Abstract
Ste6p, the a-factor transporter in Saccharomyces cerevisiae, is a multispanning membrane protein with 12 transmembrane spans and two cytosolic ATP binding domains. Ste6p belongs to the ATP binding cassette (ABC) superfamily and provides an excellent model for examining the intracellular trafficking of a complex polytopic membrane protein in yeast. Previous studies have shown that Ste6p undergoes constitutive endocytosis from the plasma membrane, followed by delivery to the vacuole, where it is degraded in a Pep4p-dependent manner, even though only a small portion of Ste6p is exposed to the vacuolar lumen where the Pep4p-dependent proteases reside. Ste6p is known to be ubiquitinated, a modification that may facilitate its endocytosis. In the present study, we further investigated the intracellular trafficking of Ste6p, focusing on the role of the ubiquitin-proteasome machinery in the metabolic degradation of Ste6p. We demonstrate by pulse-chase analysis that the degradation of Ste6p is impaired in mutants that exhibit defects in the activity of the proteasome (doa4 and pre1,2). Likewise, by immunofluorescence, we observe that Ste6p accumulates in the vacuole in the doa4 mutant, as it does in the vacuolar protease-deficient pep4 mutant. One model consistent with our results is that the degradation of Ste6p, the bulk of which is exposed to the cytosol, requires the activity of both the cytosolic proteasomal degradative machinery and the vacuolar lumenal proteases, acting in a synergistic fashion. Alternatively, we discuss a second model whereby the ubiquitin-proteasome system may indirectly influence the Pep4p-dependent vacuolar degradation of Ste6p. This study establishes that Ste6p is distinctive in that two independent degradative systems (the vacuolar Pep4p-dependent proteases and the cytosolic proteasome) are both involved, either directly or indirectly, in the metabolic degradation of a single substrate.
Our understanding of the events and cellular components required for the intracellular trafficking, targeting, and degradation of complex multispanning membrane proteins is in its infancy. The study of the life cycle of polytopic membrane proteins has become increasingly important, since improper intracellular trafficking has been implicated in a number of genetic diseases, such as cystic fibrosis, in which the misfolded cystic fibrosis conductance transmembrane regulator (CFTR) chloride channel is retained in the endoplasmic reticulum (ER) or certain forms of hypercholesterolemia in which the mutant low-density lipoprotein receptor is either ER retained or not internalized from the cell surface (1, 24, 52).
This study focuses on the intracellular trafficking of Ste6p, the a-factor mating pheromone transporter in Saccharomyces cerevisiae. Studies on yeast membrane proteins provide excellent model systems for studying general aspects of the trafficking and degradation of distinct classes of membrane proteins such as transporters, receptors, or channels. Ste6p is comprised of two homologous halves, each containing six predicted transmembrane spans and a large ATP nucleotide binding domain that is cytosolically disposed (4, 34, 35). Based on sequence homology and predicted structure, Ste6p belongs to the ATP binding cassette (ABC) superfamily of proteins, along with the multidrug resistance protein (MDR) and CFTR (4, 22). S. cerevisiae contains 30 ABC proteins (10, 51), of which 22, including the well-studied drug transporter Pdr5p that mediates pleiotropic drug resistance and exhibits several features of intracellular trafficking in common with Ste6p (12), are predicted to be membrane transporters.
Many of the major events involved in the intracellular trafficking of Ste6p have been elucidated by examining the metabolic stability of Ste6p by using pulse-chase labeling and by determining its localization by using immunofluorescence (2, 32). These studies have led to the view that Ste6p travels to the cell surface via the secretory pathway, resides at the plasma membrane only transiently, and undergoes rapid internalization by the endocytic machinery followed by delivery to the vacuole, where it is degraded. The degradation of Ste6p is dependent on Pep4p, a master protease that is involved in the proteolytic activation of a large group of intravacuolar proteases (2, 30, 32). Any mutations that block the efficient trafficking of Ste6p to the vacuole, such as those affecting the secretory pathway (sec23, sec1, and sec6) or endocytosis (end3, end4, and sac6), result in the stabilization of Ste6p, as do mutations affecting vacuolar degradation (pep4). An aspect of Ste6p intracellular trafficking that remains incompletely understood is the observation that, by immunofluorescence, the majority of Ste6p in the cell is found in an intracellular punctate compartment, speculated to correspond to the Golgi complex. In this study, we provide clear evidence by coimmunofluorescence of Ste6p with the marker Kex2p that this compartment is indeed the late Golgi complex. It is likely that this intracellular staining pattern for Ste6p and the apparent lack of cell surface staining are due to the slow rate of Ste6p exit to the cell surface, relative to its rapid rate of internalization. Whether Ste6p is functional to transport a-factor across the Golgi membrane is not known.
Additional aspects of Ste6p trafficking have emerged from the observations of Kölling and Hollenberg (32), who showed that the metabolic instability of Ste6p is significantly reduced in the ubc4Δ ubc5Δ double mutant, which is defective for a redundant pair of ubiquitin-conjugating enzymes (49). Furthermore, in an end4 mutant, compromised for endocytosis, Ste6p accumulates at the cell surface in a ubiquitinated form, suggesting that ubiquitination occurs before or during the normally transient residency of Ste6p at the cell surface (11, 32). These observations imply that the ubiquitination of Ste6p is important for its internalization, which in turn is necessary for its vacuolar degradation. It should be noted, however, that the ubiquitination of Ste6p has previously been detected only in endocytosis mutants, not in wild-type cells. Here, we demonstrate that the conjugation of ubiquitin to Ste6p also occurs in a wild-type strain, strengthening the view that ubiquitination is a normal part of the Ste6p life cycle.
The ubiquitin-mediated protein degradation pathway generally involves two distinct processes: conjugation of ubiquitin to appropriate substrates by specific recognition and conjugating enzymes, and degradation of these ubiquitinated proteins by the proteasome (for reviews, see references 14, 20, and 23). Until recently, the ubiquitination of a protein has been thought to be exclusively associated with its immediate degradation by the 26S proteasome. Known substrates for ubiquitination include aberrantly folded proteins in the cytosol or ER membrane and normally short-lived cyclins and transcription factors in the nucleus. However, certain observations have demonstrated that ubiquitination can lead to outcomes other than the immediate degradation by the proteasome (24). For example, ubiquitination of the immunoglobulin E receptor at the cell surface has been proposed to regulate its signaling function (38). Compelling evidence for a nonclassical role for ubiquitin as a signal to initiate the endocytosis of membrane proteins has also been proposed for the yeast mating pheromone receptors, Ste2p and Ste3p; neither receptor is efficiently internalized in a ubc4 ubc5Δ mutant (21, 47). In each case, a defect in ubiquitination of the receptor correlates with a defect in its endocytosis and subsequent Pep4p-dependent degradation in the vacuole (9, 21, 47). The role of ubiquitination in endocytosis may also apply to Ste6p, as ste6 mutant proteins that are not efficiently ubiquitinated accumulate at the cell surface (33). Thus, a role for ubiquitination as an endocytosis signal appears to hold true for several membrane proteins in yeast, and a decrease in their ubiquitination affects their degradation not directly, but only indirectly, by delaying their access to proteases in the vacuole.
The aim of the present study was to further probe basic features of the life cycle of Ste6p. Our first goal was to complete the characterization of Ste6p trafficking. To this end, we show by coimmunofluorescence that Ste6p exhibits colocalization with the well-characterized Golgi marker Kex2p. Thus, at any given moment, most of the Ste6p present in cells is in the late Golgi complex, presumably representing a slow step during its transit to the plasma membrane. We also demonstrate that the trafficking of Ste6p to the vacuole is blocked in a ren1-1 mutant and that Ste6p, like the pheromone receptor Ste3p (9), accumulates in the prevacuolar (class E) compartment in this strain. A second and major goal of this study was to elucidate the role of the ubiquitin-proteasome degradation pathway in the intracellular life cycle of Ste6p by assessing the consequences of mutationally blocking either ubiquitin conjugation or proteasomal activity. We find that in the doa4Δ mutant, in which deubiquitination of proteins and the activity of the proteasome are compromised (39), Ste6p is stabilized and accumulates in the vacuole, as it does in the pep4Δ mutant defective in intravacuolar proteolysis. Ste6p degradation appears to require the chymotryptic activity of the proteasome, at least in part, since in a pre1,2 mutant we found that the rate of Ste6p degradation is significantly reduced. Our results suggest that vacuolar and proteasomal proteolysis are both required and act in a synergistic manner to accomplish the degradation of Ste6p in the vacuolar membrane. A functional cooperativity between the two degradation machineries has not been reported to date for the vacuolar degradation of other membrane proteins. We discuss two models, one in which the proteasome may be directly involved in degrading Ste6p and another in which it may play an indirect role in facilitating the Pep4p-dependent degradation of Ste6p, for instance, by influencing vacuolar integrity or the deubiquitination of Ste6p. In this study, we also observed that Ste6p is localized at the cell surface in a polar pattern in a ubc4 ubc5Δ mutant, suggesting that there may normally be a redistribution step prior to the endocytosis of Ste6p which does not occur when ubiquitination of Ste6p is blocked.
MATERIALS AND METHODS
Strains, media, and growth conditions.
Yeast strains used in this study are listed in Table 1. Plate and liquid dropout media were prepared as described previously (31, 36). Yeast transformants were obtained by the plasmid transformation technique described elsewhere (13, 27). Cultures were grown at 30°C except where indicated otherwise.
TABLE 1.
Yeast strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| AAY1047 | MATa sac6::LEU2 ura3 leu2 his3 lys2 | A. Adams (University of Arizona) |
| GPY74-15C | MATa ura3-52 leu2-3,112 trp1-289 sst1-3 his4/6 | G. Payne (University of California, Los Angeles) |
| GPY385 | As GPY74-15C but pep4::LEU2 | G. Payne |
| MHY500 | MATa ura3-52 leu2-3,112 his3Δ200 trp1-1 lys2-801 | M. Hochstrasser (University of Chicago) |
| MHY508 | As MHY500 but MATα ubc4::HIS3 ubc5::LEU2 | M. Hochstrasser |
| MHY622 | As MHY500 but doa4::LEU2 | 39 |
| SM1058 | MATa ura3 leu2 his4 trp1 can1 | 36 |
| SM2544 | MATa ura3 leu2 his4 trp1 can1 ste6-Δ4 | This study |
| SM2474 | Transformant of GPY385 with pSM693 | This study |
| SM2631 | Transformant of AAY1047 with pSM693 | This study |
| SM2654 | MATa derivative of MHY508 (made with pGAL::HO) | This study |
| SM2718 | Transformant of SM2544 with pSM1361 and pBMKX22 | This study |
| SM3126 | Transformant of MHY500 with pSM683 and YEp96 | This study |
| SM3127 | Transformant of MHY500 with pSM683 and YEp105 | This study |
| SM3128 | Transformant of MHY500 with pSM192 and YEp105 | This study |
| SM3129 | Transformant of SM2654 with pSM683 and YEp105 | This study |
| SM3133 | Transformant of MHY622 with pSM683 and YEp105 | This study |
| SM3288 | Transformant of WCG4a with pSM683 | This study |
| SM3290 | Transformant of WCG4-11/21a with pSM683 | This study |
| SM3498 | Transformant of MHY500 with pSM693 | This study |
| SM3499 | Transformant of MHY500 with pSM683 | This study |
| SM3501 | Transformant of MHY622 with pSM693 | This study |
| SM3502 | Transformant of MHY622 with pSM683 | This study |
| SM3628 | Transformant of SM2654 with pSM683 | This study |
| SM3629 | Transformant of SM2654 with pSM693 | This study |
| SM3631 | Transformant of GPY74-15C with pSM683 | This study |
| SM3632 | Transformant of GPY385 with pSM683 | This study |
| SM3636 | Transformant of SY1614 with pSM693 | This study |
| SY1614 | MATa ren1-1 ura3 leu2 met14 ade2-1 ade1 his6 trp1 GAL1::STE3 | 9 |
| WCG4a | MATa ura3 leu2-3,112 his3-11,15 | 19 |
| WCG4-11/21a | MATa pre1-1 pre2-1 ura3 leu2-3,112 his3-11,15 | 19 |
The ste6 deletion allele (ste6-Δ4) was constructed by the two-step disruption method (7). Strain SM1058 was transformed with SnaBI-linearized pSM738 (YIp URA3 ste6-Δ4 [see below]). Ura+ transformants were selected on synthetic complete (SC)-Ura dropout plates. Segregants that had excised the plasmid were selected on 5-fluoro-orotic acid plates and screened for a nonmater phenotype, indicating that the deletion allele had replaced the wild-type allele in the chromosome. The deletion at the STE6 locus was confirmed by Southern analysis.
Plasmid constructions.
Plasmids used in this study are listed in Table 2. Vectors are described in reference 50. Plasmid pSM738, used to introduce the deletion ste6-Δ4 into the chromosome, was constructed by cloning a SalI-HindIII fragment containing the STE6 locus deleted for an internal SpeI fragment (encompassing nucleotide positions −368 to +843 in the STE6 gene) into pRS306, a yeast URA3 integrating vector (50).
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant markers | Source or reference |
|---|---|---|
| pBMKX22 | CEN URA3 pGAL::KEX2 | R. Fuller (University of Michigan) |
| pSM192 | CEN URA3 STE6 | 3 |
| pSM683 | CEN URA3 STE6::HA ecto | This study |
| pSM693 | 2μm URA3 STE6::HA | 2 |
| pSM738 | YIp URA3 ste6-Δ4 | This study |
| pSM1361 | CEN LEU2 pGAL::STE6::HA C-term | This study |
| YEp96 | 2μm TRP1 pCUP1 UBI | 25 |
| YEp105 | 2μm TRP1 pCUP1 UBI::myc | 25 |
To detect Ste6p by immunofluorescence, immunoprecipitation, and immunoblotting, we used strains containing plasmids that expressed the STE6 gene tagged with the triply iterated hemagglutinin epitope (HA) from influenza virus. The allele designated STE6::HA ecto in Table 2 contains the epitope in a predicted extracellular loop near the N-terminal end of Ste6p between amino acids 68 and 69 (2). The allele designated STE6::HA C-term contains the epitope at the extreme C terminus of Ste6p (5).
Plasmid pSM683 (CEN URA3 STE6::HA ecto) was generated by subcloning the SalI-NotI fragment containing the entire insert from pSM693 (2) into pRS316 (CEN URA3). Plasmid pSM1361 (pGAL::STE6::HA C-term) was constructed in several steps. First, a HindIII site was generated at nucleotide position −10, upstream of the STE6 gene, by site-directed mutagenesis of pSM351 (CEN LEU2 STE6) (3) with oligonucleotide oSM131 (5′-CAT GAC GTA GCT AAG CTT TGT TCT TTG TTT CC-3′). The resulting plasmid is pSM579. Second, the SalI-NcoI fragment of pSM579 that contains the 5′ untranslated region and about two-thirds of the STE6 gene was then exchanged with the corresponding fragment of pSM498 (CEN LEU2 STE6::HA C-term) (5) to generate pSM580, which contains a C-terminally HA-tagged version of STE6 with a HindIII site at position −10. Third, the 5-kb HindIII fragment of pSM580, containing promoterless STE6::HA, was subcloned downstream of the GAL1 promoter of pRS316GU, to generate pSM785 (CEN URA3 pGAL::STE6::HA C-term). Finally, the 2.5-kb AlwNI fragment of pSM785 containing a 5′ portion of STE6 in front of the GAL1 promoter was then exchanged with the 2.8-kb fragment of pSM498 to generate pSM1361 (CEN LEU2 pGAL::STE6::HA C-term).
Antibodies.
The mouse anti-HA monoclonal antibody 12CA5 was purchased from Babco (Richmond, Calif.) and was obtained at a concentration of 4.3 mg/ml. The rabbit anti-Kar2p antibody was a gift from M. Rose (Princeton University). The anti-Cpy antibody was a gift from E. Jones (Carnegie Mellon University). The mouse anti-Myc monoclonal antibody 9E10 was obtained from the monoclonal antibody facility at the Johns Hopkins University School of Medicine. Secondary rhodamine- or fluorescein isothiocyanate (FITC)-conjugated anti-rabbit and anti-mouse immunoglobulin G antibodies were purchased from Boehringer Mannheim (Indianapolis, Ind.).
Indirect immunofluorescence.
Cells were prepared for immunofluorescence essentially as previously described (2). For most experiments, cultures were grown overnight in SC dropout media to an optical density at 600 nm (OD600) of 0.5 to 1.0. For coimmunofluorescence in which Kex2p and Ste6p were expressed from the GAL1 promoter, cells were diluted to an OD600 of 0.1 in SC containing 2% raffinose and 2% galactose and grown to an OD600 of 0.8. Five OD600 units was harvested and resuspended in 5 ml of KP buffer (0.1 M potassium phosphate [pH 6.5]). A volume of 0.6 ml of a 37% formaldehyde solution (J. T. Baker) was added dropwise to the cell suspension, and fixation was allowed to occur for 40 min at 30°C with gentle agitation. Cells were washed twice in KP buffer and once in KPS buffer (KP buffer with 1.2 M sorbitol). For spheroplasting, 5 μl of Zymolyase (5 mg/ml) and 5 μl of β-mercaptoethanol were added to cells resuspended in 1 ml of KPS buffer. Cells were incubated 20 min at 30°C with gentle rotation. Cells were harvested gently (2,000 rpm in a Beckman TJ-6 clinical centrifuge for 3 min) and washed once in 5 ml of KPS buffer. Finally, the cells were resuspended in 1 ml of KPS–0.1% Tween 20 and left at room temperature for 15 min.
For immunodetection, an aliquot (15 μl) of the cell suspensions was applied to polylysine-coated slides and allowed to settle for 15 min. Wells were washed once with PBST buffer (0.04 M K2HPO4, 0.01 M KH2PO4, 0.15 M NaCl, 10 mg of bovine serum albumin per ml, 0.1% NaN3), 15 μl of primary antibody diluted in PBST buffer was applied, and incubation was carried out overnight at room temperature. In all experiments involving immunofluorescence, Ste6p was detected with the anti-HA antibody 12CA5 (dilution of 1:2,000), and Kar2p was detected with polyclonal rabbit anti-Kar2p antibodies (dilution of 1:1,000). Secondary incubations were performed for at least 2 h at room temperature in the dark. For the anti-HA immunofluorescence, a rhodamine-conjugated anti-mouse antibody was used; for the Kar2p immunofluorescence, an FITC-conjugated anti-rabbit antibody was used (both at a dilution of 1:500 in PBST). Wells were washed four times with PBST buffer between primary and secondary incubations.
Slides were mounted as described previously (31) and visualized by using a Zeiss Axiovert with a 100× objective. Images were captured on a Power Mac 7100, using the IP Lab software (Analytics Corp.). Images were further processed with Adobe Photoshop and printed on a dye sublimation printer (Phaser 440; Tektronix).
Metabolic labeling and immunoprecipitation of Ste6p and Cpy.
For immunoprecipitations, cultures diluted from a 2-day overnight culture in SC dropout media were grown from an OD600 of 0.2 to 0.6 to 1.0. The cultures took 8 to 10 h to reach this stage. A total of 12.5 OD600 units of cells was harvested (corresponding to 2.5 OD600 units per time point) and resuspended in 2.5 ml of SD media supplemented with appropriate amino acids. Cells were incubated with shaking at 30°C for 10 min and pulse-labeled with 25 μCi of Express 35S (DuPont, Wilmington, Del.) for 10 min. For the doa4Δ and ubc4,5Δ strains, cultures were grown at 25°C and shifted for 40 min at 30°C prior to labeling. The pre1 pre2 mutant was shifted to 37°C for 40 min prior to the labeling. In all cases, the isogenic wild-type strains were submitted to the identical growth conditions. The label was chased with 50 μl of chase mix (1 M cysteine, 1 M methionine), and 0.5-ml samples were collected at 0, 15, 30, and 60 min. The chase was terminated by mixing cells with 0.5 ml of stop mix (40 mM cysteine, 40 mM methionine, 20 mM NaN3) on ice.
Protein extracts for each time point were prepared as follows. Cells were washed once and resuspended in 1 ml of cold H2O, lysed by adding 150 μl of 2 N NaOH–1 M β-mercaptoethanol, vortexed vigorously, and incubated on ice for 15 min. Trichloroacetic acid was added to 5%, and samples were left on ice an additional 15 min. Tubes were microcentrifuged for 10 min, and protein pellets were resuspended in 50 μl of trichloroacetic acid sample buffer (3.5% sodium dodecyl sulfate [SDS], 0.5 M dithiothreitol, 80 mM Tris, 8 mM EDTA, 15% glycerol, 0.1 mg of bromophenol blue per ml).
For immunoprecipitation of Ste6p, 25 μl of extract in sample buffer was added to 0.5 ml of dilution buffer (1% Triton X-100, 150 mM NaCl, 5 mM EDTA, 50 mM Tris [pH 7.5]) and incubated on ice for 60 min. The lysate was cleared twice, and 250 μl of 12CA5 antibody dilution (final 12CA5 dilution was 1:1,500 in dilution buffer) was added to the diluted protein. Primary incubation was carried out overnight at 4°C. Immune complexes were pelleted with protein A-Sepharose beads (Pharmacia Biotech, Piscataway, N.J.). Cpy immunoprecipitation was carried out in the same way except that 10 μl of extract was used and the final dilution of anti-Cpy antibody was 1:1,500. Immunoprecipitates were dissociated from the beads by the addition of 15 μl of 2× Laemmli sample buffer and incubation at 37°C for 20 min. Immunoprecipitates were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and fluorography.
For determination of Ste6p half-life, dried gels were analyzed with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). Counts corresponding to the Ste6p signal were quantified in each lane by using the ImageQuant software (Molecular Dynamics). The 0-min chase time point was used as the 100% reference value for each time course experiment. Semilogarithmic graphs and exponential extrapolations were done with the Kaleidagraph software (Synergy Software, Reading, Pa.).
Immunoprecipitation and immunoblotting to detect Ste6p-ubiquitin conjugates.
Unlabeled protein extracts for immunoprecipitation were prepared by the β-mercaptoethanol–NaOH extraction procedure as described in the previous section except that 25 OD600 units of cells was harvested and N-ethylmaleimide (Sigma) was present at a concentration of 10 mM throughout the preparation. The anti-HA antibody was used at a dilution of 1:750 in the immunoprecipitations. Immunoprecipitates were treated as described above, analyzed by SDS-PAGE on 8% gels, and transferred to nitrocellulose. For Western blots, anti-HA and anti-Myc antibodies were used at dilutions of 1:10,000 and 1:3,000, respectively, in the presence of 0.1% Tween 20. Immunoblots were developed by using the ECL detection system (Amersham Life Sciences, Arlington Heights, Ill.).
RESULTS
Ste6p shows significant colocalization with the late Golgi marker Kex2p.
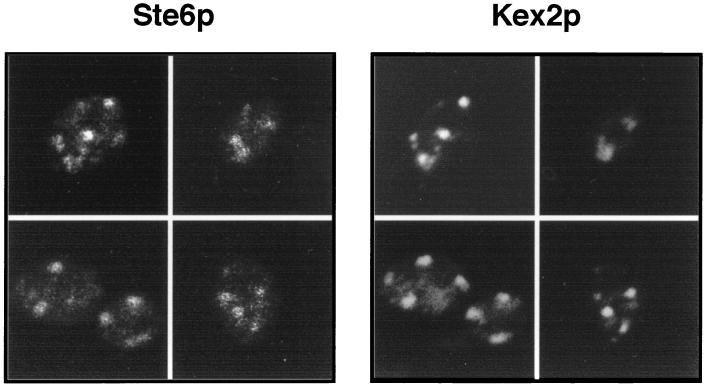
Although many aspects of the overall life cycle of Ste6p have been elucidated, the precise identity of the compartment(s) in which Ste6p predominates at steady state has not been unambiguously determined. A Golgi localization pattern for Ste6p has been proposed, based on its punctate immunofluorescence staining, which is reminiscent of several Golgi markers (2, 32), and on sucrose gradient cofractionation with dipeptidylaminopeptidase A (DPAPA) (32). Both of these findings provide a suggestive but not definitive localization for Ste6p. To better ascertain the identity of the punctate compartment in which Ste6p predominates, we performed coimmunofluorescence with the well-established Golgi marker Kex2p (15, 43). As shown in Fig. 1, in cells where both Ste6p and Kex2p were expressed from the GAL1 promoter, the immunofluorescence patterns detected for these two proteins are essentially overlapping. In most cells, the size, shape, and distribution of the dots detected for Ste6p were the same as seen for Kex2p.
FIG. 1.
Coimmunofluorescence of Ste6p and Kex2p. HA-tagged Ste6p and Kex2p were expressed from the GAL1 promoter from the centromeric-based plasmids pSM1361 and pBMKX22 (provided by R. Fuller), respectively, in strain SM2544 (MATa ste6-Δ4). Cells were processed for immunofluorescence after induction in galactose as described in Materials and Methods. Ste6p was detected with the mouse anti-HA antibody 12CA5, and Kex2p was detected with the rabbit anti-Kex2p antibody. Rhodamine-conjugated anti-mouse and FITC-conjugated anti-rabbit secondary antibodies were used to visualize the Ste6p and Kex2p staining patterns, respectively.
To quantitate the extent of the colocalization between Ste6p and Kex2p, we counted the dots that colocalized between the two markers, and the ones that failed to do so, in 100 cells where staining for both Ste6p and Kex2p could be detected. The proportion of Ste6p dots that coincided with Kex2p was 77%. The Kex2p pattern has been previously shown to correspond to the yeast equivalent of the late Golgi complex (15) or to a possible equivalent of the trans-Golgi network in yeast (44). We conclude that a significant amount of Ste6p is in the Golgi complex at any given moment, presumably on its way to the cell surface. Because Ste6p is rapidly endocytosed upon arrival at the cell surface, it is generally not visible at the plasma membrane, except in mutants blocked in endocytosis (2, 32) (see Fig. 4).
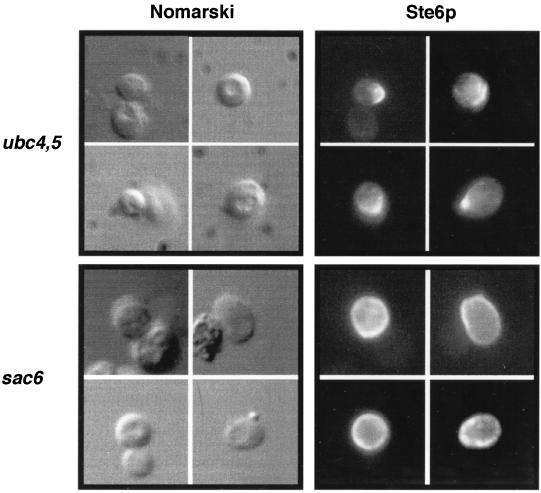
FIG. 4.
Comparison of the cell surface staining pattern of Ste6p in mutants defective in ubiquitin conjugation (ubc4,5) and endocytosis (sac6). The ubc4,5Δ and sac6 strains (SM3629 and SM2631, respectively) that bear pSM693 (2μm STE6::HA) were processed for immunofluorescence as described in Materials and Methods. Ste6p was detected with the anti-HA antibody 12CA5.
Ste6p requires REN1 for its delivery to the vacuole, as evidenced by its accumulation in the prevacuolar (class E) compartment in the ren1-1 mutant.
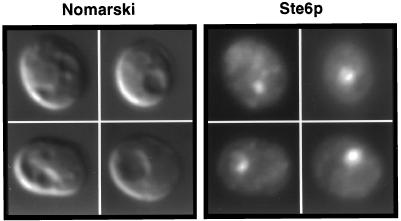
We wished to determine whether Ste6p uses the same endocytic machinery as that employed by Ste3p, the a-factor pheromone receptor, another yeast plasma membrane protein that undergoes constitutive endocytosis. Ste3p requires the REN1 gene product, which functions late in endocytosis, for its delivery to the vacuole (9). A ren1-1 mutant accumulates a distinctive compartment adjoining the vacuole (the class E compartment) that is thought to represent a late endosome or prevacuolar compartment (40, 42). Ste3p does not reach the vacuole in a ren1-1 strain but instead accumulates in the class E compartment. We examined the immunolocalization pattern of Ste6p in the ren1-1 mutant. As shown in Fig. 2, Ste6p accumulates in a single, concentrated compartment that is adjacent to and clearly distinct from the main vacuole as visualized in the Nomarski image. This pattern is precisely that of the class E prevacuolar compartment. Thus, Ste6p appears to use machinery in common with Ste3p to reach the vacuole.
FIG. 2.
Ste6p concentrates in the prevacuolar class E compartment in the ren1-1 mutant. Strain SY1614 (ren1-1) was transformed with a 2μm STE6::HA plasmid (pSM693). Cells were processed for coimmunofluorescence as described in Materials and Methods. Ste6p was detected with a mouse anti-HA antibody.
Ste6p is ubiquitinated in a Ubc4,5p-dependent manner and accumulates in a polarized pattern at the cell surface in a ubc4,5Δ mutant.
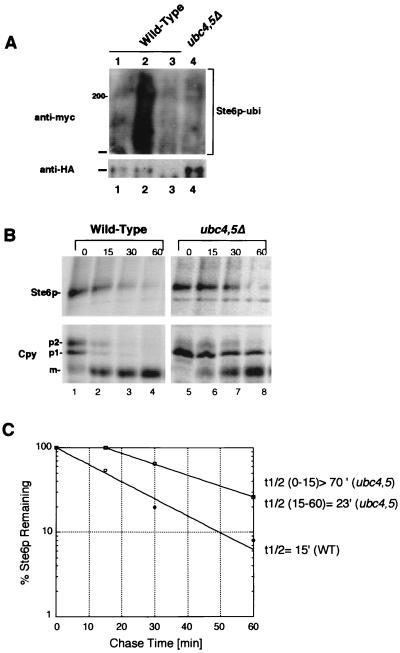
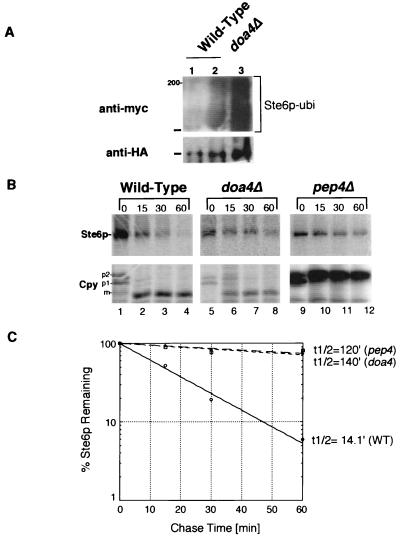
To better characterize the life cycle of Ste6p, we tested whether known components of the ubiquitin-proteasome pathway are required for the ubiquitination, trafficking, internalization, or degradation of Ste6p. While previous studies have shown that Ste6p is ubiquitinated in an end4 mutant (32, 33), a direct demonstration that Ste6p is ubiquitinated in a wild-type strain is lacking. As a starting point here, we examined whether we could detect the ubiquitination of Ste6p in a wild-type strain. We used an assay system described previously (25). Accordingly, Ste6p-HA was immunoprecipitated from a strain bearing Myc-tagged ubiquitin, and immunoprecipitates were subjected to SDS-PAGE and transferred to nitrocellulose. Ubiquitin-conjugated forms of Ste6p were detected by immunoblotting with the anti-Myc antibody (Fig. 3A, top). The total amount of Ste6p present was visualized by immunoblotting with anti-HA antibodies (Fig. 3A, bottom). In the wild-type strain, ubiquitinated forms of Ste6p were clearly detectable, ranging in size from 145 kDa (the size of full-length Ste6p) to over 210 kDa (Fig. 3, lane 2). We attribute the lack of resolution of the ubiquitinated forms of Ste6p to their aggregation during preparation, as has been suggested by others (32). The signal detected by the anti-Myc antibody in Fig. 3A is specific for Myc-tagged ubiquitinated Ste6p-HA, since it is absent in strains that bear untagged ubiquitin (Fig. 3A, lane 1) or untagged STE6 plasmids (Fig. 3A, lane 3). No detectable ubiquitination of Ste6p above background levels is seen in the ubc4,5Δ mutant (Fig. 3A, lane 4), suggesting that this pair of ubiquitin-conjugating enzymes is required for ubiquitination of Ste6p. We note that in this mutant, a higher steady-state level of Ste6p is detectable, presumably due to the increased metabolic stability of Ste6p (Fig. 3B). These results establish that Ste6p is ubiquitinated in a wild-type strain and not just in mutants in which endocytosis is defective.
FIG. 3.
Ubiquitination and metabolic stability of Ste6p in a ubc4,5Δ mutant, defective for a pair of ubiquitin-conjugating enzymes. (A) To determine the extent of ubiquitination of Ste6p in wild-type and mutant strains, unlabeled extracts were prepared from a strain containing two plasmids, pSM683 (STE6-HA) and YEp105 (UBI::myc), or the corresponding untagged versions pSM192 (CEN URA3 STE6) and YEp96 (2μm TRP1 UBI::myc). Ste6p-HA was immunoprecipitated from the unlabeled extracts with anti-HA antibodies; immunoprecipitates were subjected to SDS-PAGE and transferred to nitrocellulose. Two separate gels and filters were prepared. One filter was probed with anti-Myc antibodies to detect ubiquitin-conjugated forms of Ste6p-HA, and the other was probed with anti-HA antibodies to assess the total amount of Ste6p-HA in each extract. Extracts in panel A were prepared from SM3126 (pSTE6::HA, vector) in lane 1, SM3127 (pSTE6::HA, pUBI::myc) in lane 2, SM3128 (pUBI::myc, vector) in lane 3, and SM3129 (pSTE6::HA, pUBI::myc, in ubc4Δ ubc5Δ) in lane 4. The position at which Ste6p migrates (ca. 145 kDa) is indicated by a bar, and the 200-kDa size marker is indicated in panel A. (B) To compare the half-life of Ste6p-HA in wild-type and ubc4,5Δ mutant strains, cells were pulse-labeled for 10 min with Express 35S. Strains used were SM3624 (wild type) and SM3628 (ubc4,5Δ). The label was chased for the indicated times (minutes). Cell extracts were immunoprecipitated with anti-HA antibodies to determine the half-life of Ste6p (top) and with anti-Cpy antibodies to examine the time course of Cpy processing (bottom). The p1 (ER), p2 (Golgi), and mature (m; vacuolar) forms are indicated. The kinetics of Ste6p degradation was determined by quantitation of the Ste6p band at each time point (C). Quantitation of Ste6p degradation was performed by PhosphorImager analysis as described in Materials and Methods. Open circles and squares designate the time points for the wild-type (WT) and ubc4,5Δ mutants, respectively. The calculated half-life (t1/2) of Ste6p-HA is indicated.
We wished to examine the consequences of blocking the ubiquitination of Ste6p on its trafficking. To this end, we assessed the fate of Ste6p by pulse-chase analysis (Fig. 3B) and find, in agreement with a previous report (32), that Ste6p is considerably stabilized in the ubc4,5Δ mutant. We note that a strong stabilization occurs in the first 30 min of chase, followed by significant degradation at later time points. Additional experiments (not shown) confirmed that the metabolic degradation of Ste6p appears to be biphasic in the ubc4,5Δ mutant, perhaps representing a severe delay rather than a complete defect in degradation. To determine whether the activity of Pep4p in the ubc4,5Δ strain is normal, we monitored the processing of the vacuolar protease Cpy, which requires Pep4p for conversion from the precursor (p2) to the mature form. Overall, the kinetics of the appearance of mature Cpy is similar to that seen for the wild-type strain (Fig. 3B; compare lanes 1 to 4 to lanes 5 to 8), indicating that Pep4p activity is not defective in the ubc4,5Δ mutant and thus cannot account for the stabilization of Ste6p in this experiment (see also reference 21). However, we noted a slow step in conversion of the ER precursor species (p1) to the Golgi species (p2), suggestive of a possible delay in ER to Golgi trafficking in the ubc4,5Δ mutant strain. Such an overall cellular trafficking defect could contribute to some extent to the observed stabilization of Ste6p.
To determine whether the stabilization of Ste6p in the ubc4,5Δ mutant was truly due to a delay in reaching the vacuole, we examined the localization of Ste6p by immunofluorescence and found that Ste6p accumulates at the cell surface in the ubc4,5Δ mutant (Fig. 4, top panels). Interestingly, the cell surface staining is not uniform and instead concentrates at one edge of the cell. Although Ste6p resides at the plasma membrane when its ubiquitination is blocked, the staining pattern of Ste6p in the ubc4,5Δ mutant is distinctly different from its staining pattern in a mutant defective in endocytosis, such as end3 (37), end4 (2), or sac6Δ (Fig. 4, bottom panels), where the signal is seen as overall rim staining. The polarized cell surface staining may indicate a previously uncharacterized step involved in the cell surface distribution of Ste6p.
Ste6p is metabolically stabilized in the doa4Δ mutant, which exhibits defects in the activity of the proteasome.
The degradation of proteins in yeast is carried out by two major proteolytic systems, intravacuolar proteolysis, which is dependent on enzymes activated by Pep4p processing, and ubiquitin-mediated proteolysis, which is thought to occur in the cytoplasm and the nucleus and is proteasome dependent (reviewed in references 29 and 30). We and others have shown previously that Pep4p-dependent vacuolar proteolysis is required for Ste6p degradation in the vacuole (2, 32). Because Ste6p is ubiquitinated, we sought to determine whether the proteolytic activity of the proteasome might also be required for the turnover of Ste6p.
We analyzed the fate of Ste6p in the doa4Δ mutant by pulse-chase analysis. The DOA4 gene encodes a deubiquitinating enzyme that, when mutated, has been shown to lead to a pleiotropic phenotype, including defects both in the deubiquitination of proteins and in the proteolytic activity of the proteasome (39). Thus, proteolysis of ubiquitinated substrates is strongly inhibited in the doa4 mutant strain. We compared the metabolic stability of Ste6p in doa4 and pep4 mutant strains by pulse-chase analysis. The stabilization of Ste6p that we observe in the doa4Δ mutant is comparable to the effect seen in the pep4Δ mutant (Fig. 5B, lanes 9 to 12). The fraction of Ste6p remaining 60 min after the chase is about 70% in both cases (Fig. 5C), whereas the amount remaining in the wild-type strain is 6%. In the doa4Δ mutant, the processing of Cpy occurs normally (Fig. 5B, lanes 6 to 10), showing that the stabilization of Ste6p is not due to a defect in Pep4p activity and that the strain used is indeed PEP4+. These results suggest that, directly or indirectly, either deubiquitination or the activity of the proteasome is required for Ste6p turnover in the cell, in addition to the intravacuolar Pep4p-dependent degradation machinery. The finding that Ste6p is stabilized to a similar extent in both the pep4Δ and doa4Δ single mutants suggests that the effects on Ste6p of the ubiquitin-proteasome and Pep4p-dependent systems are not additive but interdependent.
FIG. 5.
Ste6p is metabolically stabilized to similar extents in mutants defective in vacuolar proteases (pep4Δ) or in the activity of the proteasome (doa4Δ). (A) Ubiquitinated Ste6p was detected as described in the legend to Fig. 3A. Extracts from the following strains were prepared and processed: SM3126(pSM683, YEp96) in lane 1, SM3127(pSM683, YEp105) (wild-type strain) in lane 2, and SM3133 (pSM683, YEp105) (doa4Δ mutant) in lane 3. (B) Cells were pulse-labeled for 10 min with Express 35S, and the label was chased for the indicated times (minutes). Ste6p and Cpy were immunoprecipitated with anti-HA and anti-Cpy antibodies, respectively, analyzed by SDS-PAGE, and detected by fluorography. The conversion of the glycosylated precursor (p2) form of Cpy to mature (m) is dependent on Pep4p activity in the vacuole, hence the accumulation of p2 in the pep4Δ mutant strain. Circles, wild-type (WT) strain; squares, doa4Δ mutant; diamonds, pep4Δ mutant. Strains examined are wild type (SM3631), pep4Δ (SM3632), and doa4Δ (SM3502), which all carry pSM683 (CEN STE6::HA).
We assessed the level of ubiquitination of Ste6p in the doa4Δ mutant to rule out the possibility that Ste6p is underubiquitinated in this strain, which in turn could indirectly account for the observed stabilization of Ste6p. We found instead that substantially more ubiquitinated Ste6p is present in the doa4Δ mutant than in the wild-type strain (Fig. 5A; compare lanes 2 and 3), presumably correlating with the increased steady-state levels of Ste6p in the doa4Δ mutant strain resulting from its stabilization. We conclude that ubiquitination of Ste6p was not deficient in the doa4Δ mutant.
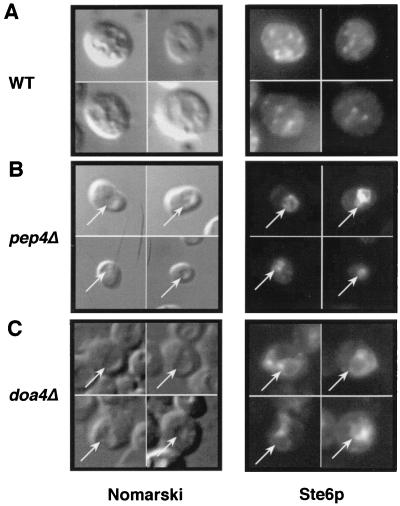
Ste6p accumulates in the vacuole in the doa4Δ mutant.
If the stabilization of Ste6p in the doa4 mutant is due solely to its lack of degradation and not to a defect in its internalization from the cell surface, we would expect Ste6p to accumulate in the vacuolar membrane in this strain. Accordingly, we examined the localization pattern of Ste6p by immunofluorescence. As shown in Fig. 6C, Ste6p localizes to the vacuole in the doa4Δ mutant, compatible with a defect in vacuolar degradation and not intracellular trafficking. The pattern seen in the doa4Δ strain is similar to that observed in the pep4Δ strain (Fig. 6B), where the signal coincides with the vacuole. The staining pattern forms a ring delimiting the vacuole, suggesting that Ste6p concentrates in the vacuolar membrane. The vacuolar localization of Ste6p was easily distinguishable from the punctate Golgi pattern of Ste6p in wild-type cells (Fig. 6A) and from an ER immunolocalization pattern (not shown). Taken together, our metabolic labeling and immunofluorescence studies imply that Pep4p-mediated degradation and Doa4p-dependent degradation are both required to achieve the degradation of Ste6p in the vacuole, at the endpoint of its life cycle. When either of these degradative machineries is compromised, Ste6p fails to be efficiently degraded.
FIG. 6.
Ste6p localizes to the vacuole in the doa4Δ mutant. Ste6p localization was analyzed by immunofluorescence in a wild-type (WT) strain (SM3498; A) a pep4Δ mutant (SM2474; B), and a doa4Δ mutant (SM3501; C), all of which contain a 2μm STE6::HA plasmid (pSM693). Ste6p was detected by using antibody 12CA5. Intracellular indentations in the Nomarski image correspond to the vacuole. The arrows point to the position of the vacuole in the Nomarski images and to the position of the Ste6p signal in the middle panels.
The degradation of Ste6p is slowed in the pre1-1 pre2-1 double mutant, defective in the chymotrypsin-like activity of the proteasome.
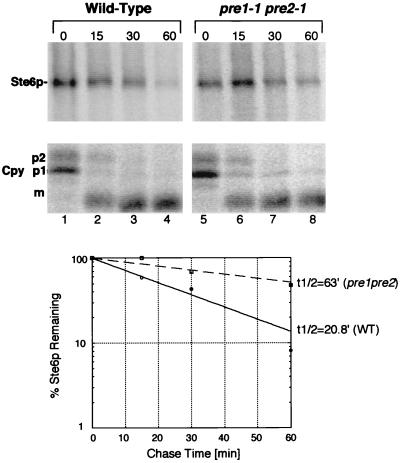
The yeast proteasome has been shown to have at least three biochemically distinct proteolytic activities: a chymotrypsin-like, a trypsin-like, and a peptidyl-prolyl-glutamyl-like activity (19). We asked if Ste6p degradation was affected in the pre1-1 pre2-1 double mutant, in which the chymotrypsin-like activity of the proteasome is reduced to 3% in vitro (18). Cells were subjected to a shift to the nonpermissive temperature (37°C) prior to labeling to impose the block in proteasome function (16). As shown in Fig. 7A, the half-life of Ste6p in the pre1-1 pre2-1 mutant is approximately three times greater than in the isogenic wild-type strain (63 min in the mutant strain and 21 min in the wild-type strain). Thus, the pre1,2 mutations result in significant stabilization of Ste6p. This level of stabilization, however, is not as dramatic as that observed in the doa4Δ mutant. The partial effect of the pre1-1 pre2-1 mutations could be explained by the contribution of other proteasomal proteolytic activity(ies) to the degradation of Ste6p or to the leakiness of the mutations under the conditions used. The processing of Cpy is normal in the pre1-1 pre2-1 mutant (Fig. 7A, bottom), showing that the stabilizing effect on Ste6p is not due to a defect in Pep4p activity. We conclude that the chymotrypsin-like activity of the proteasome is at least partially required for the metabolic degradation of Ste6p.
FIG. 7.
Ste6p is partially stabilized in a pre1-1 pre2-1 mutant, defective in the chymotrypsin-like activity of the proteasome. Metabolic pulse-chase labeling and immunoprecipitation of Ste6p and Cpy were carried out as described in the legend to Fig. 3B. Cells were shifted to 38°C for 40 min prior to metabolic labeling to impose the pre1 pre2 block. Strains used were SM3288 (PRE, pSM683) and SM3290 (pre1-1 pre2-1, pSM683). For abbreviations, see the legend to Fig. 5.
DISCUSSION
The activity of the proteasome is required, either directly or indirectly, for the degradation of Ste6p in the vacuole.
The results presented here, together with studies by others (32), provide evidence that the ubiquitin-proteasome pathway influences the life cycle of Ste6p at two different steps, each step occurring in a distinct cellular location. First, ubiquitination appears to be required for the efficient endocytosis of Ste6p from the cell surface, since the ubc4,5 mutant, defective in the conjugation of ubiquitin to Ste6p, exhibits a delay in the degradation of Ste6p and a cell surface staining pattern for Ste6p that is indicative of an endocytosis defect (Fig. 3 and 4). Second, we have demonstrated here that the metabolic instability of Ste6p, which requires the vacuolar master protease Pep4p, also requires the integrity of the proteasome; in a pre1-1 pre2-1 mutant defective in the activity of the proteasome, the degradation of Ste6p is significantly slowed. In addition, in the doa4Δ mutant, which is defective in the deubiquitination of proteins and in the activity of the proteasome, the degradation of Ste6p is impaired to the same extent as in a pep4Δ mutant. Furthermore, as in the pep4Δ mutant, Ste6p accumulates in the vacuole in the doa4Δ mutant. Because each single (doa4Δ and pep4Δ) mutant is as defective as the other in Ste6p degradation, the two degradative systems appear to both be required, directly or indirectly, for the metabolic instability of Ste6p and can be said to act in a cooperative or interdependent fashion to accomplish degradation.
The cooperation of the Pep4p- and Doa4p-mediated degradation systems was unexpected. Pep4p is a master protease in the vacuole lumen responsible for the maturation and activation of various intravacuolar proteases and is therefore required for most known vacuolar proteolytic activity (30). An assumption in the field of protein degradation has been that vacuolar proteolysis is completely independent of proteasomal proteolysis, which is believed to occur in the cytosol or possibly in the nucleus but not in the vacuole. Our results suggest either that Ste6p is an exception to the rule or that the two systems are both required in other instances as well. It is notable that in the case of catabolite inactivation of the fructose-1,6-bisphosphatase (FBPase) in yeast, the degradation of FBPase has been shown to be Pep4p dependent by one group (8) and proteasome dependent by another (48); indeed, Schork et al. (48) specifically demonstrated that under their conditions, Pep4p has no impact on the metabolic stability of FBPase. It may be that there are actually two distinct pathways for FBPase degradation, one involving delivery to the vacuole, and possibly involving an autophagic-like process followed by Pep4p-dependent degradation, and the other involving cytosolic degradation by the ubiquitin-proteasome system. The use of one versus the other of these pathways could be dictated by subtly different physiological conditions employed by the two different laboratories, thus reconciling their apparent differences. In contrast, for Ste6p, the two systems do not appear to operate in an either/or fashion. Instead, our results indicate that the integrity of the proteasome and that of the Pep4p-dependent proteases are both required for the metabolic degradation of Ste6p. A defect in either one of the proteolytic systems blocks Ste6p degradation. It is of interest that the degradation of FBPase has been postulated to involve an autophagic-like process, in which FBPase-containing vesicles are engulfed by the vacuole (26). Because we have demonstrated here using a ren1 mutant (Fig. 2) and elsewhere with end3, end4, and sac6 mutants that Ste6p travels to the vacuole via the endocytic pathway, it is unlikely that autophagy is involved in the delivery of Ste6p to the vacuole. However, the possibility of an as yet uncharacterized autophagic-like process acting after endocytosis and prior to Pep4p-mediated degradation of Ste6p cannot be excluded.
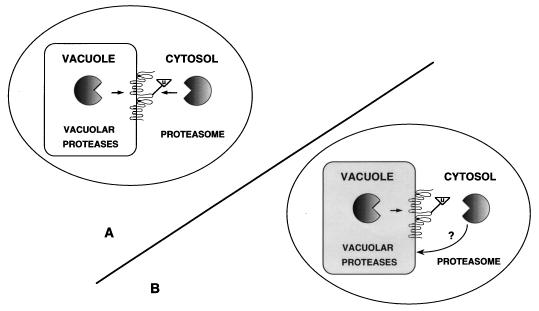
Topologically, Ste6p contains large cytosolic domains and only small lumenal regions (Fig. 8). We propose two models to account for the finding that Ste6p requires both the vacuolar and proteasomal degradation systems. According to one model (Fig. 8A), the cytosolic proteasome and vacuolar Pep4p-dependent proteolytic enzymes act directly on Ste6p to perform degradation from both sides of the vacuolar membrane. It is the proteolytic activity of the proteasome that is required for the degradation of Ste6p, and not an associated activity, because Ste6p is significantly stabilized in the pre1-1 pre2-1 mutant, defective for the chymotrypsin-like activity of the proteasome. We note that another group has reported that the metabolic stability of Ste6p was unaffected by the pre1 pre2 mutations, suggesting that the proteasome had no influence on the degradation of Ste6p (33). Because the pre1-1 mutation has been shown to compromise the proteolytic activity of the proteasome at 30°C in vitro, its in vivo phenotype is often examined at this temperature. Unfortunately, the exact conditions under which the previously reported pre1 pre2 Ste6p stability experiment was performed were not reported, nor were the data shown; it is likely that the temperature used was not fully restrictive for the mutant defect, as we also do not observe any defect in degradation of Ste6p at 30°C in the pre1,2 mutant (34a). It is relevant that the pre1,2 mutant has been shown to have an effect on the degradation of substrates in two other cases, MATα2p (46), and Sec61-2p, a mutant form of Sec61p degraded in the ER (6). In both of these studies, the cells were shifted to 38°C for the experiment. We also observe stabilization of Ste6p in a doa4 mutant, in which proteasome function is defective. Doa4p encodes a deubiquitinating enzyme which, when mutated, is thought to lead to defects in the activity of the proteasome due to accumulation of deubiquitinated substrates in the cell (39). Together, our results indicate that Ste6p is stabilized in two different mutant strains (carrying pre1,2 and doa4), both of which are compromised for the function of the proteasome.
FIG. 8.
Models for Ste6p vacuolar degradation. Two models for Ste6p degradation are shown to explain how the degradation of Ste6p in the vacuolar membrane may be mediated by two distinct sets of proteolytic machinery, the vacuolar proteolytic (PEP4-dependent) machinery and the proteasome (DOA4- and PRE1,2-dependent) machinery. Ste6p is represented in the vacuolar membrane according to its predicted topology (17, 34, 35), with the sizes of the loops drawn roughly to proportion. The ATP binding cassettes are predicted to face the cytosol. The triangle represents ubiquitin moieties attached to Ste6p. The precise sites on Ste6p that are ubiquitinated are currently unknown. According to model A, the cytosolic proteasome recognizes ubiquitinated Ste6p; this event could activate intravacuolar Pep4p-mediated degradation. Both proteolytic systems could then act synergistically to directly degrade Ste6p from each side of the membrane, since little degradation of Ste6p occurs in mutants defective in one or the other system. According to model B, the proteasome may indirectly affect the efficient Pep4p-dependent intravacuolar degradation of Ste6p, by as yet unknown mechanisms (indicated by arrows). For instance, proteasome malfunction could lead to an overall subtle defect in vacuolar integrity (indicated by the grey shading), which could in turn block the Pep4p-dependent proteolysis of Ste6p. It should be noted, however, that neither the gross overall vacuolar morphology nor the Pep4p-dependent processing of Cpy is affected in doa4 or pre1,2 mutants (Fig. 5 and 6). Alternatively, the Pep4p-dependent degradation of Ste6p may require a deubiquitinated Ste6p substrate. As discussed in the text, a functional proteasome might be indirectly required for the deubiquitination, and thus the Pep4p-dependent degradation, of Ste6p.
As shown in the model in Fig. 8A, the simplest explanation for the dependence of Ste6p degradation on both the proteasome and Pep4p-dependent proteases is that both proteolytic systems act directly to mediate the proteolysis of Ste6p from the cytoplasmic and vacuolar compartments, respectively. However, we cannot exclude the possibility that a fraction of the total cellular proteasomes may reside in the lumen of the vacuole and collaborate with Pep4p to degrade Ste6p from within the vacuole, although the lack of evidence for the presence of proteasomes inside the vacuole renders this possibility unlikely.
According to the second model for the degradation of Ste6p (Fig. 8B), the activity of the proteasome may have only an indirect effect on the Pep4p-mediated degradation of Ste6p. For instance, it is possible that the integrity of the vacuole itself, or of the Pep4p-dependent degradation of certain substrates, such as Ste6p, is defective in mutants lacking a functional proteasome. However, we do not observe a defect in overall vacuolar morphology or in Pep4p-dependent processing of Cpy in either the doa4 or pre1,2 mutant (Fig. 5 and 6), indicating that if there are vacuolar defects in these mutants, they are not global defects. Alternatively, the Pep4p-dependent degradation system might act only on a deubiquitinated Ste6p substrate. According to this view, the doa4 mutant, in which one of several of the cellular deubiquitinating enzymes is absent, might accumulate hyperubiquitinated substrates, including Ste6p, which in turn would not be efficiently degraded by Pep4p. To explain the lack of Ste6p degradation in the pre1,2 proteasome mutant it could be postulated that in the absence of a functional proteasome, hyperubiquitinated Ste6p also accumulates, although there is no direct evidence for this latter point.
Although our data cannot unambiguously enable us to distinguish between the two models shown in Fig. 8, we favor the first model, in which the two proteolytic systems act on Ste6p directly, from opposite sides of the membrane. It seems parsimonious to propose that the cytosolic proteasome recognizes the cytosolic ATP binding domains of Ste6p whereas the lumenal Pep4p-dependent enzymes act on the small lumenally disposed loops. The reason why the proteasome and intravacuolar proteases might act in an interdependent fashion, each apparently requiring the activity of the other, is not obvious. One possibility is that a shared requirement for both Pep4p and proteasomal activities acting synergistically would ensure that Ste6p be degraded only upon its arrival in the vacuole, where Pep4p-dependent proteases reside.
Ubiquitination is required for Ste6p to reach the vacuole.
We show here that Ste6p is ubiquitinated in a wild-type strain and not just in endocytosis mutants, as had been previously seen (32, 33). We also observe that ubiquitination is required for the efficient endocytosis of Ste6p, based on its hyperstability and the surface staining pattern of Ste6p in the ubc4,5Δ double mutant. These findings are in agreement with the recent report that mutant forms of Ste6p that are underubiquitinated fail to be internalized from the cell surface (33).
Interestingly, the cell surface staining pattern of Ste6p in the ubc4,5Δ mutant is different from that seen in mutants defective in endocytosis, such as end4 (2) or sac6 (this study) mutants. In the latter case, an all-around rim staining pattern is observed, while in the ubc4,5Δ mutant a nonuniform polar staining pattern is seen (Fig. 4). Thus, a defect in ubiquitination may affect the trafficking of Ste6p at a different step from that blocked in endocytosis mutants such as end3, end4, and sac6 mutants, raising the possibility of a heretofore undescribed trafficking step for Ste6p. We would speculate that conjugation of ubiquitin to Ste6p could be required for proper distribution of the molecule at the plasma membrane subsequent to polarized arrival at the cell surface.
Is only ubiquitinated Ste6p degraded in the vacuole, or is nonubiquitinated Ste6p also subject to degradation? Our results cannot directly address this question. Since ubiquitination is necessary for endocytosis, this problem will be difficult to answer in vivo. Ubiquitination could have two separable functions in the life cycle of Ste6p; it could be a signal for endocytosis from the cell surface, and it may also be required for the recognition of Ste6p by the proteasome when it arrives in the vacuolar membrane. Thus, for Ste6p, ubiquitination may not lead to immediate degradation as it does for certain cell cycle proteins or transcription factors, but instead it may lead to delayed and highly regulated degradation by the proteasome and the vacuolar proteolytic machinery.
Ste6p colocalizes with Kex2p by immunofluorescence.
Based on our colocalization with Kex2p, and the fact that Ste6p cofractionates on a sucrose gradient with the Golgi markers DPAPA (32) and Kex2p (47a), it appears that Ste6p experiences a slow step in its trafficking from the Golgi complex to the plasma membrane. However, Ste6p cannot be considered a Golgi resident such as Kex2p or DPAPA, which are both metabolically stable in the cell (55). Paradoxically, even though it is not apparent at the plasma membrane at steady state as revealed by immunofluorescence and subcellular fractionation, Ste6p is best considered a plasma membrane protein. Its cell surface localization is revealed in mutants defective for endocytosis or ubiquitin conjugation, which block Ste6p internalization from the plasma membrane, thus trapping it at the cell surface. Likewise, although Ste6p clearly traffics to the vacuole, it is also never visible in the vacuole in wild-type cells. Instead, Ste6p can be detected in the vacuole only in pep4 or doa4 mutants in which it accumulates due to a block in its metabolic degradation. All in all, the apparent Golgi localization may reflect a slow step in trafficking and metabolism and not a site of functional residency.
Ste6p: the exception or the rule?
The trafficking of several membrane proteins has been studied in yeast, and a subset of these exhibit metabolic instability, at least under certain conditions. Strikingly, it is beginning to emerge that the major aspects of the life cycle of several of these may be mediated by the same machinery as Ste6p. For instance, like Ste6p, the pheromone receptors, Ste2p and Ste3p, are known to require ubiquitination for efficient endocytosis and are degraded in the vacuole in a Pep4p-dependent manner (21, 47). However, unlike the case for Ste6p, it has been reported that proteasome mutants do not exhibit defects in the degradation of these membrane proteins. Similarly, for the uracil permease (Fur4p) (16), the maltose transporter (Mal1p) (45), and an ABC transporter involved in drug resistance (Pdr5p) (12), vacuolar trafficking requires ubiquitination, but degradation is thought to be proteasome independent. Thus, to our knowledge, Ste6p is the first membrane protein that has been shown to require the integrity of two proteolytic systems for degradation. We speculate that Ste6p may represent a special case, in which the degradation of a complex transmembrane protein must occur extremely rapidly to facilitate efficient mating-type interconversion. The topology of Ste6p, like that of other ABC proteins, could dictate a strong requirement for a cytosolically disposed proteolytic machinery such as the proteasome, because the bulk of Ste6p is inaccessible to lumenal vacuolar proteases. Notably, the 26S proteasome has also been implicated in the degradation of CFTR (28, 54). In this case, however, the fraction of CFTR that is degraded by the proteasome (about 75%) fails to exit the ER and does not reach the later compartments (53). The remaining 25% of the CFTR that is synthesized is transported to the cell surface and rapidly endocytosed (41). Therefore, the requirement for the proteasome for degradation might be a feature particular to some ABC transporters, which might have to rely on complex degradation mechanisms due to their particular topology.
ACKNOWLEDGMENTS
We thank C. Pickart, W. Schmidt, and G. Nijbroek for helpful comments on the manuscript; M. Hochstrasser, G. Sprague, and D. Wolf for strains and plasmids; M. Rose and E. Jones for antibodies; R. Fuller for precious affinity-purified anti-Kex2p antibody; and D. Murphy and W. Guggino for generous help with microscopes and computers. We thank members of the Michaelis laboratory for valuable ideas and C. Berkower for help with plasmid and strain construction.
This work was supported by National Institutes of Health grant GM51508 to S.M.
REFERENCES
- 1.Amara J F, Cheng S H, Smith A E. Intracellular protein trafficking defects in human disease. Trends Cell Biol. 1992;2:145–149. doi: 10.1016/0962-8924(92)90101-r. [DOI] [PubMed] [Google Scholar]
- 2.Berkower C, Loayza D, Michaelis S. Metabolic instability and constitutive endocytosis of STE6, the a-factor transporter of Saccharomyces cerevisiae. Mol Biol Cell. 1994;5:1185–1198. doi: 10.1091/mbc.5.11.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkower C, Michaelis S. Mutational analysis of the yeast a-factor transporter STE6, a member of the ATP binding cassette (ABC) protein superfamily. EMBO J. 1991;10:3777–3785. doi: 10.1002/j.1460-2075.1991.tb04947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkower, C., and S. Michaelis. ATP binding cassette proteins in yeast. In S. Rothman (ed.), Membrane protein transport, in press. JAI Press, Greenwich, Conn.
- 5.Berkower C, Taglicht D, Michaelis S. Functional and physical interactions between partial molecules of STE6, a yeast ATP-binding-cassette transport protein. J Biol Chem. 1996;271:22983–22989. doi: 10.1074/jbc.271.38.22983. [DOI] [PubMed] [Google Scholar]
- 6.Biederer T, Volkwein C, Sommer T. Degradation of subunits of the Sec61p complex, an integral component of the ER membrane, by the ubiquitin-proteasome pathway. EMBO J. 1996;15:2069–2076. [PMC free article] [PubMed] [Google Scholar]
- 7.Boeke J D, Trueheart J, Natsoulis G, Fink G R. 5-Fluoro-orotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;152:481–504. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 8.Chiang H L, Schekman R. Regulated import and degradation of a cytosolic protein in the yeast vacuole. Nature. 1991;350:313–318. doi: 10.1038/350313a0. [DOI] [PubMed] [Google Scholar]
- 9.Davis N G, Horecka J L, Sprague G F., Jr Cis- and trans-acting functions required for endocytosis of the yeast pheromone receptors. J Cell Biol. 1993;122:53–65. doi: 10.1083/jcb.122.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Decottignies A, Goffeau A. Complete inventory of the yeast ABC proteins. Nat Genet. 1997;15:137–145. doi: 10.1038/ng0297-137. [DOI] [PubMed] [Google Scholar]
- 11.Egner R, Kuchler K. The yeast multidrug transporter Pdr5 of the plasma membrane is ubiquitinated prior to endocytosis and degradation in the vacuole. FEBS Lett. 1996;378:177–181. doi: 10.1016/0014-5793(95)01450-0. [DOI] [PubMed] [Google Scholar]
- 12.Egner R, Mahe Y, Pandjaitan R, Kuchler K. Endocytosis and vacuolar degradation of the plasma membrane-localized Pdr5 ATP-binding cassette multidrug transporter in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:5879–5887. doi: 10.1128/mcb.15.11.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elble R. A simple and efficient procedure for transformation of yeasts. BioTechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- 14.Finley D, Chau V. Ubiquitination. Annu Rev Cell Biol. 1991;7:25–69. doi: 10.1146/annurev.cb.07.110191.000325. [DOI] [PubMed] [Google Scholar]
- 15.Franzusoff A, Redding K, Crosby J, Fuller R S, Schekman R. Localization of components involved in protein transport and processing through the yeast Golgi apparatus. J Cell Biol. 1991;112:27–37. doi: 10.1083/jcb.112.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galan J M, Moreau V, Andre B, Volland C, Haguenauer-Tsapis R. Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J Biol Chem. 1996;271:10946–10952. doi: 10.1074/jbc.271.18.10946. [DOI] [PubMed] [Google Scholar]
- 17.Geller D, Taglicht D, Edgar R, Tam A, Pines O, Michaelis S, Bibi E. Comparative topology studies in Saccharomyces cerevisiae and in Escherichia coli of the N-terminal half of the yeast ABC protein, Ste6. J Biol Chem. 1996;271:13746–13753. doi: 10.1074/jbc.271.23.13746. [DOI] [PubMed] [Google Scholar]
- 18.Heinemeyer W, Gruhler A, Mohrle V, Mahe Y, Wolf D H. PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chymotryptic activity and degradation of ubiquitinated proteins. J Biol Chem. 1993;268:5115–5120. [PubMed] [Google Scholar]
- 19.Heinemeyer W, Kleinschmidt J A, Saidowsky J, Escher C, Wolf D H. Proteinase yscE, the yeast proteasome/multicatalytic-multifunctional proteinase: mutants unravel its function in stress induced proteolysis and uncover its necessity for cell survival. EMBO J. 1991;10:555–582. doi: 10.1002/j.1460-2075.1991.tb07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hershko A, Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- 21.Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- 22.Higgins C F. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 23.Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 24.Hochstrasser M. Protein degradation or regulation: Ub the judge. Cell. 1996;84:813–815. doi: 10.1016/s0092-8674(00)81058-2. [DOI] [PubMed] [Google Scholar]
- 25.Hochstrasser M, Ellison M J, Chau V, Varshavsky A. The short-lived MATα2 transcriptional regulator is ubiquitinated in vivo. Proc Natl Acad Sci USA. 1991;88:4606–4610. doi: 10.1073/pnas.88.11.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang P-H, Chiang H-L. Identification of novel vesicles in the cytosol to vacuole protein degradation pathway. J Cell Biol. 1997;136:803–810. doi: 10.1083/jcb.136.4.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jensen T J, Loo M A, Pind S, Williams D B, Goldberg A L, Riordan J R. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- 29.Jentsch S. Ubiquitin-dependent protein degradation: a cellular perspective. Trends Cell Biol. 1992;2:98–103. doi: 10.1016/0962-8924(92)90013-d. [DOI] [PubMed] [Google Scholar]
- 30.Jones E W. Three proteolytic systems in the yeast Saccharomyces cerevisiae. J Biol Chem. 1991;266:7963–7966. . (Review.) [PubMed] [Google Scholar]
- 31.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. A Cold Spring Harbor course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 32.Kölling R, Hollenberg C P. The ABC-transporter Ste6 accumulates in the plasma membrane in a ubiquitinated form in endocytosis mutants. EMBO J. 1994;13:3261–3271. doi: 10.1002/j.1460-2075.1994.tb06627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kölling R, Losko S. The linker region of the ABC transporter Ste6 mediates ubiquitination and fast turnover of the protein. EMBO J. 1997;16:2251–2261. doi: 10.1093/emboj/16.9.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuchler K, Sterne R E, Thorner J. Saccharomyces cerevisiae STE6 gene product: a novel pathway for protein export in eukaryotic cells. EMBO J. 1989;8:3973–3984. doi: 10.1002/j.1460-2075.1989.tb08580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a.Loayza, D., and S. Michaelis. Unpublished observation.
- 35.McGrath J P, Varshavsky A. The yeast STE6 gene encodes a homologue of the mammalian multidrug resistance P-glycoprotein. Nature. 1989;340:400–404. doi: 10.1038/340400a0. [DOI] [PubMed] [Google Scholar]
- 36.Michaelis S, Herskowitz I. The a-factor pheromone of Saccharomyces cerevisiae is essential for mating. Mol Cell Biol. 1988;8:1309–1318. doi: 10.1128/mcb.8.3.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paddon C, Loayza D, Vangelista L, Solari R, Michaelis S. Analysis of the localization of STE6/CFTR chimeras in a Saccharomyces cerevisiae model for the cystic fibrosis defect CFTRΔF508. Mol Microbiol. 1996;19:1007–1017. doi: 10.1046/j.1365-2958.1996.444973.x. [DOI] [PubMed] [Google Scholar]
- 38.Paolini R, Kinet J-P. Cell surface control of the multiubiquitination and deubiquitination of high-affinity immunoglobulin E receptors. EMBO J. 1993;12:779–786. doi: 10.1002/j.1460-2075.1993.tb05712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Papa F R, Hochstrasser M. The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature. 1993;366:313–319. doi: 10.1038/366313a0. [DOI] [PubMed] [Google Scholar]
- 40.Piper R C, Cooper A A, Yang H, Stevens T H. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J Cell Biol. 1995;131:603–617. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prince L S, Workman J, Marchase R B. Rapid endocytosis of the cystic fibrosis transmembrane conductance regulator chloride channel. Proc Natl Acad Sci USA. 1994;91:5192–5196. doi: 10.1073/pnas.91.11.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raymond C K, Howald-Stevenson I, Vater C A, Stevens T H. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Redding K, Holcomb C, Fuller R S. Immunolocalization of Kex2 protease identifies a putative late Golgi compartment in the yeast Saccharomyces cerevisiae. J Cell Biol. 1991;113:527. doi: 10.1083/jcb.113.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Redding K, Seeger M, Payne G S, Fuller R S. The effects of clathrin inactivation on localization of Kex2 protease are independent of the TGN localization signal in the cytosolic signal in hte cytosolic tail of Kex2p. Mol Biol Cell. 1996;7:1667–1677. doi: 10.1091/mbc.7.11.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riballo E, Herweijer M, Wolf D H, Lagunas R. Catabolite inactivation of the yeast maltose transporter occurs in the vacuole after internalization by endocytosis. J Bacteriol. 1995;177:5622–5627. doi: 10.1128/jb.177.19.5622-5627.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richter-Ruoff B, Wolf D H, Hochstrasser M. Degradation of the yeast MATα2 transcriptional regulator is mediated by the proteasome. FEBS Lett. 1994;354:50–52. doi: 10.1016/0014-5793(94)01085-4. [DOI] [PubMed] [Google Scholar]
- 47.Roth A F, Davis N G. Ubiquitination of the yeast a-factor receptor. J Cell Biol. 1996;134:661–674. doi: 10.1083/jcb.134.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47a.Schmidt, W., and S. Michaelis. Unpublished data.
- 48.Schork S M, Thumm M, Wolf D H. Catabolite inactivation of fructose-1,6-bisphosphatase of Saccharomyces cerevisiae. J Biol Chem. 1995;270:26446–26450. doi: 10.1074/jbc.270.44.26446. [DOI] [PubMed] [Google Scholar]
- 49.Seufert W, Jentsch S. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 1990;9:543–550. doi: 10.1002/j.1460-2075.1990.tb08141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taglicht, D., and S. Michaelis. A complete catalog of Saccharomyces cerevisiae ABC proteins and their relevance to human health and disease. Methods Enzymol., in press. [DOI] [PubMed]
- 52.Thomas P J, Bao-He Q, Pedersen P. Defective protein folding as a basis of human disease. Trends Biochem Sci. 1995;20:456–459. doi: 10.1016/s0968-0004(00)89100-8. [DOI] [PubMed] [Google Scholar]
- 53.Ward C L, Kopito R R. Intracellular turnover of cystic fibrosis transmembrane conductance regulator. J Biol Chem. 1994;269:25710–25718. [PubMed] [Google Scholar]
- 54.Ward C L, Omura S, Kopito R R. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 55.Wilcox C A, Redding R, Wright R, Fuller R S. Mutation of a tyrosine localization signal in the cytosolic tail of yeast Kex2 protease disrupts Golgi retention and results in default transport to the vacuole. Mol Biol Cell. 1992;3:1353–1371. doi: 10.1091/mbc.3.12.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]