Abstract
Co-translational insertion of selenocysteine (Sec) into proteins in response to UGA codons is directed by selenocysteine insertion sequence (SECIS) elements. In known bacterial selenoprotein genes, SECIS elements are located in the coding regions immediately downstream of UGA codons. Here, we report that a distant SECIS element can also function in Sec insertion in bacteria provided that it is spatially close to the UGA codon. We expressed a mammalian phospholipid hydroperoxide glutathione peroxidase in Escherichia coli from a construct in which a natural E.coli SECIS element was located in the 3′-untranslated region (3′-UTR) and adjacent to a sequence complementary to the region downstream of the Sec UGA codon. Although the major readthrough event at the UGA codon was insertion of tryptophan, Sec was also incorporated and its insertion was dependent on the functional SECIS element in the UTR, base-pairing potential of the SECIS flanking region and the Sec UGA codon. These data provide important implications into evolution of SECIS elements and development of a system for heterologous expression of selenoproteins and show that in addition to the primary sequence arrangement between UGA codons and SECIS elements, their proximity within the tertiary structure can support Sec insertion in bacteria.
INTRODUCTION
Selenium is a trace element that is vital for many life forms. It occurs in cells in the form of small compounds and selenium-containing proteins. In most selenoproteins, selenium is present in the form of the amino acid selenocysteine (Sec). Sec is encoded by UGA codons (1) and recognized as the 21st amino acid in proteins (2) as it has its own codeword, tRNA (3–5) and a Sec-specific elongation factor (6–9). UGA also serves as a signal for termination of protein synthesis. To distinguish between these two functions of UGA, a designated RNA structure, Sec insertion sequence (SECIS) element (10,11), is present in selenoprotein mRNAs and directs Sec incorporation into selenoproteins (11,12).
In both prokaryotes and eukaryotes, SECIS elements serve as cis-elements that recognize specific trans-acting factors and direct these factors to the ribosome. SECIS elements show little primary sequence conservation, but possess common secondary structures (within each domain of life) (13,14). There is a key difference in the location of SECIS elements in bacteria versus archaea and eukaryotes. In known bacterial selenoprotein genes, SECIS elements are located in coding regions immediately downstream of UGA codons such that the distance between UGA and the apical loop of SECIS elements is ∼20–25 nt; in contrast, in archaea and eukaryotes, SECIS elements are present in 3′-untranslated regions (3′-UTRs) and the distance between the UGA codon and the SECIS element is highly variable (15). In addition, bacterial SECIS elements reversibly bind Sec-specific elongation factor SelB, which in turn recruits selenocysteyl-tRNASec and inserts Sec at the UGA codon; whereas in eukaryotes SECIS elements are associated with a SECIS binding protein (8,16), which in turn binds elongation factor EFSec/SelB. In archaea, this issue has not been resolved. Previous data suggested that archaeal SelB was similar to the eukaryotic counterpart in that it lacked the SECIS binding domain (17,18); however, a recently reported structure of an archaeal SelB suggested that this protein might actually bind SECIS directly, similarly to the bacterial SelB (19).
The differences in location and structural features of SECIS elements in the major life kingdoms might represent evolutionary preferences. To achieve Sec insertion at high efficiency, trans-acting factors must be in close proximity to the ribosome, which is provided by the presence of SECIS elements immediately downstream of UGA in bacteria. However, the coding region location of SECIS elements has disadvantages, as the nucleotide sequences downstream of UGA codons must satisfy both structural features of SECIS elements and support insertion of specific amino acids as dictated by protein function. The compromise between the coding and the SelB-binding functions of SECIS elements might limit the choice of sequences downstream of Sec as well as restrict selenoprotein diversity. In eukaryotes and archaea, this problem is addressed by having SECIS elements in 3′-UTRs. The 3′-UTR location of SECIS elements also has additional advantages, such as (i) the processivity of Sec insertion. 3′-UTR SECIS elements can stably bind SECIS binding proteins (20), increasing Sec insertion efficiency; and (ii) utilization of one stem–loop structure for insertion of multiple Sec residues. However, the UTR location of the stem–loop structure creates a different challenge as Sec insertion may be slower due to distance factor. Introduction of additional trans-acting factors that bridge ribosome, Sec insertion machinery and SECIS elements through protein–protein and protein–RNA interactions may partially alleviate this problem. The difference in transcription and translation processes between bacteria and eukaryotes is another factor that may affect evolution of SECIS elements. In bacteria, transcription and translation are coupled; therefore, a distant SECIS element might not yet be transcribed when the upstream UGA is already translated. Thus, a distant SECIS element in 3′-UTR might not be favorable in bacteria, whereas this is not an issue in eukaryotes due to separation of transcription and translation.
In this report, we expressed a mouse selenoprotein glutathione peroxidase 4 (GPx4; also known as phospholipid hydroperoxide glutathione peroxidase, PHGPX) in E.coli using a bacterial SECIS element located within the 3′-UTR. We used a construct, in which the UGA and the SECIS element were bridged by base-pair interactions between the regions adjacent to the SECIS element and the UGA codon. Our data show that the 3′-UTR SECIS element can function in Sec insertion with low efficiency in E.coli.
MATERIALS AND METHODS
Constructs
Mouse GPx4 was cloned from an expressed sequence tag (gi:390827) (Invitrogen) using a 5′ primer 5′-GGAATTCCATATGTGTGCATCCCGCGATG-3′ and four overlapping 3′ primers 5′-CCTAGTGGTGGTGGTGGTGGTGGAGATAGCACGGCAGGTCC-3′, 5′-TCAGCTAGTCGATCTGCATGCCCCTAGTGGTGGTGGTGGTGGTG-3′, 5′-GCAACCGATACGTAAACTACACTCAGCTAGTCGATCTGCATGCC-3′ and 5′-CGGGATCCATTGGTGCAGACCTGCAACCGATACGTAAACTACAC-3′. The PCR product was digested with NdeI and BamHI and ligated into a pET21b vector (Novagen), which was digested with the same pair of restriction enzymes.
All mutants were based on the GPx4 expression construct described above. The SECIS mutant, UAA mutant and cysteine mutant were obtained using QuikChange® XL Site-Directed Mutagenesis Kit (Stratagene). To generate the SECIS mutant, primers 5′-GTGTAGTTTACGTATCGGATGGAGGTCTGGACCAATGGATCC-3′ and 5′-GGATCCATTGGTCCAGACCTCCATCCGATACGTAAACTACAC-3′ were used. Primers 5′-CGTGGCCTCGCAATAAGGCAAAACTGACG-3′ and 5′-CGTCAGTTTTGCCTTATTGCGAGGCCACG-3′ were used to obtain the UAA mutant. To generate the cysteine mutant, primers 5′-CGTGGCCTCGCAATGCGGCAAAACTGACGT-3′ and 5′-ACGTCAGTTTTGCCGCATTGCGAGGCCACG-3′ were used. Additional controls that lacked the base-pairing potential, SECIS element and combination of both were generated using ExSite™ PCR-Based Site-Directed Mutagenesis Kit (Stratagene). For the former, primers 5′-p-ATCGGTTGCAGGTCTGCACCAATGG-3′ and 5′-CTAGTGGTGGTGGTGGTGGTGGAG-3′ were used. For the second, primers 5′-p-GGATCCGAATTCGAGCTCCGTCGAC-3′ and 5′-ACGTAAACTACACTCAGCTAGTCGATCTGC-3′ were used. The mutant that combined deletions of SECIS element and base pairing was generated using primers 5′-p-ATCGGTTGCAGGTCTGCACCAATGG-3′ and 5′-ACGTAAACTACACTCAGCTAGTCGATCTGC-3′.
75Se labeling
The GPx4 expression construct and all mutants were expressed in BL21(DE3) cells with or without co-expression of SelA, SelB and SelC from the plasmid pSUABC (21). In each case, 10 ml cells were grown to OD600 0.6, 0.02 mCi of freshly neutralized 75Se[selenite] was added, and the cell culture was supplemented with 50 μM IPTG and 100 mg/l l-cysteine. Cells were grown for 12 h at 30°C, collected by centrifugation and broken by sonication. Cell lysates were fractionated on a Talon™ resin (BD Biosciences). Eluted proteins were subjected to SDS–PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane, and the pattern of 75Se radioactivity was visualized using a PhosphorImager. The same membrane was also subjected to western blot analysis using anti-His-tag antibodies, then stripped and re-probed with anti-GPx4 antibodies.
Protein expression
The strain carrying GPx4-SECIS and pSUABC and the strain carrying pET21b and pSUABC were grown in 3 liters of Luria–Bertani medium, cells were collected by centrifugation and proteins purified according to The QIAexpressionist™ (Qiagen) using an Ni-NTA resin (QIAGEN). The proteins were visualized on an SDS–PAGE gel by staining with Bio-Safe™ Coomassie (Bio-Rad).
Mass spectrometry and selenium content analysis
The recombinant GPx4 expressed from a construct carrying a 3′-UTR SECIS element and purified using an affinity column was thoroughly dialyzed in 0.4 M ammonium bicarbonate and alkylated with iodoacetamide. Following tryptic digestion, peptide sequences were determined by tandem mass spectrometry sequencing at the Nebraska Redox Biology Center proteomics/metabolomics facility. The selenium content of the recombinant GPx4 was analyzed by inductively coupled plasma-emission spectrometry at the Chemical Analysis Facility at University of Georgia. Before the selenium analysis, the sample was dialyzed against phosphate-buffered saline, and the dialysis buffer was used as background metal ion content control for selenium determination.
RESULTS
Design of expression constructs
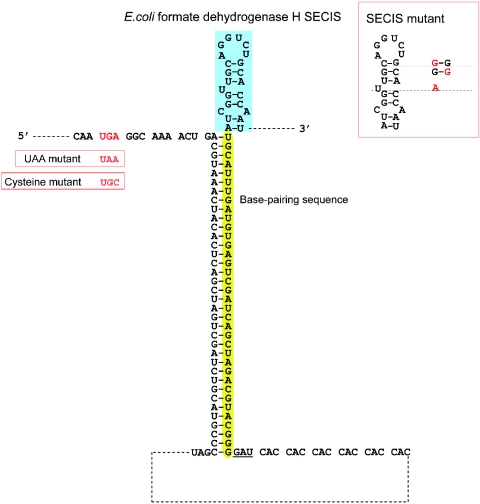
To test whether a distant SECIS element can support Sec insertion in bacteria, we developed a construct coding for a full-size, Sec-containing mouse GPx4. In this construct, we placed an E.coli formate dehydrogenase H SECIS element (highlighted in blue in Figure 1) in the 3′-UTR. This construct also contained a sequence that coded for six histidines at the C-terminus of the open reading frame (ORF), followed with a UAG stop codon (underlined in Figure 1). In the event of readthrough of the Sec UGA codon, the His-tag could be used for protein detection and purification by affinity chromatography. To bring the SECIS element in close proximity to the UGA codon (shown in red in Figure 1), we inserted a 36 nt 3′-UTR segment (highlighted in yellow in Figure 1), which was complementary to the sequence downstream of the UGA codon, between the ORF and the SECIS element. The strong base-pairing potential of the 3′-UTR 36 nt segment and the region adjacent to the UGA should position the UGA and the SECIS element next to each other, approximately as close as the UGA and the SECIS element in the wild-type formate dehydrogenase H gene (Figure 1).
Figure 1.
Design of the GPx4 expression construct containing a 3′-UTR SECIS element. The in-frame UGA codon that codes for Sec is shown in red. At the end of the ORF, six histidine codons were added, followed by the stop signal, UAG (underlined). In addition, a 36 nt segment, which was complementary to the 36 nt downstream of the Sec UGA codon, was cloned in the 3′-UTR downstream of the UAG codon (highlighted in yellow). Further downstream of this segment, the E.coli formate dehydrogenase H SECIS element was inserted (highlighted in blue). The red box on the right shows the SECIS mutant used in this study. Letters shown in red (on the right of the SECIS element) indicate mutations and their positions within the SECIS element. The UAA and cysteine mutants are shown in red boxes on the left.
We also developed a series of control constructs: (i) A SECIS mutant: this construct was identical to the construct described above except that three point mutations were introduced in the stem–loop structure (as shown in the red box in Figure 1). This mutant SECIS element was previously reported to have a very low ability to direct Sec insertion (22,23). (ii) A UAA mutant: in this construct, the Sec UGA codon was mutated to UAA. (iii) A UAA/SECIS mutant: this construct combined the mutations in constructs ‘i’ and ‘ii’. (iv) A cysteine mutant: the Sec UGA codon was mutated to a cysteine codon, UGC. (v) A construct lacking the 3′-UTR base-pairing region (highlighted in yellow in Figure 1). (vi) A construct lacking the SECIS element (highlighted in blue in Figure 1). (vii) A construct lacking both the base-pairing region and the SECIS element.
The 3′-UTR SECIS element directs Sec insertion in E.coli
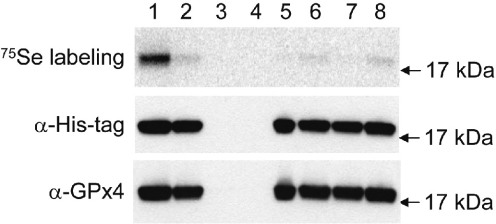
The wild-type and various mutant GPx4 constructs were transformed into E.coli cells. To monitor Sec incorporation, all samples were metabolically labeled with 75Se and recombinant proteins isolated using affinity columns (Figure 2). These procedures were performed in parallel for all samples. GPx4 expressed from the construct containing the wild-type SECIS element showed a strong 75Se signal (Figure 2, lane 1, upper panel). The 75Se-labeled protein migrated with the size that was expected of the full-size protein. The enrichment of GPx4 on the affinity resin specific for His-tag indicated that the C-terminal tag was part of the protein and that the sequences downstream of the UGA were coding. The protein that was expressed from the construct containing a mutant SECIS element showed a very weak 75Se signal (Figure 2, lane 2, upper panel), suggesting that the functional SECIS element was required for Sec insertion at the UGA codon. However, when the UGA codon was replaced with UAA (independently of whether the wild-type or mutant SECIS was used), no 75Se signal was observed, indicating that the Sec insertion was dependent on the UGA codon (Figure 2, lanes 3 and 4, upper panel). In addition, a cysteine mutant showed a very weak 75Se signal (Figure 2, lane 5, upper panel), and the deletion of the base-pairing sequence, SECIS element or both also resulted in very weak 75Se signals (Figure 2, lanes 6, 7 and 8, upper panel). These data suggested that Sec was inserted into GPx4 in a 3′-UTR SECIS element-dependent manner and that Sec insertion was supported exclusively by UGA. The weak labeling of some samples with 75Se most probably resulted from non-specific labeling of the protein by random incorporation of selenium in place of sulfur-containing amino acids. We used 100 mg/l l-cysteine in the growth medium (24), however, the non-specific labeling could not be completely blocked. Nevertheless, the non-specific labeling could easily be distinguished from the specific insertion of Sec as the latter occurred at a much higher level (compare lane 1 and lanes 2, 5–8 in Figure 2).
Figure 2.
Sec insertion directed by the 3′-UTR SECIS element. Following the induction of recombinant protein synthesis, BL21(DE3) cells containing various GPx4 expression constructs were metabolically labeled with 75Se. All cells were labeled in parallel and proteins purified on affinity columns using identical procedures. Approximately equal amounts of GPx4 were obtained in each sample, except in mutants containing an in-frame UAA (no GPx4 was detected) and the cysteine mutant (expression level of the GPx4 cysteine mutant was much higher than in other samples). The pattern of 75Se radioactivity of recombinant GPx4 (upper panel) was analyzed using a PhosphorImager on a PVDF membrane following SDS–PAGE. Lane 1, wild-type 3′-UTR SECIS element; lane 2, SECIS element mutant; lane 3, UAA mutant; lane 4, UAA/SECIS mutant; lane 5, cysteine mutant; lane 6, a mutant lacking the 3′-UTR base-pairing segment; lane 7, a mutant lacking the SECIS element; and lane 8, a mutant lacking both the 3′-UTR base-pairing segment and the SECIS element. The membrane was also subjected to immunoblot assays with antibodies specific for His-tag (middle panel) and GPx4 (lower panel). Migration of a molecular a weight standard (17 kDa) is shown on the right.
To verify that the 75Se signal indeed corresponded to the recombinant GPx4, all affinity-purified protein samples were subjected to immunoblot assays with anti-His-tag and anti-GPx4 antibodies. Both gave signals that coincided with the 75Se signal (middle and lower panels in Figure 2) except the two UAA mutants, in which no GPx4 could be detected. In contrast to the 75Se patterns, both immunoblot assays revealed a similar amount of the full-size GPx4 in the samples expressed from the constructs containing the UGA codon. As expected, the cysteine mutant of GPx4 was overexpressed (we normalized its loading on the SDS–PAGE gel shown in Figure 2). Taken together, these data suggested that there was an additional major readthrough event other than Sec insertion, which responded to UGA, but not to UAA, and which was independent of the base-pairing sequence or the SECIS element.
We repeated the 75Se labeling experiments five times, and in each case, Sec insertion was dependent on the 3′-UTR SECIS and the UGA codon. We also attempted to express the selenoprotein by co-expressing SelA (Sec synthetase), SelB (Sec-specific elongation factor) and SelC (Sec tRNA) (21), which was reported to increase Sec insertion into mammalian thioredoxin reductase 1 by 8-fold (21). Again, dependence of 75Se insertion on the 3′-UTR SECIS element was evident (data not shown).
Opal suppression accounts for the major readthrough event
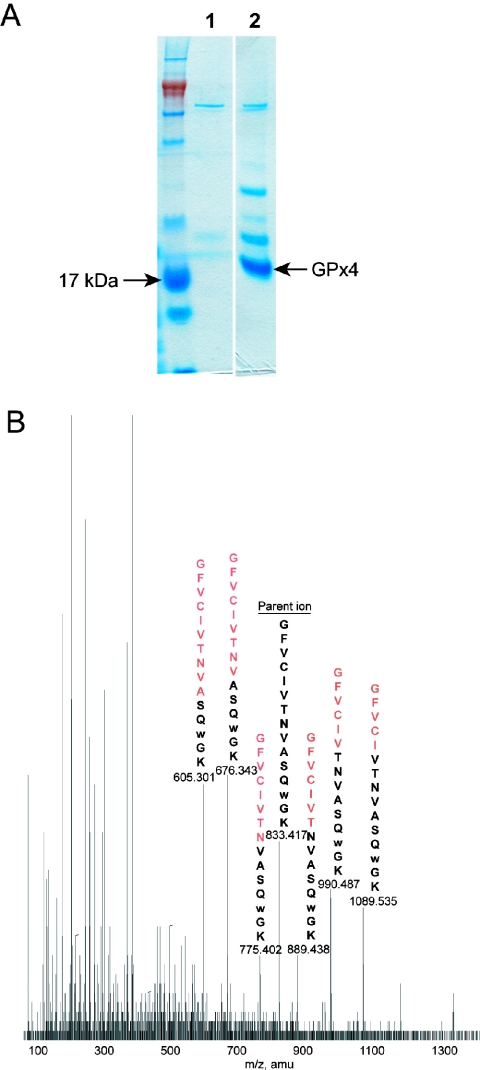
To characterize readthrough forms GPx4, we carried out a large-scale isolation of GPx4 expressed from the construct with the functional 3′-UTR SECIS element. Approximately 1 mg of the affinity-purified protein (∼60% purity, Figure 3A) was obtained from 1 liter of bacterial culture. This recombinant GPx4 was subjected to selenium quantification, which found that only ∼3% of the protein molecules had this trace element.
Figure 3.
Opal suppression is a major readthrough event. (A) SDS–PAGE analysis of the recombinant GPx4 expressed from the construct carrying the functional 3′-UTR SECIS element. Extracts prepared from BL21(DE3) cells carrying pET21b and pSUABC (lane 1) and the GPx4 construct with the functional SECIS element and pSUABC (lane 2) were applied to a His-tag affinity column and proteins bound to the column were eluted and analyzed by SDS–PAGE. (B) Mass spectrometry analysis of the readthrough product. The recombinant protein shown in (A) was cut from the SDS–PAGE gel, digested with trypsin, alkylated with iodoacetamide and subjected to tandem mass spectrometry sequencing. A double-charged fragment at 833.4 m/z was identified, which corresponded to a tryptophan-containing readthrough fragment. The ms/ms spectrum of the 833.4 parent ion is shown and the major daughter ions and the parent ion are labeled. We identified these peaks as Y, B or A ions (not shown in this figure) resulting from fragmentation of the sequence GFVCIVTNVASQWGK. The black letters above each peak indicate the amino acid sequence of the corresponding daughter ion and the red letters indicate the amino acids lost in the fragmentation.
To determine the identity of the major readthrough form, the recombinant GPx4 was subjected to tandem mass spectrometry sequencing. More than 90% of sequence coverage was obtained (data not shown); however, the fragment containing the readthrough amino acid was missing if either Sec or Cys (alkylated or unalkylated) were used to calculate peptide masses. In addition to the assigned peptides, a major double-charged fragment at 833.4 m/z was detected, which corresponded to the active site tryptic peptide if the UGA was read as tryptophan. Indeed, the mass-spectrometry fingerprint pattern of this fragment matched the W-containing peptide sequence (Figure 3B). UGA suppression by tRNATrp is known as opal suppression. It was reported previously (25) and was also observed in selenoprotein genes (26). Thus, it appears that GPx4 expression from a construct containing a functional 3′-UTR SECIS element resulted largely in Trp insertion (97% of the readthrough product, ∼1 mg protein per liter of cell culture), whereas the Sec-containing product accounted for ∼3% of the readthrough protein.
DISCUSSION
It has long been known that bacterial SECIS elements are located immediately downstream of the UGA codon (27–30), whereas in eukaryotes and archaea, these structures reside in the 3′-UTR and the UGA codon/SECIS element distance is not critical (12). In this study, we expressed a mouse selenoprotein, GPx4, in bacteria with the help of a 3′-UTR SECIS element that was distant to the UGA codon. The expression construct also contained a 3′-UTR sequence, which was complementary to the sequence downstream of the UGA. This resulting base-pairing interaction was expected to bring the Sec codon and the SECIS element near each other within the mRNA structure (Figure 1). We found that this 3′-UTR SECIS element directed Sec insertion into GPx4 in E.coli. Various controls revealed that the Sec insertion was dependent on the UGA codon, base-pairing region and the SECIS element.
One important question regarding Sec evolution is whether SECIS elements in the three domains of life have a common origin or they evolved independently. The similarities in Sec biosynthesis (SelD/SPS2) and insertion (tRNASec, SelB/EFSec) components among bacteria, archaea and eukaryotes strongly argue that all systems have a common origin. Interestingly, the situation in archaea might represent an interesting evolutionary mid-point as most archaeal selenoproteins have homologs exclusively in bacteria (31), whereas their SECIS elements are located in the 3′-UTRs (17) as in eukaryotes. It is possible that the SECIS elements of selenoproteins specific for bacteria and archaea have the same ancestors. However, when and how SECIS elements might have relocated, and whether these events were linked to changes in the Sec insertion machinery, is not clear. Our results show that bacterial SECIS elements may be functional in the 3′-UTR provided that these structures remain spatially close to UGA codons. Although the method that we used to preserve the proximity between the SECIS element and the UGA codon might not necessarily be the one adopted in evolution, the data show a principal possibility that the relocation can be achieved without any changes in the rest of the Sec insertion machinery.
We detected weak 75Se signals of the recombinant GPx4 when the SECIS element was mutated or deleted, the base-pairing sequence was deleted or both the SECIS element and the base-pairing sequence were deleted. In addition, a weak 75Se signal was observed in the cysteine mutant. It appears that these 75Se signals were due to non-specific labeling of GPx4 with 75Se. It is known that selenium can enter sulfur pathways and be inserted non-specifically into proteins as Sec and selenomethionine (24).
The efficiency of Sec insertion from our construct was low. This is not unexpected since (i) the primary sequence constraints that organize the interaction between the SECIS element and the UGA through Sec insertion machinery and translation apparatus would certainly be more efficient than an interaction relying on the secondary structure; (ii) the orientation of the SECIS element relative to the UGA codon as well as translating ribosomes was likely not optimal; and (iii) the coupled transcription and translation in bacteria might make the SECIS element unavailable for translation of some polypeptides synthesized from the GPx4 mRNA. The low efficiency of Sec insertion by the 3′-UTR SECIS element suggests that the relocation of the SECIS element into the 3′-UTR is not favorable in bacteria. However, the data also show that such an evolutionary event is possible.
We found that the opal suppression by tRNATrp in response to UGA occurred at a rate much higher than the rate of Sec insertion. The nonsense codon suppression has long been known in both bacteria and eukaryotes (32,33). The context of the sequence that flanks UGA as well as the secondary structures of the mRNA determine the efficiency of opal suppression (26,34). The high efficiency of opal suppression in our study suggests that it efficiently competed with the translation termination process. It would be interesting to determine what percentage of the opal suppression is observed in natural selenoproteins in vivo and what the functional consequences of opal suppression in selenoproteins are.
Expression of heterologous proteins in E.coli has become a routine procedure in biochemistry and molecular biology. However, the incompatibility of SECIS elements and their distinct locations in eukaryotes and archaea on one hand, and bacteria on the other hand, generally preclude heterologous expression of selenoproteins in bacteria. A bacterial SECIS element can be designed downstream of UGA in selenoproteins, but it requires changes in amino acid sequences and is feasible only if Sec is located close to the C-terminus (35–37). Since Sec is often found in the enzyme active sites (12,38,39), changes in the flanking sequences are not desirable for functional characterization of these proteins. Our study shows that, in principle, it is possible to utilize SECIS elements in non-coding regions to direct Sec insertion in E.coli. Such a technique may find applications in studies of mammalian and other selenoproteins. Although, at present, the low efficiency of Sec insertion and opal suppression pose challenges, these problems might be alleviated if the technique is improved. For example, positioning a SECIS element within the 5′-UTR might increase the efficiency of Sec incorporation as it solves the transcription/translation coupling problem. At least one archaeal SECIS element is located in the 5′-UTR (40). In addition, the tryptophan insertion could potentially be suppressed (26) and orientation and spatial proximity between SECIS element and UGA adjusted.
Acknowledgments
We thank Donna Driscoll for providing anti-GPx4 antibodies and Elias Arnér for providing pSUABC plasmid. We also thank Jiong Yu and the Redox Biology Center mass-spectrometry facility for help with mass spectrometry. This work was supported by NIH grant GM061603. Funding to pay the Open Access publication charges for this article was provided by GM061603.
Conflict of interest statement. None declared.
REFERENCES
- 1.Chambers I., Frampton J., Goldfarb P., Affara N., McBain W., Harrison P.R. The structure of the mouse glutathione peroxidase gene: the selenocysteine in the active site is encoded by the ‘termination’ codon, TGA. EMBO J. 1986;5:1221–1227. doi: 10.1002/j.1460-2075.1986.tb04350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Söll D. Genetic code: enter a new amino acid. Nature. 1988;331:662–663. doi: 10.1038/331662a0. [DOI] [PubMed] [Google Scholar]
- 3.Leinfelder W., Zehelein E., Mandrand-Berthelot M.A., Böck A. Gene for a novel tRNA species that accepts l-serine and cotranslationally inserts selenocysteine. Nature. 1988;331:723–725. doi: 10.1038/331723a0. [DOI] [PubMed] [Google Scholar]
- 4.Lee B.J., Worland P.J., Davis J.N., Stadtman T.C., Hatfield D.L. Identification of a selenocysteyl-tRNA(Ser) in mammalian cells that recognizes the nonsense codon, UGA. J. Biol. Chem. 1989;264:9724–9727. [PubMed] [Google Scholar]
- 5.Sturchler C., Westhof E., Carbon P., Krol A. Unique secondary and tertiary structural features of the eucaryotic selenocysteine tRNA(Sec) Nucleic Acids Res. 1993;21:1073–1079. doi: 10.1093/nar/21.5.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forchhammer K., Leinfelder W., Böck A. Identification of a novel translation factor necessary for the incorporation of selenocysteine into protein. Nature. 1989;342:453–456. doi: 10.1038/342453a0. [DOI] [PubMed] [Google Scholar]
- 7.Fagegaltier D., Hubert N., Yamada K., Mizutani T., Carbon P., Krol A. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 2000;19:4796–4805. doi: 10.1093/emboj/19.17.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Copeland P.R., Fletcher J.E., Carlson B.A., Hatfield D.L., Driscoll D.M. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 2000;19:306–314. doi: 10.1093/emboj/19.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tujebajeva R.M., Copeland P.R., Xu X.M., Carlson B.A., Harney J.W., Driscoll D.M., Hatfield D.L., Berry M.J. Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Rep. 2000;1:158–163. doi: 10.1093/embo-reports/kvd033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zinoni F., Heider J., Böck A. Features of the formate dehydrogenase mRNA necessary for decoding of the UGA codon as selenocysteine. Proc. Natl Acad. Sci. USA. 1990;87:4660–4664. doi: 10.1073/pnas.87.12.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berry M.J., Banu L., Chen Y.Y., Mandel S.J., Kieffer J.D., Harney J.W., Larsen P.R. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature. 1991;353:273–276. doi: 10.1038/353273a0. [DOI] [PubMed] [Google Scholar]
- 12.Hatfield D.L., Gladyshev V.N. How selenium has altered our understanding of the genetic code. Mol. Cell. Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin G.W., III, Harney J.W., Berry M.J. Selenocysteine incorporation in eukaryotes: insights into mechanism and efficiency from sequence, structure, and spacing proximity studies of the type 1 deiodinase SECIS element. RNA. 1996;2:171–182. [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z., Reches M., Groisman I., Engelberg-Kulka H. The nature of the minimal ‘selenocysteine insertion sequence’ (SECIS) in Escherichia coli. Nucleic Acids Res. 1998;26:896–902. doi: 10.1093/nar/26.4.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kryukov G.V., Castellano S., Novoselov S.V., Lobanov A.V., Zehtab O., Guigo R., Gladyshev V.N. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 16.Copeland P.R., Driscoll D.M. Purification, redox sensitivity, and RNA binding properties of SECIS-binding protein 2, a protein involved in selenoprotein biosynthesis. J. Biol. Chem. 1999;274:25447–25454. doi: 10.1074/jbc.274.36.25447. [DOI] [PubMed] [Google Scholar]
- 17.Rother M., Resch A., Wilting R., Böck A. Selenoprotein synthesis in archaea. Biofactors. 2001;14:75–83. doi: 10.1002/biof.5520140111. [DOI] [PubMed] [Google Scholar]
- 18.Böck A. Selenium metabolism in bacteria. In: Hatfield D.L., editor. Selenium: Its Molecular Biology and Role in Human Health. Norwell: Kluwer Academic Publishers; 2001. p. 7. [Google Scholar]
- 19.Leibundgut M., Frick C., Thanbichler M., Böck A., Ban N. Selenocysteine tRNA-specific elongation factor SelB is a structural chimaera of elongation and initiation factors. EMBO J. 2005;24:11–22. doi: 10.1038/sj.emboj.7600505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zavacki A.M., Mansell J.B., Chung M., Klimovitsky B., Harney J.W., Berry M.J. Coupled tRNA(Sec)-dependent assembly of the selenocysteine decoding apparatus. Mol. Cell. 2003;11:773–781. doi: 10.1016/s1097-2765(03)00064-9. [DOI] [PubMed] [Google Scholar]
- 21.Arnér E.S., Sarioglu H., Lottspeich F., Holmgren A., Böck A. High-level expression in Escherichia coli of selenocysteine-containing rat thioredoxin reductase utilizing gene fusions with engineered bacterial-type SECIS elements and co-expression with the selA, selB and selC genes. J. Mol. Biol. 1999;292:1003–1016. doi: 10.1006/jmbi.1999.3085. [DOI] [PubMed] [Google Scholar]
- 22.Heider J., Baron C., Böck A. Coding from a distance: dissection of the mRNA determinants required for the incorporation of selenocysteine into protein. EMBO J. 1992;11:3759–3766. doi: 10.1002/j.1460-2075.1992.tb05461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandman K.E., Tardiff D.F., Neely L.A., Noren C.J. Revised Escherichia coli selenocysteine insertion requirements determined by in vivo screening of combinatorial libraries of SECIS variants. Nucleic Acids Res. 2003;31:2234–2241. doi: 10.1093/nar/gkg304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müller S., Heider J., Böck A. The path of unspecific incorporation of selenium in Escherichia coli. Arch. Microbiol. 1997;168:421–427. doi: 10.1007/s002030050517. [DOI] [PubMed] [Google Scholar]
- 25.Sambol A.R., Dubes G.R. Phenotypic suppression of opal mutants by l-tryptophan. Microbios. 1984;39:19–27. [PubMed] [Google Scholar]
- 26.Sandman K.E., Noren C.J. The efficiency of Escherichia coli selenocysteine insertion is influenced by the immediate downstream nucleotide. Nucleic Acids Res. 2000;28:755–761. doi: 10.1093/nar/28.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Böck A., Forchhammer K., Heider J., Leinfelder W., Sawers G., Veprek B., Zinoni F. Selenocysteine: the 21st amino acid. Mol. Microbiol. 1991;5:515–520. doi: 10.1111/j.1365-2958.1991.tb00722.x. [DOI] [PubMed] [Google Scholar]
- 28.Hüttenhofer A., Heider J., Böck A. Interaction of the Escherichia coli fdhF mRNA hairpin promoting selenocysteine incorporation with the ribosome. Nucleic Acids Res. 1996;24:3903–3910. doi: 10.1093/nar/24.20.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suppmann S., Persson B.C., Böck A. Dynamics and efficiency in vivo of UGA-directed selenocysteine insertion at the ribosome. EMBO J. 1999;18:2284–2293. doi: 10.1093/emboj/18.8.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stadtman T.C. Selenocysteine. Annu. Rev. Biochem. 1996;65:83–100. doi: 10.1146/annurev.bi.65.070196.000503. [DOI] [PubMed] [Google Scholar]
- 31.Kryukov G.V., Gladyshev V.N. The prokaryotic selenoproteome. EMBO Rep. 2004;5:538–543. doi: 10.1038/sj.embor.7400126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gesteland R.F., Atkins J.F. Recoding: dynamic reprogramming of translation. Annu. Rev. Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- 33.Namy O., Rousset J.P., Napthine S., Brierley I. Reprogrammed genetic decoding in cellular gene expression. Mol. Cell. 2004;13:157–168. doi: 10.1016/s1097-2765(04)00031-0. [DOI] [PubMed] [Google Scholar]
- 34.Miller J.H., Albertini A.M. Effects of surrounding sequence on the suppression of nonsense codons. J. Mol. Biol. 1983;164:59–71. doi: 10.1016/0022-2836(83)90087-6. [DOI] [PubMed] [Google Scholar]
- 35.Arnér E.S. Recombinant expression of mammalian selenocysteine-containing thioredoxin reductase and other selenoproteins in Escherichia coli. Methods Enzymol. 2002;347:226–235. doi: 10.1016/s0076-6879(02)47022-x. [DOI] [PubMed] [Google Scholar]
- 36.Bar-Noy S., Moskovitz J. Mouse methionine sulfoxide reductase B: effect of selenocysteine incorporation on its activity and expression of the seleno-containing enzyme in bacterial and mammalian cells. Biochem. Biophys. Res. Commun. 2002;297:956–961. doi: 10.1016/s0006-291x(02)02314-8. [DOI] [PubMed] [Google Scholar]
- 37.Kim H.Y., Gladyshev V.N. Methionine sulfoxide reduction in mammals: characterization of methionine-R-sulfoxide reductases. Mol. Biol. Cell. 2004;15:1055–1064. doi: 10.1091/mbc.E03-08-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gladyshev V.N., Hatfield D.L. Selenocysteine-containing proteins in mammals. J. Biomed. Sci. 1999;6:151–160. doi: 10.1007/BF02255899. [DOI] [PubMed] [Google Scholar]
- 39.Behne D., Kyriakopoulos A. Mammalian selenium-containing proteins. Annu. Rev. Nutr. 2001;21:453–473. doi: 10.1146/annurev.nutr.21.1.453. [DOI] [PubMed] [Google Scholar]
- 40.Wilting R., Schorling S., Persson B.C., Böck A. Selenoprotein synthesis in archaea: identification of an mRNA element of Methanococcus jannaschii probably directing selenocysteine insertion. J. Mol. Biol. 1997;266:637–641. doi: 10.1006/jmbi.1996.0812. [DOI] [PubMed] [Google Scholar]