative model, given that elongin B contains a ubiquitin-like domain, is that elongin B itself becomes covalently linked to certain proteins, such as has been observed for the ubiquitinlike protein named either GMP1, SUMO-1, or Sentrin (20, 33). In either case, pVHL competes with elongin A in vitro for binding to elongins B and C (6). Furthermore, we have thus far been unable to detect complexes containing elongin A and Cul2 in native cell extracts (data not shown). Therefore, one might hypothesize that the function of elongins B and C, when bound to pVHL and Cul2, is different from their function when they are bound to elongin A (Fig. 10). As a corollary, these data raise the possibility that the previously described ability of pVHL to inhibit transcriptional elongation by elongin A in vitro is not responsible for its ability to suppress tumor growth in vivo.
What proteins are the likely downstream targets of Cul2? Levy and coworkers identified RNA-binding proteins that interact specifically with the 3′ untranslated regions of hypoxia-inducible mRNAs (29, 30). Significantly, these proteins appear to stabilize these mRNAs. In VHL (+/+) cells, these proteins are detectable with RNA gel shift assays under hypoxic conditions but not normoxic conditions. In contrast, these proteins are detectable in VHL (−/−) cells under both hypoxic and normoxic conditions, perhaps accounting for the failure of VHL (−/−) cells to properly degrade the mRNAs under normoxic conditions (29). Thus, Cul2 may, directly or indirectly, affect the stability or function of these mRNA-binding proteins. There is precedence for complexes containing Cdc53 and Cdc34 playing a role in nutritional or environmental adaptation. For example, under nutritionally replete conditions, Cdc34 is required for the ubiquitin-dependent proteolysis of the transcription factor Gcn4, which is required for amino acid biosynthesis (25). Under conditions of nutritional starvation, however, Gcn4 becomes stabilized (25).
A loss of cul1 in Caenorhabditis elegans leads to hyperplasia in certain cell lineages, perhaps due to a failure to exit the cell cycle (23). To our knowledge, the phenotype of cul2 −/− C. elegans has not been reported. Given the apparent role of Cdc53 in the regulation of Cln2 and Sic1, one might speculate that cul2 also plays a role in cell cycle control (15, 19). pVHL does not grossly affect the cell cycle control of renal carcinoma cells in vitro (11, 18). Whether it might do so under certain experimental conditions, however, remains to be determined. In any event, control of the cell cycle and responses to environmental stress clearly need not be mutually exclusive activities.
pVHL inhibits both the stability of hypoxia-inducible mRNAs and tumor cell growth in vivo. For the first time, we have now linked the former to a biochemical activity, namely, the ability to form complexes which contain elongin B, elongin C, and Cul2. Clearly, determining the degree to which tumor suppression likewise relies upon this activity is an important question for the future. Similarly, it will be important to determine if Cul2, as suspected, acts as an E3 protein. If so, it will be essential to identify its downstream targets and to ascertain how its behavior is modulated by complex formation with pVHL.
ACKNOWLEDGMENTS
We thank Stephen Elledge, Sushant Kuman, and Pengbo Zhou for critical reading of the manuscript and for useful discussions and Edward Kipreos for the human Cul2 partial cDNA. In particular, we thank Stephen Elledge for bringing the similarity of Skp1 to elongin C and the presence of an F box in elongin A to our attention. We thank our colleagues in the Kaelin laboratory for many helpful comments.
This work was supported by grants from the National Institutes of Health (to W.G.K. and J.W.C.), the Oklahoma Medical Research Foundation (to J.W.C.), the MRC of Canada (to M.O.), the VHL Family Alliance and the Murray Foundation (to O.I.), the H. A. and Mary K. Chapman Trust (to J.W.C.), and Novartis Pharmaceuticals (to W.G.K.). J.W.C. is an Associate Investigator of the Howard Hughes Medical Institute.
Kim M. Lonergan and Othon Iliopoulos contributed equally to this work.
REFERENCES
- 1.Aso T, Haque D, Barstead R, Conaway R, Conaway J. The inducible elongin A elongation activation domain: structure, function and interaction with elongin BC complex. EMBO J. 1996;15:101–110. [PMC free article] [PubMed] [Google Scholar]
- 2.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 3.Berse B, Brown L, Livingston V, Dvorak H, Senger D. Vascular permeability factor (vascular endothelial growth factor) gene is expressed differentially in normal tissues, macrophages, and tumors. Mol Biol Cell. 1992;3:211–220. doi: 10.1091/mbc.3.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conaway R, Reines D, Garrett K, Powell W, Conaway J. Purification of RNA polymerase II general transcription factors from rat liver. Methods Enzymol. 1996;273:194–207. doi: 10.1016/s0076-6879(96)73020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Conaway, R. C., J. W. Conaway, and W. G. Kaelin, Jr. Unpublished data.
- 5.Corless C L, Kibel A, Iliopoulos O, Kaelin W G. Immunostaining of the von Hippel-Lindau gene product (pVHL) in normal and neoplastic human tissues. Hum Pathol. 1997;28:459–464. doi: 10.1016/s0046-8177(97)90035-6. [DOI] [PubMed] [Google Scholar]
- 6.Duan D R, Pause A, Burgress W, Aso T, Chen D Y T, Garrett K P, Conaway R C, Conaway J W, Linehan W M, Klausner R D. Inhibition of transcriptional elongation by the VHL tumor suppressor protein. Science. 1995;269:1402–1406. doi: 10.1126/science.7660122. [DOI] [PubMed] [Google Scholar]
- 7.Foster K, Prowse A, van den Berg A, Fleming S, Hulsbeek M M F, Crossey P A, Richards F M, Cairns P, Affara N A, Ferguson-Smith M A, Buys C H C M, Maher E R. Somatic mutations of the von Hippel-Lindau disease tumor suppressor gene in non-familial clear cell renal carcinoma. Hum Mol Genet. 1994;3:2169–2173. doi: 10.1093/hmg/3.12.2169. [DOI] [PubMed] [Google Scholar]
- 8.Garrett K, Aso T, Bradsher J, Foundling S, Lane W, Conaway R, Conaway J. Positive regulation of general transcription factor SIII by a tailed ubiquitin homolog. Proc Natl Acad Sci USA. 1995;92:7172–7176. doi: 10.1073/pnas.92.16.7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gnarra J R, Duan D R, Weng Y, Humphrey J S, Chen D Y T, Lee S, Pause A, Dudley C F, Latif F, Kuzmin I, Schmidt L, Duh F-M, Stackhouse T, Chen F, Kishida T, Wei M H, Lerman M I, Zbar B, Klausner R D, Linehan W M. Molecular cloning of the von Hippel-Lindau tumor suppressor gene and its role in renal cell carcinoma (review) Biochim Biophys Acta. 1996;1242:201–210. doi: 10.1016/0304-419x(95)00012-5. [DOI] [PubMed] [Google Scholar]
- 10.Gnarra J R, Tory K, Weng Y, Schmidt L, Wei M H, Li H, Latif F, Liu S, Chen F, Duh F-M, Lubensky I, Duan D R, Florence C, Pozzatti R, Walther M M, Bander N H, Grossman H B, Brauch H, Pomer S, Brooks J D, Isaacs W B, Lerman M I, Zbar B, Linehan W M. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nature Genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- 11.Gnarra J R, Zhou S, Merrill M J, Wagner J, Krumm A, Papavassiliou E, Oldfield E H, Klausner R D, Linehan W M. Post-transcriptional regulation of vascular endothelial growth factor mRNA by the VHL tumor suppressor gene product. Proc Natl Acad Sci USA. 1996;93:10589–10594. doi: 10.1073/pnas.93.20.10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harlow E, Crawford L V, Pim D C, Williamson N M. Monoclonal antibodies specific for simian virus 40 tumor antigens. J Virol. 1981;39:861–869. doi: 10.1128/jvi.39.3.861-869.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harlow E, Lane D. Antibodies—a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 14.Herman J G, Latif F, Weng Y, Lerman M I, Zbar B, Liu S, Samid D, Duan D-S R, Gnarra J R, Linehan W M, Baylin S B. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci USA. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hopkin K. Regulating cellular processes: the power of protein degradation. J NIH Res. 1997;9:36–42. [Google Scholar]
- 16.Iliopoulos O, Jiang C, Levy A P, Kaelin W G, Goldberg M A. Negative regulation of hypoxia-inducible genes by the von Hippel-Lindau protein. Proc Natl Acad Sci USA. 1996;93:10595–10599. doi: 10.1073/pnas.93.20.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iliopoulos O, Kaelin W G. The molecular basis of von Hippel-Lindau disease. Mol Med. 1997;3:289–293. [PMC free article] [PubMed] [Google Scholar]
- 17a.Iliopoulos, O., and W. G. Kaelin, Jr. Unpublished data.
- 18.Iliopoulos O, Kibel A, Gray S, Kaelin W G. Tumor suppression by the human von Hippel-Lindau gene product. Nature Med. 1995;1:822–826. doi: 10.1038/nm0895-822. [DOI] [PubMed] [Google Scholar]
- 19.Jackson P K. Cell cycle: cull and destroy. Curr Biol. 1996;6:1209–1212. doi: 10.1016/s0960-9822(96)00697-5. [DOI] [PubMed] [Google Scholar]
- 20.Kamitani T, Nguyen H P, Yeh E T H. Preferential modification of nuclear proteins by a novel ubiquitin-like molecule. J Biol Chem. 1997;272:14001–14004. doi: 10.1074/jbc.272.22.14001. [DOI] [PubMed] [Google Scholar]
- 21.Kanno H, Kondo K, Ito S, Yamamoto I, Fujii S, Torigoe S, Sakai N, Hosaka M, Shuin T, Yao M. Somatic mutations of the von Hippel-Lindau tumor suppressor gene in sporadic central nervous system hemangioblastomas. Cancer Res. 1994;54:4845–4847. [PubMed] [Google Scholar]
- 22.Kibel A, Iliopoulos O, DeCaprio J D, Kaelin W G. Binding of the von Hippel-Lindau tumor suppressor protein to elongin B and C. Science. 1995;269:1444–1446. doi: 10.1126/science.7660130. [DOI] [PubMed] [Google Scholar]
- 23.Kipreos E T, Lander L E, Wing J P, He W W, Hedgecock E M. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- 24.Kishida T, Stackhouse T M, Chen F, Lerman M I, Zbar B. Cellular proteins that bind the von Hippel-Lindau disease gene product: mapping of binding domains and the effect of missense mutations. Cancer Res. 1995;55:4544–4548. [PubMed] [Google Scholar]
- 25.Kornitzer D, Raboy B, Kulka R G, Fink G R. Regulated degradation of the transcription factor Gcn4. EMBO J. 1994;13:6021–6030. doi: 10.1002/j.1460-2075.1994.tb06948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koura A N, Liu W, Kitadai Y, Singh R K, Radinsky R, Ellis L M. Regulation of vascular endothelial growth factor expression in human colon carcinoma cells by cell density. Cancer Res. 1996;56:3891–3894. [PubMed] [Google Scholar]
- 27.Latif F, Tory K, Gnarra J, Yao M, Duh F-M, Orcutt M L, Stackhouse T, Kuzmin I, Modi W, Geil L, Schmidt L, Zhou F, Li H, Wei M H, Chen F, Glenn G, Choyke P, Walther M M, Weng Y, Duan D-S R, Dean M, Glavac D, Richards F M, Crossey P A, Ferguson-Smith M A, Pasiler D L, Chumakov I, Cohen D, Chinault A C, Maher E R, Linehan W M, Zbar B, Lerman M I. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science. 1993;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 28.Lee S, Chen D Y T, Humphrey J S, Gnarra J R, Linehan W M, Klausner R D. Nuclear/cytoplasmic localization of the von Hippel-Lindau tumor suppressor gene product is determined by cell density. Proc Natl Acad Sci USA. 1996;93:1770–1775. doi: 10.1073/pnas.93.5.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levy A P, Levy N S, Goldberg M A. Hypoxia-inducible protein binding to vascular endothelial growth factor mRNA and its modulation by the von Hippel-Lindau protein. J Biol Chem. 1996;271:25492–25497. doi: 10.1074/jbc.271.41.25492. [DOI] [PubMed] [Google Scholar]
- 30.Levy A P, Levy N S, Iliopoulos O, Jiang C, Kaelin W G, Goldberg M A. Regulation of vascular endothelial growth factor by hypoxia and its modulation by the von Hippel-Lindau tumor suppressor gene. Kidney Int. 1997;51:575–578. doi: 10.1038/ki.1997.82. [DOI] [PubMed] [Google Scholar]
- 31.Los M, Jansen G H, Kaelin W G, Lips C J M, Blijham G H, Voest E E. Expression pattern of the von Hippel-Lindau protein in human tissues. Lab Invest. 1996;75:231–238. [PubMed] [Google Scholar]
- 32.Lubensky I A, Gnarra J R, Bertheau P, Walther M M, Linehan W M, Zhuang Z. Allelic deletions of the VHL gene detected in multiple microscopic clear cell renal lesions in von Hippel-Lindau disease patients. Am J Pathol. 1996;149:2089–2094. [PMC free article] [PubMed] [Google Scholar]
- 33.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 34.Marsilio E, Cheng S H, Schaffhausen B, Paucha E, Livingston D M. The T/t common region of simian virus 40 large T antigen contains a distinct transformation-governing sequence. J Virol. 1991;65:5647–5652. doi: 10.1128/jvi.65.10.5647-5652.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKusick V A. Mendelian inheritance in man. Baltimore, Md: The Johns Hopkins University Press; 1992. [Google Scholar]
- 36.Morii K, Tanaka R, Washiyama K, Kumanishi T, Kuwano R. Expression of vascular endothelial growth factor in capillary hemangioblastoma. Biochem Biophys Res Commun. 1993;194:749–755. doi: 10.1006/bbrc.1993.1885. [DOI] [PubMed] [Google Scholar]
- 37.Mukhopadhyay D, Knebelmann B, Cohen H, Ananth S, Sukhatme V. The von Hippel-Lindau tumor suppressor gene product interacts with Sp1 to repress vascular endothelial growth factor promoter activity. Mol Cell Biol. 1997;17:5629–5639. doi: 10.1128/mcb.17.9.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pause A, Lee S, Worrell R A, Chen D Y T, Burgess W H, Linehan W M, Klausner R D. The von Hippel-Lindau tumor-suppressor gene product forms a stable complex with human CUL-2, a member of the Cdc53 family of proteins. Proc Natl Acad Sci USA. 1997;94:2156–2161. doi: 10.1073/pnas.94.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sato K, Terada K, Sugiyama T, Takahashi S, Saito M, Moriyama M, Kakinuma H, Suzuki Y, Kato M, Kato T. Frequent overexpression of vascular endothelial growth factor gene in human renal cell carcinoma. Tohoku J Exp Med. 1994;173:355–360. doi: 10.1620/tjem.173.355. [DOI] [PubMed] [Google Scholar]
- 40.Shuin T, Kondo K, Torigoe S, Kishida T, Kubota Y, Hosaka M, Nagashima Y, Kitamura H, Latif F, Zbar B, Lerman M I, Yao M. Frequent somatic mutations and loss of heterozygosity of the von Hippel-Lindau tumor suppressor gene in primary human renal cell carcinomas. Cancer Res. 1994;54:2852–2855. [PubMed] [Google Scholar]
- 41.Siemeister G, Weindel K, Mohrs K, Barleon B, Martiny-Baron G, Marme D. Reversion of deregulated expression of vascular endothelial growth factor in human renal carcinoma cells by von Hippel-Lindau tumor suppressor protein. Cancer Res. 1996;56:2299–2301. [PubMed] [Google Scholar]
- 42.Takagi Y, Conaway R, Conaway J. Characterization of elongin C functional domain required for interaction with elongin B and activation of elongin A. J Biol Chem. 1996;271:1–7. doi: 10.1074/jbc.271.41.25562. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi A, Sasaki H, Kim S, Tobisu K, Kakizoe T, Tsukamoto T, Kumamoto Y, Sugimura T, Terada M. Markedly increased amounts of messenger RNAs for vascular endothelial growth factor and placenta growth factor in renal cell carcinoma associated with angiogenesis. Cancer Res. 1994;54:4233–4237. [PubMed] [Google Scholar]
- 44.Tsuchiya H, Iseda T, Hino O. Identification of a novel protein (VBP-1) binding to the von Hippel-Lindau (VHL) tumor suppressor gene product. Cancer Res. 1996;56:2881–2885. [PubMed] [Google Scholar]
- 45.Whaley J M, Naglich J, Gelbert L, Hsia Y E, Lamiell J M, Green J S, Collins D, Neumann H P H, Laidlaw J, Li F P, Klein-Szanto A J P, Seizinger B R, Kley N. Germ-line mutations in the von Hippel-Lindau tumor suppressor gene are similar to somatic von Hippel-Lindau aberrations in sporadic renal cell carcinoma. Am J Hum Genet. 1994;55:1092–1102. [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson I A, Haft D H, Getzoff E D, Tainer J A, Lerner R A, Brenner S. Identical short peptide sequences in unrelated proteins can have different conformations: a testing ground for theories of immune recognition. Proc Natl Acad Sci USA. 1985;82:5255–5259. doi: 10.1073/pnas.82.16.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wizigmann-Voos S, Breier G, Risau W, Plate K. Up-regulation of vascular endothelial growth factor and its receptors in von Hippel-Lindau disease-associated and sporadic hemangioblastomas. Cancer Res. 1995;55:1358–1364. [PubMed] [Google Scholar]
- 48.Zbar B, Kishida T, Chen F, Schmidt L, Maher E R, Richards F M, Crossey P A, Webster A R, Affara N A, Ferguson-Smith M A, Brauch H, Glavac D, Neumann H P H, Tisherman S, Mulvihill J J, Gross D, Shuin T, Whaley J, Seizinger B, Kley N, Olschwang S, Boisson C, Richard S, Lips C H M, Linehan M W, Lerman M. Germline mutations in the von Hippel-Lindau (VHL) gene in families from North America, Europe, and Japan. Hum Mutat. 1996;8:348–357. doi: 10.1002/(SICI)1098-1004(1996)8:4<348::AID-HUMU8>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 49.Zhuang Z, Bertheau P, Emmert-Buck M, Liotta L, Gnarra J, Linehan W, Lubensky I. A microscopic dissection technique for archival DNA analysis of specific cell populations in lesions < 1mm in size. Am J Pathol. 1995;146:620–625. [PMC free article] [PubMed] [Google Scholar]