Abstract
The number of antral follicles is considered an important fertility trait because animals with a high follicle count (HFC) produce more oocytes and embryos per cycle. Identification of these animals by genetic markers such as single nucleotide polymorphisms (SNPs) can accelerate selection of future generations. The aim of this study was to perform a genome wide association study (GWAS) on Nelore and Angus heifers with HFC and low (LFC) antral follicle counts. The groups HFC and LFC for genotyping were formed based on the average of total follicles (≥ 3 mm) counted in each breed consistently ± standard deviation. A total of 72 Nelore heifers (32 HFC and 40 LFC) and 48 Angus heifers (21 HFC and 27 LFC) were selected and the DNA was extracted from blood and hair bulb. Genotyping was done using the Illumina Bovine HD 770K BeadChip. The GWAS analysis showed 181 and 201 SNPs with genotype/phenotype association (P ≤ 0.01) in Nelore and Angus heifers, respectively. Functional enrichment analysis was performed on candidate genes that were associated with SNPs. A total of 97 genes were associated to the 181 SNPs in the Nelore heifers and the functional analysis identified genes (ROBO1 and SLIT3) in the ROBO-SLIT pathway that can be involved in the control of germ cell migration in the ovary as it is involved in lutheal cell migration and fetal ovary development. In the Angus heifers, 57 genes were associated with the 201 SNPs, highlighting Fribilin 1 (FBN1) gene, involved in regulation of growth factors directly involved in follicle activation and development. In summary, GWAS for Nelore and Angus heifers showed SNPs associated with higher follicle count phenotype. Furthermore, these findings offer valuable insights for the further investigation of potential mechanism involved in follicle formation and development, important for breeding programs for both breeds.
Keywords: Bovine HD 770 K SNP, candidate genes, follicle, reproduction, SNPs
Introduction
Reproductive performance is an important trait in livestock production. The number of antral follicles (AFC) greater than 3 mm has been associated with fertility in dairy cows. Dairy cows with low follicle counts have lower pregnancy rates at first service, a longer interval from calving to conception, and a higher number of services during the breeding season compared to cows with high follicle counts (HFC; Mossa et al., 2012). In addition, cows with a HFC have a higher number of total oocyte recovery by ultrasound-guided follicular aspiration (OPU) and consequently a greater number of viable embryos produced in vitro (Ireland et al., 2007).
Indicine animals (Brahman and Nelore) have been shown to have a greater number of antral follicles, whereas taurine animals (Angus) have a greater number of preantral follicles (Cushman et al., 2019; Favoreto et al., 2019). In addition, a microarray study showed that the gene expression profile differs between these breeds. Several genes associate with osteoblast differentiation (BMP4, IGF1, IGFBP3 and IGFBP5), cell proliferation (BMP4, SATB1, EMX2, IGF1, MAP7, EMPEP and FABP7) and bone development (BMP4, COL13A1, IGF1, IGFBP3 and IGFBP5) showed higher expression in follicles of Nelore heifers. The genes are related to follicle activation and development (Favoreto et al., 2019). Moreover, genomic studies have shown that Indian and Taurine cattle differ in the expression of genes associated with important traits (Liu et al., 2021), SNPs (Dar et al., 2021; Verardo et al., 2021), gene ontologies (Cortez et al., 2022), and chromatin and methylation profiles (Powell et al., 2023; Capra et al., 2023). Even though there are studies showing that zebu and taurine cattle trace back to a common ancestor, Bos primigenius (Perez-Pardal et al., 2010; Pitt et al., 2018).
In the past years, the use of genomic information has been widely employed as a strategy to improve selection for phenotypes such as increased milk production (Chamberlain et al., 2012), fertility (Huang et al., 2010; Peñagaricano et al., 2012), and other traits of economic interest in cattle. Genome-wide association studies (GWAS) are a method in genetics that aims to identify specific genetic variations, known as single nucleotide polymorphisms (SNPs). The studies are made based on a population that has a specific trait, these animals will have their genotype searched for a common SNPs that can be associated with the phenotype (Dehghan, 2018). Single nucleotide polymorphisms (SNP) enabled the identification of a subset of markers that can explain important portions of the variation in these traits. An alternative approach to identifying SNPs associated with a phenotype of interest is a candidate gene approach, in which individual genes are selected as candidates based on their known function (Naukkarinen et al., 2010).
Identification of SNPs and possible genes responsible for genetic variation in Nelore and Angus with HFC and LFC may improve understanding of the biological pathways involved in the AFC phenotype.
In summary, animals that are considered HFC are associated with better fertility as they allow more oocytes per cycle and thus more embryos in in vivo and in vitro embryo production (Ireland et al., 2007). The identification of these animals by genetic markers could improve the fertility of future generations. Therefore, the aim of the present work was to perform a GWAS study in Nelore and Angus heifers with HFC and LFC using the high-density SNP array to identify genetic variants and possible genes associated with these phenotypes. The main hypothesis was that HFC animals have genetic markers associated with follicular development that could be used as biomarkers for genomic selection.
Material and methods
Animals selection and phenotypic data
Animals were maintained according to the Bioethics Committee of the Faculty of Veterinary Medicine (N° 439) and Animal Sciences of the São Paulo State University (Botucatu, São Paulo, Brazil) and in accordance with the specific guidelines and standards of the SSR.
To pre-select animals, a group of 155 Nelore and 132 Angus heifers of similar body weight (Nelore = 442.93 ± 6.97 kg and Angus = 466.23 ± 10,13) and age (25.57 ± 2,05) were examined by ultrasound (US; Mindray Vet DPS 2200, São Paulo, Brazil) with a 7.5-MHz probe on a random day of the estrous cycle. Only heifers that were cyclic and did not have a follicle greater than 5 mm were selected. In this initial evaluation, the total number of follicles ≥ 3 mm in both ovaries was counted and the mean follicle number for each breed was determined. These heifers were injected with two doses of PGF2 alpha 11 days apart to initiate luteolysis and synchronize the occurrence of ovulation. The ovaries of each animal were examined one day after ovulation in three consecutive estrous cycles to determine the high (HFC) and low (LFC) follicle count groups. Groups were formed based on the average of total follicles counted (≥ 3 mm) for each breed consistently ± standard deviation (Table 1). A total of 72 Nelore heifers were selected and classified into the groups: 32 animals were classified as HFC (40 ≥ follicles) and 40 animals were classified as LHC (20 ≤ follicles). For Angus heifers, 48 animals were selected and assigned to the groups: 21 animals were classified as HFC (20 ≥ follicles) and 27 as LFC (≤ 10 follicles).
Table 1. Numbers of animals, follicle average count and standard deviation for Nelore and Angus heifers with HFC and LFC.
| Nelore | Angus | ||||
|---|---|---|---|---|---|
| Number of animals | 43 | 35 | 22 | 27 | |
| Fol average count | 49 | 15 | 25 | 6 | |
| Standard deviation | 9.3 | 3.9 | 5.1 | 2.3 | |
Sample and DNA extraction
During ultrasound examination a sample of blood from the tail vein or capillary bulbs from the tail hair were collected from all selected animals for DNA extraction. For the blood samples, DNA was extracted using the MiniPrep kit (Axygen bioscience, Union City, New Jersey, USA), while the capillary bulb was extracted using the NucleoSpin Tissue Kit (Macherey-Nagel, Duren, Germany) according to the manufacturer’s instructions. After extraction, the quality of the DNA samples was assessed by determining the A260/280 ratio in a biophotometer. Samples were accepted if the values were between 1.8 and 2.0. DNA was quantified and diluted to a concentration between 50 ng/µL and 150 ng/µL for subsequent genotyping.
Genotyping and SNPs quality control
Genotyping was performed by DEOXI BIOTECNOLGIA LTDA, Araçatuba, São Paulo, Brazil, using the Illumina Bovine HD 770 K BeadChip (Infinium BeadChip, Illumina, San Diego, CA) according to the protocol of the manufacturer’s instructions. Genotypes were determined in Illumina A/B allele format and used to represent a covariate value at each locus coded as 0, 1, or 2 indicating the number of B alleles. Initial data analysis and visualization was performed using GenomeStudio Data Analysis Software (Illumina, 2023). For GWAS, SNPs were quality controlled using only autosomal SNPs with known genomic coordinates according to UMD 3.1 Bovine Genome. Samples with a call rate (IDCR) of less than 90% were removed from the study. SNPs were removed if they had a minor allele frequency ≤ 0.02, a call rate ≤ 0.98, and a P value for the Fisher exact test for Hardy-Weindberg equilibrium ≤ 1 x 0.00001. After filtering, 538.575 SNPs were used for the GWAS study.
GWAS test and gene enrichment analysis
The Cochran-Armitage test has been used for genomic association between HFC and LFC in Nelore and Angus heifers. This test is commonly used as a genotype-based test for candidate gene association. It uses a range of scores that can be obtained as an efficient score test for logistic regression (Clarke et al., 2011). In this analysis, it was used for comparisons as a case-control study (GenABEL package; Aulchenko et al., 2007) with LFC as the control group in both subspecies. Significant SNPs (P ≤ 0.001) were mapped to corresponding or nearby genes by linkage disequilibrium (LD) using the software PLINK (Bush et al., 2009; Bohmanova et al., 2010). We considered only the genes with r2 > 0.8 significant for functional analysis. The genes associated with SNPs directly or indirectly by linkage disequilibrium were subjected to the functional enrichment analyzes on DAVID (Database for Annotation, Visualization and Integrated Discovery; (Dennis et al., 2003) to determine gene ontologies (GO), functions, and pathways in which the genes were overrepresented at P < 0.05.
Results
After quality control, a total of 538.575 SNPs distributed on 29 chromosomes were used to study the association between phenotype and genotype. The profiles of p-values (in terms of -log[p]) of all tested SNPs for Nelore and Angus heifers are shown in Figures 1 and 2. A total of 181 SNPs for Nelore and 201 SNPs for Angus heifers showed an association (p ≤ 0.001) between the HFC and LFC groups.
Figure 1. Genome-wide association analysis for HFC comparing with LFC in Nelore heifers. The Manhattan plot demonstrates the results of association after correction for population structure. The horizontal red line indicates the whole-genome with significance threshold [-log (p ≤ 10E-3)].
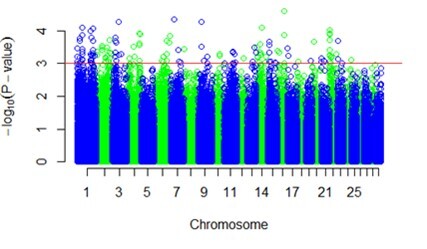
Figure 2. Genome-wide association analysis for HFC comparing with LFC in Angus heifers. The Manhattan plot demonstrates the results of association after correction for population structure. The horizontal red line indicates the whole-genome with significance threshold [-log (p ≤ 10E-3)].
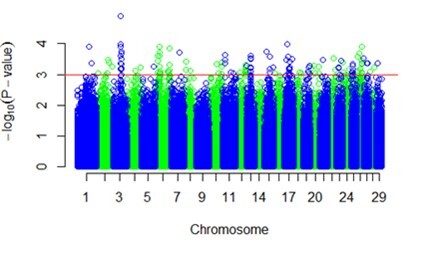
The 181 SNPs of Nelore heifers were mapped on 23 different chromosomes, and the most significant SNPs (p ≤ 0.00098) were located on chromosomes 1, 3, 7, 14, 16, and 22 (Table 2). In Angus heifers, all 201 were mapped on 29 chromosomes, and the most significant SNP (p = 0.000124) is located on chromosome 3 (Table 2).
Table 2. Description of the most significant SNPs for Nelore e Angus heifers.
| Chromosome | SNPs | ||
|---|---|---|---|
| 1 | rs136289764 (8.11E-04) | ||
| 1 | rs109443367 (8.47E-04) | ||
| 3 | rs43704025 (5.39E-04) | rs43338364 (1.24E-04) | |
| 7 | rs110807077 (4.53E-04) | ||
| 9 | rs136692332 (5.39E-04) | ||
| 14 | rs132707253 (8.01E-04) | ||
| 14 | rs41730052 (8.01E-04) | ||
| 14 | rs110253276 (8.01E-04) | ||
| 16 | rs109100442 (2.47E-04) | ||
| 22 | rs133905094 (9.38E-04) | ||
SNPs Identification: see Ensembl (2023).
GWAS revealed a total of 97 different genes associated directly or indirectly via LD (r2> 0.8) with all 181 SNPs of Nelore heifers (Table 3). Functional enrichment analysis was applied to all 97 genes associated in the integrated network using the DAVID Functional Annotation Tool. In Nelore heifers, a total of 5 gene ontologies (GO) were significantly overrepresented in 2 categories: biological process and cellular component (Table 5). In Angus heifers, 52 genes were associated with all 201 SNPs (Table 4), and functional enrichment analysis showed that 18 genes were significantly GO (p < 0.05) overrepresented in the 3 categories (biological process, cellular component, and molecular function; Table 5).
Table 3. Associated SNPs with respective candidate genes for Nelore heifers.
| Gene | Chromosome | SNP | Gene | Chromosome | SNP | Gene | Chromosome | SNP |
|---|---|---|---|---|---|---|---|---|
| RSRC1 | 1 | BTB-01568926 | DC1I1 | 4 | BovineHD0400003849 | SGK3 | 14 | Hapmap49131-BTA-34531 |
| PLCH1 | 1 | BovineHD0100031992 | AGMO | 4 | BovineHD0400007019 | PREX2 | 14 | BovineHD1400009858 |
| ANKUB1 | 1 | BovineHD0100033727 | A0JNG1 | 4 | BovineHD0400001266 | KCNB2 | 14 | BTA-107899-no-rs |
| WWTR1 | 1 | BovineHD0100033813 | RERG | 5 | BovineHD0500027036 | JPH1 | 14 | BovineHD1400011391 |
| AT1B3 | 1 | BovineHD0100036157 | BST1 | 6 | BovineHD0600032820 | SNTB1 | 14 | BovineHD1400023903 |
| EPHB1 | 1 | BovineHD0100038454 | NPNT | 6 | BovineHD0600005674 | ZC3H12C | 15 | BovineHD1500005193 |
| CEP63 | 1 | BTB-00063883 | A0JN38 | 6 | BovineHD0600019557 | F1N2Z9 | 15 | BovineHD1500005842 |
| APC13 | 1 | BovineHD0100038705 | EPHA5 | 6 | BovineHD0600022781 | E1BA24 | 15 | BovineHD1500009535 |
| ENTK | 1 | BovineHD0100005454 | ART3 | 6 | BovineHD0600025443 | STIM1 | 15 | BTA-114838-no-rs |
| C21orf63 | 1 | BovineHD0100000686 | 11-Sep | 6 | BovineHD0600025874 | LOC790886 | 16 | BovineHD1600001673 |
| ROBO1 | 1 | BovineHD0100007707 | FRAS1 | 6 | BovineHD0600026271 | RALGPS2 | 16 | BovineHD1600017259 |
| E1BJS9 | 1 | BovineHD0100010534 | SGCD | 7 | BovineHD0700020503 | ANGL1 | 16 | BovineHD1600017298 |
| A5PKG1 | 1 | BovineHD0100022104 | HEM2 | 8 | BovineHD0800031076 | FBXW8 | 17 | BovineHD1700017212 |
| SMARCAL1 | 2 | BovineHD0200030269 | CLCN3 | 8 | BovineHD0800000483 | ADAMTS18 | 18 | BovineHD1800001383 |
| F1MTX0 | 2 | BovineHD0200040258 | TMEM2 | 8 | BovineHD0800014467 | F1MX91 | 18 | ARS-BFGL-NGS-59215 |
| KIAA1486 | 2 | BovineHD0200033024 | RIMS1 | 9 | BovineHD0900002983 | DOCK2 | 20 | BovineHD2000000529 |
| NMI | 2 | BovineHD0200013025 | ZNF292 | 9 | Hapmap54718-rs29022960 | RNF180 | 20 | BovineHD2000021261 |
| MYO7B | 2 | BovineHD0200001364 | EML5 | 10 | BovineHD1000029437 | FAM196B | 20 | BovineHD2000000555 |
| IWS1 | 2 | BovineHD0200001420 | A6QLI2 | 10 | BTA-114684-no-rs | SLIT3 | 20 | BovineHD2000000133 |
| DNAH7 | 2 | BovineHD0200024162 | A7YWN4 | 11 | ARS-BFGL-NGS-10436 | F1MJV4 | 20 | BovineHD2000018883 |
| ORC2 | 2 | BovineHD0200025582 | LBH | 11 | BovineHD1100019791 | CCDC33 | 21 | BovineHD2100021281 |
| NRP2 | 2 | BovineHD0200027119 | FAM179A | 11 | BovineHD1100020297 | LYZL4 | 22 | BovineHD2200004477 |
| PIK3R3 | 3 | BTA-20822-no-rs | IFT172 | 11 | BovineHD1100020629 | F2Z4H7 | 22 | BovineHD2200005456 |
| NHRF3 | 3 | BovineHD0300006834 | Q3SZR7 | 11 | BovineHD1100022452 | AZI2 | 22 | BovineHD2200000741 |
| SYWM | 3 | BovineHD0300007564 | HS1BP3 | 11 | BovineHD4100009043 | ZCWPW2 | 22 | BovineHD4100015404 |
| TBX15 | 3 | BovineHD0300007604 | XRN2 | 13 | BovineHD1300011855 | RBMS3 | 22 | BTB-00830411 |
| SPAG17 | 3 | BovineHD0300007956 | C20orf123 | 13 | BovineHD1300021958 | ATP2B2 | 22 | ARS-BFGL-NGS-14331 |
| MAN1A2 | 3 | BovineHD0300035509 | KIAA0146 | 14 | BovineHD1400005988 | EGFR | 22 | BovineHD2200000246 |
| ATP1A1 | 3 | BovineHD0300036028 | PRKDC | 14 | BovineHD1400006081 | DCDC2 | 23 | BovineHD2300009744 |
| Q2KI63 | 3 | BovineHD0300002619 | A6QLA9 | 14 | BovineHD4100011332 | TTC39C | 24 | BTB-01623856 |
| ECHD2 | 3 | BovineHD0300027093 | A4IFV2 | 14 | BovineHD4100011399 | RB27B | 24 | BovineHD2400015606 |
| AT1A2 | 3 | BovineHD0300003116 | TRIM55 | 14 | BovineHD1400009278 | CTNNA3 | 28 | BTB-00980953 |
| F1MY54 | 4 | BovineHD0400033064 |
Table 5. Gene Ontology terms related to biological process, cellular component and molecular function of the genes associated directly or indirectly with SNPs in the HFC from Nelore and Angus heifers.
| Category | GO ~ Term | Count | % | P Value | Genes | |
|---|---|---|---|---|---|---|
| Nelore | GOTERM_BP_FAT | GO:0006928 ~ cell motion | 5 | 11.4 | 0.02 | ROBO1, PRKDC, DCDC2, DNAH7, SLIT3 |
| GOTERM_BP_FAT | GO:0048870 ~ cell motility | 4 | 9.1 | 0.03 | ROBO1, PRKDC, DCDC2, DNAH7 | |
| GOTERM_BP_FAT | GO:0051674 ~ localization of cell | 4 | 9.1 | 0.03 | ROBO1, PRKDC, DCDC2, DNAH7 | |
| GOTERM_CC_FAT | GO:0044459 ~ plasma membrane part | 11 | 25.0 | 0.01 | EPHA5, ART3, CLCN3, ROBO1, KCNB2, SNTB1, SGCD, STIM1, ATP1A1, RIMS1, CTNNA3 | |
| GOTERM_CC_FAT | GO:0016010 ~ dystrophin-associated glycoprotein complex | 2 | 4.5 | 0.03 | SNTB1, SGCD | |
| Angus | GOTERM_BP_FAT | GO:0001822 ~ kidney development | 3 | 8.3 | 0.01 | PKHD1, FBN1, NID1 |
| GOTERM_BP_FAT | GO:0001655 ~ urogenital system development | 3 | 8.3 | 0.02 | PKHD1, FBN1, NID1 | |
| GOTERM_CC_FAT | GO:0043228 ~ non-membrane-bounded organelle | 12 | 33.3 | 0.00 | FNTB, TNS3, PARN, PKHD1, MYO16, RBM19, WDR1, MYH14, DNAH2, SHANK1, XRN2, CBS | |
| GOTERM_CC_FAT | GO:0043232 ~ intracellular non-membrane-bounded organelle | 12 | 33.3 | 0.00 | FNTB, TNS3, PARN, PKHD1, MYO16, RBM19, WDR1, MYH14, DNAH2, SHANK1, XRN2, CBS | |
| GOTERM_CC_FAT | GO:0005604 ~ basement membrane | 3 | 8.3 | 0.01 | FRAS1, FBN1, NID1 | |
| GOTERM_CC_FAT | GO:0044420 ~ extracellular matrix part | 3 | 8.3 | 0.02 | FRAS1, FBN1, NID1 | |
| GOTERM_CC_FAT | GO:0044430 ~ cytoskeletal part | 6 | 16.7 | 0.03 | FNTB, PKHD1, MYO16, MYH14, DNAH2, SHANK1 | |
| GOTERM_CC_FAT | GO:0005730 ~ nucleolus | 5 | 13.9 | 0.04 | TNS3, PARN, RBM19, XRN2, CBS | |
| GOTERM_CC_FAT | GO:0005856 ~ cytoskeleton | 7 | 19.4 | 0.04 | FNTB, PKHD1, MYO16, WDR1, MYH14, DNAH2, SHANK1 | |
| GOTERM_MF_FAT | GO:0004532 ~ exoribonuclease activity | 2 | 5.6 | 0.02 | PARN, XRN2 | |
| GOTERM_MF_FAT | GO:0016896 ~ exoribonuclease activity, producing 5'-phosphomonoesters |
2 | 5.6 | 0.02 | PARN, XRN2 | |
| GOTERM_MF_FAT | GO:0003779 ~ actin binding | 4 | 11.1 | 0.02 | MYO16, WDR1, MYH14, MYLK | |
| GOTERM_MF_FAT | GO:0005516 ~ calmodulin binding | 3 | 8.3 | 0.03 | ADCY1, MYH14, MYLK | |
| GOTERM_MF_FAT | GO:0003774 ~ motor activity | 3 | 8.3 | 0.03 | MYO16, MYH14, DNAH2 | |
| GOTERM_MF_FAT | GO:0016796 ~ exonuclease activity, active with either ribo- or deoxyribonucleic acids and producing 5'-phosphomonoesters | 2 | 5.6 | 0.03 | PARN, XRN2 | |
| GOTERM_MF_FAT | GO:0046872 ~ metal ion binding | 14 | 38.9 | 0.04 | FRAS1, ADCY1, RBM20, FBN1, SYT9, PRKCH, NID1, FNTB, TNS3, PARN, EBF1, XRN2, MYLK, CBS | |
| GOTERM_MF_FAT | GO:0043169 ~ cation binding | 14 | 38.9 | 0.04 | FRAS1, ADCY1, RBM20, FBN1, SYT9, PRKCH, NID1, FNTB, TNS3, PARN, EBF1, XRN2, MYLK, CBS | |
| GOTERM_MF_FAT | GO:0043167 ~ ion binding | 14 | 38.9 | 0.04 | FRAS1, ADCY1, RBM20, FBN1, SYT9, PRKCH, NID1, FNTB, TNS3, PARN, EBF1, XRN2, MYLK, CBS |
Table 4. Associated SNPs with respective candidate genes for Angus heifers.
| Gene | Chromosome | SNP | Gene | Chromosome | SNP | Gene | Chromosome | SNP |
|---|---|---|---|---|---|---|---|---|
| Q32KQ4 | 1 | BovineHD0100028647 | FBN1 | 10 | BovineHD1000017884 | SHANK1 | 18 | ARS-BFGL-NGS-25117 |
| CBS | 1 | BovineHD0100041801 | PRKCH | 10 | BovineHD1000021003 | DNAH2 | 19 | BovineHD1900008265 |
| MYLK | 1 | BovineHD0100019384 | FNTB | 10 | BovineHD1000022127 | LEPREL4 | 19 | BovineHD1900012163 |
| A8E661 | 2 | BovineHD0200026032 | A8E641 | 11 | BovineHD1100020762 | PSA4 | 21 | ARS-BFGL-NGS-24797 |
| PALMD | 3 | BovineHD0300013319 | MYO16 | 12 | BovineHD1200025736 | PTPRG | 22 | BovineHD2200011275 |
| IFI44 | 3 | BovineHD0300019648 | RALGAPA2 | 13 | BovineHD1300011643 | ATG7 | 22 | BovineHD2200016071 |
| KAD5 | 3 | BovineHD0300019991 | KIZ | 13 | BovineHD1300011819 | CRISP1 | 23 | BovineHD2300005886 |
| ACM2 | 4 | BovineHD0400028468 | XRN2 | 13 | BovineHD1300011851 | PKHD1 | 23 | BovineHD2300006404 |
| TNS3 | 4 | BovineHD0400020885 | A6QQD3 | 15 | BTB-01820462 | FAM59A | 24 | BovineHD2400006778 |
| ADCY1 | 4 | BovineHD0400021263 | SYT9 | 15 | BovineHD1500013002 | TNFRSF11A | 24 | BovineHD2400017755 |
| F19A5 | 5 | BovineHD0500034919 | A6QNL3 | 16 | BovineHD1600012798 | PARN | 25 | BovineHD2500003745 |
| PPP2R2C | 6 | BovineHD0600029214 | F262 | 16 | BovineHD1600001371 | LIPA | 26 | BTA-62081-no-rs |
| SLC2A9 | 6 | BovineHD0600030984 | CD045 | 17 | BovineHD1700011280 | SORCS1 | 26 | BovineHD2600007572 |
| WDR1 | 6 | BovineHD0600031016 | CUX2 | 17 | BovineHD1700016280 | RBM20 | 26 | BovineHD2600008410 |
| A7YWG6 | 6 | BovineHD0600008622 | RBM19 | 17 | BovineHD1700017987 | F1MX91 | 18 | ARS-BFGL-NGS-59215 |
| FRAS1 | 6 | BovineHD0600026329 | LRBA | 17 | BovineHD1700002101 | DOCK2 | 20 | BovineHD2000000529 |
| EBF1 | 7 | BovineHD0700021323 | MYH14 | 18 | ARS-BFGL-NGS-3584 | RNF180 | 20 | BovineHD2000021261 |
| A1A4J5 | 8 | ARS-BFGL-NGS-1787 | FAM196B | 20 | BovineHD2000000555 |
Discussion
Genome-wide association studies (GWAS) were conducted to identify quantitative trait loci (QTL) and candidate genes associated with fertility in cattle. The most common traits that affect reproduction and are candidates for genetic selection are age at first calving (Hutchison et al., 2017; Mota et al., 2020), conception rate (Galvão et al., 2013), and daughter pregnancy rate (Parker Gaddis et al., 2014).
In Nelore and Angus heifers, the GWAS study showed no genomic region associated with variation in antral follicle number across all 29 autosomal chromosomes, which is shown in the Manhattan plots (Figures 1 and 2). The mean/low peaks observed in the figures can be attributed to two factors: Many SNPs have a small effect on phenotype and variable antral follicle number in cattle is regulated by many genes. GWAS identified several SNPs with association and candidate genes in animals with HFC in Nelore and Angus heifers when analyzed separately.
In Nelore heifers, the epidermal growth factor receptor (EGFR) gene was associated with the SNP BovineHD2200000246 on chromosome 22 (Table 3). EGFR is a transmembrane glycoprotein consisting of an extracellular ligand-binding domain and a cytoplasmic segment with tyrosine kynase activity, which is central to the cell proliferative effects of EGF. EGF has been shown to play a role in spermatogenesis, oocyte development and maturation (Wald, 2005; Richani and Gilchrist, 2018). In spermatogenesis, EGF enhances the effect of gonadotropin on testicular testosterone production by modulating the activity of enzymes involved in the biosynthetic pathway of testosterone and by increasing the availability of cholesterol for mitochondrial steroidogenesis (Wald, 2005). In follicular development, the EGF network is an essential component of the ovulatory cascade by relaying the signal LH from the periphery of the follicle to the cumulus-oocyte complex (COC). Although the EGF network act in the late stages of follicle development, a new concept to emerge is that cumulus cell acquisition of EGF receptor responsiveness in early stages of development represents a developmental hallmark in folliculogenesis. (Richani and Gilchrist, 2018; Ritter et al., 2015).
Nominal associated gene analyzes using DAVID bioinformatics resources identified several functional categories that differed in the HFC group compared with the LFC group in Nelore and Angus heifers. Notably, cell motion (ROBO1, PRKDC, DCDC2, DNAH7, and SLIT3), cell motility (ROBO1, PRKDC, DCDC2, and DNAH7), and localization (ROBO1, PRKDC, DCDC2, and DNAH7) were the GO associated with biological process in Nelore heifers. The Roundabout (ROBO) transmembrane proteins constitute a conserved family of receptors that includes ROBO1, ROBO2, ROBO3, and ROBO4, which together with their repellent ligand SLIT (SLIT1, SLIT2, and SLIT3) play an important role in regulating axon guidance decisions (Devine and Key, 2008). However, there is evidence that the SLIT/ROBO pathway also plays a role in cellular processes outside the nervous system (Wong et al., 2002). In a study of adult human ovaries, the expression of SLIT2, SLIT3, and ROBO2 was increased during the luteal phase and was negatively regulated by hCG and cortisol (Dickinson et al., 2008). In addition, blocking the activity of SLIT-ROBO decreased apoptosis and increased migration in luteal cells (Dickinson et al., 2008). In ovarian development, the SLIT-ROBO signaling pathway may be involved in controlling germ cell migration. Gene expression of SLIT2, SLIT3, ROBO1, ROBO2, and ROBO4 was detected in the ovine fetal ovary at days 50, 60, 70, and 80 of gestation, the time when follicles are formed. In addition, at the time of increased SLIT-ROBO expression, there was a significant reduction in the proliferation of oocytes in the developing ovary (Dickinson et al., 2010). Although GWAS showed that the ROBO1 (BovineHD0100007707) and SLIT3 (BovineHD2000000133) genes are associated with HFC in Nelore, the influence of the SLIT-ROBO pathway on follicle formation in this breed remains to be confirmed by other functional experiments. Nevertheless, this opens a new field for studies on how and whether the SLIT-ROBO pathway affects AFC in Nelore heifers.
A hapFLK study of the Nelore bull genome identified 83,326 SNPs. Most of these are in Chr1, Chr2, Chr5, Chr11, Chr13, and Chr17. The is region in Chr11 is located near genes just between the FSHR and NRXN1 genes. FSHR is required for ovarian follicle development. The authors also highlighted the ‘Roundabout signaling pathway’ involving the ROBO1 and SLIT protein genes (Maiorano et al., 2022). This signaling pathway is important for physiological adaptation of grazing cattle (Muroya et al., 2015).
Two GO terms related to biological processes, kidney development (PKHD1, FBN1, and NID1) and urogenital system development (PKHD1, FBN1, and NID1), were significantly associated with Angus heifers. Fibrillins (FBN1, 2, and 3) are glycoproteins found in connective tissues (Zhang et al., 1995) and form the backbone structure of small diameter microfibrils (Sakai et al., 1991). In addition, fibrillins have regulatory functions by binding and sequestering growth factors. Fibrillin contributes to the extracellular regulation of endogenous TGFB activity by providing a structural platform that controls the diffusion, storage, presentation, and release (Ramirez and Sakai, 2010). In addition, they also interact with the prodomains of bone morphogenic proteins (BMPs) and growth and differentiation factors (GDFs; Sengle et al., 2008), which are members of the TGFB superfamily and have been described as important for ovarian follicular development and function (Dong et al., 1996; McNatty et al., 2000; Yan et al., 2001; Knight and Glister, 2006).
Taurine animals have been shown to have more preantral follicles than indicine animals. On the other hand, indicine animals have more antral follicles counted by ultrasound and histology (Cushman et al., 2019; Favoreto et al., 2019). In addition, follicles from Nelore heifers show higher expression of IGF, BMP, and TGFbeta family genes (Favoreto et al., 2019). These genes are known to be associated with follicle activation and development. The fact that Angus heifers have a polymorphism associated with fibrillin, which regulates these important growth factors, may be one of the reasons for the difference in preantral and antral follicle density in these animals.
Understanding how candidate genes act in modeling the phenotype is a major challenge that is further complicated by the fact that the environment can influence the phenotype. However, the functional information obtained in the present GWAS has focused the study of biological processes and may contribute to future research aimed at validating these genes and elucidating the mechanisms involved in the phenotype. The trait age at first calving in heifers from Nellore exposed to different environmental conditions, especially rainfall and feeding, is associated with regions showing environmental dependence. In animals reared under different environmental conditions, the genomic region on BTA14 acts on metabolic substrates, and these variations are directly involved in the control of reproductive pathways that are essential for early maturity (Mota et al., 2020). BTA 14 also has significant SNPs associated with reproductive function here and in other studies (Kneeland et al., 2004; Buzanskas et al., 2017).
In summary, the GWAS in Nelore and Angus heifers showed SNPs associated with the higher follicle count phenotype. In addition, the HFC heifers had SNPs associated with follicle formation and development.
Acknowledgements
The authors thank São Paulo Research Foundation (FAPESP) for funding (Grant #2011/50964-0) and scholarships for Favoreto, Loureiro, Ereno, Pupulim and Queiroz.
Funding Statement
Financial support: Received funding for this research from Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP). Grant: #2011/50964-0.
Footnotes
Financial support: Received funding for this research from Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP). Grant: #2011/50964-0.
How to cite: Loureiro B, Ereno RL, Pupulim AGR, Tramontana MCVB, Tabosa HP, Barros CM, Favoreto MG. Genome-wide association study of Nelore and Angus heifers with low and high ovarian follicle counts. Anim Reprod. 2024;21(1):e20230110. https://doi.org/10.1590/1984-3143-AR2023-0110
References
- Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23(10):1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- Bohmanova J, Sargolzaei M, Schenkel FS. Characteristics of linkage disequilibrium in North American Holsteins. BMC Genomics. 2010;11(1):421. doi: 10.1186/1471-2164-11-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush WS, Chen G, Torstenson ES, Ritchie MD. LD-spline: mapping SNPs on genotyping platforms to genomic regions using patterns of linkage disequilibrium. BioData Min. 2009;2(1):7. doi: 10.1186/1756-0381-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzanskas ME, Grossi DA, Ventura RV, Schenkel FS, Chud TCS, Stafuzza NB, Rola LD, Meirelles SLC, Mokry FB, Mudadu MA, Higa RH, Silva MVGB, Alencar MM, Regitano LCA, Munari DP. Candidate genes for male and female reproductive traits in Canchim beef cattle. J Anim Sci Biotechnol. 2017;8(1):67. doi: 10.1186/s40104-017-0199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra E, Lazzari B, Milanesi M, Nogueira GP, Garcia JF, Utsunomiya YT, Ajmone-Marsan P, Stella A. Comparison between indicine and taurine cattle DNA methylation reveals epigenetic variation associated to differences in morphological adaptive traits. Epigenetics. 2023;18(1):2163363. doi: 10.1080/15592294.2022.2163363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain AJ, Hayes BJ, Savin K, Bolormaa S, McPartlan PJ, van de Jagt C, MacEachern S, Goddard ME. Validation of single nucleotide polymorphisms associated with milk production traits in dairy cattle. J Dairy Sci. 2012;95(2):864–875. doi: 10.3168/jds.2010-3786. [DOI] [PubMed] [Google Scholar]
- Clarke GM, Anderson CA, Pettersson FH, Cardon LR, Morris AP, Zondervan KT. Basic statistical analysis in genetic case-control studies. Nat Protoc. 2011;6(2):121–133. doi: 10.1038/nprot.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez T, Montenegro H, Coutinho LL, Regitano LCA, Andrade SCS. Molecular evolution and signatures of selective pressures on Bos, focusing on the Nelore breed (Bos indicus) PLoS One. 2022;17(12):e0279091. doi: 10.1371/journal.pone.0279091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman RA, Soares EM, Yake HK, Patterson AL, Rosasco SL, Beard JK, Northrop EJ, Rich JJJ, Miles JR, Chase CC, Jr, Gonda MG, Perry GA, McNeel AK, Summers AF. Brangus cows have ovarian reserve parameters more like Brahman than Angus cows. Anim Reprod Sci. 2019;209:106170. doi: 10.1016/j.anireprosci.2019.106170. [DOI] [PubMed] [Google Scholar]
- Dar MR, Singh M, Thakur S, Verma A. Exploring the relationship between polymorphisms of leptin and IGF-1 genes with milk yield in indicine and taurine crossbred cows. Trop Anim Health Prod. 2021;53(4):413. doi: 10.1007/s11250-021-02866-1. [DOI] [PubMed] [Google Scholar]
- Dehghan A. Genome-wide association studies. Methods Mol Biol. 2018;1793:37–49. doi: 10.1007/978-1-4939-7868-7_4. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- Devine CA, Key B. Robo-Slit interactions regulate longitudinal axon pathfinding in the embryonic vertebrate brain. Dev Biol. 2008;313(1):371–383. doi: 10.1016/j.ydbio.2007.10.040. [DOI] [PubMed] [Google Scholar]
- Dickinson RE, Hryhorskyj L, Tremewan H, Hogg K, Thomson AA, McNeilly AS, Duncan WC. Involvement of the SLIT/ROBO pathway in follicle development in the fetal ovary. Reproduction. 2010;139(2):395–407. doi: 10.1530/REP-09-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson RE, Myers M, Duncan WC. Novel regulated expression of the SLIT/ROBO pathway in the ovary: possible role during luteolysis in women. Endocrinology. 2008;149(10):5024–5034. doi: 10.1210/en.2008-0204. [DOI] [PubMed] [Google Scholar]
- Dong J, Albertini DF, Nishimori K, Kumar TR, Lu N, Matzuk MM. Growth differentiation factor-9 is required during early ovarian folliculogenesis. Nature. 1996;383(6600):531–535. doi: 10.1038/383531a0. [DOI] [PubMed] [Google Scholar]
- Ensembl Ensembl. 2023. [cited 2023 Jul 25]. Internet. Available from: http://www.ensembl.org/index.html .
- Favoreto MG, Loureiro B, Ereno RL, Pupulim AG, Queiroz V, Silva NA, Barros CM. Follicle populations and gene expression profiles of Nelore and Angus heifers with low and high ovarian follicle counts. Mol Reprod Dev. 2019;86(2):197–208. doi: 10.1002/mrd.23095. [DOI] [PubMed] [Google Scholar]
- Galvão KN, Federico P, De Vries A, Schuenemann GJ. Economic comparison of reproductive programs for dairy herds using estrus detection, timed artificial insemination, or a combination. J Dairy Sci. 2013;96(4):2681–2693. doi: 10.3168/jds.2012-5982. [DOI] [PubMed] [Google Scholar]
- Huang W, Kirkpatrick BW, Rosa GJ, Khatib H. A genome-wide association study using selective DNA pooling identifies candidate markers for fertility in Holstein cattle. Anim Genet. 2010;41(6):570–578. doi: 10.1111/j.1365-2052.2010.02046.x. [DOI] [PubMed] [Google Scholar]
- Hutchison JL, VanRaden P, Null D, Cole J, Bickhart D. Genomic evaluation of age at first calving. J Dairy Sci. 2017;100(8):6853–6861. doi: 10.3168/jds.2016-12060. [DOI] [PubMed] [Google Scholar]
- Illumina . GenomeStudio Data Analysis Software. San Diego: 2023. [Google Scholar]
- Ireland JJ, Ward F, Jimenez-Krassel F, Ireland JL, Smith GW, Lonergan P, Evans AC. Follicle numbers are highly repeatable within individual animals but are inversely correlated with FSH concentrations and the proportion of good-quality embryos after ovarian stimulation in cattle. Hum Reprod. 2007;22(6):1687–1695. doi: 10.1093/humrep/dem071. [DOI] [PubMed] [Google Scholar]
- Kneeland J, Li C, Basarab J, Snelling WM, Benkel B, Murdoch B, Hansen C, Moore SS. Identification and fine mapping of quantitative trait loci for growth traits on bovine chromosomes 2, 6, 14, 19, 21, and 23 within one commercial line of Bos taurus. J Anim Sci. 2004;82(12):3405–3414. doi: 10.2527/2004.82123405x. [DOI] [PubMed] [Google Scholar]
- Knight PG, Glister C. TGF-beta superfamily members and ovarian follicle development. Reproduction. 2006;132(2):191–206. doi: 10.1530/rep.1.01074. [DOI] [PubMed] [Google Scholar]
- Liu R, Tearle R, Low WY, Chen T, Thomsen D, Smith TPL, Hiendleder S, Williams JL. Distinctive gene expression patterns and imprinting signatures revealed in reciprocal crosses between cattle sub-species. BMC Genomics. 2021;22(1):410. doi: 10.1186/s12864-021-07667-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiorano AM, Cardoso DF, Carvalheiro R, Fernandes GA, Jr, Albuquerque LG, de Oliveira HN. Signatures of selection in Nelore cattle revealed by whole-genome sequencing data. Genomics. 2022;114(2):110304. doi: 10.1016/j.ygeno.2022.110304. [DOI] [PubMed] [Google Scholar]
- McNatty KP, Fidler AE, Juengel JL, Quirke LD, Smith PR, Heath DA, Lundy T, O’Connell A, Tisdall DJ. Growth and paracrine factors regulating follicular formation and cellular function. Mol Cell Endocrinol. 2000;163(1-2):11–20. doi: 10.1016/S0303-7207(99)00235-X. [DOI] [PubMed] [Google Scholar]
- Mossa F, Walsh SW, Butler ST, Berry DP, Carter F, Lonergan P, Smith GW, Ireland JJ, Evans AC. Low numbers of ovarian follicles ≥3 mm in diameter are associated with low fertility in dairy cows. J Dairy Sci. 2012;95(5):2355–2361. doi: 10.3168/jds.2011-4325. [DOI] [PubMed] [Google Scholar]
- Mota LFM, Lopes FB, Fernandes GA, Jr, Rosa GJM, Magalhães AFB, Carvalheiro R, Albuquerque LG. Genome-wide scan highlights the role of candidate genes on phenotypic plasticity for age at first calving in Nellore heifers. Sci Rep. 2020;10(1):6481. doi: 10.1038/s41598-020-63516-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muroya S, Ogasawara H, Hojito M. Grazing affects exosomal circulating MicroRNAs in cattle. PLoS One. 2015;10(8):e0136475. doi: 10.1371/journal.pone.0136475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naukkarinen J, Surakka I, Pietiläinen KH, Rissanen A, Salomaa A, Ripatti V, Yki-Järvinen H, van Duijn CM, Wichmann HE, Kaprio J, Taskinen MR, Peltonen L. Use of genome-wide expression data to mine the “Gray Zone” of GWA studies leads to novel candidate obesity genes. PLoS Genet. 2010;6(6):e1000976. doi: 10.1371/journal.pgen.1000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker Gaddis KL, Cole J, Clay J, Maltecca C. Genomic selection for producer-recorded health event data in US dairy cattle. J Dairy Sci. 2014;97(5):3190–3199. doi: 10.3168/jds.2013-7543. [DOI] [PubMed] [Google Scholar]
- Peñagaricano F, Weigel KA, Khatib H. Genome-wide association study identifies candidate markers for bull fertility in Holstein dairy cattle. Anim Genet. 2012;43(Suppl. 1):65–71. doi: 10.1111/j.1365-2052.2012.02350.x. [DOI] [PubMed] [Google Scholar]
- Pérez-Pardal L, Royo LJ, Beja-Pereira A, Chen S, Cantet RJC, Traore A, Curik I, Sölkner J, Bozzi R, Fernández I, Alvarez I, Gutiérrez JP, Gómez E, Ponce de León FA, Goyache F. Multiple paternal origins of domestic cattle revealed by Y-specific interspersed multilocus microsatellites. Heredity. 2010;105(6):511–519. doi: 10.1038/hdy.2010.30. [DOI] [PubMed] [Google Scholar]
- Pitt D, Sevane N, Nicolazzi EL, MacHugh DE, Park SDE, Colli L, Martinez R, Bruford MW, Orozco-terWengel P. Domestication of cattle: two or three events? Evol Appl. 2018;12(1):123–136. doi: 10.1111/eva.12674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell J, Talenti A, Fisch A, Hemmink JD, Paxton E, Toye P, Santos I, Ferreira BR, Connelley TK, Morrison LJ, Prendergast JGD. Profiling the immune epigenome across global cattle breeds. Genome Biol. 2023;24(1):127. doi: 10.1186/s13059-023-02964-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F, Sakai LY. Biogenesis and function of fibrillin assemblies. Cell Tissue Res. 2010;339(1):71–82. doi: 10.1007/s00441-009-0822-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richani D, Gilchrist RB. The epidermal growth factor network: role in oocyte growth, maturation and developmental competence. Hum Reprod Update. 2018;24(1):1–14. doi: 10.1093/humupd/dmx029. [DOI] [PubMed] [Google Scholar]
- Ritter LJ, Sugimura S, Gilchrist RB. Oocyte induction of EGF responsiveness in somatic cells is associated with the acquisition of porcine oocyte developmental competence. Endocrinology. 2015;156(6):2299–2312. doi: 10.1210/en.2014-1884. [DOI] [PubMed] [Google Scholar]
- Sakai LY, Keene DR, Glanville RW, Bächinger HP. Purification and partial characterization of fibrillin, a cysteine-rich structural component of connective tissue microfibrils. J Biol Chem. 1991;266(22):14763–14770. doi: 10.1016/S0021-9258(18)98752-1. [DOI] [PubMed] [Google Scholar]
- Sengle G, Charbonneau NL, Ono RN, Sasaki T, Alvarez J, Keene DR, Bächinger HP, Sakai LY. Targeting of bone morphogenetic protein growth factor complexes to fibrillin. J Biol Chem. 2008;283(20):13874–13888. doi: 10.1074/jbc.M707820200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verardo LL, Silva FF, Machado MA, Panetto JCC, Faza DRLR, Otto PI, Regitano LCA, Silva LOC, Egito AA, Albuquerque MSM, Zanella R, Silva MVGB. Genome-wide analyses reveal the genetic architecture and candidate genes of indicine, taurine, synthetic crossbreds, and locally adapted cattle in Brazil. Front Genet. 2021;12:702822. doi: 10.3389/fgene.2021.702822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wald M. Eidermal growth factor and spermatogenesis. J Urol. 2005;174(6):2089–2090. doi: 10.1097/01.ju.0000187410.24477.cf. [DOI] [PubMed] [Google Scholar]
- Wong K, Park HT, Wu JY, Rao Y. Slit proteins: molecular guidance cues for cells ranging from neurons to leukocytes. Curr Opin Genet Dev. 2002;12(5):583–591. doi: 10.1016/S0959-437X(02)00343-X. [DOI] [PubMed] [Google Scholar]
- Yan C, Wang P, DeMayo J, DeMayo FJ, Elvin JA, Carino C, Prasad SV, Skinner SS, Dunbar BS, Dube JL, Celeste AJ, Matzuk MM. Synergistic roles of bone morphogenetic protein 15 and growth differentiation factor 9 in ovarian function. Mol Endocrinol. 2001;15(6):854–866. doi: 10.1210/mend.15.6.0662. [DOI] [PubMed] [Google Scholar]
- Zhang H, Hu W, Ramirez F. Developmental expression of fibrillin genes suggests heterogeneity of extracellular microfibrils. J Cell Biol. 1995;129(4):1165–1176. doi: 10.1083/jcb.129.4.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]


