Summary
Growing evidence indicates that transposable elements (TEs) play important roles in evolution by providing genomes with coding and non-coding sequences. Identification of TE-derived functional elements, however, has relied on TE annotations in individual species, which limits its scope to relatively intact TE sequences. Here, we report a novel approach to uncover previously unannotated degenerate TEs (degTEs) by probing multiple ancestral genomes reconstructed from hundreds of species. We applied this method to the human genome and achieved a 10.8% increase in coverage over the most recent annotation. Further, we discovered that degTEs contribute to various cis-regulatory elements and transcription factor binding sites, including those of a known TE-controlling family, the KRAB zinc-finger proteins. We also report unannotated chimeric transcripts between degTEs and human genes expressed in embryos. This study provides a novel methodology and a freely available resource that will facilitate the investigation of TE co-option events on a full scale.
Keywords: transposable elements, transposon, gene regulation, genome evolution, KRAB zinc-finger protein, endogenous retrovirus, cis-regulatory element, promoter, enhancer, splice variant
Graphical abstract
Highlights
-
•
A transposon annotation pipeline through probing reconstructed ancestral genomes
-
•
Identification of previously unannotated degenerate transposable elements (degTEs)
-
•
DegTEs contribute various regulatory elements and transcription factor binding sites
-
•
DegTEs form non-canonical chimeric transcripts with the host genes
Transposable elements (TEs) are selfish genetic elements and have invaded our genomes throughout evolution. Matsushima et al. developed a computational method to annotate TEs by probing reconstructed ancestral genomes and discovered that a large number of functional sequences are derived from previously unannotated ancient and highly degenerate TEs.
Introduction
Transposable elements (TEs) are genetic units capable of mobilizing their sequence within the host genome. In the most recent telomere-to-telomere complete human genome assembly, 54% of genomic DNA is estimated to be derived from TEs and repetitive elements, consisting mostly of long terminal repeats (LTRs), long and short interspersed nuclear elements (LINEs and SINEs, respectively), and DNA transposons.1 However, the vast majority of the sequences in the human genome annotated as TEs are transposition incompetent due to mutations including insertions/deletions accumulated during evolution, and only an estimated 1:1,000 integrants belonging to L1, Alu, SVA, or HERV-K remain active.2
It has become apparent that transposition-incompetent TEs are not just “fossils” of parasitic sequences. Indeed, TEs are frequently co-opted as regulatory elements (reviewed in Fueyo et al.3) and, at times, even give rise to new genes (reviewed in Jangam et al.4 and Modzelewski et al.5). TEs are also spliced into genic transcripts, resulting in TE-gene chimeric transcripts, or transpochimeric gene transcripts (TcGTs), often produced in specific cell types as alternatives to canonical transcripts.6,7 Since each lineage has acquired distinct TE subfamilies during evolution, TEs contribute to lineage-specific genomic innovations.
Currently, the gold-standard method to annotate TEs in an assembled genome is repository-based annotation, best exemplified by RepeatMasker.8 This software scans a genome and identifies loci that significantly resemble one of the consensus sequences registered on a TE repository such as Repbase9 (https://www.girinst.org/repbase/) or Dfam10 (https://www.dfam.org/). Thus, this methodology is inherently limited to discovery of TEs that still retain a high sequence similarity to the consensus and might miss highly degenerate TEs, typically evolutionarily old elements.
Recent efforts have provided whole-genome sequences from an increasing number of species, including more than 600 vertebrates.11,12 Armstrong et al. described a scalable method to align large numbers of genomes through the reconstruction of their ancestral sequences.13 These reconstructed ancestral genomes (RAGs) are the best proxy to the genomes of extinct common ancestors, which are otherwise challenging to obtain because of difficulties in tracing back lineages and assembling sequences obtained from fossilized materials. Since RAGs are generated correcting for later-occurring mutational changes based on phylogeny, the sequences of their TEs are, in theory, closer to the consensus. This was in part confirmed for the L1PA6 subfamily13 but has not been verified for other TEs.
Here, we present a novel method that utilizes multiple RAGs to identify TEs that escape common annotation techniques. It allowed us to discover previously unannotated integrants belonging to all the major TE classes in the human genome, extending its total TE coverage by 10.8%. We further found these newly unearthed TEs to contribute various cis-regulatory elements (CREs) as well as transcription factor binding sites (TFBSs) and to form chimeric transcripts with human genes expressed at specific embryonic stages.
Design
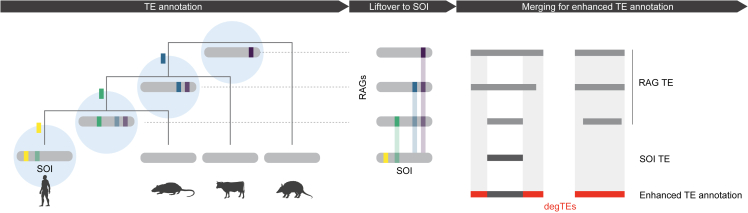
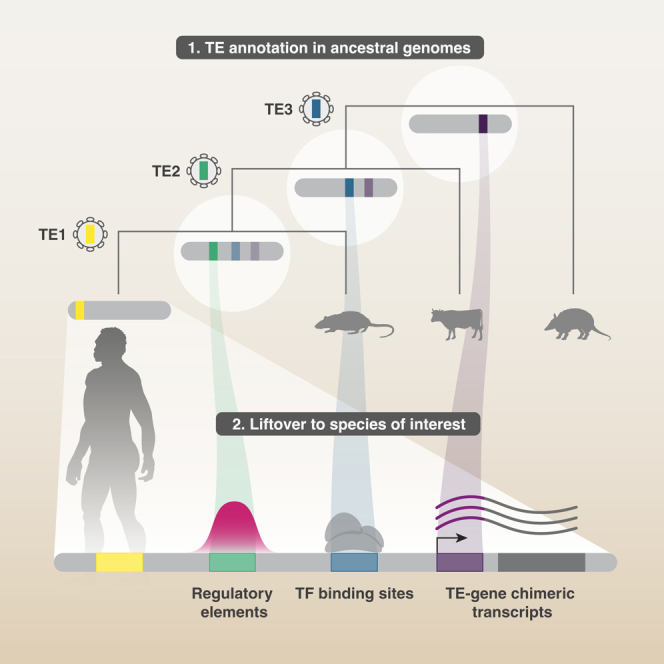
We present a versatile method to identify degTEs using multiple RAGs (Figure 1). While TE integrants detected in older ancestral genomes are expected to be closer to the consensus because of a shorter divergence time since the initial emergence of the corresponding TE subfamilies, RAGs are devoid of TE subfamilies that appeared later in evolution (e.g., the primate ancestral genome does not harbor human-specific TEs). Thus, to recover TEs that emerged at wide-ranged evolutionary time points, multiple RAGs ancestral to a species of interest (SOI) are used. First, TEs are annotated in the RAGs and in the SOI. The RAG TEs are then lifted over to the SOI genome to find their corresponding sites. Finally, the lifted-over TEs are compared against the SOI TEs. If they do not overlap, the lifted-over TEs are added as novel elements, and the ones with a partial overlap are merged to extend existing integrants in the SOI genome.
Figure 1.
Overview of the RAG-enhanced TE annotation
Schematic representation of the workflow to achieve enhanced TE annotation by utilizing multiple RAGs. Firstly, TEs are annotated in the species of interest (SOI) as well as in RAGs (blue circles). TEs annotated in RAGs are then lifted over to corresponding regions in SOI. Finally, lifted-over TE annotations not overlapping TEs in SOI (degTEs) are either merged to extend existing annotations (left) or are added as new integrants (right). TEs inserted at different evolutionary time points are shown as colored boxes, with transparency indicating their divergence level from the consensus.
Results
TE annotation in RAGs
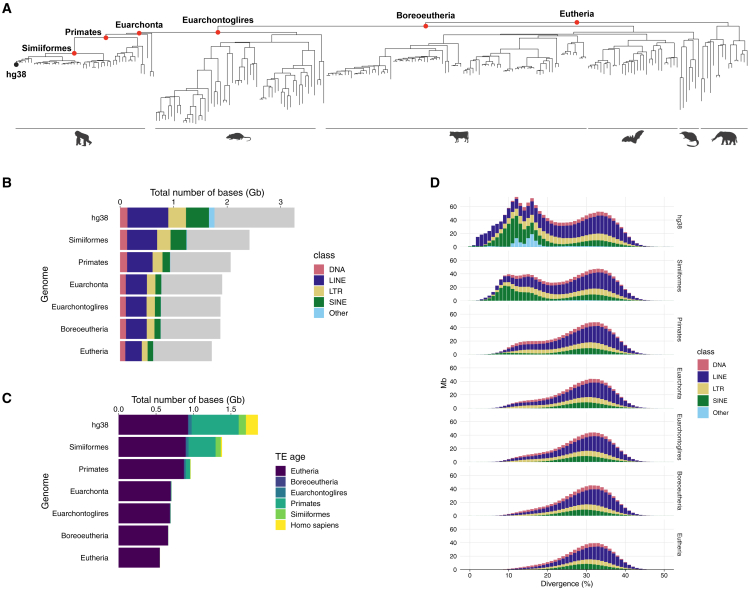
As a proof of principle, we enhanced TE annotation in the human genome assembly GRCh38 (hg38). We chose six RAGs ranging between the oldest available, Eutheria, and the simian common ancestor, namely Simiiformes, primates, Euarchonta, Euarchontoglires, Boreoeutheria, and Eutheria common ancestors (Figure 2A). Employing the same RepeatMasker parameters, we annotated TEs in hg38 (hereafter, hg38 TE) and in the RAGs, 30%–50% of which were attributed to TEs (Figure 2B). We found that the vast majority of the TEs identified in each RAG were older than their host genome as anticipated (Figure 2C). Furthermore, TE divergence profiles revealed that a peak (approximately 10%–20% divergence) corresponding mostly to evolutionarily young L1, Alu, and SVA elements was only seen in hg38 and the simian RAG and was absent in the older RAGs (Figure 2D; Data S1). Together, these results suggest that reconstruction and annotation of TEs were achieved in the RAGs without a high degree of false positives.
Figure 2.
Characterizations of TEs annotated in the RAGs
(A) A phylogenetic tree of 241 species used to reconstruct the ancestral genomes (red points) for enhanced hg38 TE annotation.
(B) Coverage of TE classes detected in each RAG and in hg38. Gray bars represent the non-TE regions in each genome.
(C) TE age proportions in hg38 and the RAGs. TEs that emerged outside of these six age categories were classified into the next younger age category (e.g., TEs born between primates and Simiiformes common ancestors were classified as Simiiformes).
(D) Distribution of divergence levels of TE integrants in hg38 and the RAGs. The divergence level was rounded to a nearest integer.
See also Data S1.
RAGs enhanced human TE annotation by unbiased identification of degTEs
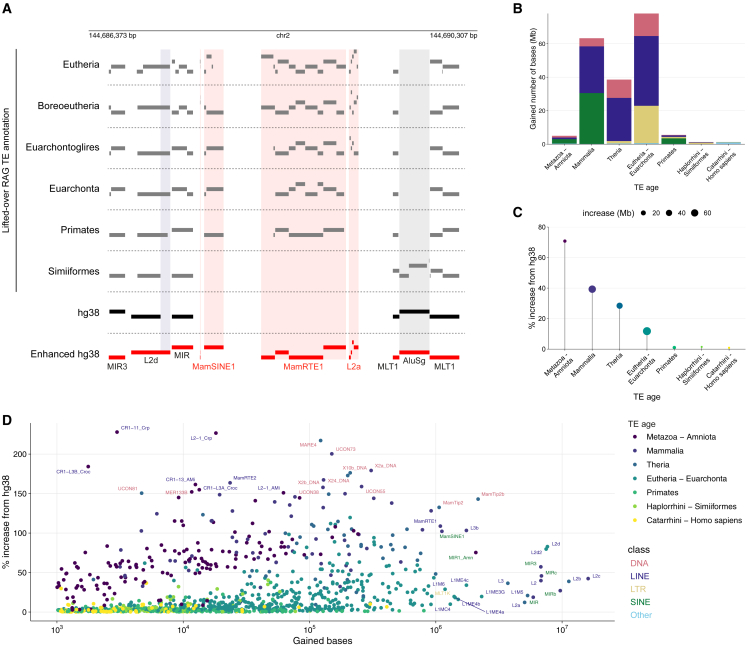
For the TE annotation of each RAG, corresponding genomic loci in hg38 were sought using halLiftover14 (Figure S1A). Of the TEs found in each RAG, between 60 and 130 Mb corresponded to genomic sequences unassigned to TEs in hg38, which, when combined, amounted to 191 Mb additional sequences annotated as TEs (Figure S1B). We will refer to such newly identified degenerate TE-derived elements in hg38 as degTEs for simplicity (Figure 1). As shown in Figure 3A as an example genomic locus, degTEs either represent newly discovered integrants (MamSINE1, MamRTE1, and L2a) or extend already annotated inserts (L2d). Also, each integrant was discovered in distinct sets of RAGs. The MamRTE1 integrant was detected in a relatively young RAG, the primate common ancestor, while the L2a insert was identified only when going up to the Euarchontoglires common ancestor, presumably because genetic drift erased their signature features after that. Conversely, the primate-specific AluSg integrant was not annotated in the RAGs older than the simian common ancestor, which suggests that it inserted in the middle of the MLT1 element between the primate and simian common ancestors.
Figure 3.
Summary statistics of degTEs
(A) A representative genomic locus where degTEs were identified. Red bars represent enhanced TE annotation produced by merging hg38 TE (black bars) and lifted-over RAG TE (gray bars) annotations. Newly identified integrants are highlighted in red shades, and those extending existing hg38 TE annotations are shown in blue shades. The gray shade represents an integrant belonging to a primate-specific subfamily, AluSg. TE segments are visualized in multiple rows to indicate breaks of the segments.
(B and C) Evolutionary age distribution of degTEs (B) and their percentage of increase from the already annotated TEs in hg38 (C).
(D) For the TE subfamilies that gained more than 1 kb by the enhanced annotation, the number of bases gained and percentage of increase relative to the hg38 annotation are plotted with a color representing the TE age. The subfamilies that gained either more than 1 Mb or 150% are labeled with their subfamily name in a color of a corresponding TE class.
See also Figures S1–S5 and Data S2.
The enhanced annotation, a union of degTEs and hg38 TEs, increased TE coverage in the human genome by 10.8% without major changes in the proportions of TE classes (Figure S1C). The vast majority of degTEs were found in either intronic or intergenic regions, similarly to TEs already annotated in hg38 (Figure S1D). They are distributed across all chromosomes (Figure S1E, with a weak but significant bias (p < 2.2e−16) toward regions harboring fewer TEs already annotated in hg38 (Figure S1F). We further estimated a false positive rate (FPR) of our method by defining genomic regions that are unlikely to be of TE origin as negative control regions and by calculating how often such regions are falsely called degTEs with our method (Figure S2A). With this, we estimated the FPR to be 0.56%, which is only 0.16% higher than that of RepeatMasker alone15 (Figure S2B). Additionally, by using the frequency of degTEs not matching the age of RAGs in which they were found (Figure S2C), we approximated the false discovery rate (FDR) to be 1.6% (Figure S2D). In both cases, LTR elements significantly contribute to false positives (p < 0.001; Figures S2E, and S2F), which suggests that care must be taken when analyzing LTR degTEs.
97% of the degTEs are derived from TE subfamilies that emerged sometime between the mammalian and Eutherian common ancestors (Figure 3B; Data S1), and the enhanced annotation achieved 10%–70% increase in coverage for these ancient TEs from hg38 (Figure 3C). In particular, the L2 and MIR subfamilies that emerged during this evolutionary time window significantly contribute to the new annotation, each gaining approximately an additional 10 Mb (Figure 3D; Data S2). Noteworthily, several ancient DNA transposon subfamilies increased by more than 150% in the enhanced annotation relative to the hg38 TE annotation.
Next, we tested how much sequence similarity each degTE retained compared to the corresponding inserts found in the RAGs. For this, we searched degTE sequences across the Dfam TE library and calculated homology scores without the inclusion threshold imposed when annotation was performed with RepeatMasker. This analysis revealed that more than 20% of the degTE integrants displayed significant homology to the same subfamily as the one found for the corresponding sequences in the RAGs, whereas the remaining elements were either assigned to a different TE subfamily or showed no significant homology to any TEs (Figure S3A). The homology scores obtained for degTEs were expectedly lower than those of the corresponding TEs in the RAGs (Figure S3B). Also, we observed that degTE L2c elements exhibited similar coverage to those already annotated in hg38. The observed 5′ truncation is known to be a typical feature for LINE integrants16 (Figure S3C).
Finally, we tested the robustness of the pipeline by running it with a different TE annotation tool and by applying it on other species. Instead of RepeatMasker, we used another reference-based annotation tool specialized in SINE annotation, SINEBase,17 to enhance hg38 TE annotation. While it correctly identified abundant Alu elements in hg38 (Figure S4A), it added SINE degTEs, which are mostly evolutionarily old Ther-1 (MIR) and Ther-2 (MIR3), each gaining 3–5 times the coverage in hg38 (Figure S4B). The vast majority of these degTEs were also found with RepeatMasker (Figure S4A). To examine the applicability of the method to other genomes, we performed the enhancement on three species from distinct taxa, namely mouse (Mus musculus), dog (Canin lupus familialis), and armadillo (Dasypus novemcinctus), using the same RAG (Figures S5A and S5B). Intriguingly, the enhancements achieved in the three additional species were higher than in the human genome, ranging between a 23% and 33% increase in coverage, and were proportional neither to the genome sizes nor to the already annotated TE coverage in the genome (Figure S5C).
Together, these results show that our novel method robustly achieved enhanced TE annotation by recovering evolutionarily ancient TEs that exhibited higher homology to the consensus in the RAGs with low FPRs and FDRs. Importantly, while some (∼20%) degTEs may be discovered by simply running RepeatMasker with a lower threshold, the remaining elements will not, presumably because of more extensive mutational changes.
degTEs contribute regulatory elements and TFBSs
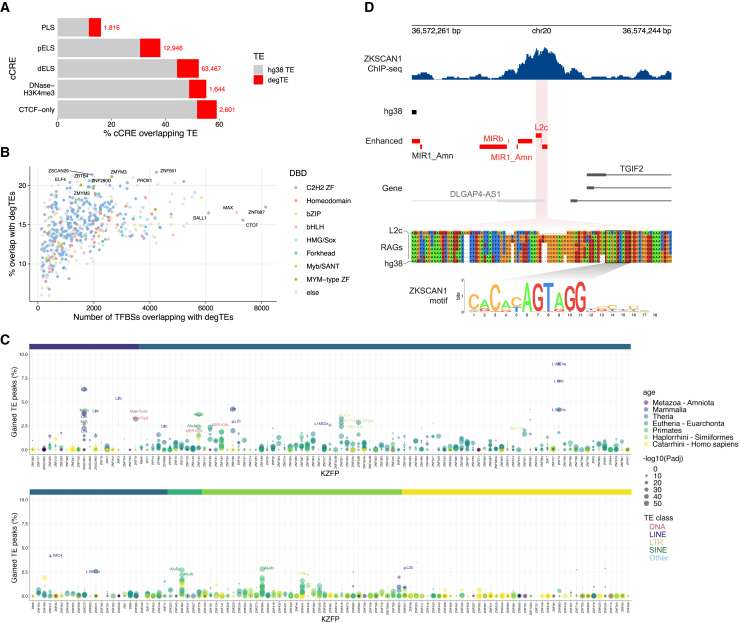
Given that TE-derived CREs are prevalent and play crucial gene regulatory roles, we next tested if the discovered degTEs contribute CREs to the human genome. We crossed the enhanced hg38 TE annotation with the genome-wide candidate CRE (cCRE) annotation, which was defined based on epigenetic signatures in multiple cell types.18 In addition to the cCREs overlapping with previously annotated hg38 TEs, we identified 82,474 cCREs associated with degTEs, which corresponds to ∼8% of each cCRE group (Figure 4A).
Figure 4.
cCREs and TFBSs derived from degTEs
(A) Percentage of cCREs overlapping the hg38 TE and degTEs. The red numbers indicate cCREs that overlap with one or more degTEs. PLS, promoter-like signatures; pELS, proximal enhancer-like signatures; dELS, distal enhancer-like signatures.
(B) ENCODE TFBSs overlapping with one or more degTEs. The colors represent DNA-binding domains of the TFs.
(C) A summary of degTE-overlapping KZFP binding sites shown in percentage relative to the total number of peaks. Each point represents a TE subfamily and its color and size indicate its evolutionary age and an adjusted enrichment p-value, respectively. The same color scale is used for the ribbon above to indicate the ages of the KZFPs.
(D) A representative genomic region where a degTE-derived KZFP binding site was found. A multiple sequence alignment is shown for the newly found L2c element in the middle of the ZKSCAN1 peak and its corresponding sequences in the RAGs together with the L2c consensus. The sequence matching the ZKSCAN1 binding motif is highlighted.
Next, to see if the degTEs contribute TFBSs, we crossed the enhanced TE annotation with the ENCODE TFBS data. We identified ∼6,000 degTE-associated TFBSs for each TF (Figure 4B; Data S3). Among the TFs with the highest proportion of degTE-derived TFBSs, we found the co-repressor of repressor element-1 silencing transcription (CoREST) complex components ZMYM2 and its paralog, ZMYM3.19 ZMYM2 has been reported to be involved in TE silencing through its interaction with TE DNA,20,21 which may explain its frequent association with degTEs.
KRAB-domain-containing zinc-finger proteins (KZFPs) are major TFs that recognize TEs, usually binding TE subfamilies of a matching evolutionary age.22,23 We thus tested if some of the degTEs are also bound by KZFPs. We compared degTE annotation with the chromatin immunoprecipitation sequencing (ChIP-seq) peaks from 210 KZFPs displaying significant enrichment for one of the TE subfamilies in the human genome.23 In line with the previously observed association between the ages of KZFPs and their target TEs,23 degTE-overlapping peaks are more prevalent among evolutionarily old KZFPs (Figure S6). Moreover, we found that the binding sites of KZFPs that were previously identified as targeting evolutionarily old TE subfamilies overlap often with degTEs belonging to the same subfamilies23 (Figure 4C). To further probe the evolution of the KZFP-bound degTEs, we focused on a KZFP, ZKSCAN1, which significantly binds elements belonging to the L2 family (adjusted p < 2.2e−308). We found a ZKSCAN1 binding site centered on a L2c degTE (Figure 4D). A multiple sequence alignment of the degTE and corresponding RAG sequences revealed that the latter were indeed closer to the L2c consensus. However, interestingly, the region corresponding to the ZKSCAN1-binding motif is highly conserved across the RAGs and hg38, suggesting that it has been under purifying selection more than the rest of this TE sequence.
Thus, the recovery of degTEs through the analysis of RAGs revealed the previously unknown association of cCREs and TFBSs with TEs.
degTEs contribute to chimeric transcripts
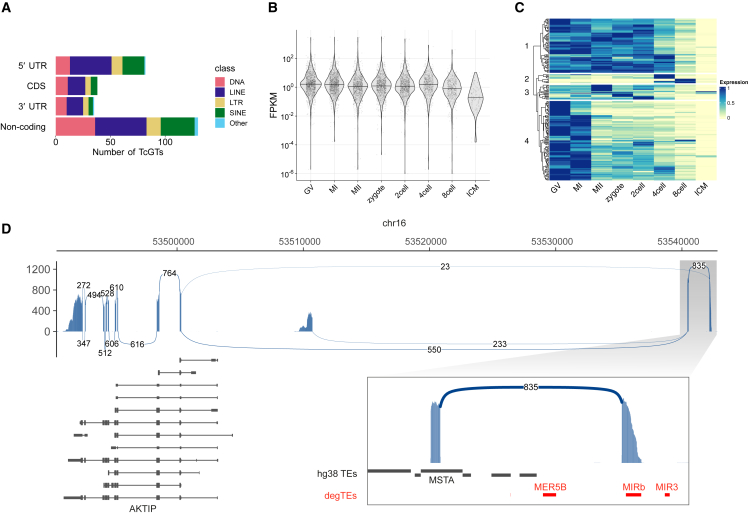
In addition to regulatory elements discussed above, TEs, including transposition-incompetent integrants, are known to contribute novel sequences to transcripts of the host. Non-canonical chimeric transcripts between TEs and genes, or TcGTs, have been detected in various cell types.24 A TE-derived sequence in a TcGT can initiate transcription,25 insert a non-canonical coding sequence,26 and serve as a microRNA target.27 Especially in the mammalian embryos, TEs have been reported to define stage-specific expression of the host genes by forming TE-driven TcGTs.6,7 To test if degTEs are also involved in such TcGTs expressed in the human embryos, we sought for non-canonical transcripts involving degTEs using previously published RNA sequencing data from human embryos,28 which led us to identify 222 non-canonical exons that involve at least one degTE, most of which contribute non-coding exons (Figures 5A and S7A; Data S4). Many TcGTs are highly expressed until the 8-cell stage (Figures 5B and 5C) and constitute major (>50%) isoforms in their host genes (Figure S7B). In agreement with previous reports, degTE-associated TcGTs exhibit distinct expression in the developing embryo (Figure 5C). Figure 5D shows an example TcGT that initiates AKTIP transcription from a non-canonical exon derived from a MIRb degTE located 37 kb upstream of the first annotated exon of this gene. This indicates that some degTEs also form TcGTs by either driving their expression or being included as a non-canonical exon.
Figure 5.
DegTE-gene chimeric transcripts expressed during human embryonic development
(A) Summary of relative positions of degTE-derived exons in TcGTs.
(B) The expression distributions of degTE-associated TcGTs during human embryonic development. GV, germinal vesicle oocyte; MI, metaphase I oocyte; MII, metaphase II oocyte; ICM, inner cellular mass.
(C) A heatmap summarizing expression changes of degTE-associated TcGTs during embryonic development. Expression levels are normalized by their maximum expression level. The heatmap was split into four clusters (the numbers shown on the left) based on k-mer clustering.
(D) A Sashimi plot indicating the number of splicing events observed for a gene, AKTIP, where a chimeric transcript with a degTE, MIRb, was found. Splice junctions with more than 10 supporting reads are shown.
Discussion
Upon discovering TEs in the middle of the 20th century, Barbara McClintock coined the term controlling elements for the regulatory potential of TEs,29 and two decades later, Eric Davidson and Roy Britten postulated that they play fundamental roles in building regulatory networks30,31. Accumulating evidence has now validated this hypothesis, demonstrating that TE-derived proteins, non-coding transcripts, and various cis-acting elements are key to the evolution of genome regulation and architecture (reviewed in Fueyo et al.3 and Jangam et al.4).
To study these TE co-option events, TE annotation plays a central role. TE annotation can be either repository based or performed de novo (reviewed in Goerner-Potvin and Bourque32). The former most commonly uses a software called RepeatMasker, which annotates TEs by looking for sequences that resemble those cataloged in a reference repeat database such as Repbase or Dfam. Thus, inherently, this method can only detect TEs with a reasonably high similarity to the consensus sequences. Also, since sequences are annotated based solely on a given genome, mutations gained over evolution are not taken into consideration, which may result in false classifications. On the other hand, de novo annotation is generally considered to be more robust to sequence variations from the consensus and thus is more likely to discover a higher load of TEs than repository-based annotation for a given genome. However, it is also prone to a high FPR. A report estimates that P-clouds,33 which predicted the highest number of elements in the human genome, may entail an FPR of ∼60%, which is much higher than the estimated 0.4% FPR of RepeatMasker.15
In this study, we describe a novel method to obtain a more complete TE annotation by incorporating information from multiple RAGs while still utilizing a commonly used repository-based annotation method. This allows for not only more sensitive recovery of degTEs but also more interpretable results of newly annotated TEs since corresponding sequences in RAGs are available with nucleotide-resolution alignment to the consensus sequences. Our method may also be useful in updating already-annotated TE integrants based on the annotation of corresponding sequences in RAGs, as these are closer to the ancestral state of the integrants. Importantly, we estimated infrequent false positives of the method (FPR = 0.56% and FDR = 1.6%; Figure S2).
Ancestral reconstructions of several TE subfamilies have been achieved using multiple integrants either within or across species.34,35,36 Campitelli et al. took TE reconstruction to the next level by employing ancestral genome reconstruction and achieved the reconstruction of the full-length L1 progenitor sequence.37 However, to our knowledge, our study is the first attempt where whole RAGs were used to mine degTEs in a genome of interest. We demonstrated that our approach is applicable to different TE annotation methods (Figure S4) and species (Figure S5), though high-quality RAGs would be critical in achieving accurate degTE annotation. Thus, although we provide a pipeline that employs RepeatMasker for TE annotation and six RAGs ranging between primate and Eutherian common ancestors, these should be adjusted to the research questions asked. For instance, if enhanced annotation of a certain TE class is desired, then there are several other software tools that are optimized for this purpose.32,38 Also, if evolutionary trajectories of TE subfamilies at a higher resolution in a specific clade are sought (e.g., Alu subfamilies in primates), then more RAGs within the clade should be screened.
The largest increase in the enhanced annotation was seen for the L2 subfamilies, each gaining approximately 10 Mb degTEs corresponding to ∼90% from the current hg38 annotation (Figure 3D). Multiple studies have reported an enrichment of L2 elements in various CREs, including promoters, enhancers, and DNA loop anchors.39,40 Also, L2 elements have been reported to be a source of microRNAs and their target sites.41 Thus, newly annotated L2 elements in our enhanced annotation will help achieve a full grasp of the L2-mediated gene regulatory system.
Our approach identified a large number of cCREs, TFBSs, and TcGTs to be derived from old TEs, which has not been achieved with current hg38 TE annotation, further reinforcing the validity of Britten and Davidson’s original hypothesis on the role played by mobile genetic elements in the genome-wide dissemination of response-mediating sequences. In addition, our enhanced TE annotation method as well as the resulting improved human TE annotation would open up wide-ranged applications. For example, since this approach achieves locus-level reconstruction of each element, it will facilitate defining how individual TE subfamilies have propagated across genomes and how various TE-derived elements have evolved over time. Another potential approach to RAG-assisted enhanced TE annotation is to construct TE libraries de novo probing RAGs. This may discover previously unidentified TE clades that have long been extinct and are highly degenerate in many present-day genomes. With the ever-increasing number of sequenced genomes, RAGs will become more accurate, and older RAGs will be reconstructed, which will further extend the breadth of applications of this method.
Limitations
Our method relies on two major factors: reconstruction of ancestral genomes and TE annotation with RepeatMasker. For the former, although the ancestral genomes used were reconstructed from up to 241 genomes, the result still is only a prediction of the ancestral states, which might involve a certain degree of error and might affect the TE prediction accuracy at specific genomic loci. For the latter, any TE annotation software applies a threshold to call elements. Although RepeatMasker is no different and may report some false positives, the major benefit of our method is that the TEs in the SOI were also called with the same method and threshold. In other words, TEs found in RAGs and SOI are called with the same confidence.
We present a reference-based TE annotation in RAGs, as we believe that this provides the most comprehensible annotation of TEs. However, the use of consensus sequences constructed based on present-day genomes to probe ancestral genomes may in itself harbor limitations. It is likely that the consensus sequences generated this way contain mutations that are prevalent in modern genomes because they were important for the domestication of particular TEs but do not necessarily reflect their original sequences. This may explain why so few “young” or less-diverged TEs were discovered in the RAGs in our study (Figure 2D). This might be because the reconstructed TE sequences either appeared diverged despite being closer to the original state or were missed because they deviate significantly from the consensus. Ultimately, what might be required to further improve the quality of annotation in RAGs is reconstruction of full-length TE progenitor sequences for a number of TE families, as was done for L1.37
While our method is purely computational, relying only on non-commercial software, it requires a high-performance computing cluster to complete the analysis within a reasonable time frame. Especially, whole-genome TE annotation with RepeatMasker is computationally intensive, and it took us approximately ∼72 h with 24 CPUs (∼1,728 CPU h) for each genome, though this may change proportional to genome sizes.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| 241-way mammalian HAL alignment | Armstrong et al.13 | https://cglgenomics.ucsc.edu/data/cactus/ |
| Dfam family database (version 3.6) | Wheeler et al.10 | https://www.dfam.org/ |
| Enhanced TE annotations | This paper | Zenodo: https://doi.org/10.5281/zenodo.7716408 |
| RepeatMasker outputs | This paper | Zenodo: https://doi.org/10.5281/zenodo.7716408 |
| SINEBase | Vassetzky el al.17 | https://sines.eimb.ru |
| The human candidate cis-regulatory elements (cCREs) data (registry V3) | ENCODE Project Consortium18 | https://screen.encodeproject.org/index/cversions |
| ENCODE TF ChIP-seq peaks | ENCODE Project Consortium42 | Data S3 |
| Human embryo RNA-seq data | Zou et al.28 | GEO: GSE197265 |
| Software and algorithms | ||
| Code for enhancing TE annotation of a Eutherian genome | This paper | Zenodo: https://doi.org/10.5281/zenodo.7716408 |
| RepeatMasker (version 4.1.2-p1) | Smit et al.8 | http://www.repeatmasker.org; RRID:SCR_012954 |
| HAL tools (version 2.1) | Hickey et al.14 | https://github.com/ComparativeGenomicsToolkit/hal |
| BLAST+ (version 2.14.0) | Camacho et al.43 | https://blast.ncbi.nlm.nih.gov/Blast.cgi; RRID:SCR_004870 |
| bedtoolsr (version 2.30.0-1) | Patwardhan et al.44 | https://github.com/PhanstielLab/bedtoolsr |
| HMMER (version 3.3.2) | Wheeler et al.45 | http://hmmer.org; RRID:SCR_005305 |
| bedtools (version 2.30.0) | Quinlan et al.46 | https://bedtools.readthedocs.io/en/latest/; RRID:SCR_006646 |
| MAFFT (version 7.508) | Katoh et al.47 | https://mafft.cbrc.jp/alignment/software/; RRID:SCR_011811 |
| Jalview (version 2.11.2.6) | Waterhouse et al.48 | https://www.jalview.org/; RRID:SCR_006459 |
| pyTEnrich (version 0.6) | Alexandre Coudray | https://alexdray86.github.io/pyTEnrich/build/html/index.html |
| STAR (version 2.7.10b) | Dobin et al.49 | https://github.com/alexdobin/STAR; RRID:SCR_004463 |
| StringTie (version 2.2.1) | Pertea et al.50 | https://github.com/gpertea/stringtie; RRID:SCR_016323 |
| TransDecoder (version 5.7.1) | Haas51 | https://github.com/TransDecoder/TransDecoder; RRID:SCR_017647 |
| ggsashimi (version 1.1.5) | Garrido-Martín et al.52 | https://github.com/guigolab/ggsashimi |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Didier Trono (didier.trono@epfl.ch).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
(1)
RepeatMasker outputs and enhanced TE annotations have been deposited at Zenodo and are publicly available as of the date of publication. DOIs are listed in the key resources table. This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
-
(2)
All original code has been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
(3)
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Method details
TE annotation
Cactus genomic alignment HAL file was downloaded from the Cactus alignment project website13 (https://cglgenomics.ucsc.edu/data/cactus/). All the reconstructed ancestral genomes and the human genome were isolated from the HAL file using the hal2fasta script from HAL tools14 (version 2.1) (https://github.com/ComparativeGenomicsToolkit/hal). The corresponding node names of the RAGs used in this study are summarised in Table S1. Dfam family database10 (Dfam-p1_curatedonly.h5.gz, version 3.6) was downloaded from Dfam website (https://www.dfam.org/) and was used as a reference. Search engine HMMER and species “human” together with the options “-s -a -nolow” were used to run RepeatMasker (version 4.1.2-p1) for all the genomes used. RepeatMasker output files were converted to the BED file format for downstream analyses.
The computational pipeline to enhance hg38 TE annotation is shown in Figure S8 and its associated scripts are shared on Zenodo:https://doi.org/10.5281/zenodo.7716408. TE annotation for each ancestral genome was lifted over to the human genome using halLiftover from HAL tools (version 2.1). For the enrichment analyses shown below, to prevent inflated significant scores, the fragmented annotation after the halLiftover was merged. Specifically, TE annotations that are on the same strand, are within 100 bp from each other, and belong to the same subfamily were merged into a single continuous annotation. For the other statistics, the unmerged fragmented annotation was used.
For the TE annotation with SINEBase, we downloaded the SINE consensus FASTA file from the website (https://sines.eimb.ru). We first built database files of the consensus sequences with makeblastdb application (blast version 2.14.0).43 Using the database, we then scanned hg38 as well as the Eutherian RAG with blastn (blast version 2.14.0) with an option of “-culling_limit 1” to keep only a best-matching annotation for overlapping features. For the rest, it was run with the default parameters. The downstream enhancement was conducted in the same manner as described above.
For the enhanced TE annotation on multiple Eutherian genomes, TEs annotated in the Eutherian RAG was lifted over to human (Homo sapiens, GRCh38), mouse (Mus musculus, GRCm38), dog (Canin lupus familialis, canFam3), and armadillo (Dasypus novemcinctus, dasNov3). Then, the lifted-over annotation was compared against the TE annotation obtained as RepeatMasker output files available on the UCSC Genome Browser website (http://genome-euro.ucsc.edu/), and non-overlapping novel TEs were identified as degTEs. The combined enhanced TE annotations of these species were also shared on Zenodo: https://doi.org/10.5281/zenodo.7716408.
TE characterization
The estimated age of each TE subfamily was obtained from Dfam_curatedonly.embl file downloaded from the Dfam website (https://dfam.org/releases/Dfam_3.6/). In cases where a subfamily is associated with multiple nested clades, the oldest clade was assigned (e.g., “Eutheria; Primates” was reclassified as “Eutheria”).
To annotate genomic loci degTEs fall into, BED files of genic regions, UTRs, introns, and coding exons, were downloaded from the UCSC Table Browser (http://genome-euro.ucsc.edu/). The intergenic regions were defined as regions outside the genic regions.
TE and degTE distribution analysis
All the manipulations were done with the R package bedtoolsr44 (version 2.30.0-1). The human genome was split into 100-kb bins with bt.makewindows, and the coverage calculation for TE/degTE in each bin were performed by bt.coverage. The hg38 centromere annotation was obtained using the UCSC Table Browser (http://genome-euro.ucsc.edu/).
degTE homology analysis
A degTE was defined as a genomic region where no TE was annotated in hg38 but was assigned a TE in one of the corresponding loci in the RAGs. The homology of the degTEs to TE sequences was determined by running the nhmmscan function from HMMER software45 (version 3.3.2) with the default parameters against the same Dfam database (version 3.6) as in the RepeatMasker annotation.
L2c alignment to the consensus
The sequences annotated as L2c were obtained with bedtools46 getfasta (version 2.30.0) command by running it on hg38 and the Eutherian RAG. 1,000 elements were randomly chosen from each set and were aligned to the consensus sequences obtained from Dfam (version 3.6) with MAFFT47 –addfragments command (version 7.508). The resulting alignment was visualised with Jalview48 (version 2.11.2.6).
degTE overlap with cCREs and TFBSs
The human candidate cis-regulatory elements (cCREs) data (registry V3) were downloaded from the SCREEN website (https://screen.encodeproject.org/index/cversions). To simplify the categories, the CTCF-bound cCRE categories were merged into other cCRE categories (e.g., both “PLS” and “PLS,CTCF-bound” were collapsed into a single category “PLS”). Overlaps between hg38 TE/degTEs and cCREs were defined by “bedtools intersect -f 0.5 -F 0.5 -e” (version 2.30.0), meaning an overlap length must be longer than 50% of either the TE/degTEs or cCRE annotation. With this, we defined cCREs that do not overlap with hg38 TE but overlap with degTEs.
For the TFBS analysis, we downloaded TF ChIP-seq data generated by the Richard Myers group from the ENCODE website42 (https://www.encodeproject.org/). ENCODE dataset ID can be found in the “id” column of Data S3. We defined TFBSs that do not overlap with hg38 TE but overlap with degTEs by bedtools using the same parameters used for cCREs as shown above.
Chimeric transcripts in human embryos
FASTQ files of RNA-seq on human embryos were obtained from GEO: GSE197265.28 The reads were mapped to hg38 using STAR49 (version 2.7.10b) with the following parameters “--outFilterMultimapNmax 20 --alignSJoverhangMin 10 --alignSJDBoverhangMin 1 --outFilterMismatchNmax 999 --outFilterMismatchNoverReadLmax 0.04 --alignIntronMin 20 --alignIntronMax 1000000 --alignMatesGapMax 1000000”. Using the obtained alignment, de novo transcriptome assembly was performed with StringTie50 (version 2.2.1) with the following parameters “-c 1 -f 0.01 -p 1 -j 1 -a 10”. Of the resulting transcripts, exons that were identified in all the biological replicates for each embryonic stage were kept for downstream analyses. The filtered exon annotation obtained from the de-novo identified transcripts were then crossed with degTEs that do not overlap with the canonical exons. Per-transcript expression quantification was also performed with StringTie.
For the prediction of coding regions of the de novo transcript assembly, DNA sequences of the transcripts were obtained with gtf_genome_to_cdna_fasta.pl script and the resulting FASTA file was used to predict coding regions with TransDecoder.LongOrfs script with an option “--complete_orfs_only”. These scripts are from TransDecoder51 (version 5.7.1) package.
To visualise the splicing event of the RNA-seq data, ggsashimi52 was used.
Quantification and statistical analysis
TE enrichment in KZFP binding sites
KZFP binding sites were taken from ref. 23 Statistical test on the enrichment levels of hg38 TE in the KZFP binding sites was performed with pyTEnrich software with the default parameters (https://alexdray86.github.io/pyTEnrich/build/html/index.html).
Acknowledgments
We thank the Trono lab members for constructive discussions. Most computational works were conducted on the high-performance computing cluster developed and maintained by Scientific IT and Application Support (SCITAS) at EPFL. This work was supported by grants from the EMBO Postdoctoral Fellowship (ALTF 1287-2020) and the JSPS Overseas Research Fellowship (no. 202360326) to W.M. and the European Research Council (KRABnKAP, no. 268721; Transpos-X, no. 694658), and the Swiss National Science Foundation (310030_152879 and 310030B_173337) to D.T.
Author contributions
Conceptualization, W.M.; methodology, W.M.; software, W.M.; formal analysis, W.M.; investigation, W.M. and E.P.; resources, W.M.; writing—original draft, W.M.; writing—review & editing, W.M., E.P., and D.T.; visualization, W.M.; supervision, D.T.; funding acquisition, W.M. and D.T.
Declaration of interests
The authors declare no competing interests.
Published: January 30, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xgen.2024.100497.
Contributor Information
Wayo Matsushima, Email: wayo.matsushima@epfl.ch.
Didier Trono, Email: didier.trono@epfl.ch.
Supplemental information
Data S1. Divergence levels of the TEs found in the RAGs, Related to Figure 2
Data S2. Summary statistics of degTEs, Related to Figure 3
Data S3. Summary statistics of ENCODE TFBSs overlapping with degTEs, Related to Figure 4
Data S4. List of degTE-derived non-canonical exons forming TcGTs, Related to Figure 5
References
- 1.Nurk S., Koren S., Rhie A., Rautiainen M., Bzikadze A.V., Mikheenko A., Vollger M.R., Altemose N., Uralsky L., Gershman A., et al. The complete sequence of a human genome. Science. 2022;376:44–53. doi: 10.1126/science.abj6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mills R.E., Bennett E.A., Iskow R.C., Devine S.E. Which transposable elements are active in the human genome? Trends Genet. 2007;23:183–191. doi: 10.1016/j.tig.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Fueyo R., Judd J., Feschotte C., Wysocka J. Roles of transposable elements in the regulation of mammalian transcription. Nat. Rev. Mol. Cell Biol. 2022;23:481–497. doi: 10.1038/s41580-022-00457-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jangam D., Feschotte C., Betrán E. Transposable Element Domestication As an Adaptation to Evolutionary Conflicts. Trends Genet. 2017;33:817–831. doi: 10.1016/j.tig.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Modzelewski A.J., Gan Chong J., Wang T., He L. Mammalian genome innovation through transposon domestication. Nat. Cell Biol. 2022;24:1332–1340. doi: 10.1038/s41556-022-00970-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peaston A.E., Evsikov A.V., Graber J.H., de Vries W.N., Holbrook A.E., Solter D., Knowles B.B. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Göke J., Lu X., Chan Y.-S., Ng H.-H., Ly L.-H., Sachs F., Szczerbinska I. Dynamic Transcription of Distinct Classes of Endogenous Retroviral Elements Marks Specific Populations of Early Human Embryonic Cells. Cell Stem Cell. 2015;16:135–141. doi: 10.1016/j.stem.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Smit A.F.A., Hubley R., Green P. 2021. RepeatMasker Open-4.0; pp. 2013–2015. [Google Scholar]
- 9.Jurka J. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- 10.Wheeler T.J., Clements J., Eddy S.R., Hubley R., Jones T.A., Jurka J., Smit A.F.A., Finn R.D. Dfam: a database of repetitive DNA based on profile hidden Markov models. Nucleic Acids Res. 2013;41:D70–D82. doi: 10.1093/nar/gks1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zoonomia Consortium A comparative genomics multitool for scientific discovery and conservation. Nature. 2020;587:240–245. doi: 10.1038/s41586-020-2876-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng S., Stiller J., Deng Y., Armstrong J., Fang Q., Reeve A.H., Xie D., Chen G., Guo C., Faircloth B.C., et al. Dense sampling of bird diversity increases power of comparative genomics. Nature. 2020;587:252–257. doi: 10.1038/s41586-020-2873-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Armstrong J., Hickey G., Diekhans M., Fiddes I.T., Novak A.M., Deran A., Fang Q., Xie D., Feng S., Stiller J., et al. Progressive Cactus is a multiple-genome aligner for the thousand-genome era. Nature. 2020;587:246–251. doi: 10.1038/s41586-020-2871-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hickey G., Paten B., Earl D., Zerbino D., Haussler D. HAL: a hierarchical format for storing and analyzing multiple genome alignments. Bioinformatics. 2013;29:1341–1342. doi: 10.1093/bioinformatics/btt128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caballero J., Smit A.F.A., Hood L., Glusman G. Realistic artificial DNA sequences as negative controls for computational genomics. Nucleic Acids Res. 2014;42:e99. doi: 10.1093/nar/gku356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ostertag E.M., Kazazian H.H., Jr. Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 17.Vassetzky N.S., Kramerov D.A. SINEBase: a database and tool for SINE analysis. Nucleic Acids Res. 2013;41:D83–D89. doi: 10.1093/nar/gks1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.ENCODE Project Consortium. Moore J.E., Purcaro M.J., Pratt H.E., Epstein C.B., Shoresh N., Adrian J., Kawli T., Davis C.A., Dobin A., et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature. 2020;583:699–710. doi: 10.1038/s41586-020-2493-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hakimi M.-A., Dong Y., Lane W.S., Speicher D.W., Shiekhattar R. A candidate X-linked mental retardation gene is a component of a new family of histone deacetylase-containing complexes. J. Biol. Chem. 2003;278:7234–7239. doi: 10.1074/jbc.M208992200. [DOI] [PubMed] [Google Scholar]
- 20.Graham-Paquin A.-L., Saini D., Sirois J., Hossain I., Katz M.S., Zhuang Q.K.-W., Kwon S.Y., Yamanaka Y., Bourque G., Bouchard M., et al. ZMYM2 Is Essential for Methylation of Germline Genes and Active Transposons in Embryonic Development. bioRxiv. 2022 doi: 10.1101/2022.09.13.507699. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Owen D., Aguilar-Martinez E., Ji Z., Li Y., Sharrocks A.D. ZMYM2 controls transposable element transcription through distinct co-regulatory complexes. bioRxiv. 2023 doi: 10.1101/2023.01.24.525372. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolf D., Goff S.P. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458:1201–1204. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imbeault M., Helleboid P.-Y., Trono D. KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature. 2017;543:550–554. doi: 10.1038/nature21683. [DOI] [PubMed] [Google Scholar]
- 24.Faulkner G.J., Kimura Y., Daub C.O., Wani S., Plessy C., Irvine K.M., Schroder K., Cloonan N., Steptoe A.L., Lassmann T., et al. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 25.Swergold G.D. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol. Cell Biol. 1990;10:6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makałowski W., Mitchell G.A., Labuda D. Alu sequences in the coding regions of mRNA: a source of protein variability. Trends Genet. 1994;10:188–193. doi: 10.1016/0168-9525(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 27.Smalheiser N.R., Torvik V.I. Alu elements within human mRNAs are probable microRNA targets. Trends Genet. 2006;22:532–536. doi: 10.1016/j.tig.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 28.Zou Z., Zhang C., Wang Q., Hou Z., Xiong Z., Kong F., Wang Q., Song J., Liu B., Liu B., et al. Translatome and transcriptome co-profiling reveals a role of TPRXs in human zygotic genome activation. Science. 2022;378:abo7923. doi: 10.1126/science.abo7923. [DOI] [PubMed] [Google Scholar]
- 29.Mcclintock B. Controlling elements and the gene. Cold Spring Harb. Symp. Quant. Biol. 1956;21:197–216. doi: 10.1101/sqb.1956.021.01.017. [DOI] [PubMed] [Google Scholar]
- 30.Britten R.J., Davidson E.H. Gene regulation for higher cells: a theory. Science. 1969;165:349–357. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- 31.Davidson E.H., Britten R.J. Regulation of gene expression: possible role of repetitive sequences. Science. 1979;204:1052–1059. doi: 10.1126/science.451548. [DOI] [PubMed] [Google Scholar]
- 32.Goerner-Potvin P., Bourque G. Computational tools to unmask transposable elements. Nat. Rev. Genet. 2018;19:688–704. doi: 10.1038/s41576-018-0050-x. [DOI] [PubMed] [Google Scholar]
- 33.de Koning A.P.J., Gu W., Castoe T.A., Batzer M.A., Pollock D.D. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivics Z., Hackett P.B., Plasterk R.H., Izsvák Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 35.Miskey C., Izsvák Z., Plasterk R.H., Ivics Z. The Frog Prince: a reconstructed transposon from Rana pipiens with high transpositional activity in vertebrate cells. Nucleic Acids Res. 2003;31:6873–6881. doi: 10.1093/nar/gkg910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dewannieux M., Harper F., Richaud A., Letzelter C., Ribet D., Pierron G., Heidmann T. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 2006;16:1548–1556. doi: 10.1101/gr.5565706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campitelli L.F., Yellan I., Albu M., Barazandeh M., Patel Z.M., Blanchette M., Hughes T.R. Reconstruction of full-length LINE-1 progenitors from ancestral genomes. Genetics. 2022;221 doi: 10.1093/genetics/iyac074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petri R., Brattås P.L., Sharma Y., Jönsson M.E., Pircs K., Bengzon J., Jakobsson J. LINE-2 transposable elements are a source of functional human microRNAs and target sites. PLoS Genet. 2019;15 doi: 10.1371/journal.pgen.1008036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ou S., Su W., Liao Y., Chougule K., Agda J.R.A., Hellinga A.J., Lugo C.S.B., Elliott T.A., Ware D., Peterson T., et al. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol. 2019;20:275. doi: 10.1186/s13059-019-1905-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao Y., Chen G., Wu G., Zhang X., McDermott J., Chen X., Xu C., Jiang Q., Chen Z., Zeng Y., et al. Widespread roles of enhancer-like transposable elements in cell identity and long-range genomic interactions. Genome Res. 2019;29:40–52. doi: 10.1101/gr.235747.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roller M., Stamper E., Villar D., Izuogu O., Martin F., Redmond A.M., Ramachanderan R., Harewood L., Odom D.T., Flicek P. LINE retrotransposons characterize mammalian tissue-specific and evolutionarily dynamic regulatory regions. Genome Biol. 2021;22:62. doi: 10.1186/s13059-021-02260-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo Y., Hitz B.C., Gabdank I., Hilton J.A., Kagda M.S., Lam B., Myers Z., Sud P., Jou J., Lin K., et al. New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res. 2020;48:D882–D889. doi: 10.1093/nar/gkz1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architecture and applications. BMC Bioinf. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patwardhan M.N., Wenger C.D., Davis E.S., Phanstiel D.H. Bedtoolsr: An R package for genomic data analysis and manipulation. J. Open Source Softw. 2019;4 doi: 10.21105/joss.01742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wheeler T.J., Eddy S.R. nhmmer: DNA homology search with profile HMMs. Bioinformatics. 2013;29:2487–2489. doi: 10.1093/bioinformatics/btt403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katoh K., Misawa K., Kuma K.-I., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pertea M., Pertea G.M., Antonescu C.M., Chang T.-C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haas B.J.T.D. TransDecoder Source. https://github.com/TransDecoder/TransDecoder
- 52.Garrido-Martín D., Palumbo E., Guigó R., Breschi A. ggsashimi: Sashimi plot revised for browser- and annotation-independent splicing visualization. PLoS Comput. Biol. 2018;14 doi: 10.1371/journal.pcbi.1006360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Divergence levels of the TEs found in the RAGs, Related to Figure 2
Data S2. Summary statistics of degTEs, Related to Figure 3
Data S3. Summary statistics of ENCODE TFBSs overlapping with degTEs, Related to Figure 4
Data S4. List of degTE-derived non-canonical exons forming TcGTs, Related to Figure 5
Data Availability Statement
-
(1)
RepeatMasker outputs and enhanced TE annotations have been deposited at Zenodo and are publicly available as of the date of publication. DOIs are listed in the key resources table. This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
-
(2)
All original code has been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
(3)
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.