Abstract
We have studied the functional importance of the N terminus of mouse Sos1 (mSos1), a ubiquitously expressed Ras-specific guanine nucleotide exchange factor whose C-terminal sequences bind Grb-2. Consistent with previous reports, addition of a myristoylation signal to mSos1 (MyrSos1) rendered it transforming for NIH 3T3 cells and deletion of the mSos C terminus (MyrSos1-ΔC) did not interfere with this activity. However, an N-terminally deleted myristoylated mSos1 protein (MyrSos1-ΔN) was transformation defective, although the protein was stable and localized to the membrane. Site-directed mutagenesis was used to examine the role of the Dbl and pleckstrin homology (PH) domains located in the N terminus. When mutations in the PH domain were introduced into two conserved amino acids either singly or together in MyrSos1 or MyrSos1-ΔC, the transforming activity was severely impaired. An analogous reduction in biological activity was seen when a cluster of point mutations was engineered into the Dbl domain. The mitogen-activation protein (MAP) kinase activities induced by the various Dbl and PH mutants of MyrSos1 correlated with their biological activities. When NIH 3T3 cells were transfected with a myristoylated Sos N terminus, their growth response to epidermal growth factor (EGF), platelet-derived growth factor, lysophosphatidic acid or serum was greatly impaired. The dominant inhibitory biological activity of the N terminus correlated with its ability to impair EGF-dependent activation of GTP-Ras and of MAP kinase, as well with the ability of endogenous Sos to form a stable complex with activated EGF receptors. The N terminus with mutations in the Dbl and PH domains was much less inhibitory in these biological and biochemical assays. In contrast to wild-type Sos1, nonmyristoylated versions of Sos1-ΔN and Sos1-ΔC did not form a stable complex with activated EGF receptors. We conclude that the Dbl and PH domains are critical for Sos function and that stable association of Sos with activated EGF receptors requires both the Sos N and C termini.
The ras genes encode membrane-associated proteins which transduce a variety of extracellular signals that regulate diverse biological effects, including cell growth and differentiation (27). Ras proteins function as molecular switches that cycle between an active GTP-bound state (GTP-Ras) and an inactive GDP-bound state (GDP-Ras). Activated Ras transmits its signal to several downstream targets, the best characterized being the Raf–mitogen-activated protein kinase (MAPK) pathway (2, 16, 22, 24, 28).
GTP-Ras is negatively regulated by guanine nucleotide-activating proteins (Ras GAPs), which hydrolyze GTP-Ras to GDP-Ras, while GDP-Ras is activated by Ras-specific guanine nucleotide exchange factors (Ras GNEFs), which catalyze the exchange of GDP for GTP on Ras. One of the best-studied Ras GNEFs in mammalian cells is Sos1 (3, 6, 16). Sos was first identified in Drosophila melanogaster, where it was placed genetically between a receptor protein tyrosine kinase (Sev) and Ras (5, 37). The central region of the protein, which contains the Ras-specific exchange activity and has been designated the catalytic region, is flanked by several hundred N-terminal and C-terminal amino acids.
In Caenorhabditis elegans, an adapter protein (Sem-5) was shown to lie genetically between a receptor protein tyrosine kinase and Sos (13), and a mammalian Sem-5 homolog, Grb-2, has been shown to bind to the C terminus of mammalian Sos1 (7, 11, 17, 19, 25). While Sos-1 is largely cytoplasmic and inactive in quiescent cells, activation of receptor protein tyrosine kinases such as the epidermal growth factor (EGF) receptor (EGFR) has been shown to lead to the Grb-2-dependent binding of a Sos1 complex to the activated receptor, and a concomitant increase in GTP-Ras (7, 19). Furthermore, the addition of a membrane-targeting signal to Sos1 renders the protein constitutively active in vivo, leading to Ras-dependent focal transformation of established rodent fibroblasts (1, 35). The membrane-targeted protein does not require the Sos C terminus, which suggests that the membrane-targeting signal substitutes for this function. This series of observations has led to a still widely accepted model in which activation of wild-type Sos depends entirely on its C terminus (30).
However, several lines of evidence suggest that this model of wild-type Sos activation may require at least some modification. Studies of Drosophila Sos in flies have shown that its C terminus is dispensable for Sev-dependent Sos function (23). Furthermore, a premature termination mutant of mammalian Sos1, which therefore lacks the Grb-2 binding site, has been shown to be more active in vivo than full-length Sos1 (40), and the N terminus of mammalian Sos1 has recently been shown to interfere with EGF-dependent signaling in mammalian cells (8).
The foregoing studies therefore suggest that the N terminus of Sos1 may contribute significantly to Sos1 function. Two potentially important motifs in the N terminus are a Dbl homology domain and a pleckstrin homology (PH) domain. Dbl is a protein that has been shown to have GNEF activity for the human homolog of Cdc42 and RhoA, which are members of the Rho GTPase family (10, 20). Domains with homology to the catalytic region of Dbl have been identified in a variety of signaling molecules (10). In most of these proteins, including Sos, the function of the Dbl homology region has not been established. PH domains are also present in many signaling molecules (36). In most well-studied examples (26, 33, 34, 39, 42), they contribute to membrane association and have functional importance.
To examine the role of the Sos1 N terminus, we have carried out a mutational analysis of the Dbl and PH domains in the context of full-length Sos1 and C-terminally truncated Sos1 with or without a membrane-targeting sequence. We report that membrane-targeted Sos1 depends on both the Dbl and PH domains for full biological activity, implicating a role for them that is independent of membrane association. Furthermore, the N terminus of Sos is required for the stable association between Sos and activated EGFR.
MATERIALS AND METHODS
DNA constructs.
The mouse Sos1 (mSos1) cDNA was fused at its N-terminal coding region with a fragment encoding the Src myristoylation sequence (MGSSKSPKDPSQRRM) (15, 31) and at its C-terminal coding region with a c-Myc coding epitope (EEQKLISEEDLL) followed by a unique XhoI site for subcloning convenience. A PCR primer-mediated silent point mutation at codons 1048 and 1049 of mSos1 (GGAACC→GGTACC) resulted in the creation of a KpnI site. MyrSos1-ΔC (truncated at residue 1050) was then generated by deleting the sequence between the KpnI and XhoI sites and inserting a linker sequence encoding the c-Myc epitope tag. MyrSos1-ΔN constructs were generated by deleting a 1.7-kb fragment between BglI and BamHI sites in the coding region of mSos1 by partial digestion and inserting a small linker fragment in frame (TGGCGG/AAAGCTGGGATC, encoding WR20/K595AGI), resulting in the elimination of the Sos1 N-terminal codons between residues 21 and 594. MyrSos1-NT was made by deleting the sequence between PstI and XhoI, resulting in a truncation at E562. A sequence encoding the hemagglutinin (HA1) epitope tag (YPYDVPDYASL) (32) was fused in frame at the C termini of both MyrSos1-ΔN and MyrSos1-NT.
Mutated versions of MyrSos1, MyrSos1-ΔC, and MyrSos-NT carrying one or two point mutations in the PH domain and/or a cluster of mutations in the Dbl homology domain were generated by four-primer PCR. Two initial amplification reactions were performed; one used a sense mutagenic oligonucleotide and a 3′ antisense primer, while the second used an antisense mutagenic oligonucleotide and a 5′ sense primer. The two overlapping PCR products were then gel purified, diluted, and mixed for the second round of amplification. Both 3′ and 5′ primers were used to amplify the mutagenic fragment in the second-round reaction. The fragment containing the point mutation R459C or W537F was used to replace the wild-type sequence by subcloning, respectively, between NheI and SphI sites or SphI and PstI sites of the Sos PH domain in the Bluescript vector. The R459C and W537F mutations in the PH domain are designated by the symbols ∗ and #, respectively. The PCR products with the cluster mutation at 351-IIIRDII-357 substituted for wild-type 351-LHYFELL-357 in the Dbl homology domain were generated by successive mutagenesis and replaced the wild-type sequence by subcloning into NdeI and NheI site of the Sos1 cDNA construct. The cluster mutation is designated Dbl∼. All of the deletion and point mutation in the Sos constructs were confirmed by sequence analysis and in vitro translation. The ability of the in vitro-translated proteins encoded by the Sos deletion mutants to bind Grb-2 in vitro was assayed by coprecipitation with 5 μg of glutathione S-transferase (GST)–Grb-2 protein (Upstate Biotechnology Inc.).
The myristoylated Sos1 cDNAs were cloned into pGV16, a retroviral vector that places the sos gene under control of a murine retroviral long terminal repeat and the Neor gene under control of an internal simian virus 40 promoter (41). The nonmyristoylated Sos1 cDNAs were cloned into an internal ribosome entry site (IRES) vector (a generous gift from Susan E. Kane, City of Hope Medical Center, Duarte, Calif.) which expresses Sos and Neor from the same mRNA. The cDNA encoding HA-tagged MAPK was provided by Silvio Gutkind, National Institute of Dental Research.
The v-rasH expression plasmid (pPA90) was constructed as follows. A 1,329-bp fragment containing the promoter for the human elongation factor 1a gene (38) was excised from pEBG (a gift of Silvio Gutkind) by using HindIII and BspEI and cloned into pGFP1 that had been digested with the same restriction enzymes to yield pEFP1. Subsequently, a v-rasH DNA fragment was obtained by PCR from the v-rasH-containing plasmid pBW1423 (41) as the template and the following oligonucleotides: 5′CCGGATCCACCATGACAGAATACAAGCTTGTGG and 5′AAGGGCCCTCAGGACAGCACACACTTGCAGC. This fragment was cloned between the BamHI site and SmaI site of pEFP2, a version of pEFP1 with a modified polylinker.
Cell culture, transfection, and focus formation assays.
NIH 3T3 and COS-7 cells were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C in a humidified 5% CO2 atmosphere. Then 0.5 μg of wild-type or mutant mSos1 cDNA construct was transfected into NIH 3T3 (clone 7), using calcium phosphate precipitation as described previously (41). Foci were counted after 14 days. Clones stably expressing Sos were generated by G418 selection.
Ligand-dependent colony growth assay.
NIH 3T3 cells were transfected with empty Neor IRES vector, IRES MyrSos1-NT, or IRES MyrSos1-NT (Dbl∼PH∗/#), using calcium phosphate. Transfected NIH 3T3 cells were replated in multiple 10-cm-diameter dishes the next day and cultured in 1% FBS–DMEM containing geneticin and various kinds of ligands: EGF (Life Technologies, Inc.), 10 ng/ml; platelet-derived growth factor (PDGF; Life Technologies, Inc.), 10 ng/ml; and lysophosphatidic acid (LPA; Sigma), 1 μM. The media were changed twice weekly for 2 weeks. Growth efficiency was determined by counting the total number of G418-resistant colonies composed of more than 30 cells.
Subcellular fractionation.
Stably transfected clones expressing wild-type or mutant MyrSos1 proteins were grown to confluence. Either unlabeled cells or cells that had been metabolically labeled with [35S]methionine as described previously (9) were scraped and resuspended in hypotonic buffer as described previously (7) except that various phosphatase inhibitors were omitted. After homogenization and separation of nuclei by centrifugation at 15,000 rpm, the soluble cytosol and membrane pellet were fractionated by ultracentrifugation at 100,000 × g. Fractions of equal amounts of protein or trichloroacetic acid-precipitable counts were analyzed by immunoprecipitation with anti-Sos antibodies (Santa Cruz Biotechnology) followed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). The [35S]methionine-labeled samples were detected by autoradiography. Alternatively, the nonlabeled samples were analyzed by immunoblotting with an anti-Sos monoclonal antibody (Transduction Laboratory). Immunocomplexes on nitrocellulose membrane were visualized by enhanced chemiluminescence (ECL), using an ECL kit from Amersham.
MAPK assay.
COS-7 or NIH 3T3 cells were transiently transfected with pcDNA3.HA-MAPK (14) along with the empty vector or various Sos constructs, using LipofectAMINE (Life Technologies, Inc.) according to the manufacturer’s instructions. MAPK was assayed 48 h after the transfection. Serum-starved cells, with or without ligand treatment, were lysed with radioimmunoprecipitation assay RIPA buffer (20 mM Tris [pH 8.0], 137 mM NaCl, 10% glycerol, 1% Nonidet P-40, 1% SDS, 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, aprotinin [10 μg/ml], leupeptin [10 μg/ml], 5 mM Na3VO4). Equal amounts of protein from cell extracts were immunoprecipitated with anti-HA antibody (Babco). The immunocomplexes were washed once with radioimmunoprecipitation assay buffer, three times with phosphate-buffered saline containing 1% Nonidet P-40 and 2 mM Na3VO4, and once with kinase buffer (30 mM HEPES [pH 7.4], 10 mM MgCl2, 1 mM dithiothreitol, 5 μM ATP). The HA-MAPK complexes were then incubated with 50 μl of reaction buffer containing 5 μCi of [γ-32P]ATP (NEN-Dupont) and 2 μg of myelin basic protein (MBP; Upstate Biotechnology Inc.). After incubation for 20 min at 30°C, kinase reactions were terminated by the addition of 2× Laemmli sample buffer. The samples were then resolved by SDS-PAGE, and the phosphorylated MBP was visualized by autoradiography. The relative intensities of MBP phosphorylation were scanned with the AMBIS radioanalytic imaging system. The exogenous HA-MAPK proteins were examined by immunoblotting using anti-HA antibody (Babco).
In vivo Ras-GTP and Ras-GDP.
Subconfluent NIH 3T3 cells stably overexpressing MyrSos1NT, MyrSos1-NT(Dbl∼PH∗/#), or empty vector were starved in phosphate-free DMEM for 6 h. Cells were then labeled with medium containing 32Pi (0.4 mCi/ml) and 1.5% dialyzed FBS for 10 h. After treatment with or without EGF (100 ng/ml) for 5 min, cells were lysed and analyzed for Ras-GTP and Ras-GDP as described previously (43).
Immunoprecipitation of Sos1 and EGF receptors.
Subconfluent NIH 3T3 cells stably expressing nonmyristoylated Sos1 or mutated derivatives were starved in serum-free DMEM for ∼16 h. After treatment with or without EGF (50 ng/ml) for 5 min at 37°C, cells were lysed with nondenaturing buffer (20 mM Tris [pH 7.4], 150 mM NaCl, 1% Triton, 1 mM EDTA, 1 mM Na3VO4, 10 mM NaF, 1 mM phenylmethylsulfonyl fluoride, aprotinin, [10 μg/ml], leupeptin [10 μg/ml]), clarified by centrifugation, and diluted with lysis buffer to a protein concentration of 1.2 mg/ml. Endogenous Sos1 and exogenous Sos1 proteins were immunoprecipitated with anti-Sos and anti-epitope tag antibodies, respectively. After 3 h incubation on ice, 50 μl of 50% protein A-Sepharose (Sigma) was added, and the mixture was rotated for 2 h at 4°C. Immunocomplexes were washed twice with nondenaturing buffer, washed once with phosphate-buffered saline, and denatured in Laemmli sample buffer. Following resolution by SDS-PAGE and transfer to nitrocellulose membranes, immunoblotting was performed to detect tyrosine-phosphorylated EGFR, using antiphosphotyrosine antibody 4G10 (Upstate Biotechnology Inc.) at a 1:10,000 dilution. The EGFR immunoprecipitated by Sos was quantitated with the NIH Image 1.61 program. The blots were later stripped as instructed by the manufacturer (Amersham) and reprobed with anti-Sos antibody (Santa Cruz Biotechnology) at a 1:1,000 dilution. For each blot, horseradish peroxidase-conjugated anti-mouse or anti-rabbit immunoglobulin G (Amersham) was used for the second reaction at a 1:10,000 dilution. Immunocomplexes on nitrocellulose were visualized by ECL.
RESULTS
A membrane-localized, N-terminally deleted Sos protein is not biologically active.
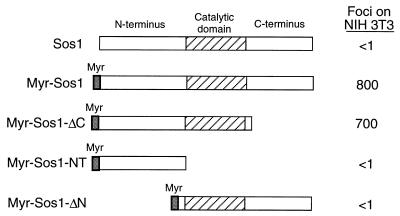
As noted in the introduction, addition of a membrane-targeting signal to full-length Sos renders the protein constitutively active, leading to Ras-dependent transformation of NIH 3T3 cells (1, 31, 35). We first confirmed that adding the Src myristoylation signal to the N terminus of wild-type mSos1 (creating MyrSos1) localized the protein to the membrane and converted it from one that lacks transforming activity in NIH 3T3 cells under our standard assay conditions to one that efficiently induces focal transformation (Fig. 1). A myristoylated Sos1 mutant which lacked the C terminus (MyrSos1-ΔC) was also highly transforming, as expected (Fig. 1).
FIG. 1.
The Sos N terminus is required for transformation by myristoylated Sos1. The structures of wild-type Sos1, myristoylated Sos1 (MyrSos1), and the derived deletion mutants are shown. These Sos1 constructs were transfected into NIH 3T3 cells and assayed for focal transformation as described in Materials and Methods. Data represent mean number of foci per 0.5 μg of DNA from five separate experiments.
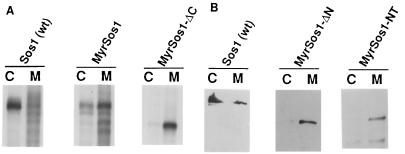
However, a myristoylated Sos1 mutant from which sequences that lie N terminal to the catalytic region had been deleted (MyrSos1-ΔN) was found unexpectedly to be defective for transformation (Fig. 1). Analysis of the protein encoded by MyrSos1-ΔN indicated that it was stable in NIH 3T3 cells and localized mainly in the membrane fraction (Fig. 2); its distribution was similar to that of the biologically active MyrSos1 and distinct from that of (nonmyristoylated) wild-type Sos, which localized mainly in the cytosolic fraction (Fig. 2). In addition, in vitro-translated MyrSos1-ΔN was able to bind an in vitro-translated GST–Grb-2 fusion protein as efficiently as MyrSos1, in contrast to MyrSos1-ΔC, which did not bind (data not shown). Taken together, the results strongly suggested that, contrary to expectation, membrane localization of a Sos protein that contained only the catalytic and C-terminal regions was not sufficient to render it biologically active.
FIG. 2.
Subcellular fractionation of cells expressing of myristoylated Sos mutant proteins. Stable clones of NIH 3T3 cells expressing control wild-type (wt) Sos1, full-length MyrSos1, or deleted versions of MyrSos1 proteins (see Fig. 1 for structures) were metabolically labeled with [35S]methionine (A) or unlabeled (B) and fractionated as described in Materials and Methods. The cytosolic (lanes C) and membrane (lanes M) fractions were immunoprecipitated with anti-Sos antibodies. The labeled samples (A) were analyzed by autoradiography after SDS-PAGE. Alternatively, to avoid the interference of background signals near the region of the gel in which the deleted MyrSos1 proteins migrated, the nonlabeled samples (B) were subjected to SDS-PAGE and analyzed by immunoblotting using anti-Sos antibodies. Wild-type Sos1 served as a control in each panel. Similar results were seen in two separate experiments.
Analysis of Dbl and PH substitution mutants.
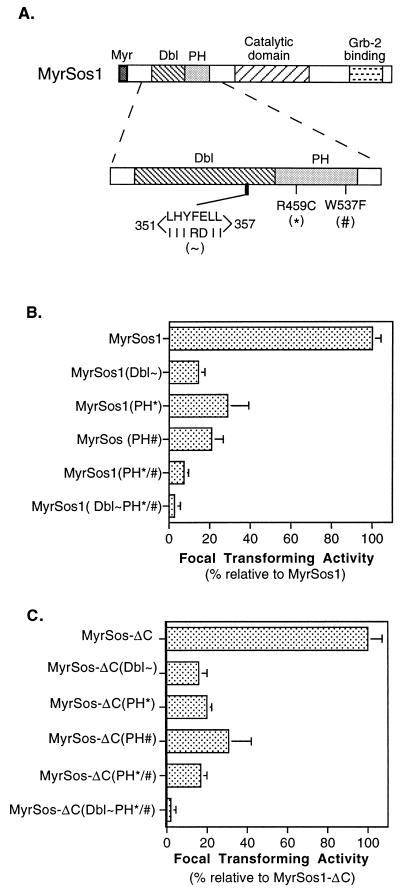
It remained possible that the negative results with MyrSos-ΔN resulted from an inadvertently induced conformational distortion of Sos catalytic and/or C-terminal domains as a consequence of the deletion of the N-terminal sequences. To rule out this possibility and to examine the potential role of specific motifs within the N terminus, we used MyrSos1 as our starting clone and engineered substitution mutations in the Dbl and PH domains located in the N terminus (Fig. 3A). We constructed a cluster of seven substitution mutations in a conserved region of the Dbl domain (351-IIIRDII-357 for amino acid residues 351-LHYFELL-357) (designated Dbl∼). A similar cluster of substitution mutations was reported to inactivate the in vitro exchange activity and in vivo biological activity of the Dbl oncoprotein (21). Two point mutations in the PH domain were constructed singly and together: R459C, located in the second β sheet of the domain; and W537F, in the α-helical region (designated by the symbols ∗ and #, respectively). Analogous mutations in the PH domain of the Bruton’s tyrosine kinase were reported to reduce its biological activity and membrane association (26, 39).
FIG. 3.
Reduced transforming activity of MyrSos1 and MyrSos1-ΔC with substitution mutations in the Dbl and PH domains. (A) Structures of substitution mutations in Dbl and PH domains. A seven-residue cluster mutation was introduced into the Sos Dbl homology domain (indicated as Dbl∼ in this and other figures). A single-point mutation was introduced into the N-terminal or C-terminal region of the Sos PH domain, (PH∗ or PH#, respectively). PH∗/# represents the presence of both point mutations in the PH domain, and Dbl∼PH∗/# stands for the combination of all mutations. (B and C) The full-length MyrSos1 and C-terminally truncated MyrSos1-ΔC carrying the designated substitution mutations were transfected into NIH 3T3 cells. Panel B shows the focal transforming efficiency of MyrSos1 and its derivatives; panel C shows that of MyrSos1-ΔC and its derivatives. Data shown are summarized from four separate experiments (four readings per experiment). For each mutant, the bar shows the relative transforming activity and standard error compared with that of MyrSos1 (B) or MyrSos1-ΔC (C).
The focus-forming activity of each mutant was reproducibly less than that of MyrSos1 (Fig. 3B). For the PH domain mutants, the R459C mutant, MyrSos1(PH∗), was about 1/3 as active, the W537F mutant, MyrSos1(PH#), was about 1/5 as active, and the double mutant, MyrSos1(PH∗/#), was about 1/10 as active. The Dbl homology domain mutant, MyrSos1(Dbl∼), was less than one-fifth as active, and the mutant containing the Dbl cluster mutation and the PH double mutation, MyrSos1(Dbl∼PH∗/#), was 2 orders of magnitude less efficient than MyrSos1. As with the MyrSos1 deletion mutants, the proteins encoded by the MyrSos1 substitution mutants involving the Dbl and PH domains were stable, membrane associated, and expressed at comparable levels (data not shown). These results indicated that the Dbl and PH domains each made an independent contribution to the biological activity of the myristoylated Sos protein.
We also examined the biological activity of the Dbl and PH mutants in the context of MyrSos1-ΔC, the myristoylated Sos protein lacking the C terminus (Fig. 3C). The relative activities of the Dbl and PH mutants, when the mutations were engineered into MyrSos1-ΔC, were qualitatively similar to their activities in the full-length MyrSos1. The results indicate that under standard growth conditions, the biological activity of myristoylated Sos is heavily dependent on the Dbl and PH domains whether or not the C terminus, which contains the Grb-2 binding site, is present.
The biological activities of the mutants correlate with their abilities to activate MAPK.
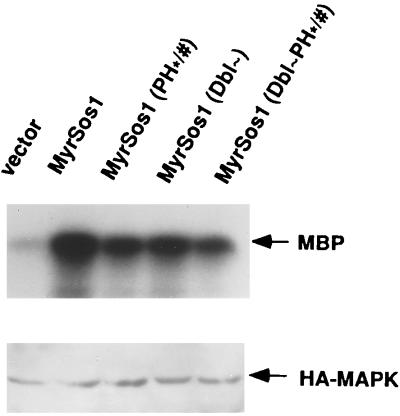
As noted in the introduction, Sos activation leads to the Ras-dependent activation of MAPK. To confirm that the lower biological activities of the MyrSos1 Dbl and PH mutants correlated with a decrease in MAPK activity, we evaluated the abilities of the MyrSos1 mutants to activate MAPK in transient transfection assays. COS-7 and NIH 3T3 cells were cotransfected with the MyrSos1 mutants and a plasmid encoding an epitope-tagged MAPK. Forty-eight hours later, the activity of the transfected MAPK was measured in a standard assay using MBP as the substrate (Fig. 4). As expected, the MAPK activity induced by the mutants was substantially lower than that induced by MyrSos1 (data shown for COS-7 cells).
FIG. 4.
Reduced MAPK activity of MyrSos1 Dbl and PH mutants in COS-7 cells. Cells were transiently cotransfected with HA-MAPK and either empty vector, MyrSos1, or the indicated MyrSos1 substitution mutants. The basal HA-MAPK activity was determined following anti-HA immunoprecipitation using MBP as the substrate (top panel). Compared with MyrSos1, the amounts of MBP were 39% for the PH∗/# mutant, 42% for the Dbl∼ mutant, 26% for the Dbl-PH∗/# mutant, and 5% for the empty vector. The expression of exogenous HA-MAPK was examined by anti-HA immunoblotting (bottom panel). Data shown are representative for three separate experiments.
The Sos N terminus, through the Dbl and PH domains, can act as a dominant interfering mutant.
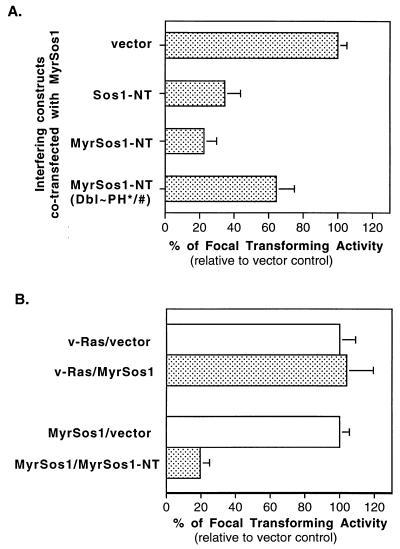
The requirement for the Sos N terminus for transformation by MyrSos1 raised the possibility that the isolated Sos N terminus might interfere with the biological activity of MyrSos1. To test this experimentally, the Sos N terminus, with a myristoylation signal (MyrSos1-NT) or without one (Sos1-NT), was cotransfected with MyrSos1 (Fig. 5A). Compared with the empty vector, Sos1-NT reduced the transforming activity of MyrSos1 threefold, while MyrSos1-NT was even more inhibitory, reducing the activity four- to fivefold, which suggested that the functional elements in the N terminus were more accessible in the myristoylated version. To determine if the Dbl and PH domains contributed to the inhibitory activity of MyrSos1-NT, we tested the effect of an isogenic construct N terminus containing the Dbl and PH mutations, MyrSos1-NT(Dbl∼PH∗/#), when cotransfected with MyrSos1. This mutant inhibited focus formation less than twofold, implying that a substantial proportion of the inhibitory activity of MyrSos1-NT was attributable to its Dbl and PH domains.
FIG. 5.
The Sos1 N terminus interferes with MyrSos1-induced focal transformation. (A) MyrSos1 was cotransfected either with vector or interfering DNA constructs as indicated. The resultant focus-forming activity from MyrSos1-vector (mean number of foci per 0.5 μg of DNA, as in Fig. 1) was designated 100%. The relative (bar, standard error) focus-forming activities from cotransfection of MyrSos1 with the interfering DNAs relative to vector control represent the average from five separate experiments. (B) MyrSos1 or plasmid pPA90, which contains v-rasH, was cotransfected with either vector or MyrSos1-NT. The resultant focus-forming activities from MyrSos1-vector and pPA90-vector were each designated 100%. The relative (bar, standard error) focus-forming activities from cotransfection of pPA90 or MyrSos1 with MyrSos1-NT compared with their activities with the vector control represent the average from three separate experiments. The mean numbers of foci induced by the pPA90-vector and MyrSos1-vector were 1,860/0.5 μg of DNA and 920/0.5 μg of DNA, respectively.
To confirm that the inhibition was specific, we examined the ability of MyrSos-NT to affect the transforming activity of a mutationally activated ras gene (v-rasH), which should be Sos independent (Fig. 5B). MyrSos1-NT did not inhibit transformation induced by the activated ras gene, in contrast to its effects on MyrSos1.
The Sos N terminus can interfere with normal cell growth.
The foregoing experiments analyzed the biological and biochemical activities of exogenous myristoylated Sos and the ability of the myristoylated N terminus to interfere with these effects. To examine the role of the N terminus in a more physiologic context, the ability of the myristoylated N terminus to interfere with normal cell growth was evaluated. MyrSos1-NT and the isogenic MyrSos1-NT(Dbl∼PH∗/#) mutant were used in these studies.
NIH 3T3 cells were transfected with the N-terminal coding sequences linked to Neor, and cells were grown under four different conditions (in medium containing EGF, PDGF, LPA, or 10% FCS) in the presence of G418. Under all conditions, the number of G418-resistant colonies was substantially lower than with control cells that received the empty vector (Fig. 6). The degree of inhibition was greatest for EGF and serum, followed by PDGF and LPA. The role of the Dbl and PH domains was evaluated by testing the activity of an N terminus containing the Dbl and PH mutations. In each instance, the degree of inhibition of cell growth was much less than with the wild-type N terminus. These results strongly suggest that cell growth under each of these conditions depends on Sos, that the N terminus of Sos is essential for Sos activity, and that the Dbl and PH domains make critical contributions to the effects of the N terminus.
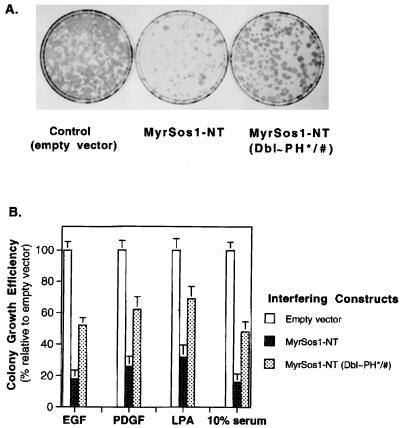
FIG. 6.
Inhibition of mitogen-dependent growth by a myristoylated Sos N-terminal fragment: importance of Dbl and PH domains. NIH 3T3 cells transfected with either vector or interfering DNA constructs were grown in low serum containing G418 and various mitogens as described in Materials and Methods. After 2 weeks, the dishes were stained with hematoxylin, and the colonies were counted. (A) Stained dishes obtained after growth in EGF. (B) Quantitation of colony formation. For each mitogen (EGF, PDGF, LPA, and 10% serum), the mean number of colonies (a colony contained >30 cells) obtained from vector-transfected cells was designated 100%. The relative colony-forming efficiencies of cells expressing MyrSos1-NT and MyrSos1-NT(Dbl∼PH∗/#) are shown. The data represent the average of three independent experiments.
Effects of the Sos N-terminus on EGF-dependent activation of Ras and MAPK.
To study the mechanism of inhibition by the myristoylated Sos N terminus, we directed our attention to the EGF-dependent phenomena. The effect of the myristoylated Sos N terminus on EGF-dependent activation of MAPK activity was studied in NIH 3T3 cells that were transiently cotransfected with MyrSos1-NT and a plasmid encoding epitope-tagged MAPK, stimulated with EGF, and analyzed for activity of the transfected MAPK (Fig. 7). As with the EGF-dependent growth of NIH 3T3 cells, the EGF-dependent MAPK activity of cells transfected with MyrSos1-NT was lower than that in cells transfected with the control plasmid vector alone, while the MAPK activity in cells that received the MyrSos1-NT(Dbl∼PH∗/#) mutant was greater than with MyrSos1-NT. The MAPK data therefore paralleled the biological results obtained in the NIH 3T3 cells in Fig. 6.
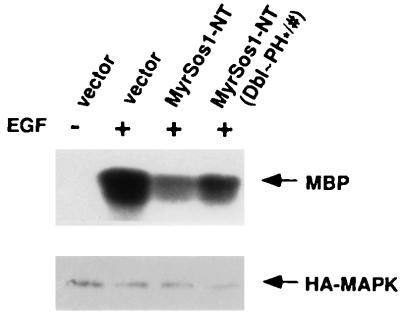
FIG. 7.
The myristoylated Sos N terminus inhibits EGF-induced MAPK activity in NIH 3T3 cells; importance of Dbl and PH domains. NIH 3T3 cells transiently cotransfected with HA-MAPK and empty vector, MyrSos1-NT, or MyrSos1-NT(Dbl∼PH∗/#) were serum starved and then treated with EGF (50 ng/ml) for 5 min at 37°C. The immunocomplexes were formed and analyzed for in vitro MAPK assays as described in Materials and Methods and in the legend to Fig. 4. Compared with the vector alone (100%), the amounts of MBP were 48% for MyrSos1-NT and 71% for MyrSos1-NT(Dbl∼PH∗/#). Expression of the exogenous HA-MAPK in these transiently transfected cells was confirmed by anti-HA blotting. Similar results were obtained in two other experiments.
To examine the influence of MyrSos1-NT on EGF-dependent activation of Ras, an NIH 3T3 cell line that stably expressed MyrSos-NT linked to Neor was derived by G418 selection (Fig. 8, inset, lane C). A companion cell line that expressed the MyrSos1-NT(Dbl∼PH∗/#) mutant was also derived (Fig. 8, inset, lane B). The cell lines were serum starved and then stimulated with EGF and analyzed for the percentage of GTP-Ras with and without EGF (Fig. 8). EGF stimulation of an NIH 3T3 cell line containing the vector control resulted in an increase in GTP-Ras from about 4 to 20%, as expected. By contrast, the line expressing MyrSos-NT rose from 5% GTP-Ras to only 10% in response to EGF. The response of the line expressing the MyrSos1-NT(Dbl∼PH∗/#) mutant was much closer to that of the vector control, rising from 7 to 20%.
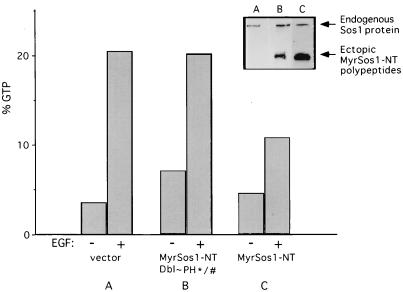
FIG. 8.
The myristoylated Sos N terminus inhibits EGF-induced Sos-dependent Ras activation; importance of Dbl and PH domains. The proportion of Ras in the GTP-bound form in cell cultures stably transfected with empty vector (A), MyrSos1-NT(Dbl∼PH∗/#) (B), and MyrSos1-NT (C) was determined following a 5-min treatment with or without EGF as indicated. In vivo levels of Ras-GTP were quantitated as described previously (43). Sos1 immunoblot (insert) of corresponding cellular lysates of each cell line demonstrates endogenous level of Sos1 and ectopic of MyrSos1 polypeptides.
Stable EGF-dependent association of Sos with the EGFR requires the Sos N terminus and C terminus.
The foregoing results indicated that the myristoylated Sos N terminus could interfere with EGF-dependent growth, with MAPK activity, and with Ras activation and that the Dbl and PH domains contributed to the inhibition. One possibility to explain these findings was that the Sos N terminus, in addition to its C terminus, might be required for the stable association of Sos with the activated EGFR.
To test this hypothesis experimentally, NIH 3T3 cell lines stably expressing nonmyristoylated versions of the Sos deletion mutants described in Fig. 1 were examined for the ability of EGF to induce complex formation between the Sos mutants and the activated endogenous EGFR. Preliminary experiments showed that all lines contained similar levels of endogenous EGFR, as determined by immunoprecipitation with an anti-EGFR antibody (data not shown).
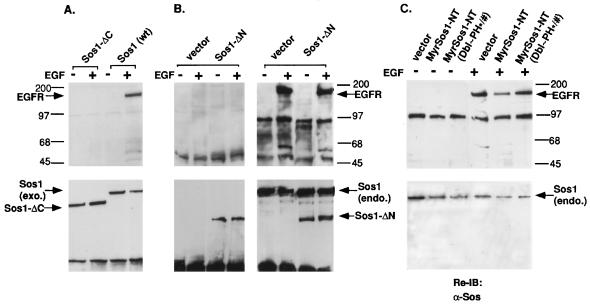
As expected, cells expressing wild-type Sos1, tagged with a c-Myc epitope, bound to the EGFR in an EGF-dependent manner (Fig. 9A). By contrast, cells expressing similar levels of c-Myc epitope-tagged Sos1-ΔC, which lacks the Sos1 C-terminus, did not bind the EGFR after EGF stimulation (Fig. 9A). This result confirms that the Sos C terminus, which binds Grb-2, is required for stable complex formation with the activated EGFR. Under the same conditions, cells expressing an HA-tagged Sos1-ΔN, which lacks the Sos1 N terminus, behaved similarly to Sos1-ΔC and did not bind to the activated EGFR (Fig. 9B, left), although the endogenous Sos did bind the receptors (Fig. 9B, right). These results indicate that stable complex formation between activated EGFR and Sos requires the Sos N terminus as well as its C terminus. In addition, expression of Sos1-ΔN did not lead to an increase in the association between the endogenous Sos and the EGFR compared with the vector control cells (Fig. 9B, right), further supporting the conclusion that Sos1-ΔN made no detectable contribution to the active signaling complex.
FIG. 9.
EGF-induced association of Sos1 with the activated EGFR requires the Sos N terminus and C terminus. (A and B) Complex formation between exogenous (exo.) Sos proteins and the EGFR. Extracts from NIH 3T3 cells stably transfected with the designated epitope-tagged Sos fragment were serum deprived and treated with EGF for 5 min (+) or left untreated (−). Cell extracts were prepared and immunoprecipitated with antitag antibodies (anti-c-Myc [9E10] [A] and anti-HA [B, left panel]). The endogenous (endo.) Sos1 protein was also immunoprecipitated with anti-Sos antibodies to serve as an internal control (B, right panel). The immunoprecipitates were subjected to SDS-PAGE and transferred to membranes for immunoblotting. The membranes were first probed with antiphosphotyrosine antibody (top panels) and then reprobed with anti-Sos antibodies after stripping (bottom panels). (C) Association between endogenous Sos1 proteins with EGFR. Cell extracts from EGF-treated or untreated control cells and stable NIH 3T3 transfected clones expressing MyrSos1-NT or MyrSos1-NT(Dbl∼PH∗/#) (Fig. 8) were immunoprecipitated with anti-Sos1 antibody follows by antiphosphotyrosine immunoblotting (top). The stripped membranes were then reprobed with anti-Sos antibody (bottom). All data shown are representative of three independent experiments.
To determine if the Dbl and PH domains participate in this requirement for the N terminus, the NIH 3T3 lines used for Fig. 8, which express stably MyrSos1-NT or the MyrSos1-NT(Dbl∼PH∗/#) mutant, were treated with EGF (Fig. 9C). As expected from the data obtained with Sos1-ΔN (Fig. 9B, left panel) and the ability of MyrSos1-NT to act as a dominant interfering mutant, endogenous Sos1 associated less efficiently with the activated EGFR in cells expressing MyrSos1-NT compared with cells expressing the control vector; only 30% as much EGFR was precipitated in the cells expressing MyrSos1-NT compared with the cells containing the vector control. By contrast, endogenous Sos1 in the line expressing the MyrSos1-NT(Dbl∼PH∗/#) mutant associated with the EGFR similarly to that of the vector control (89% as much EGFR as with the vector control). These results imply that the Dbl and PH domains contribute to the role of the N terminus in forming a stable complex with the activated EGFR.
DISCUSSION
In this study, we used a genetic approach to establish that the N-terminal portion of Sos plays a significant role in mediating Sos activity, and we identified the Dbl and PH domains as motifs that are critical to the function of the N terminus. Furthermore, we found that the Sos N terminus, which can function as a dominant inhibitory mutant that impairs Ras and MAPK activation, is required for the stable association between Sos and an activated EGFR. Our experiments confirm aspects of previous reports which have implied a role for the N terminus in Sos activity and lead to important additional conclusions.
In our initial experiments, we observed unexpectedly that in NIH 3T3 cells, membrane association of a Sos protein that carries intact catalytic and C-terminal domains but lacks the N terminus (MyrSos1-ΔN) is not sufficient for Sos to be biologically active. The results obtained with the Dbl and PH mutants indicated that each of these domains contributes to the function of a Sos protein that has been successfully targeted to the membrane via an N-terminal myristoylation signal. The Dbl- and PH-dependent functions have been established biologically and biochemically through the ability of myristoylated Sos to induce focal transformation and to activate MAPK, respectively. Thus, our results indicate that each motif has a function in myristoylated Sos, and presumably also in wild-type Sos, that is independent of membrane association per se. However, the precise molecular roles of the Dbl and PH domains in the context of the myristoylated Sos remain to be determined.
The PH domain isolated from human Sos1 has recently been shown to associate with the plasma membrane in a serum-dependent manner (12). Interestingly, the PH domain isolated in that study localized preferentially to the leading edges of motile cells, rather than being randomly distributed in the membrane. These observations together with our findings lead us to speculate that the Sos PH domain helps to position the myristoylated Sos protein within the membrane to increase the efficiency with which the protein signals to Ras, and possibly to other hypothetical targets. The reduced MAPK activity seen with the PH domain double mutant indicates a role for the PH domain of myristoylated Sos in promoting the MAPK-dependent activity.
Our data also implicate the Dbl domain in the biological and biochemical activities of Sos. It is unclear whether this domain functions primarily to increase an activity that is Ras dependent or Ras independent. One possibility is that the Dbl domain may activate one or more members of the Rho family of GTPases (10). However, this may not be the case for Ras-GRF, which is a calcium-responsive 140-kDa brain-specific Ras-specific GNEF that also contains adjacent Dbl and PH domains in addition to the Ras catalytic domain (18). In that system, mutation of the Dbl or PH domain leads to a protein that is less responsive to calcium, although the Dbl domain did not seem to be a GNEF for the Rho GTPase family members, and the mutations did not alter the membrane association of GRF.
Our results with the PH domain of mammalian Sos are consistent with the in vivo results of Karlovich et al., who showed that the PH domain of Drosophila Sos could interfere with Sos-dependent eye development in the fly (23). However, in apparent contrast to the results reported here for the substitution mutations in the Dbl or PH domain, the same group reported that deletion of either the Dbl or PH domain from wild-type Drosophila Sos did not inhibit its ability to activate the fos promoter in mammalian cells (29). In addition, Chen et al. (12) found that a double mutation (KR to EE) in the second β sheet of the human Sos1 PH domain interfered with serum-dependent MAPK activity as efficiently as did the wild-type PH domain. Although the R459C mutation examined in the present study mutated the corresponding arginine residue of mSos1, this PH single mutant and the W537F mutant were each less active than the wild type; the PH double mutant was even more impaired, perhaps because the W537F mutation is in a second region, the α-helical region, of the PH domain. Differences in the various assays and/or the mutants may account for the apparent discrepancies.
Byrne et al. (8) recently found that the N terminus of human Sos1 could interfere with serum-dependent DNA synthesis of NIH 3T3 cells and the EGF-dependent activation of ERK2 in COS-7 cells. Our results have confirmed that the N terminus of mSos1 behaves as a dominant inhibitory mutant. In addition to showing that a myristoylated Sos N terminus can inhibit the growth of NIH 3T3 cells when such growth depended on serum, EGF, PDGF, or LPA, our results also indicate that much of the inhibitory activity could be abrogated by mutations in the Dbl and PH domains. It remains to be determined whether the remaining inhibitory activity of the myristoylated N terminus with the Dbl and PH mutants is attributable to residual activity in one or both of these domains or represents an activity that resides elsewhere in the N terminus.
Our demonstration that the Sos N terminus is required for EGF to induce the formation of a stable complex between Sos and the activated EGFR provides a new mechanistic function for this region. Our data directly implicate the Dbl and/or PH domains in this function. It is likely that the PH domain makes an important contribution to this function, given its known role in other systems (26, 33, 34, 37, 42). Further analysis is required to establish this point experimentally and to examine the potential role of the Dbl domain in this process.
The Sos premature termination mutant lacking the C terminus was also unable to bind the activated EGFR. Based on the experimental findings, we propose a model in which stable binding between the activated EGFR and Sos requires cooperation between the N terminus and C terminus of Sos, with stable complex formation being required for the efficient activation of Sos. As in the usual model, the C-terminus-dependent binding presumably depends on Grb-2-bound Sos to bind directly or indirectly via Shc to the EGFR (4). Additional experimentation will be required to establish whether the region(s) of contact within the Sos N terminus involves the EGFR itself or, more likely, an adjacent region of the membrane.
Although our analysis of Sos binding has been confined to EGF-dependent activation of Sos, our dominant inhibitory studies with the myristoylated Sos N terminus make it likely that the Sos N terminus plays an analogous role in many situations in which Sos is activated. Given the diverse types of extracellular signals that activate Sos, it may prove interesting to determine whether specific ligands activate Sos primarily through the N terminus, the C terminus, or both regions, as well as to define in more detail the molecular mechanisms by which the N terminus contributes to Sos activation.
ACKNOWLEDGMENTS
We thank Silvio Gutkind and Susan E. Kane for providing plasmids and Marc Symons for helpful discussion.
REFERENCES
- 1.Aronheim A, Engelberg D, Li N X, Al-Alawi N, Schlessinger J, Karin M. Membrane targeting of the nucleotide exchange factor Sos is sufficient for activating the Ras signaling pathway. Cell. 1994;78:949–961. doi: 10.1016/0092-8674(94)90271-2. [DOI] [PubMed] [Google Scholar]
- 2.Avruch J, Zhang X F, Kyriakis J M. Raf meets Ras: completing the framework of a signal transduction pathway. Trends Biochem Sci. 1994;19:279–283. doi: 10.1016/0968-0004(94)90005-1. [DOI] [PubMed] [Google Scholar]
- 3.Bar-Sagi D. The Sos (son of sevenless) protein. Trends Endocrinol Metab. 1994;5:165–169. doi: 10.1016/1043-2760(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 4.Batzer A G, Rotin D, Urena J M, Skolinik E Y, Schlessinger J. Hierarchy of binding sites for Grb2 and Shc on the epidermal growth factor receptor. Mol Cell Biol. 1994;14:5192–5201. doi: 10.1128/mcb.14.8.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonfini L, Karlovich C A, Dasgupta C, Banerjee U. The son of sevenless gene product—a putative activator of Ras. Science. 1992;255:603–606. doi: 10.1126/science.1736363. [DOI] [PubMed] [Google Scholar]
- 6.Bowtell D, Fu P, Simon M, Senior P. Identification of murine homologues of the Drosophila son of sevenless gene—potential activators of ras. Proc Natl Acad Sci USA. 1992;89:6511–6515. doi: 10.1073/pnas.89.14.6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buday L, Downward J. Epidermal growth factor regulates p21(Ras) through the formation of a complex of receptor, GRB2 adapter protein, and Sos nucleotide exchange factor. Cell. 1993;73:611–620. doi: 10.1016/0092-8674(93)90146-h. [DOI] [PubMed] [Google Scholar]
- 8.Byrne J L, Paterson H F, Marshall C J. p21Ras activation by the guanine nucleotide exchange factor Sos, requires the Sos/Grb2 interaction and a second ligand-dependent signal involving the Sos N-terminus. Oncogene. 1996;13:2055–2065. [PubMed] [Google Scholar]
- 9.Cen H, Papageorge A G, Vass W C, Zhang K, Lowy D R. Regulated and constitutive activity by CDC25(Mm) (GRF), a Ras-specific exchange factor. Mol Cell Biol. 1993;13:7718–7724. doi: 10.1128/mcb.13.12.7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cerione R A, Zheng Y. The Dbl family of oncogenes. Curr Opin Cell Biol. 1996;8:216–222. doi: 10.1016/s0955-0674(96)80068-8. [DOI] [PubMed] [Google Scholar]
- 11.Chardin P, Camonis J H, Gale N W, Van Aelst L, Schlessinger J, Wigler M H, Bar-Sagi D. Human sos1—a guanine nucleotide exchange factor for Ras that binds to GRB2. Science. 1993;260:1338–1343. doi: 10.1126/science.8493579. [DOI] [PubMed] [Google Scholar]
- 12.Chen R H, Corbalan G S, Bar-Sagi D. The role of the PH domain in the signal-dependent membrane targeting of Sos. EMBO J. 1997;16:1351–1359. doi: 10.1093/emboj/16.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark S G, Stern M J, Horvitz H R. C. elegans cell-signalling gene sem-5 encodes a protein with SH2 and SH3 domains. Nature. 1992;356:340–344. doi: 10.1038/356340a0. [DOI] [PubMed] [Google Scholar]
- 14.Crespo P, Xu N, Daniotti J L, Troppmair J, Rapp U R, Gutkind J S. Signaling through transforming G protein-coupled receptors in NIH 3T3 cells involves c-Raf activation. Evidence for a protein kinase C-independent pathway. J Biol Chem. 1994;269:21103–21109. [PubMed] [Google Scholar]
- 15.DeClue J E, Vass W C, Papageorge A G, Lowy D R, Willumsen B M. Inhibition of cell growth by lovastatin is independent of ras function. Cancer Res. 1991;51:712–717. [PubMed] [Google Scholar]
- 16.Downward J. Control of ras activation. Cancer Surv. 1996;27:87–100. [PubMed] [Google Scholar]
- 17.Egan S E, Giddings B W, Brooks M W, Buday L, Sizeland A M, Weinberg R A. Association of Sos Ras exchange protein with Grb2 is implicated in tyrosine kinase signal transduction and transformation. Nature. 1993;363:45–51. doi: 10.1038/363045a0. [DOI] [PubMed] [Google Scholar]
- 18.Freshney N W, Goonesekera S D, Feig L A. Activation of the exchange factor Ras-GRF by calcium requires an intact Dbl homology domain. FEBS Lett. 1997;407:111–115. doi: 10.1016/s0014-5793(97)00309-8. [DOI] [PubMed] [Google Scholar]
- 19.Gale N W, Kaplan S, Lowenstein E J, Schlessinger J, Bar-Sagi D. Grb2 mediates the EGF-Dependent activation of guanine nucleotide exchange on ras. Nature. 1993;363:88–92. doi: 10.1038/363088a0. [DOI] [PubMed] [Google Scholar]
- 20.Hart M J, Eva A, Evans T, Aaronson S A, Cerione R A. Catalysis of guanine nucleotide exchange on the CDC42Hs protein by the dbl oncogene product. Nature. 1991;354:311–314. doi: 10.1038/354311a0. [DOI] [PubMed] [Google Scholar]
- 21.Hart M J, Eva A, Zangrilli D, Aaronson S A, Evans T, Cerione R A, Zheng Y. Cellular transformation and guanine nucleotide exchange activity are catalyzed by a common domain on the dbl oncogene product. J Biol Chem. 1994;269:62–65. [PubMed] [Google Scholar]
- 22.Herrmann C, Nassar N. Ras and its effectors. Prog Biophys Mol Biol. 1996;66:1–41. doi: 10.1016/s0079-6107(96)00015-6. [DOI] [PubMed] [Google Scholar]
- 23.Karlovich C A, Bonfini L, McCollam L, Rogge R D, Daga A, Czech M P, Banerjee U. In vivo functional analysis of the Ras exchange factor son of sevenless. Science. 1995;268:576–579. doi: 10.1126/science.7725106. [DOI] [PubMed] [Google Scholar]
- 24.Katz M E, McCormick F. Signal transduction from multiple Ras effectors. Curr Opin Genet Dev. 1997;7:75–79. doi: 10.1016/s0959-437x(97)80112-8. [DOI] [PubMed] [Google Scholar]
- 25.Li N, Batzer A, Daly R, Yajnik V, Skolnik E, Chardin P, Bar-Sagi D, Margolis B, Schlessinger J. Guanine-nucleotide-releasing factor hSos1 binds to Grb2 and links receptor tyrosine kinases to ras signalling. Nature. 1993;363:85–88. doi: 10.1038/363085a0. [DOI] [PubMed] [Google Scholar]
- 26.Li T, Tsukada S, Satterthwaite A, Havlik M H, Park H, Takatsu K, Witte O N. Activation of Bruton’s tyrosine kinase (BTK) by a point mutation in its pleckstrin homology (PH) domain. Immunity. 1995;2:451–460. doi: 10.1016/1074-7613(95)90026-8. [DOI] [PubMed] [Google Scholar]
- 27.Lowy D R, Willumsen B M. Function and regulation of Ras. Annu Rev Biochem. 1993;62:851–891. doi: 10.1146/annurev.bi.62.070193.004223. [DOI] [PubMed] [Google Scholar]
- 28.Marshall C J. Ras effectors. Curr Opin Cell Biol. 1996;8:197–204. doi: 10.1016/s0955-0674(96)80066-4. [DOI] [PubMed] [Google Scholar]
- 29.McCollam L, Bonfini L, Karlovich C A, Conway B R, Kozma L M, Banerjee U, Czech M P. Functional roles for the pleckstrin and Dbl homology regions in the Ras exchange factor Son-of-sevenless. J Biol Chem. 1995;270:15954–15957. doi: 10.1074/jbc.270.27.15954. [DOI] [PubMed] [Google Scholar]
- 30.Migliaccio E, Mele S, Salcini A E, Pelicci G, Lai K M, Superti F G, Pawson T, Di F P, Lanfrancone L, Pelicci P G. Opposite effects of the p52shc/p46shc and p66shc splicing isoforms on the EGF receptor-MAP kinase-fos signalling pathway. EMBO J. 1997;16:706–716. doi: 10.1093/emboj/16.4.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nielsen K H, Papageorge A G, Vass W C, Willumsen B M, Lowy D R. The Ras-specific exchange factors mouse Sos1 (mSos1) and mSos2 are regulated differently: mSos2 contains ubiquitination signals absent in mSos1. Mol Cell Biol. 1997;17:7132–7138. doi: 10.1128/mcb.17.12.7132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niman H L, Houghten R A, Walker L E, Reisfeld R A, Wilson I A, Hogle J M, Lernor R A. Generation of protein-reactive antibodies by short peptides is an event of high frequency: implications for the structural basis of immune recognition. Proc Natl Acad Sci USA. 1983;80:4949–4953. doi: 10.1073/pnas.80.16.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paterson H F, Savopoulos J W, Perisic O, Cheung R, Ellis M V, Williams R L, Katan M. Phospholipase C delta 1 requires a pleckstrin homology domain for interaction with the plasma membrane. Biochem J. 1995;312:661–666. doi: 10.1042/bj3120661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pitcher J A, Touhara K, Payner E S, Lefkowitz R J. Pleckstrin homology domain-mediated membrane association and activation of the β-adreergic receptor kinase requires coordinate interaction wiht Gβγ subunits and lipid. J Biol Chem. 1995;270:11707–11710. doi: 10.1074/jbc.270.20.11707. [DOI] [PubMed] [Google Scholar]
- 35.Quilliam L A, Huff S Y, Rabun K M, Wei W, Park W, Broek D, Der C J. Membrane-targeting potentiates guanine nucleotide exchange factor CDC25 and SOS1 activation of Ras transforming activity. Proc Natl Acad Sci USA. 1994;91:8512–8516. doi: 10.1073/pnas.91.18.8512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaw G. The pleckstrin homology domain: an intriguing multifunctional protein module. Bioessays. 1996;18:35–46. doi: 10.1002/bies.950180109. [DOI] [PubMed] [Google Scholar]
- 37.Simon M A, Bowtell D D L, Dodson G S, Laverty T R, Rubin G M. Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell. 1991;67:701–716. doi: 10.1016/0092-8674(91)90065-7. [DOI] [PubMed] [Google Scholar]
- 38.Uetsuki T, Naito A, Nagata S, Kaziro Y. Isolation and characterization of the human chromosomal gene for polypeptide chain elongation factor-1α. J Biol Chem. 1989;264:5791–5798. [PubMed] [Google Scholar]
- 39.Vihinen M, Zvelebil M J, Zhu Q, Brooimans R A, Ochs H D, Zegers B J, Nilsson L, Waterfield M D, Smith C I. Structural basis for pleckstrin homology domain mutations in X-linked agammaglobulinemia. Biochemistry. 1995;34:1475–1481. doi: 10.1021/bi00005a002. [DOI] [PubMed] [Google Scholar]
- 40.Wang W, Fisher E M, Jia Q, Dunn J M, Porfiri E, Downward J, Egan S E. The Grb2 binding domain of mSos1 is not required for downstream signal transduction. Nat Genet. 1995;10:294–300. doi: 10.1038/ng0795-294. [DOI] [PubMed] [Google Scholar]
- 41.Willumsen B M, Vass W C, Velu T J, Papageorge A G, Schiller J, Lowy D R. The BPV E5 oncogene can cooperate with ras: identification of a p21 amino acid segment required for transformation by c-rasH but not v-rasH. Mol Cell Biol. 1991;9:6026–6033. doi: 10.1128/mcb.11.12.6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yenush L, Makati K J, Smith H J, Ishibashi O, Myers M J, White M F. The pleckstrin homology domain is the principal link between the insulin receptor and IRS-1. J Biol Chem. 1996;271:24300–24306. doi: 10.1074/jbc.271.39.24300. [DOI] [PubMed] [Google Scholar]
- 43.Zhang K, Papageorge A G, Lowy D R. Mechanistic aspects of signalling through Ras in NIH 3T3 cells. Science. 1992;257:671–674. doi: 10.1126/science.1496380. [DOI] [PubMed] [Google Scholar]