Abstract
Objectives.
The ongoing risk of emerging infectious disease has renewed calls for understanding the origins of zoonoses and identifying future zoonotic disease threats. Given their close phylogenetic relatedness and geographic overlap with humans, non-human primates (NHPs) have been the source of many infectious diseases throughout human evolution. NHPs harbor diverse parasites, with some infecting only a single host species while others infect species from multiple families.
Materials and Methods.
We applied a novel link-prediction method to predict undocumented instances of parasite sharing between humans and NHPs. Our model makes predictions based on phylogenetic distances and geographic overlap among NHPs and humans in six countries with high NHP diversity: Columbia, Brazil, Democratic Republic of Congo, Madagascar, China and Indonesia.
Results.
Of the 899 human parasites documented in the Global Infectious Diseases and Epidemiology Network (GIDEON) database for these countries, 12% were shared with at least one other NHP species. The link prediction model identified an additional 54 parasites that are likely to infect humans but were not reported in GIDEON. These parasites were mostly host generalists, yet their phylogenetic host breadth varied substantially.
Discussion.
As human activities and populations encroach on NHP habitats, opportunities for parasite sharing between human and non-human primates will continue to increase. Our study identifies specific infectious organisms to monitor in countries with high NHP diversity, while the comparative analysis of host generalism, parasite taxonomy, and transmission mode provides insights to types of parasites that represent high zoonotic risk.
Keywords: surveillance, zoonoses, host generalism, parasite sharing, spillover
Introduction
Infectious diseases have been a major force in shaping human history (Snowden, 2019), inequality and health disparities (Bonds, Keenan, Rohani, & Sachs, 2010), and the human genome (Fumagalli et al., 2011; Karlsson, Kwiatkowski, & Sabeti, 2014). These effects are not simply a part of our past, with outbreaks over the past 20 years renewing interest in disease ecology and evolution and catalyzing discussions on best practices for managing future disease threats (Petrovan et al., 2021). Global connectivity facilitates the rapid spread of highly transmissible diseases across international borders, and the international community has demonstrated low preparedness in the face of pandemic threats (Coccia, 2021). Controlling disease outbreaks at the local level before they escalate to a pandemic is a major challenge that requires an intimate understanding of risk factors that promote disease emergence.
Most major pandemics and epidemics in humans originated as zoonoses, and many of these can be traced back to one or a small number of zoonotic spillover events (Devaux, Mediannikov, Medkour, & Raoult, 2019; Ellwanger & Chies, 2021; Pépin, 2021; Piret & Boivin, 2020; Plowright et al., 2017). A successful cross-species spillover requires the alignment of ecological, physiological, and immunological conditions involving the host(s) and a parasite, which we define ecologically as any organism that benefits from living within or on a host at the host’s expense (Plowright et al., 2017). In a viral spillover, for example, a person must come into contact with infectious material from another host species, which requires ecological overlap of humans and infected animals in space and time. The virus must also be capable of entering the new host’s cells, replicating in those cells, and then successfully transmitting to new hosts, all before the host immune system clears the infection. Greater phylogenetic relatedness between a pair of hosts increases the probability of cross-species transmission (Cooper, Griffin, Franz, Omotayo, & Nunn, 2012; Davies & Pedersen, 2008; Poulin, 2010), while a higher mutation rate provides more opportunities for a parasite to adapt to a novel host (Loverdo & Lloyd-Smith, 2013; Woolhouse, Haydon, & Antia, 2005). The multiple factors that lead to spillover and then sustained transmission in human populations are complex, making it extremely challenging to predict zoonotic outbreaks (Plowright et al., 2017). However, their frequency is increasing as global anthropogenic change exerts mounting pressure on wildlife (Daszak, Cunningham, & Hyatt, 2001; Marani, Katul, Pan, & Parolari, 2021).
Disease surveillance and risk mitigation at the expanding human-wildlife interface are essential for early-stage control of zoonotic pathogens, yet resources for such efforts are limited. Prioritizing hosts and parasites for targeted sampling remains a major challenge. Existing data on host-parasite interactions can inform surveillance priorities, but a paucity of data on the vast majority of parasite species leads to uncertainty in inferences drawn from current datasets (Carlson, Dallas, Alexander, Phelan, & Phillips, 2020; Gibb et al., 2021; Poulin & Keeney, 2008).
A variety of modeling approaches have emerged in recent years to predict the zoonotic risk and epidemic potential of understudied parasites. These approaches, termed “zoonotic risk technology” by Carlson et al. (2021), include link prediction models—a suite of network imputation methods that can be used to identify likely host-parasite interactions that are absent from current datasets (Bartomeus et al., 2016; Dallas, Park, & Drake, 2017; Elmasri, Farrell, Davies, & Stephens, 2020; Farrell, Elmasri, Stephens, & Davies, 2022; Morales-Castilla, Matias, Gravel, & Araújo, 2015). Such absences are often thought to occur when a parasite (i.e., a link) is undocumented due to insufficient sampling (Nunn, Altizer, Jones, & Sechrest, 2003; Teitelbaum et al., 2020). Link prediction methods can help fill in these absences and highlight host-parasite combinations that have a high probability of occurring in the future. This framework could predict missing links to humans, with links representing infectious organisms in wildlife that are currently undocumented in humans or have not yet successfully transmitted to humans with onward spread in human populations.
Link prediction models can derive probabilities of parasite occurrence in a new species based on various predictors. The structure of the network itself can inform the addition of missing host-parasite links, but this method can exacerbate sampling bias if network properties are influenced by uneven sampling (Becker et al., 2022; Elmasri et al., 2020; Farrell et al., 2022). Alternatively, link prediction models can use ecological traits as predictors, based on the assumption that ecological similarity of host species increases the likelihood of transmission between them or via shared habitats or behaviors (Becker et al., 2022; Cooper, Kamilar, & Nunn, 2012; Dallas et al., 2017). Such models have shown high accuracy in imputing parasites of small mammals (Dallas et al., 2017) and bats (Becker et al., 2022), but they depend on the availability of extensive ecological trait data. In the absence of ecological data, the evolutionary relatedness of hosts, measured by phylogenetic distance, has strong predictive power in models of host-parasite interactions (Elmasri et al., 2020; Farrell et al., 2022; Morales-Castilla et al., 2015). Geographic overlap has also been shown to predict parasite sharing, as expected because spillover requires spatial overlap of organisms (Davies & Pedersen, 2008). Though the covariates on which they rely may vary, all link prediction models are affected by data limitations, and evaluation of prediction results is necessary to arrive at relevant zoonotic risk assessments that can inform surveillance priorities (Carlson et al., 2021).
Here, we apply a novel link-prediction method to identify undocumented parasites shared by humans and non-human primates (NHPs). In addition to rodents and bats, NHPs are at high risk for harboring emerging diseases that may infect humans (Alexander et al., 2018; Devaux et al., 2019). Environments that facilitate consistent spatiotemporal overlap between humans and NHPs, including urban areas and tourist destinations, can increase opportunities for disease-relevant contact (Ahmed et al., 2019; Devaux et al., 2019). In addition, communities in some parts of rural Africa, Asia, and South America traditionally hunt NHPs, which provides an essential source of protein but places these human populations on the front lines of zoonotic disease transmission (Booth et al., 2021; Wolfe, Daszak, Kilpatrick, & Burke, 2005). Notable instances of disease transmission from NHPs to humans include yellow fever virus, which is maintained in sylvatic and urban NHP populations and transmitted via the vector Aedes aegypti (Carrington & Auguste, 2013; Hanley et al., 2013), and the human immunodeficiency viruses (HIV-1 Group M and HIV-2), which evolved from simian immunodeficiency viruses (SIVs) found in Pan troglodytes and Cercocebus atys, respectively (Gao et al., 1999; Hahn, Shaw, De Cock, & Sharp, 2000; Pépin, 2021).
The novel link-prediction method presented here builds upon the model presented by Elmasri et al. (2020) and Farrell et al. (2022) by incorporating both phylogenetic and geographic predictors to estimate undocumented NHP-parasite interactions relevant to human health. We apply this model at a regional scale to bipartite networks consisting of (1) humans and NHP species known to have high rates contact with humans due to hunting, tourism, and urban cohabitation, and (2) their parasites, where parasite refers to both micro-parasites, such as viruses, bacteria, fungi and protozoa, and macro-parasites, such as helminths and arthropods. Farrell et al. (2022) applied phylogeographic link prediction to a global network, but we focus on regional networks to obtain a manageable list of missing links, allowing us to evaluate each imputed parasite by searching the literature for existing evidence of zoonotic and epidemic risk. Finally, we investigate taxonomic and phylogenetic host generalism, parasite type, and transmission mode of these parasites to better understand traits that increase risk of spillover to humans.
We predict that the majority of parasites imputed to infect humans through link prediction will be generalists that are observed in multiple host families or broader (a taxonomically defined measure of host generalism) and infect more distantly-related species than expected by a null distribution (a phylogenetically defined measure of generalism). Previous studies in NHPs have found that their viruses infect a wider range of hosts than other groups of parasites, such as protozoa or helminths (Pedersen et al. 2005; Cooper et al. 2012). Thus, we predict that humans and NHPs will be more likely to share viruses. We also expect that vector-transmission will characterize parasites imputed to infect humans, again based on previous findings that these vector-transmission modes predict a wider host range in parasites of NHPs (Pedersen, Altizer, Poss, Cunningham, & Nunn, 2005) and of mammals (Park et al., 2018). Conversely, we predict a lower likelihood of shared helminth parasites and parasites transmitted via intermediate hosts due to the relatively host-specific nature of these groups (Cooper, Griffin, et al., 2012; Pedersen et al., 2005).
Methods
Parasite Data Collection
NHP parasite data were downloaded from the Global Mammal Parasite Database (GMPD), a resource of parasites documented in wild NHP hosts (Nunn & Altizer, 2005; Stephens et al., 2017). We focused on NHPs with the highest risk of transmitting disease to humans by including NHP species that met one of three criteria. First, all great apes were included due to their close phylogenetic relationship with humans and popularity in tourist attractions (Sharp, Rayner, & Hahn, 2013). Second, we included all NHP species that dwell in urban habitats alongside dense populations of humans (Santini et al., 2019). Third, we included all NHP species subject to routine hunting and trapping pressure according to the IUCN (2021), which we identified by searching iucnredlist.org for NHP species under the threat category “Hunting and trapping of terrestrial animals” and the use and trade categories “Food” and “Medicine.” We chose to focus on NHP species subject to the highest anthropogenic pressure in order to prioritize the parasites of hosts for which spillover to humans is a more imminent risk.
Parasite data for high-risk NHPs were included if the NHP and parasite were identified to the species level. We ensured that NHP species names in the GMPD matched the IUCN taxonomy reported by Estrada et al. (2017). We filtered out any records in which a parasite was searched for but not found (i.e., prevalence = 0). Parasite attribute data on taxonomic host specificity, type, and transmission mode were also downloaded from the GMPD. Parasites were classified to broad taxonomic categories of bacteria, protozoa, viruses, arthropods, fungi, or helminths. Though we included all parasite types in our analysis, bacteria, arthropods, and fungi are severely under sampled in primates (Cooper & Nunn, 2013); this should be considered when interpreting results for these parasite groups.
Transmission mode was classified as close contact, sexual contact, non-close (environmental), vector, and/or via an intermediate host (Pedersen et al., 2005). Multiple transmission modes were possible for each parasite species. Close contact transmission describes parasites that require close proximity for transmission, including through grooming, biting, scratching, or other physical contact. Sexual transmission describes close-contact transmission that specifically involves sexual contact. Non-close transmission involves transmission through shared environments, including by fomites, water, or soil. Vector-borne parasites are transmitted through biting arthropods that actively search for hosts, such as mosquitoes, fleas, lice, or ticks. Transmission by intermediate hosts describes parasites with complex life cycles (excluding vector-borne parasites) or trophic transmission.
Host generality, defined as the breadth of hosts that a parasite infects, can be measured in multiple ways (Park et al., 2018). We used three metrics to investigate the host generality of parasites found in NHPs. First, we used taxonomic host generality scores from the GMPD that were developed by Pedersen et al. (2005). These integer scores represent the maximum taxonomic breadth of all vertebrate hosts in which evidence of a parasite was detected and range from one through five, representing whether a parasite is documented in (1) a single host species, (2) hosts in one genus, (3) hosts in one family, (4) hosts in one order, or (5) hosts in multiple mammalian orders and possibly non-mammals. Generality scores are regularly revised to incorporate new data on a parasite’s host range, with current scores assigned in 2020 or later.
As a second measure of host generality, we used the full GMPD dataset to calculate the mean phylogenetic distance between all NHP hosts that the parasite infects (MPD). The MPD of each parasite represents the average phylogenetic “distance” between its hosts in millions of years, with smaller MPD values characterizing parasites that mostly infect closely-related hosts; this measure avoids problems with variation in the phylogenetic ages of different parasite taxonomic groups. To calculate MPD, we downloaded a posterior distribution of 100 phylogenetic trees containing all NHP species from VertLife, which provides a Bayesian inference of phylogeny for multiple groups of vertebrates (Upham, Esselstyn, & Jetz, 2019). From this posterior distribution of phylogenies we used the phytools package to obtain a consensus phylogeny by computing the mean length of an edge across all trees, with absent edges receiving a value of zero (Revell, 2012). We then calculated the MPD of each parasite using the picante package (Kembel et al., 2010) in R.
As a final measure of host generality, we controlled for varying host abundances in the GMPD and calculated a standardized metric to enable comparison of MPD values across parasites. We used picante to calculate the standard effect sizes for the mean phylogenetic distances (SES.MPD; Kembel et al., 2010). SES.MPD values were obtained by comparing the raw PD values between host pairs to the expected PDs obtained from a null distribution, which was generated by randomizing the host-parasite interaction matrix 1000 times with an independent swap algorithm (Gotelli, 2000). Host and parasite occurrence frequencies were maintained in the null distribution, and the PD of each host pair was weighted by their relative frequencies in the GMPD. The resulting z-scores indicate whether a parasite’s MPD is significantly different from the null expectation, in which case the parasite is considered a generalist (z > 1.96) or a specialist (z < −1.96).
Human parasite data were compiled from the Global Infectious Disease and Epidemiology Network (GIDEON), a database for infectious diseases in humans at a global scale (Yu & Edberg, 2005). Zoonotic link-predictions are often conducted on global host-parasite datasets, yet effective prevention and containment of emerging infectious disease should occur at the regional level, ideally before an outbreak reaches epidemic or pandemic status. In order to make targeted predictions relevant to smaller-scale efforts, we identified six countries on multiple continents that contain a high diversity of NHP species by overlaying a map of World Countries (Esri 2019) with NHP geographic range polygons (IUCN 2021) in ArcMap 10.7 and determining total NHP species richness within each country. These countries included Madagascar (54 NHP spp.), Brazil (39 NHP spp.), Indonesia (30 NHP spp.), Democratic Republic of the Congo (DRC, 25 NHP spp.), China (17 NHP spp.), and Colombia (17 NHP spp.). We examined all references under the “Relevant Diseases’’ section of the GIDEON profile for each country. GIDEON organizes data by disease rather than the organism that causes disease. Thus, we excluded disease-only data and retained only records in which the parasite causing disease was identified to the species-level. All parasite names were corrected to match those in the GMPD. If a parasite was not found in the GMPD, we used the NCBI taxonomy browser (Sayers et al., 2019; Schoch et al., 2020) and the ICTV taxonomy for viruses (Lefkowitz et al., 2017) to assign the most widely accepted species name.
Link Prediction Model
To predict undocumented parasite sharing between human and NHPs, we constructed a Bayesian phylogeographic link-prediction model using the HPpredictions package in R (see Elmasri et al. 2020). The phylogeny-only version of this model takes a binary host-parasite interaction matrix and a phylogenetic distance matrix and uses phylogenetic relatedness to predict parasite sharing. We used an extended version of this model that adds a geographic distance matrix and predicts host-parasite sharing with phylogenetic relatedness and geographic overlap (Figure 1; S1), both of which are known predictors of parasite sharing in NHPs (Davies & Pedersen, 2008). We did not include ecological traits as predictors in the model because ecological trait data are subject to uneven sampling in NHP datasets, and key traits, such as population density, are difficult to generalize to humans.
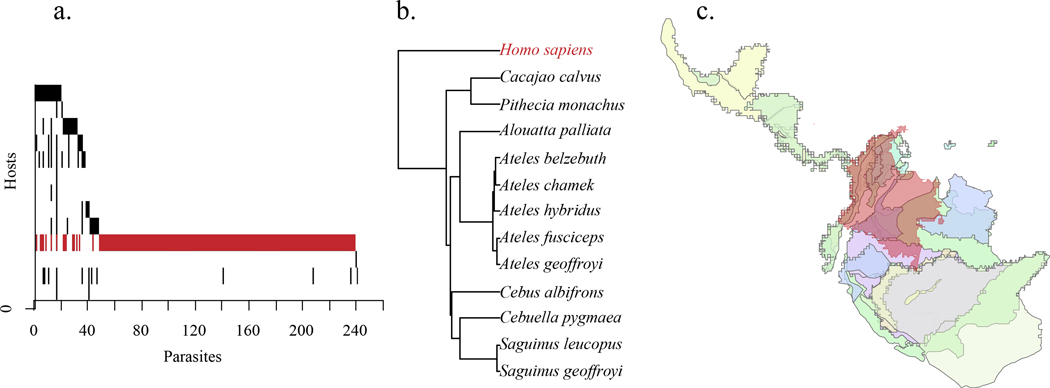
Figure 1. Imputing missing interactions using link prediction.
Our updated Elmasri et al. (2020) phylogeographic model takes (a) a binary matrix of observed host-parasite interactions and uses (b) phylogenetic relatedness and (c) geographic range overlap to identify the probability of undocumented interactions. Depicted here are the parasites, phylogeny, and geographic ranges of humans (indicated in red) and NHPs found in Colombia. The human range is limited to Columbia, while the NHP ranges often include neighboring countries. Geographic overlap with humans is the area of the NHP range that overlaps the country of interest, while overlap amongst NHPs makes use of their entire ranges, including outside the country
We obtained pairwise phylogenetic distances from the consensus phylogeny of humans and NHPs, as described above (Upham et al., 2019). Geographic range shapefiles for each NHP were downloaded from the IUCN spatial database (IUCN 2021). The “geographic range” for humans was defined by the boundaries of each country (Esri). We used the sp package in R to project NHP ranges into the EPSG 3035 projection, which preserves polygon areas across the globe (Bivand, Pebesma, & Gomez-Rubio, 2013). We then created a geographic overlap matrix for each country by calculating the area of intersection between that country and the ranges of all NHPs found within its boundaries, as well as the intersection between the entire range of each pair of NHPs. By incorporating range overlap into our phylogeographic model, we assume that parasites found in a host are evenly distributed throughout its range. Phylogenetic distances and area of geographic overlap occur on different scales; thus, to incorporate them simultaneously into the prediction model, we standardized the values in each matrix to z-scores with a mean of zero and standard deviation of one. Area of geographic overlap is a measure of similarity, but the Elmasri et al. (2020) model requires a matrix of non-negative distances. We transformed the similarity values into distances by subtracting the z-score of each value from one. To meet the requirements of having only positive elements in the distance matrix, we added a constant to ensure all values remained positive, with a minimum value of 0.01.
We scaled the phylogenetic distance matrix to account for branch-length uncertainty by applying a single-parameter, early-burst evolutionary model (Elmasri et al., 2020; Harmon et al., 2010). Model performance was measured by the area under the receiver operating characteristic (ROC) curve (AUC; Elmasri et al. 2020). We ran the network imputation model with 2,000 iterations and implemented five-fold cross validation to test the posterior predictive strength of the model. We obtained a ROC curve by averaging the true positive and false positive rates across all folds (Elmasri et al., 2020). We also derived the ranked posterior distributions for the relative probabilities of each host-parasite interaction. The model assigned a cutoff relative probability threshold that maximized the AUC and produced an output dataset containing all imputed edges with a probability value above this threshold.
From the imputation results list, we pulled out parasites shared by humans and NHPs. To corroborate model performance and further evaluate risk of individual parasites to human populations within each country, we searched the literature for existing evidence beyond the GIDEON database on the zoonotic threat level of each parasite predicted to infect humans. We conducted a topic search of the PubMed and ScienceDirect for publications from 1900 to the present. We used a combination of key words that included the genus and species of each parasite (including synonyms), the disease name, and “human infection OR primate infection”. We also consulted https://www.cdc.gov/ Center for Disease Control and Prevention (CDC) and Beran et al. (2019) for additional information.
To understand spillover risk more broadly, we investigated whether certain traits were more likely to occur among NHP parasites predicted to infect humans. We examined the taxonomic and phylogenetic host generality of predicted parasites with the expectation that parasites with wider host ranges would represent a greater spillover risk to humans. We also investigated the associations between parasite taxonomic categories and transmission modes to determine whether certain parasite types (e.g. viruses) and transmission modes (e.g. vector-borne) were more likely to occur among model predictions.
Results
We compiled data on 98 high-risk primate species and 1,131 parasite species in the six countries of interest. As expected given greater effort to characterize parasites of humans, most of these parasites (899) were documented in humans. Of these human parasites, 12.2% were shared with at least one other NHP species. Humans shared the highest number of parasite species with NHPs in the DRC (46 parasites) and the lowest in Madagascar (5 parasites).
The link prediction model imputed a total of 54 parasites that are undocumented in GIDEON in the six countries. Performance of the imputation model varied across datasets for the six countries, as did the number of imputed interactions above the threshold cutoff values (Table 1). DRC had the highest number of parasites imputed to infect humans (25 parasites), followed next by China (18 parasites). The AUC values indicated high model accuracy for all countries except China (Table 1). Only 3.6% of the 560 parasites reported in China were observed in more than one host species, and this lack of observed parasite sharing resulted in lower predictive power. The model output for China indicates the limitations of applying link prediction to such sparsely sampled datasets; therefore, the results for China should be interpreted with caution.
Table 1.
The maximum area under the ROC curve (AUC) for each imputation model, calculated via five-fold cross-validation, along with the number of imputed edges above the probability threshold (for all primates) and a list of the subset of those imputed to humans.
| country | max AUC | number of observed interactions | number of imputed interactions | parasites imputed to humans |
|---|---|---|---|---|
| Brazil | 0.99 | 616 | 387 |
Trypanosoma minasense
Dipetalonema gracile Prosthenorchis elegans |
| China | 0.57 | 582 | 4,458 |
Plasmodium inui
Plasmodium eylesi Plasmodium jefferyi Plasmodium youngi Strongyloides fuelleborni Colobenterobius presbytis Cytomegalovirus Macacine betaherpesvirus 3 Enterobius macaci Dirofilaria macacae Nochtia nochti Oesophagostomum bifurcum Plasmodium cynomolgi Polyomavirus Simian agent virus 40 Rubulavirus Parainfluenza virus 5 Simplexvirus Cercopithecine herpesvirus 1 Spumavirus Simian foamy virus Sandnema digitatum |
| Colombia | 0.98 | 295 | 96 |
Trypanosoma minasense
Mansonella marmosetae |
| Congo DRC | 0.93 | 463 | 636 |
Strongylouides fuelleborni
Hepatocystis kochi Alphavirus Semliki forest virus Alphavirus Chikungunya virus Staphylococcus aureus Dientamoeba fragilis Trypanosoma primatum Lentivirus Simian immunodeficiency virus Deltaretrovirus Primate T-lymphotrophic virus 3 Oesophagostomum bifurcum Oesophagostomum stephanostomum Encephalitozoon hellem Trypanosoma vivax Balantidium coli Physaloptera caucasia Enterobius antrhopopitheci Mansonella rodhaini Mastadenovirus Human mastadenovirus E Pediculus schaeffi Plasmodium gaboni Entamoeba chattoni Bertiella studeri Necator americanus Orthobunyavirus Bunyamwera orthobunyavirus Spumavirus Simian foamy virus |
| Madagascar | 0.95 | 243 | 358 |
Haemaphysalis lemuris
Trichuris lemuris Trichophilopterus babakotophilus Liponysella madagascariensis Dipetalonema petteri Lemuralges intermedius Ixodes lemuris Pararhabdonema longistriata |
| Indonesia | 0.96 | 253 | 182 |
Oesophagostomum aculeatum
Plasmodium inui Tanjong Rabok orthobunyavirus |
Parasites predicted to infect humans were mostly taxonomic generalists, with 71% observed to infect hosts across taxonomic families or broader (Figure 2a). Viruses comprised the majority of imputed parasites infecting the broadest taxonomic host range (taxonomic generality score = 5, i.e. documented in multiple taxonomic orders), while helminths dominated the imputed parasites infecting NHP hosts from multiple families (generality score = 4).
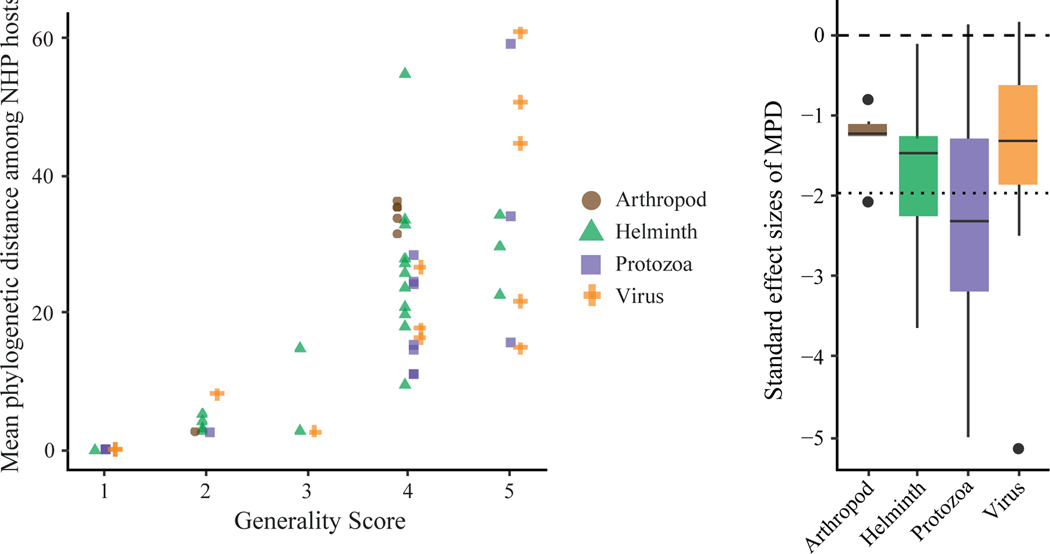
Figure 2. Host specificity of NHP parasites predicted to infect humans.
(a) Each imputed parasite is plotted along two metrics of host specificity. Parasites in the GMPD were assigned a taxonomic generality score indicating whether it is (1) species-specific, (2) documented to infect members of a single genus, (3) documented to infect members of a single family, (4) documented to infect members of a single order, or (5) documented in multiple orders of mammals and potentially some non-mammals. The phylogenetic generality of each parasite represents the mean phylogenetic distance (MPD), in millions of years, between all hosts that a parasite infects. (b) Standard effect sizes of MPD (SES.MPD) were calculated relative to a null model. Parasites with significant, negative SES.MPD values (z score < −1.96, indicated by the dotted line) are considered phylogenetic host specialists.
The mean phylogenetic distance between all NHP hosts of a given parasite (MPD) varied among the generalist parasites (Figure 2a). The average MPD of all imputed parasites was 20.7 mya, which roughly corresponds to the phylogenetic distance between the primate genera Pan and Pongo or Eulemur and Hapalemur (Upham et al., 2019). In contrast to the taxonomic scores, the SES.MPD values indicated that none of the parasites predicted to infect humans were considered generalists when compared to a null model (Figure 2b). The average SES.MPD of all imputed parasites was −1.84. According to this metric, the host specificity of most parasites did not differ from the null expectation, though 36% could be considered phylogenetic specialists. Most phylogenetic specialists were protozoa and helminths but also included two sexually transmitted viruses.
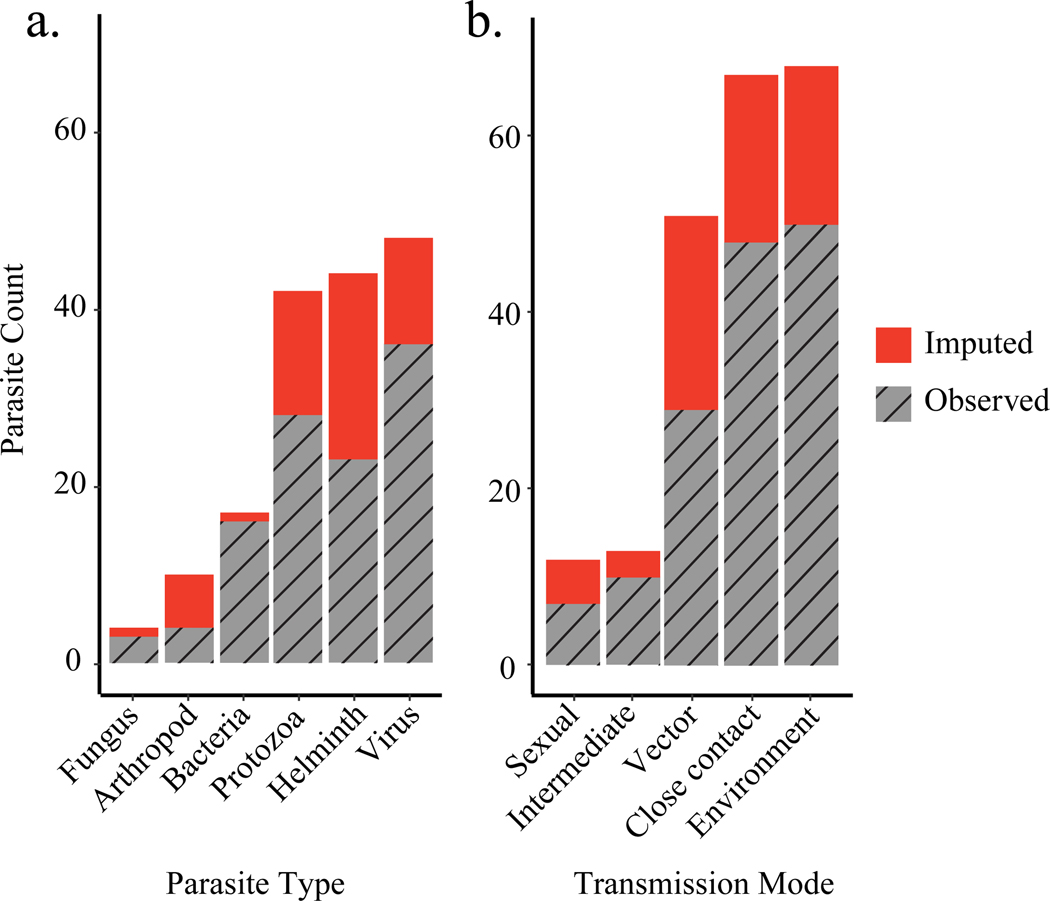
Of the observed and imputed parasites shared between humans and NHPs, the majority were viruses (29%), helminths (27%) and protozoa (25%; Figure 3a). Compared to parasites transmitted sexually and through intermediate hosts, parasites transmitted through close contact, the environment, and vectors were more commonly observed and imputed among humans and NHPs.
Figure 3. (a) Parasite type and (b) transmission mode of observed and imputed parasites shared by NHPs and humans.
Of the 54 NHP parasites predicted to have high spillover risk, we found evidence from other sources that 47% have naturally infected humans (S2). Some of these parasites pose known threats to human health but have not yet been detected in the country of interest, such as Chikungunya virus in DRC. Some known zoonotic parasites are widespread but neglected and underreported (e.g. Dientamoeba fragilis), and others have been documented in humans only rarely (e.g. Trypanosoma vivax). In addition, some zoonotic viruses have been documented in humans, but evidence of disease is lacking (e.g. Simian foamy virus and Primate T-lymphotropic virus 3).
We did not find evidence of natural human infection for 53% of the predicted parasites. However, experimental studies indicate that several of these parasites can infect human cell lines, including Parainfluenza virus 5 and Macacine betaherpesvirus 3. Other predicted parasites are not confirmed to infect humans in any setting and may pose varying levels of spillover and disease risk. These include ectoparasites of lemurs (e.g. Haemaphysalis lemuris), pinworms of Old World monkeys and apes (e.g. Enterobius anthropopitheci) and viruses that circulate in Old World monkeys (e.g. Tanjong Rabok virus).
Discussion
The threat of infectious disease transmission between humans and other mammals underscores the need for effective predictive frameworks to guide disease surveillance at the growing human-animal interface. This is especially true for NHPs, which have been the source of many novel parasites throughout human evolution due to their close phylogenetic relatedness to humans, coupled with geographic and ecological overlap that provides opportunities for spillover. To investigate these threats, we used a phylogeographic model to identify previously undocumented host-parasite interactions between humans and NHPs in six countries with high primate diversity. We found that imputations varied by country, with the DRC, China, and Indonesia having the highest percentage of imputed edges between a known parasite in NHPs and humans. Overall, the models suggest that 54 unique parasites in NHPs represent threats to humans. These findings therefore provide country-specific targets for surveillance efforts aimed at preventing zoonotic spillover events, including surveillance in NHPs in close proximity to human populations.
Literature searches supported model predictions by revealing evidence that transmission of certain NHP parasites to humans is both biologically possible (Cormier, 2016) and spatially likely (Barratt, Harkness, Marriott, Ellis, & Stark, 2011; Zhang et al., 2016). In fact, several of the predicted parasites have already been documented to naturally infect humans outside of our countries of interest (Beran, 2019; Kotepui, Masangkay, Kotepui, & Milanez, 2021). For instance, the model identified Plasmodium cynomolgi as a threat to humans in China and Plasmodium inui in China and Indonesia. Macaque-to-human and human-to-human transmission of P. cynomolgi and P. inui have been demonstrated via accidental laboratory exposures and experiments performed on imprisoned populations (Cormier, 2016), and the first natural infections were recently reported in Malaysia (Liew et al., 2021; Ta et al., 2014). P. cynomolgi and P. inui are present at high prevalence in macaques known to share space with humans (Zhang et al., 2016) and represent continued risks for spillover into human populations (Kotepui et al., 2021; Yap et al., 2021). Also included among predictions is Dientamoeba fragilis, a known enteric protozoan with a global distribution (Barratt et al., 2011). D. fragilis remains neglected due to its historically debated pathogenicity, though studies have linked it to gastrointestinal illness (Norberg, Nord, & Evengård, 2003; Stark et al., 2010). This protozoan parasite was reported in humans in all countries except the DRC, where the model filled in the undocumented interaction. Such examples corroborate model performance while calling attention to underreported parasites with known zoonotic risk.
Our findings also provide more general insights to the nature of zoonotic threats beyond the six focal countries investigated here. One important general conclusion is that taxonomic generalism is a stronger predictor of spillover likelihood than parasite type or transmission mode. Thus, as expected, taxonomic generality scores revealed that parasites predicted to infect humans were mostly generalists that infect hosts of different families, orders, or even classes. We expected that phylogenetic generaltiy metrics would correlate with taxonomic scores, but mean phylogenetic distances (MPD) between NHP hosts were highly variable. The standardized effect sizes of the MPD values (SES.MPD) indicated that even parasites with a wide taxonomic host breadth (including non-primates) were, on average, constrained to closely-related NHP hosts. Large discrepancies between taxonomic and phylogenetic generality may highlight the opposing biases of both metrics; taxonomic generality scores can exaggerate host generalism given their simplicity and emphasis on maximum host breadth, while MPD and SES.MPD can overstate host specialism in sparsely sampled datasets like the GMPD (Park et al., 2018). Examining both phylogenetic and taxonomic generality metrics together presents a more complete picture of a parasite’s host range and its potential to infect additional hosts.
We found less support for the effects of parasite taxonomy or transmission mode. We hypothesized that parasites predicted to occur in humans would be dominated by viruses and by vector-transmitted parasites, as previous work based on taxonomic specificity indicated that these traits predict greater generalism among primate hosts (Cooper, Griffin, et al., 2012; Pedersen et al., 2005). However, we found that viruses, protozoa, and helminths were evenly represented among both the NHP parasites observed in humans and those predicted to infect humans. In addition, rather than a predominance of vector transmission in the observed and imputed parasites, we found evidence for a wider range of transmission modes, including close contact, environmental, and vector transmission.
Although roughly two-thirds of the parasites predicted to infect humans were protozoans and helminths, host immunity (i.e. to Trypanosoma spp.; Vanhollebeke, Lecordier, Perez-Morga, Amiguet-Vercher, Pays, 2007) and low human-to-human transmission potential (i.e. for Physaloptera spp.; Makki et al., 2017) may limit the likelihood of widespread outbreaks originating from these parasites. The presence of helminths in the imputation results was particularly surprising given that prior studies suggest that helminth parasites of NHPs are more likely to be host specialists (Cooper, Griffin, et al., 2012; Pedersen et al., 2005). Most of the specialist helminths in the GMPD have complex life-cycles, which limits the ecological likelihood that they will adapt to new hosts (Wells, Gibson, & Clark, 2019). Consistent with this association, we found that parasites requiring an intermediate host were least likely to occur among the imputed human-parasite interactions, while the majority of predicted helminths were transmitted environmentally or through vectors. Our results thus suggest that taxonomic generalism is a more consistent indicator of spillover likelihood than parasite type or transmission mode.
The rapid generation time of viruses, especially RNA viruses, allows them to adapt quickly to novel hosts, successfully transmit between hosts, and evade natural or vaccine-induced immunity (Duffy, 2018). Hence, global surveillance efforts aimed at curbing the next pandemic focus heavily on novel viruses (Carroll et al., 2018). Our model identified undocumented or underreported viruses that may present a significant threat to communities at the human-wildlife interface and beyond (S2). For example, Simian Foamy Virus (SFV) was predicted to occur in humans in DRC and China, yet has not been documented in GIDEON. A study of 305 people living in South and Southeast Asia detected SFV in 2.6% of participants, most of whom reported bites or scratches from macaques in urban and tourist settings (Jones-Engel et al., 2008). Although no evidence of disease in humans has been reported, the evolution of a pathogenic strain and subsequent outbreak remains a possibility (Khan, 2009; Pinto-Santini, Stenbak, & Linial, 2017). By identifying potential pathways of viral spillover, the phylogeographic link-prediction model can guide targeted surveillance efforts at the human-NHP interface.
Some viruses predicted by the phylogeographic model have not been documented in humans, yet evidence suggests that they pose significant spillover risk (S2). For example, Tanjong Rabok virus (TRV) of the Bakau bunyavirus group is predicted to infect humans in Indonesia. TRV has been isolated from Culex mosquitoes in Malaysia, and antibodies have been detected in a broad range of hosts (Beran, 1994). Though it has been suspected of causing disease in Southeast Asia, a naturally-acquired TRV infection has never been confirmed in humans (Beran, 1994). The Peribunyaviridae family, of which TRV is part, contains many emergent and understudied RNA viruses (Contigiani, Diaz, & Tauro, 2017). Emerging bunyaviruses may circulate asymptomatically in susceptible communities or cause symptoms indistinguishable from those of other etiological agents (Wolfe et al., 2005). The lack of evidence for human infection by TRVs and other predicted viruses does not necessarily indicate low spillover risk; rather, surveillance initiatives and experimental studies should prioritize these viruses to further assess their zoonotic potential.
The use of a phylogeographic model allowed us to better account for the dynamic processes that influence parasite sharing across the differing primate assemblages in each country. The DRC is home to the closest evolutionary relatives of humans – Pan troglodytes and Pan paniscus – and the model predicted a high degree of parasite sharing between these species and humans. However, in the case of China, the majority of parasites predicted to infect humans were imputed from Macaca mulatta rather than their closest relatives (the lesser apes). This likely reflects the large geographic range of Macaca mulatta, resulting in greater ecological opportunities for spillover. Thus, both phylogeny and geography contributed to the predictions.
We also found cases in which phylogeny and range overlap did not adequately capture risk of transmission to humans, such as in Madagascar. The primates of Madagascar are all lemurs, and as a distinct radiation from monkeys and apes, all lemurs are equally related to humans. They are also all endemic to Madagascar, so every NHP species overlapped with humans along its entire range. Parasites of lemurs are sparsely sampled compared to those of other NHPs, resulting in lower predictive power for this group. These factors resulted in some unexpected predictions for parasites from NHP in Madagascar to infect humans. Consider, for example, the high probabilities assigned to mites and lice, which are some of the only widely-sampled parasites among lemurs. Mites and lice tend to coevolve with their hosts, so a successful host switch to humans seems improbable (Hafner, Demastes, Spradling, & Reed, 2003). Systematic sampling for parasites in understudied NHPs is needed before their role in zoonotic transmission can be accurately assessed.
Variation in sampling effort has long been recognized as a challenge to understanding parasitism (Gregory, 1990; Walther, Cotgreave, Price, Gregory, & Clayton, 1995), including in NHPs (Nunn et al., 2003; Teitelbaum et al., 2020). Cooper and Nunn (2013) identified the scale of sampling gaps and the factors that drive sampling biases, finding that even for the best studied NHPs, nearly one-half of their parasites have yet to be sampled. Bacterial parasites in particular are underrepresented, making it difficult to draw general conclusions about bacteria host range and transmission risk in NHPs. In contrast, humans are much better sampled for parasites, with an estimated 2107 species, which is likely an underestimate (Dunn, Nunn, & Horvath, 2017). This estimate for humans is two orders of magnitude higher than the number of parasites documented in other primates, suggesting that humans might be super-parasitized due to epidemiological transitions associated with the agricultural transition, the rise of high-density cities, and globalization (Barrett, Kuzawa, McDade, & Armelagos, 1998). A recent comparative study (Amoroso & Nunn, 2021) failed to find strong support for this hypothesis of super-parasitism within individual countries; when pooled across countries, however, humans do appear to have more viruses than expected based on comparative patterns in NHPs. Thus, even the exceptional number of parasites in humans is largely due to more intensive sampling, rather than ecological factors.
Uneven sampling in the GMPD may give rise to biases in the link prediction results presented here. Relatively well-studied species in the GMPD tend to be those that have the largest geographic ranges and closest phylogenetic relatedness to humans, as expected given that traits of these hosts, including larger body size, terrestriality, and the ability to inhabit human-altered environments, are also conducive to accessibility for sampling (Cooper & Nunn, 2013). The elevated risk of zoonotic spillover from species like Pan troglodytes and Macaca mulatta may be exaggerated due to the higher number parasites observed in these hosts. In addition, link prediction cannot impute parasites that have yet to be documented, which is a major limitation given that only a fraction of parasite vertebrate diversity has been described (Carlson et al., 2020). We cannot expect the model to make ecologically relevant predictions of zoonotic spillover from lemurs, for instance, when entire parasite groups (i.e. viruses) are not described for lemurs in the GMPD.
The method presented here could theoretically be used to impute parasites of humans that may be undocumented in wild NHPs, thus providing useful information for NHP health and conservation. However, the vast difference in the number of reported parasites between humans and NHPs limits the feasibility of applying our method for this purpose; humans are so “oversampled” that their parasites would automatically comprise the majority of those imputed in NHPs. Link prediction models can serve as a valuable tool for illuminating aspects of zoonotic risk and host-parasite ecology, but their limitations highlight the continued need for systematic sampling of parasites in wildlife hosts.
In conclusion, we used a novel link-prediction approach to identify undocumented zoonotic parasites and reveal gaps in global preparedness for emerging disease. We applied this method to a narrowed dataset of high-risk NHP species in selective diversity hotspots, allowing us to manually evaluate all results and make targeted, relevant predictions for zoonotic and epidemic risk. Efforts to predict NHP parasites with high spillover risk must be continuously updated to reflect both ongoing parasite discovery and global change. The ranges of NHP species and their parasites will continue to shift under the synergistic effects of habitat destruction, climate change, and biodiversity loss (Carlson et al., 2017; Estrada et al., 2017). Models that predict future parasite-sharing risk should consider shifts in geographic ranges, as demonstrated by Morales-Castilla et al. (2021) in ungulate hosts, and future extinction scenarios, as explored by Herrera et al. (2021) in NHPs. In addition, the use of link-prediction models to identify human parasites that may spillover to NHPs should be explored as documentation of NHP parasite communities improves. Future link-prediction models applied at regional scales could incorporate more comprehensive data on parasite distributions, host behavior patterns, and contact probabilities between humans and wildlife, and should eventually move beyond binary, presence-absence predictions to better quantify parasite prevalence and transmission risk among at-risk communities. Finally, the application of link-prediction models to larger-scale host-parasite networks remains an important strategy for investigating broader patterns of parasite sharing, and will become increasingly useful as gaps in host-parasite databases are filled.
Supplementary Material
Acknowledgements
We would like to thank Dr. Meredith Spence Beaulieu, Tony Goldberg, and an anonymous reviewer for helpful feedback. Participants in the Evolutionary Medicine Summer Institute - held by the Triangle Center for Evolutionary Medicine - provided additional feedback.
Funding
Funding was provided by the joint NIH-NSF-NIFA Ecology and Evolution of Infectious Disease award 1R01-TW011493-01. This research was facilitated by the Triangle Center for Evolutionary Medicine (TriCEM), which is organized by Duke University, University of North Carolina, North Carolina State University, and North Carolina Central University.
Footnotes
Data Accessibility
All code and data required to run the phylogeographic link prediction model presented in this manuscript are included in the Supplementary material. Additional information on the HPpredictions package, including a vignette, can be found at https://github.com/melmasri/HPprediction. Raw data on host-parasite interactions, primate phylogeny, and primate geographic ranges are available via databases referenced in-text.
The authors have no conflicts of interest.
References
- Ahmed S, Dávila JD, Allen A, Haklay MM, Tacoli C, & Fèvre EM (2019). Does urbanization make emergence of zoonosis more likely? Evidence, myths and gaps. Environment and Urbanization, 31(2), 443–460. doi: 10.1177/0956247819866124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander KA, Carlson CJ, Lewis BL, Getz WM, Marathe MV, Eubank SG, . . . Blackburn JK (2018). The Ecology of Pathogen Spillover and Disease Emergence at the Human-Wildlife-Environment Interface. In Advances in Environmental Microbiology (pp. 267–298): Springer International Publishing. doi: 10.1007/978-3-319-92373-4_8 [DOI] [Google Scholar]
- Amoroso CR, & Nunn CL (2021). Epidemiological transitions in human evolution and the richness of viruses, helminths, and protozoa. Evolution, Medicine, and Public Health, 9(1), 139–148. doi: 10.1093/emph/eoab009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt JLN, Harkness J, Marriott D, Ellis JT, & Stark D (2011). A review ofDientamoeba fragiliscarriage in humans: Several reasons why this organism should be considered in the diagnosis of gastrointestinal illness. Gut Microbes, 2(1), 3–12. doi: 10.4161/gmic.2.1.14755 [DOI] [PubMed] [Google Scholar]
- Barrett R, Kuzawa CW, McDade T, & Armelagos GJ (1998). Emerging and Re-emerging Infectious Diseases: The Third Epidemiologic Transition. Annual Review of Anthropology, 27(1), 247–271. doi: 10.1146/annurev.anthro.27.1.247 [DOI] [Google Scholar]
- Bartomeus I, Gravel D, Tylianakis JM, Aizen MA, Dickie IA, & Bernard‐Verdier M (2016). A common framework for identifying linkage rules across different types of interactions. Functional Ecology, 30(12), 1894–1903. doi: 10.1111/1365-2435.12666 [DOI] [Google Scholar]
- Becker DJ, Albery GF, Sjodin AR, Poisot T, Bergner LM, Chen B, Cohen LE, Dallas TA, Eskew EA, Fagre AC, Farrell MJ, Guth S, Han BA, Simmons NB, Stock MS, Teeling EC, Carlson CJ (2022). Optimising predictive models to prioritise viral discovery in zoonotic reservoirs. The Lancet Microbe. doi: 10.1016/S2666-5247(21)00245-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beran GW (1994). Handbook of Zoonoses, Section B: Viral Zoonoses: CRC Press. [Google Scholar]
- Beran GW (2019). Handbook of Zoonoses, Second Edition (Beran GWEd. 2 ed.). United Kingdom: CRC Press. [Google Scholar]
- Bivand RS, Pebesma EJ, & Gomez-Rubio V (2013). Applied spatial data analysis with R, Second edition. NY: Springer. [Google Scholar]
- Bonds MH, Keenan DC, Rohani P, & Sachs JD (2010). Poverty trap formed by the ecology of infectious diseases. Proceedings of the Royal Society B: Biological Sciences, 277(1685), 1185–1192. doi: 10.1098/rspb.2009.1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth H, Clark M, Milner-Gulland EJ, Amponsah-Mensah K, Antunes AP, Brittain S, Castilho LC, Campos-Silva JV, Constantino PAL, Li Y, Mandoloma L, Nneji LM, Iponga DM, Moyo B, McNarama J, Rakotonarivo O,S, Shi J, Tagne CTK, van Velden J, Willians DR (2021). Investigating the risks of removing wild meat from global food systems. Current Biology, 31(8). doi: 10.1016/j.cub.2021.01.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CJ, Burgio KR, Dougherty ER, Phillips AJ, Bueno VM, Clements CF, . . . Getz WM (2017). Parasite biodiversity faces extinction and redistribution in a changing climate. Science Advances, 3(9), e1602422. doi: 10.1126/sciadv.1602422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CJ, Dallas TA, Alexander LW, Phelan AL, & Phillips AJ (2020). What would it take to describe the global diversity of parasites? Proceedings of the Royal Society B: Biological Sciences, 287(1939), 20201841. doi: 10.1098/rspb.2020.1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CJ, Farrell MJ, Grange Z, Han BA, Mollentze N, Phelan AL, Rasmussen AL, Albery GF, Bett B, Brett-Major DM, Cohen LE, Dallas T, Eskew EA, Fagre AC, Forbes KM, Gibb R, Halabi S, Hammer CC, Katz R, Kindrachuk J, Muylaert RL, Nutter FB, Ogola J, Olival KJ, Rourke M, Ryan SJ, Ross N, Seifert SN, Sironen T, Standley CJ, Taylor K, Venter M, Webala PW (2021). The future of zoonotic risk prediction. Philosophical Transactions of the Royal Society B-Biological Sciences, 376. doi: 10.1098/rstb.2020.0358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington CVF, & Auguste AJ (2013). Evolutionary and ecological factors underlying the tempo and distribution of yellow fever virus activity. Infection, Genetics and Evolution, 13, 198–210. doi: 10.1016/j.meegid.2012.08.015 [DOI] [PubMed] [Google Scholar]
- Carroll D, Daszak P, Wolfe ND, Morel CM, Morzaria S, Pablos-Mendez A, Tomori O, Mazet JA (2018). The Global Virome Project. Science, 359(6378).doi: 10.1126/science.aap7463 [DOI] [PubMed] [Google Scholar]
- Coccia M (2021). Preparedness of Countries To Constrain COVID-19 Pandemic Crisis and Support Timely Vaccinations: Analysis of the Performance and Underlying Structural Factors. Environmental Research, 203(111678). doi: 10.1016/j.envres.2021.111678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contigiani MS, Diaz LA, & Tauro LB (2017). Bunyaviruses. In Marcondes C (Ed.), Arthropod Borne Diseases: Springer. [Google Scholar]
- Cooper N, Griffin R, Franz M, Omotayo M, & Nunn CL (2012). Phylogenetic host specificity and understanding parasite sharing in primates. Ecology Letters, 15(12), 1370–1377. doi: 10.1111/j.1461-0248.2012.01858.x [DOI] [PubMed] [Google Scholar]
- Cooper N, Kamilar JM, & Nunn CL (2012). Host Longevity and Parasite Species Richness in Mammals. Plos One, 7(8), e42190. doi: 10.1371/journal.pone.0042190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper N, & Nunn CL (2013). Identifying future zoonotic disease threats. Evolution, Medicine, and Public Health, 2013(1), 27–36. doi: 10.1093/emph/eot001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier LA (2016). The Ten-Thousand Year Fever: Rethinking Human and Wild-Primate Malarias: Taylor & Francis. [Google Scholar]
- Dallas T, Park AW, & Drake JM (2017). Predicting cryptic links in host-parasite networks. PLOS Computational Biology, 13(5), e1005557. doi: 10.1371/journal.pcbi.1005557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daszak P, Cunningham AA, & Hyatt AD (2001). Anthropogenic environmental change and the emergence of infectious diseases in wildlife. Acta Tropica, 78(2), 103–116. doi: 10.1016/s0001-706x(00)00179–0 [DOI] [PubMed] [Google Scholar]
- Davies TJ, & Pedersen AB (2008). Phylogeny and geography predict pathogen community similarity in wild primates and humans. Proceedings of the Royal Society B: Biological Sciences, 275(1643), 1695–1701. doi: 10.1098/rspb.2008.0284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux CA, Mediannikov O, Medkour H, & Raoult D (2019). Infectious Disease Risk Across the Growing Human-Non Human Primate Interface: A Review of the Evidence. Frontiers in Public Health, 7(305). doi: 10.3389/fpubh.2019.00305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S (2018). Why are RNA virus mutation rates so damn high? PLOS Biology, 16(8), e3000003. doi: 10.1371/journal.pbio.3000003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn RR, Nunn CL, & Horvath JE (2017). The Global Synanthrome Project: A Call for an Exhaustive Study of Human Associates. Trends in Parasitology, 33(1), 4–7. doi: 10.1016/j.pt.2016.09.007 [DOI] [PubMed] [Google Scholar]
- Ellwanger JH, & Chies JAB (2021). Zoonotic spillover: Understanding basic aspects for better prevention. Genetics and Molecular Biology, 44(1). doi: 10.1590/1678-4685-GMB-2020-0355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmasri M, Farrell MJ, Davies JT, & Stephens DA (2020). A Hierarchical Bayesian Model for Predicting Ecological Interactions Using Scaled Evolutionary Relationships. The Annals of Applied Statistics, 14(1). doi: 10.1214/19-AOAS1296 [DOI] [Google Scholar]
- Esri. “World Countries (Generalized)” [basemap]. Scale Not Given. November 2019. https://hub.arcgis.com/datasets/esri::world-countries-generalized/about. [Google Scholar]
- Estrada A, Garber PA, Rylands AB, Roos C, Fernandez-Duque E, Di Fiore A, Nekaris KA, Nijman V, Heymann EW, Lambert JE, Rovero F, Barelli C, Setchell JM, Gillespie TR, Mittermeier RA, Arregoitia LV, De Guinea M, Gouveia S, Dobrovolski R, Shanee S, Shanee N, Boyle SA, Fuentes A, Mackinnon KC, Amato KR, Meyer ALS, Wich S, Sussman RW, Pan R, Kone I, & Li B (2017). Impending extinction crisis of the world’s primates: Why primates matter. Science Advances, 3(1), e1600946. doi: 10.1126/sciadv.1600946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell MJ, Elmasri M, Stephens D, & Davies JT (2022). Predicting missing links in global host-parasite networks. Journal of Animal Ecology, 91(4). doi: 10.1111/1365-2656.13666 [DOI] [PubMed] [Google Scholar]
- Fumagalli M, Sironi M, Pozzoli U, Ferrer-Admettla A, Pattini L, & Nielsen R (2011). Signatures of Environmental Genetic Adaptation Pinpoint Pathogens as the Main Selective Pressure through Human Evolution. PLoS Genetics, 7(11), e1002355. doi: 10.1371/journal.pgen.1002355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, Cummins LB, Arthur LO, Peeters M, Shaw GM, Sharp PM, & Hahn BH (1999). Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature, 397(6718), 436–441. doi: 10.1038/17130 [DOI] [PubMed] [Google Scholar]
- Gibb R, Albery GF, Mollentze NF, Eskew EA, Brierley L, Ryan SJ, Seifert SN, & Carlson CJ (2021). Mammal virus diversity estimates are unstable due to accelerating discovery effort. bioRxiv. doi: 10.1101/2021.08.10.455791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotelli NJ (2000). Null Model Analysis of Species Co-occurrence Patterns. Ecology, 81(9), 2606–2621. doi: 10.1890/0012-9658(2000)081[2606:nmaosc]2.0.co;2 [DOI] [Google Scholar]
- Gregory RD (1990). Parasites and Host Geographic Range as Illustrated by Waterfowl. Functional Ecology, 4(5), 645. doi: 10.2307/2389732 [DOI] [Google Scholar]
- Hafner MS, Demastes JW, Spradling TA, & Reed DL (2003). Cophylogeny Between Pocket Gophers and Chewing Lice. In Page RDM (Ed.), Tangled Trees: Phylogeny, Cospeciation, and Coevolution. Chicago, IL, USA: University of Chicago Press. [Google Scholar]
- Hahn BH, Shaw GM, De Cock KM, & Sharp PM (2000). AIDS as a Zoonosis: Scientific and Public Health Implications. Sciene, 287(5453), 607–614. doi: 10.1126/science.287.5453.607 [DOI] [PubMed] [Google Scholar]
- Hanley KA, Monath TP, Weaver SC, Rossi SL, Richman RL, & Vasilakis N (2013). Fever versus fever: The role of host and vector susceptibility and interspecific competition in shaping the current and future distributions of the sylvatic cycles of dengue virus and yellow fever virus. Infection, Genetics and Evolution, 19, 292–311. doi: 10.1016/j.meegid.2013.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon LJ, Losos JB, Davies TJ, Gillespie RG, Gittleman JL, Jennings WB, Kozak KH, McPeek MA, Moreno-Roark F, Near TJ, Purvis A, Ricklefs RE, Schluter D, Schulte II JA, Seehausen O, Sidlauskas BL, Torres-Carvajal O, Weir JT, & Mooers AØ (2010). Early Bursts of Body Size and Shape Evolution are Rare in Comparative Data. Evolution: International Journal of Organic Evolution, 64(8). doi: 10.1111/j.1558-5646.2010.01025.x [DOI] [PubMed] [Google Scholar]
- IUCN (International Union for Conservation of Nature). 2021. Primata (spatial data). The IUCN Red List of Threatened Species. Version 2021–3. https://www.iucnredlist.org. [Google Scholar]
- Jones-Engel L, May CC, Engel GA, Steinkraus KA, Schillaci MA, Fuentes A, Rompis A, Chalise MK, Aggimarangsee N, Feeroz MM, Grant R, Allan JS, Putra A, Wandia IN, Watanabe R, Kuller L, Thongsawat S, Chaiwarith R, Kyes RC, & Linial ML (2008). Diverse Contexts of Zoonotic Transmission of Simian Foamy Viruses in Asia. Emerging Infectious Diseases, 14(8), 1200–1208. doi: 10.3201/eid1408.071430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson EK, Kwiatkowski DP, & Sabeti PC (2014). Natural selection and infectious disease in human populations. Nature Reviews Genetics, 15(6), 379–393. doi: 10.1038/nrg3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, . . . Webb CO (2010). Picante: R tools for integrating phylogenies and ecology. Bioinformatics, 26(11), 1463–1464. doi: 10.1093/bioinformatics/btq166 [DOI] [PubMed] [Google Scholar]
- Khan A (2009). Simian foamy virus infection in humans: prevalence and management. Expert Review of Anti-infective Therapy. 7( 5). doi: 10.1586/eri.09.39 [DOI] [PubMed] [Google Scholar]
- Kotepui M, Masangkay FR, Kotepui KU, & Milanez GDJ (2021). Preliminary review on the prevalence, proportion, geographical distribution, and characteristics of naturally acquired Plasmodium cynomolgi infection in mosquitoes, macaques, and humans: a systematic review and meta-analysis. BMC Infectious Diseases, 21(1). doi: 10.1186/s12879-021-05941-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefkowitz EJ, Dempsey DM, Hendrickson RC, Orton RJ, Siddel SG, & Smith DB (2017). Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Research, 46(D1), D708–D717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew JWK, Bukhari FDM, Jeyaprakasam NK, Phang WK, Vythilingam I, & Lau YL (2021). Natural Plasmodium inui Infections in Humans and Anopheles cracens Mosquito, Malaysia. Emerging Infectious Diseases, 27(10), 2700–2703. doi: 10.3201/eid2710.210412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loverdo C, & Lloyd-Smith JO (2013). Evolutionary Invasion and Escape in the Presence of Deleterious Mutations. Plos One, 8(7), e68179. doi: 10.1371/journal.pone.0068179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marani M, Katul GG, Pan WK, & Parolari AJ (2021). Intensity and frequency of extreme novel epidemics. PNAS, 118(35). doi: 10.1073/pnas.2105482118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Castilla I, Matias MG, Gravel D, & Araújo MB (2015). Inferring biotic interactions from proxies. Trends in Ecology & Evolution, 30(6), 347–356. doi: 10.1016/j.tree.2015.03.014 [DOI] [PubMed] [Google Scholar]
- Norberg A, Nord CE, & Evengård B (2003). Dientamoeba fragilis—a protozoal infection which may cause severe bowel distress. Clinical Microbiology and Infection, 9(1), 65–68. doi: 10.1046/j.1469-0691.2003.00459.x [DOI] [PubMed] [Google Scholar]
- Nunn CL, Altizer S, Jones KE, & Sechrest W (2003). Comparative Tests of Parasite Species Richness in Primates. The American Naturalist, 162(5), 597–614. doi: 10.1086/378721 [DOI] [PubMed] [Google Scholar]
- Nunn CL, & Altizer SM (2005). The global mammal parasite database: An online resource for infectious disease records in wild primates. Evolutionary Anthropology: Issues, News, and Reviews, 14(1), 1–2. doi: 10.1002/evan.20041 [DOI] [Google Scholar]
- Park AW, Farrell MJ, Schmidt JP, Huang S, Dallas TA, Pappalardo P, Drake JM, Stephens PR, Poulin R, Nunn CL, & Davies TJ (2018). Characterizing the phylogenetic specialism–generalism spectrum of mammal parasites. Proceedings of the Royal Society B: Biological Sciences, 285(1874), 20172613. doi: 10.1098/rspb.2017.2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen AB, Altizer S, Poss M, Cunningham AA, & Nunn CL (2005). Patterns of host specificity and transmission among parasites of wild primates. International Journal for Parasitology, 35(6), 647–657. [DOI] [PubMed] [Google Scholar]
- Pépin J (2021). The Origin of AIDS (2nd ed.). Cambridge: Cambridge University Press. [Google Scholar]
- Petrovan SO, Aldridge DC, Bartlett H, Bladon AJ, Booth H, Broad S, Broom DM, Burgess ND, Cleaveland S, Cunningham AA, Ferri M, Hinsley A, Hua F, Hughes AC, Jones K, Kelly M, Mayes G, Radakovic M, Ugwu CA, Uddin N, Verissimo D, Walzer C, White TB, Wood JL, Sutherland WJ (2021). Post COVID‐19: a solution scan of options for preventing future zoonotic epidemics. Biological Reviews. doi: 10.1111/brv.12774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto-Santini DM, Stenbak CR, & Linial ML (2017). Foamy virus zoonotic infections. Retrovirology, 14(1). doi: 10.1186/s12977-017-0379-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piret J, & Boivin G (2020). Pandemics Throughout History. Frontiers in Microbiology, 11(631736). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowright RK, Parrish CR, Mccallum H, Hudson PJ, Ko AI, Graham AL, & Lloyd-Smith JO (2017). Pathways to zoonotic spillover. Nature Reviews Microbiology, 15(8), 502–510. doi: 10.1038/nrmicro.2017.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin R (2010). Decay of similarity with host phylogenetic distance in parasite faunas. Parasitology, 137(4). doi: 10.1017/S0031182009991491 [DOI] [PubMed] [Google Scholar]
- Poulin R, & Keeney DB (2008). Host specificity under molecular and experimental scrutiny. Trends in Parasitology, 24(1). doi: 10.1016/j.pt.2007.10.002 [DOI] [PubMed] [Google Scholar]
- Revell LJ (2012). phytools: an R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution, 3(2), 217–223. doi: 10.1111/j.2041-210x.2011.00169.x [DOI] [Google Scholar]
- Santini L, González‐Suárez M, Russo D, Gonzalez‐Voyer A, Hardenberg A, & Ancillotto L (2019). One strategy does not fit all: determinants of urban adaptation in mammals. Ecology Letters, 22(2), 365–376. doi: 10.1111/ele.13199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, Cavanaugh M, Clark K, Ostell J, Pruitt KD, & Karsch-Mizrachi I (2019). GenBank. Nucleic Acids Research, 47(D1), D94–D99. doi: 10.1093/nar/gky989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch CL, Ciufo S, Domrachev M, Hotton CL, Kannan S, Khovanskaya R, Leipe D, Mcveigh R, O’Neill K, Robbertse B, Sharma S, Soussov V, Sullivan JP, Sun L, Turner S, & Karsch-Mizrachi I (2020). NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database, 2020. doi: 10.1093/database/baaa062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Rayner JC, & Hahn BH (2013). Great Apes and Zoonoses. Science, 340(6130). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden FM (2019). Epidemics and society: Yale University Press. [Google Scholar]
- Stark D, Barratt J, Ellis J, Roberts T, Marriott D, & Harkness J (2010). A Review of the Clinical Presentation of Dientamoebiasis. The American Journal of Tropical Medicine and Hygiene, 82(4), 614–619. doi: 10.4269/ajtmh.2010.09-0478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PR, Pappalardo P, Huang S, Byers JE, Farrell MJ, Gehman A, Ghai RR, Haas SE, Han B, Park AW, Schmidt JP, Altizer S, Ezenwa VO, & Nunn CL (2017). Global Mammal Parasite Database version 2.0. Ecology, 98(5), 1476–1476. doi: 10.1002/ecy.1799 [DOI] [PubMed] [Google Scholar]
- Ta TH, Hisam S, Lanza M, Jiram AI, Ismail N, & Rubio JM (2014). First case of a naturally acquired human infection with Plasmodium cynomolgi. Malaria Journal, 13(1), 68. doi: 10.1186/1475-2875-13-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitelbaum CS, Amoroso CR, Huang S, Davies TJ, Rushmore J, Drake JM, Stephens PR, Byers JE, Majewska AA, & Nunn CL (2020). A comparison of diversity estimators applied to a database of host–parasite associations. Ecography, 43(9), 1316–1328. doi: 10.1111/ecog.05143 [DOI] [Google Scholar]
- Upham NS, Esselstyn JA, & Jetz W (2019). Inferring the mammal tree: Species-level sets of phylogenies for questions in ecology, evolution, and conservation. PLOS Biology, 17(12), e3000494. doi: 10.1371/journal.pbio.3000494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther BA, Cotgreave P, Price RD, Gregory RD, & Clayton DH (1995). Sampling Effort and Parasite Species Richness. Parasitology Today, 11(8), 306–310. doi: 10.1016/0169-4758(95)80047-6 [DOI] [PubMed] [Google Scholar]
- Wells K, Gibson DI, & Clark NJ (2019). Global patterns in helminth host specificity: phylogenetic and functional diversity of regional host species pools matter. Ecography, 42(3), 416–427. doi: 10.1111/ecog.03886 [DOI] [Google Scholar]
- Wolfe ND, Daszak P, Kilpatrick AM, & Burke DS (2005). Bushmeat Hunting, Deforestation, and Prediction of Zoonotic Disease. Emerging Infectious Diseases, 11(12), 1822–1827. doi: 10.3201/eid1112.040789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse MEJ, Haydon DT, & Antia R (2005). Emerging pathogens: the epidemiology and evolution of species jumps. Trends in Ecology & Evolution, 20(5), 238–244. doi: 10.1016/j.tree.2005.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap NJ, Hossain H, Nada-Raja T, Ngui R, Muslim A, Hoh B-P, Khaw LT, Kadir KA, Divis PCS, Vythilingam I, Singh B, & Lim YA-LL (2021). Natural Human Infections with Plasmodium cynomolgi, P. inui, and 4 other Simian Malaria Parasites, Malaysia. Emerging Infectious Diseases, 27(8), 2187–2191. doi: 10.3201/eid2708.204502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu VL, & Edberg SC (2005). Global Infectious Diseases and Epidemiology Network (GIDEON): a worldwide Web-based program for diagnosis and informatics in infectious diseases. Clinical Infectious Diseases, 40(1), 123–126. [DOI] [PubMed] [Google Scholar]
- Zhang X, Kadir KA, Quintanilla-Zariñan LF, Villano J, Houghton P, Du H, Singh B, & Smith DG (2016). Distribution and prevalence of malaria parasites among long-tailed macaques (Macaca fascicularis) in regional populations across Southeast Asia. Malaria Journal, 15(1). doi: 10.1186/s12936-016-1494-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.