Abstract
Like multisubunit RNA polymerases (RNAPs), T7 RNAP frequently releases its transcript over the initial 8-12 transcribed nucleotides, when it still contacts the promoter. This abortive cycling, which is most prominent with initial sequences that deviate from those of T7 late genes, eventually compromises productive transcription. Starting from an in vivo situation where transcription of a target gene by T7 RNAP is virtually abolished because of extensive abortive cycling, we have selected a mutation in RNAP that restores target gene expression. In vitro, this mutation (P266L) weakens promoter binding but markedly reduces abortive cycling over a variety of initial sequences by stabilizing the transcription complex at nucleotides 5-8. Other substitutions of P266 have similar effects. X-ray data show that during the transition from initial to elongation complex, the N-terminal region undergoes a major structural switch of which P266 constitutes one of the hinges. How the mutation might facilitate this switch is tentatively discussed. On the practical side, the mutation can significantly improve in vitro transcription, particularly from templates carrying unfavorable initial sequences.
Keywords: abortive transcription, in vitro RNA synthesis
Like multisubunit RNA polymerases (RNAPs), the monomeric RNAP from bacteriophage T7 recognizes a cognate promoter and, after locally melting the DNA to produce an “open complex,” uses a mononucleotide to prime template-directed RNA synthesis. The contacts with the promoter are maintained during the transcription of the first ≈10 nucleotides [this DNA region is referred to here as the initially transcribed sequence (ITS), whereas the corresponding transcription complex is the initial complex (IC)]. Upon clearing the promoter, the IC undergoes a major structural rearrangement, yielding the elongation complex (EC) (1, 2). The IC is very unstable compared with the EC, and it frequently dissociates before the switch. Therefore, small abortive transcripts are repeatedly released until eventually the polymerase becomes engaged in productive transcription. One important parameter determining the extent of abortive cycling is ITS sequence. Whereas cycling is low when the purine-rich ITS from late T7 genes is used, it increases when pyrimidines, and particularly Ts, are present (3, 4). Another important parameter is the enzyme catalytic turnover: the lower this turnover, the longer it takes to transcribe the ITS and the higher the probability of IC dissociation during this time. Catalytic turnover can be decreased by active site mutations or nucleotide shortage (5). However, mutations unrelated to catalytic activity also have been found to favor abortive cycling, presumably by specifically hampering the transition from IC to EC (6).
Whereas the main features of transcription initiation, including abortive cycling, are common to all RNAPs, a distinctive feature of the T7 enzyme is that the steps leading to open complex formation are fast and easily reversible (7, 8). It follows that promoter clearance can become rate-limiting in productive transcription, particularly when abortive cycling is favored (noncognate ITS sequence, low enzyme turnover). Consistently, these conditions have been found to decrease the yield of productive transcripts in vivo or in vitro and to increase the ratio of abortive to productive transcripts in vitro (4, 9). Eventually, the combination of a low enzyme turnover and an unfavorable ITS promotes cycling to the point that the enzyme is indefinitely trapped in the abortive mode.
T7 RNAP is a popular tool for producing RNA in vitro (10), but the need of using ITSs resembling those from late T7 genes for optimal transcription limits its versatility. In this work, we describe a mutant RNAP that is less sensitive to abortive cycling so that this constraint is partially relieved.
Materials and Methods
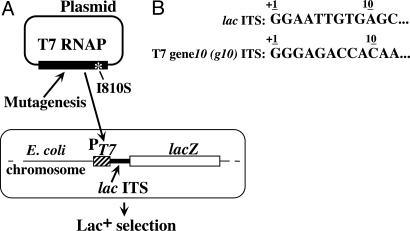
Strains and Plasmids. Strains ENS0134 and ENS0134T, which carry the lac and g10 ITS, respectively, inserted between the T7 φ10 promoter (PT7) and the lacZ gene (Fig. 1), and plasmids pT7LacZ-Arg 5 and pMAMA5-Arg 5 (Fig. 2A), which bear the same constructs, have been described (9). The pBR322-based plasmid pAR1219 (11) and its derivative encoding the I810S T7 RNAP (5, 12) carry the T7 RNAP gene under the control of the lacUV5 promoter.
Fig. 1.
An in vivo screen for T7 RNAP with improved initial processivity. (A) Overview of the experimental system. The large rectangle represents the Escherichia coli cell; its chromosome carries the T7 gene 10 promoter (PT7; hatched box), followed by the lac or g10 ITS (closed box; cf. B), and a truncated lac operon encompassing the lacZ gene and part of the lacY gene (open box). “Plasmid” stands for plasmid pAR1219 (11), which carries the T7 RNAP gene, or a derivative carrying the I810S mutation (*) in the active site. The situation illustrated here (lac ITS, I810S mutation) has been used for the isolation of T7 RNAP mutants with increased initial processivity (see text). (B) Sequence of the lac or g10 ITS.
Fig. 2.
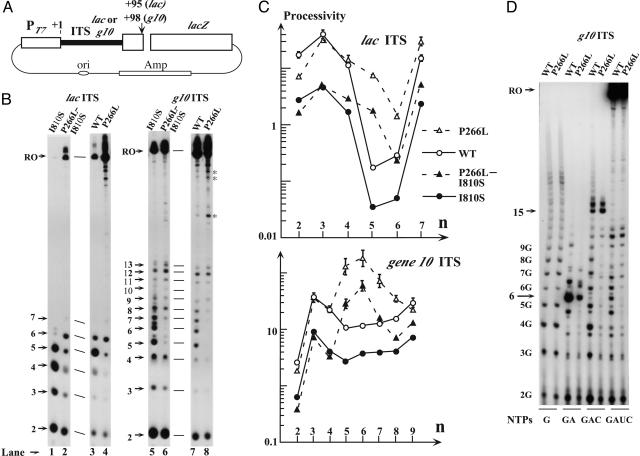
In vitro transcription assays. (A) Schematic representation of the plasmids used as templates. These plasmids, which carry the same PT7-lacZ fusions as the chromosome of the E. coli strains used in Fig. 1, are linearized with HindIII at +95 or +98 (vertical arrow). (B) Pattern of [α-32P]GTP-labeled transcripts generated by different T7 RNAPs in the presence of the lac (left) or g10 (right) ITS. The T7 RNAPs used, designated by their mutated residues, are indicated above the corresponding lanes. The positions of the productive transcripts (RO, runoff) or abortive transcripts (numbers refer to transcript length in nt) are arrowed next to the corresponding panel. Bands marked with an asterisk were not consistently observed and are presumably degradation products. (C) Processivity of the WT or mutant T7 RNAP over individual positions (n) of the lac or g10 ITS. For the definition of processivity used here, see the text. Higher processivity values are comparatively less precise because they rely on the quantification of faint bands; in this case, uncertainties have been indicated by error bars. (D) Same as B, except that only transcription from the g10 ITS by the WT and P266L RNAPs is illustrated, and that subsets of the four NTPs (as indicated below the corresponding lanes) are used. The positions of the runoff transcripts synthesized in the presence of GTP + ATP (+6), GTP + ATP + CTP (+15), or all four NTPs (RO) are shown by arrows. Also indicated are the positions of oligo(G) (4G, 5G, etc.) synthesized when T7 RNAP stalls at position +3.
Mutant Isolation and Protein Purification. For mutagenesis, the pAR1219 derivative encoding I810S T7 RNAP was incubated with aqueous hydroxylamine (13) until transformation efficiency on XL1 cells dropped 100-fold. The dialyzed plasmid was split into 11 parts, which were independently used to transform XL1 cells. After growth in batch, plasmids from each of the 11 cultures were used to transform ENS0134 cells, which were finally plated on LB-ampicillin plates containing isopropyl β-d-thiogalactoside and X-Gal. Passage through XL1 cells was necessary because direct transformation of ENS0134 with the mutagenized plasmid was inefficient. Lac+ (β-galactosidase-positive) colonies were recovered from five transformation plates. The corresponding plasmids were isolated, and fragments were subcloned into the unmutagenized plasmid to map the mutation responsible for the Lac+ phenotype, which was finally characterized by sequencing. To introduce the P266L mutation in WT T7 RNAP, the AlwNI-DraIII fragment carrying the mutation was replaced into the parent pAR1219 plasmid. For mutant protein purification, plasmids encoding His-tagged mutant T7 RNAPs were constructed by inserting the pAR1219 AlwNI-AlwNI fragment carrying most of the mutant RNAP coding sequence into the same sites of pBH161 (14); however, pBH161 derivatives encoding the P266A, P266S, and P266Y mutants were obtained by direct mutagenesis of pBH161 with the QuikChange XL mutagenesis kit (Stratagene). His-tagged RNAPs were purified, quantified, and stored as described in ref. 14.
In Vitro Transcription. Analytical transcription in the presence of [α-32P]GTP and transcript analysis on 25% acrylamide/6 M urea gels (Figs. 2 and 4C) were essentially as described in ref. 9. For preparative transcription (Fig. 4 A and B), template (15 nM) and T7 RNAP (1 μM) were incubated as in Promega technical manual 016 (Riboprobe System; synthesis of large amounts of RNA), in the presence of [α-32P]ATP [5 μCi (1 Ci = 37 GBq)/50 μl]. At timed intervals, aliquots (5 μl) were added on ice to 4 μl of stop solution (95% formamide/6 mM EDTA) and analyzed as for analytical transcription. Radioactive spots were quantified with a FLA3000 Imager (Fuji).
Fig. 4.
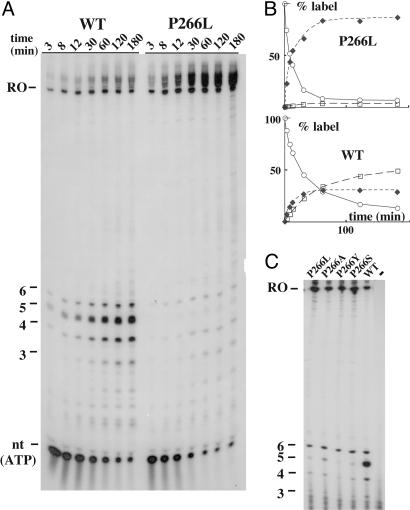
In vitro RNA synthesis. (A) Comparison of the ability of the WT or P266L enzyme to synthesize large amounts of RNA carrying an unfavorable ITS as a function of time. The same plasmidic 6.5-kb template shown in Fig. 2 A (lac ITS) was transcribed in vitro with either polymerase in the presence of [α-32P]ATP. Free ATP (nt), abortive, runoff (RO; 95 nt) and extended runoff products are visualized. (B) Quantification of the data. The fraction of the label present as free ATP (○), abortive transcripts (□), and runoff plus extended products (♦) is shown as a function of time. (C) Synthesis of abortive and productive transcripts from the same plasmid shown in Fig. 2 A (lac ITS) in the presence of various T7 RNAP mutants and [α-32P]GTP (see Materials and Methods). The nature of the mutation is shown above each lane.
AseI Protection Assays. To compare the ability of the WT or P266L enzyme to protect the PT7 promoter against AseI digestion (Fig. 3), plasmid pMAMA5-Arg 5 was first linearized with DraIII. DNA (6.6 nM) was then incubated with 0.2 AseI unit/μl in 20 mM NaCl/6 mM MgCl2/10 mM Tris·HCl, pH 7.9/10 mM DTT/0.5 mM spermidine/5% glycerol and, when appropriate, 1 μM T7 RNAP and 0.5 mM GTP or ATP (low-salt conditions; Fig. 3B). High-salt conditions (Fig. 3C) were identical, except that NaCl was 70 mM. After incubation at 37°C for 30 min, cleavage products were separated on a native 6% polyacrylamide gel in 0.5× TBE (1× TBE = 89 mM Tris/89 mM boric acid/20 mM EDTA, pH 8.3) and stained with ethidium bromide.
Fig. 3.
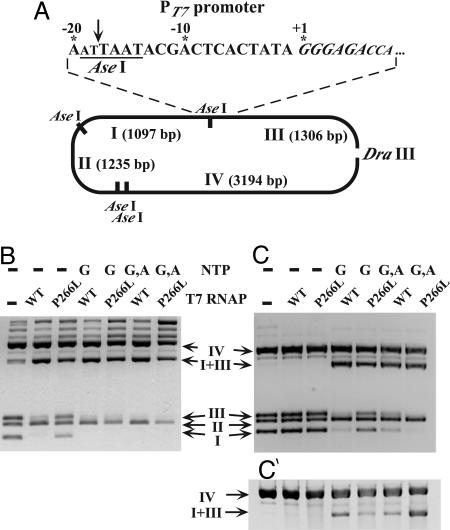
Enzymatic test for assessing the binding of T7 RNAP to the promoter. (A) Position of AseI sites in the plasmid shown in Fig. 2A (g10 ITS). The PT7 promoter used here (enlarged) encompasses an AseI site (underlined; cleavage is shown by arrow) that can be shielded by T7 RNAP binding (see B and C). The ITS is italicized; the nucleotides that are not strictly conserved among T7 late promoters are in small capitals. Before the assay, the plasmid was linearized with DraIII (see text). The size of the AseI and AseI-DraIII fragments is indicated. (B-C′) AseI digest of the plasmid shown in A in the presence of no RNAP or WT or P266L T7 RNAP, as indicated above each lane. The disappearance of fragments I and III and the accumulation of the uncut fragment I+III are diagnostic of promoter protection. The assay has been done without NTPs (-) or in the presence of either GTP (G), or GTP + ATP (G,A), which allow transcription to nucleotides +3 and +6, respectively. B and C correspond to low- and high-salt conditions, respectively; C′ is similar to C, except that AseI digestion was slightly longer, making variations in the intensity of the I+III band more visible.
Results
A Genetic Screen for Improving Initial Processivity of T7 RNAP. Previously, we described an experimental tool for studying the influence of ITS sequence or T7 RNAP turnover on the expression of target genes in vivo (9, 12). A variant of this system has been used here (Fig. 1). Briefly, two Escherichia coli strains carrying a chromosomal copy of the lacZ gene under the control of the T7 late promoter (PT7) were used. In one of them, PT7 is followed by the T7 late g10 ITS, whereas in the other, the ITS is derived from the genuine lacZ gene ITS (lac ITS). Of note, the lac ITS carries three Ts at positions 5, 6, and 8 that are expected to favor abortive cycling (Fig. 1B). The cells also contain a plasmid encoding T7 RNAP, with or without an active site mutation (I810S) that decreases the catalytic turnover ≈6-fold (5) (Fig. 1 A). Whereas the combination of the WT enzyme and g10 ITS resulted in a high β-galactosidase expression, the use of the I810S mutant or lac ITS individually decreased this expression 3.5- and 6.0-fold, respectively, and their combination caused a >1,000-fold decrease (Table 1). To clarify the basis for this decrease, the purified WT or I810S RNAP was used in vitro to transcribe linearized plasmids carrying the same constructs as in vivo under conditions where both productive and abortive transcripts could be visualized (Fig. 2 A and B). In all cases, the yield of productive transcript (Fig. 2B, lanes 1, 3, 5, and 7) paralleled the β-galactosidase expression observed in vivo (Table 1): in particular, when the lac ITS and I810S RNAP were used together, productive transcription was almost abolished (Fig. 2B, lane 1). In the meantime, the synthesis of abortive transcripts was unabated or even stimulated. Altogether, these observations fit the view that the use of either the lac ITS or of the I810S enzyme depress productive transcription by favoring abortive cycling, and that, when both are used together, the enzyme is trapped in the abortive mode. Interestingly, under these conditions, β-galactosidase expression in vivo is so low that cells are phenotypically Lac- on X-Gal plates. Because rare Lac+ colonies are easily scored within a Lac- population, this system is suitable for isolating new T7 RNAP mutants with a reduced propensity for cycling.
Table 1. β-Galactosidase expression observed with the WT or mutant T7 RNAP and with the lac or g10 ITS.
| RNAP | ITS | β-Galactosidase, activity* |
|---|---|---|
| WT | g10 | 19,000 |
| I810S | g10 | 5,300 |
| WT | lac | 3,400 |
| I810S | lac | <15 |
| I810S P266L | lac | 420 |
Given in nmoles ortho-nitrophenyl-β-d thiogalactopyranoside (ONPG) hydrolized per min per mg of protein.
The plasmid encoding the I810S RNAP was then mutagenized in vitro with hydroxylamine and introduced back into cells carrying the lac ITS, yielding Lac+ colonies with a frequency of 10-4 to 10-5. Five of them, resulting from independent mutational events, were studied; all contained a modified T7 RNAP that was at least 25- to 30-fold more efficient than the parent I810S RNAP for synthesizing β-galactosidase from the lac ITS. Sequencing of these plasmids revealed that all but one carried the same nucleotide change at codon 810 of T7 RNAP, causing the further replacement of Ser by Asn (AGT → AAT; this change is the most probable one at this codon, because hydroxylamine causes mainly C → T and G → A transitions). Presumably, this replacement alleviates the active site defect of the I810S mutant, thereby reducing abortive cycling. In only one mutant was codon 810 unaffected; it carried a transition at codon 266 (CCG → CTG), resulting in the replacement of Pro by Leu (P266L). Interestingly, P266, which is well conserved among phage or mitochondrial RNAPs (15), is located away from the active site. Our aim here is to describe the unusual properties of this mutant.
P266L, a Mutation That Stabilizes the IC over the ITS, Particularly at Nucleotides 5-8. To study how the P266L mutation stimulates β-galactosidase synthesis from the I810S enzyme, the double-mutant P266L I810S was purified and assayed in vitro as above. For completeness, the P266L single mutant also was constructed and included in the assay. Whatever the ITS and the residue present at position 810, the P266L mutation greatly diminished the synthesis of abortive transcripts. In the meantime, the P266L mutation generally favored runoff transcription (Fig. 2B). In particular, when used in combination with the I810S mutation, it allowed significant productive transcription even from the lac ITS (compare lanes 1 and 2 in Fig. 2B), as it allows β-galactosidase synthesis from this ITS in vivo (Table 1). We conclude that, in all cases, the P266L mutation diminishes abortive cycling, facilitating productive transcription. As the only exception, when introduced into the WT enzyme, the P266L mutation slightly decreased the very high level of productive transcription from the g10 ITS. It is possible that a step different from promoter clearance (e.g., enzyme recycling) then becomes limiting in transcription, and that it is slightly hampered rather than favored by the P266L mutation. However, the synthesis of abortive transcripts was reduced even more, so that even in this case the relative yield of productive to abortive transcripts was increased. This increase is presumably characteristic of the P266L mutation because it was observed with a variety of ITS beyond lac and g10 (Fig. 6, which is published as supporting information on the PNAS web site).
To assess quantitatively the effect of the P266L mutation on abortive cycling, individual transcripts in the different lanes of Fig. 2B were quantified, and the processivity of T7 RNAP was evaluated from the data. We define here processivity as the ratio of the probabilities that the transcript is extended, rather than released, at any particular position (n). This ratio, which varies from 0 to infinity, is obtained by dividing the molar abundance of all species ≥n by the molar abundance of species n. Formally, it can be expressed as kel/kdiss, where kel and kdiss are the apparent first-order rate constants for the forward motion or the dissociation, respectively, of the transcription complex at position n (16). The I810S mutation caused a 5- to 10-fold decrease in processivity all over the ITS (Fig. 2C), as expected because it decreases the enzyme turnover (i.e., kel) ≈6-fold (5). Also as expected, the replacement of the g10 ITS by the lac ITS dramatically reduced processivity at nucleotides 5 and 6, after uridine incorporation (Fig. 1B). As concerns the 266L mutation, it had little effect down to +4 but prominently increased processivity at nucleotides 5 and 6 whatever the ITS (Figs. 2C and 6). For the lac ITS, the effect of the mutation was still large at +7 but could not be assessed further downstream because of the fading of abortive transcripts. Whereas the same holds for several other ITS (Fig. 6), with the g10 ITS, transcripts could be observed down to +13. In this case, the increase in processivity due to the P266L mutation vanishes at +8-9 [no attempt was made to evaluate processivity further downstream, because species 11-13 nt long are presumably dead-end products unrelated to the normal EC → IC transition rather than genuine abortive transcripts (17)]. Further assays with templates several kilobases long evidenced no effect of P266L on processivity in the elongation mode (not shown). Of note, with both the lac ITS and g10 ITS, the processivity increase due to the P266L mutation was independent of the I810S context in which it was first isolated. Therefore, only the P266L single mutant was retained for further studies.
The local processivity increase due to the P266L mutation may reflect either an increase in kel or a decrease in kdiss at the corresponding positions. The former explanation is unattractive because the mutation, being located away from the active site, is unlikely to affect kel, which reflects enzyme turnover. The following argument further supports the view that it acts by lowering kdiss, i.e., by stabilizing the complex. When transcription is interrupted within the ITS by the absence of the next NTP, the steady-state production of the short transcript is limited by the rate of complex breakdown, i.e., by kdiss (7). The same linearized template used above (g10 ITS) was transcribed in vitro with the WT or P266L enzymes by using incomplete NTP sets. When only GTP is present, the template-directed transcription is limited to GGG, but in practice, a ladder of oligo(G) is observed because of reiterated enzyme slippage over the C stretch of the template. The P266L mutation has little effect on this synthesis (Fig. 2D, G lanes). In contrast, when both GTP and ATP were present, allowing transcription to proceed to +6, the transcript yield from the P266L mutant was markedly depressed compared with WT, consistent with a decreases in kdiss at this position (Fig. 2D, GA lanes).
When CTP was also present, synthesis proceeded to +15. The WT and P266L enzymes differed little in this synthesis, consistent with the view that the mutation decreases kdiss over a subregion of the ITS only (Fig. 2D, GAC lanes). Interestingly, with the WT enzyme, an oligo(G) ladder also was observed, reflecting the presence of a second RNAP molecule trapped in the slippage mode by the obstructing RNAP (18). The mutant failed to synthesize this ladder, even though it is capable of slippage synthesis (compare above). One possibility is that it binds the promoter more weakly than the WT enzyme, so that in the presence of an obstructing polymerase, binding is abrogated. This issue was examined further.
P266L Decreases the T7 RNAP Affinity for Promoter. To investigate possible differences in promoter affinity between the WT and P266L enzymes, we used a recently described endonuclease protection assay (19). Briefly, the T7 promoter used here carries a cleavage site for endonuclease AseI (nucleotides -14 to -19; Fig. 3A), which can be protected by promoter-bound T7 RNAP. If then the P266L enzyme binds the promoter less tightly than the WT enzyme, it also should be less efficient in this protection. One of the plasmids used above for in vitro transcription (g10 ITS) was chosen for this assay. Because supercoiling affects the T7 RNAP-promoter interaction (20), the plasmid was first linearized with a single-cutter (DraIII). Promoter protection should cause the disappearance of two fragments (I and III) and the appearance of a new one (I+III) in the AseI digestion profile (Fig. 3A). Under low-salt conditions (20 mM NaCl) that favor promoter binding but not AseI cleavage, the promoter was well protected by WT T7 RNAP but less so by the P266L enzyme (compare lanes 2 and 3 in Fig. 3B), indicating weaker binding. Interestingly, when GTP or GTP + ATP were present, allowing transcription to proceed to +3 or +6, respectively (see Fig. 3A), complete protection was observed even with the P266L enzyme (Fig. 3B, lanes 4-7). That promoter binding is tightened by the presence of GTP is well documented (21).
The use of a higher ionic strength should favor AseI cleavage and inhibit RNAP binding. Consistently, when the NaCl concentration was raised to 70 mM, neither the WT nor the P266L enzyme provided significant protection by itself (Fig. 3C, lanes 2 and 3). In contrast, in the presence of nucleotides, protection was still observed. With the WT enzyme, protection was moderate and nearly alike in the presence of either GTP or GTP + ATP. With the P266L enzyme, protection was comparatively weaker in the presence of GTP but almost complete in the presence of GTP + ATP (Fig. 3C). A straightforward interpretation is that the mutant differs from the WT enzyme in dissociating much more slowly when stalled at +6 rather than at +3, so that the promoter is now occupied most of the time. This result further supports the view that the P266L mutation stabilizes the transcription complex at position +6.
The P266L Enzyme in Preparative RNA Synthesis. To test how the P266L mutant compares with the WT enzyme under conditions of preparative RNA synthesis, the two enzymes were used to synthesize the same 95-nt transcript as in Fig. 2B (lac ITS), except that synthesis was run for 3 h by using a protocol of commercial origin. At timed intervals, aliquots were analyzed under conditions where free nucleotides, abortive products, and runoff transcripts could all be visualized and quantified (Fig. 4A). Transcripts were labeled with [α-32P]ATP, because ATP should be incorporated faster than other NTPs (A occurs 33-fold in the 95-nt transcript and 2-fold in the abundant 4- to 6-mer abortive products). Initially, the runoff transcripts accumulated 3-fold faster with the mutant than with the WT enzyme. Upon standing, the 95-nt runoff transcript was itself slowly extended by T7 RNAP, as often observed with transcripts that can self-anneal and are present in large excess over template (22). With both enzymes, ATP was almost quantitatively incorporated over time. With the WT enzyme, this incorporation was slow (15% free ATP remaining after 3 h); initially, the runoff (and extended) products predominated over abortive transcripts, but they leveled off first as the nucleotide concentration dropped, so that they never represented more than one third of the total input (Fig. 4B). With P266L, free ATP dropped much more rapidly (<10% after 1 h) and was virtually quantitatively incorporated into runoff (and extended) products. Thus, P266L appears superior to the WT enzyme from the viewpoint of rate of synthesis, maximal product yield, and low contamination with abortive products.
The Effect of the P266L Mutation Is Due to the Loss of Proline-266. The effect of P266L on processivity might reflect either the removal of the proline or the introduction of a leucine at position 266. To settle this point, we constructed three more mutants (P266A, P266S, and P266Y) and compared their processivity over the lac ITS to that of the WT and P266L enzymes (Fig. 4C). Although some differences were noted among mutants, they were all far more processive than the WT enzyme, showing that the removal of the conserved proline is mainly responsible for the observed processivity changes.
Discussion
A Mutation That Facilitates Promoter Clearance. To synthesize defined transcripts, all RNAPs must bind DNA in two successive modes, i.e., specific during promoter recognition and nonspecific during elongation. The transition from one mode to the other occurs over the ITS and it is characterized by a very low processivity, causing the repeated release of abortive transcripts before the enzyme eventually switches to elongation (3, 23, 24). For T7 RNAP, structural work has shown that a large region of the N-terminal domain (amino acids 44-266), which contributes most to promoter binding in the IC, undergoes a major structural rearrangement in the EC, where it interacts in particular with nascent RNA and DNA-RNA hybrid (1, 2, 25) (Fig. 5). By randomly mutagenizing the T7 RNAP gene and selecting in vivo for facilitated promoter clearance, we have isolated a mutation (P266L) that increases processivity over a subregion of the ITS (from +5 to about +8). Other substitutions for the conserved proline-266 appear to have the same effect. To our knowledge, no mutation with a similar phenotype has been described for a monomeric RNAP, and our efforts to isolate additional ones were largely frustrating. Using a PCR technique, we randomly mutagenized a fragment encoding amino acids 113-350 of T7 RNAP, including P266, and applied the same in vivo screen as above to search for mutants with facilitated promoter clearance. This strategy yielded three more mutants, all of which were doubly mutated (I117V V134A, I119V D147N, and H230R R291C) (J.G., unpublished data). However, in no cases did the effect of these mutations on β-galactosidase synthesis exceed one-third that of P266L (see Table 1); likewise, the increase of in vitro processivity was far more modest.
Fig. 5.
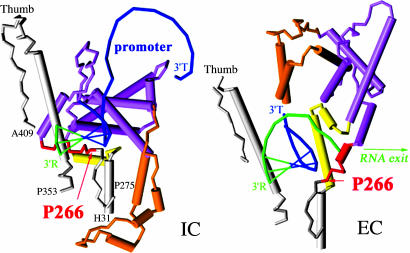
Structural differences between the IC with a 3-nt transcript (Left) and the EC (Right). The view is from the active site and toward the N-terminal domain, which rearranges in the IC → EC transition. Both IC and EC structures are seen as an α-carbon array, with α-helical regions shown as cylinders. Only amino acids 31-275 plus 353-409 (thumb) are shown. Sequences shown in gray (amino acids 31-43, 267-275, and 353-409), as well as regions not represented, are almost superposable in the two structures. The N-terminal rigid body (amino acids 72-151 and 206-257; purple), which prevents further extension of the 3-nt RNA-DNA hybrid in the IC, undergoes extensive displacement but no refolding in the EC, whereas the region separating the two parts of the rigid body (amino acids 152-205, orange) and the two linkers that connect it to invariant regions (amino acids 44-71, yellow; amino acids 258-266, red) undergo both displacement and refolding. The positions of a few selected amino acids, including P266 at the end of the C-terminal linker, are shown. Nucleic acids appear as phosphate backbones. Template strand (T) is shown in blue and nascent RNA (R) is shown in green. Nontemplate strand is omitted. Base pairings are shown as filled bars. Drawings were generated from the Protein Data Bank files 1QLN and 1MSW by using rasmol v.2 and pov-ray 3.1.r1 software (www.openrasmol.org and www.povray.org).
Although unprecedented in T7 RNAP, mutations that facilitate promoter clearance have been isolated in E. coli RNAP (26). In particular, two mutations in the σ70 subunit (P504L and S506F) that reduce abortive transcription and favor productive transcription from the highly abortive gal P2 promoter have been characterized (27). However, it is unlikely that these mutations are functionally equivalent to P266L. Both mutations are located in a region of σ that obstructs the RNA exit channel in the open complex, and they presumably act by alleviating this obstruction (27). In contrast, at nucleotides 5-8 where the effect of P266L is observed, the RNA is still fully hybridized to the template (28). In fact, given its location on the structure, the P266L mutation may act by facilitating the major rearrangement of the T7 RNAP N-terminal region (see below), which might have no counterpart in multisubunit RNAPs (1).
Structural Considerations. Besides increasing the initial processivity, the P266L mutation also decreases the affinity of the enzyme for the promoter (Fig. 3). By favoring early promoter release, this lowered affinity may be the cause of the increased initial processivity. Although this interpretation cannot be ruled out at this time (but see ref. 3), the very location of P266 on the structure also suggests an alternate interpretation. P266 is the last residue of the “C-linker,” which connects the invariant C-terminal domain (amino acids 267-883) to a large structural element (amino acids 72-151 and 206-257) that contacts the promoter in the IC and moves as a rigid body in the IC → EC rearrangement (refs. 1 and 2; Fig. 5). Although the detailed pathway of this rearrangement is not known, it is clear that it is a multistep process. Indeed, the IC structure must start rearranging at +4 to avoid a steric clash between the growing DNA-RNA hybrid and the N-terminal rigid body (29), yet biochemical data show that contacts with the promoter are maintained to position about +8 (30). Intriguingly, these positions match approximatively the boundaries of the region where P266L affects processivity. The likely pathway of the rearrangement at this stage has been modeled (2, 31). In particular, a recent study suggests that a simple translation of the rigid N-terminal body away from the active site can allow transcription to proceed to at least +6 while retaining enzyme-promoter contacts. During this process, the C-linker would reorient extensively (supplementary material in ref. 31). Possibly, the replacement of P266 by another residue adds flexibility there, facilitating the reorientation and ultimately stabilizing the complex during this step. Interestingly, it has been observed that, conversely, two mutants carrying extra prolines in this region (G259P A260P G263P and G259P A260P G264A) were inactive (W. T. McAllister, personal communication). However, although we find the comparison between modeling studies and P266L effects to be suggestive, it remains speculative at this time; understanding the effect of the mutation will presumably require more biochemical and structural work.
An Improved Enzyme for Preparing RNA in Vitro. With T7 RNAP, open complex formation is fast and reversible and promoter clearance usually constitutes the slow step in transcription when the ITS is noncognate (see Introduction). The rate of productive transcription will then reflect both the thermodynamics of open complex formation and the rate of promoter clearance. By disfavoring the former and increasing the latter, the P266L mutation may therefore affect the rate of productive transcription in two opposite ways. In practice, when the ITS is prone to abortive cycling at positions 5-8, the net effect of the mutation is to increase this rate in vitro (compare Figs. 2B, 4B, and 6). Presumably, given the relatively large enzyme concentrations used in these assays (0.1-1 μM), the positive effect on promoter clearance predominates. In contrast, when clearance is favored (g10 ITS), the mutation brings no increase or even a slight decrease in transcription rate (Fig. 2B and data not shown). These results suggest that the rate of productive transcription will generally be far less ITS-dependent with the P266L mutant than with the WT enzyme. The mutant also should increase the maximum transcript yield by reducing the fraction of NTPs wasted in abortive products (Fig. 4A), a valuable feature when this fraction is high (short runoff RNAs and noncognate ITS). Finally, in all cases, the mutant should facilitate product purification by reducing contaminating abortive products.
Supplementary Material
Acknowledgments
We thank Dr. Rui Sousa for valuable comments and Dr. W. T. McAllister for permission to cite unpublished results. This work is supported by the Centre National de la Recherche Scientifique, the École Normale Supérieure, the Association pour la Recherche sur le Cancer (Grant 4633), and grants from the Ministère de l'Éducation Nationale, de la Recherche et de la Technologie (Action Concertées Incitatives Microbiologie and Dynamique et Réactivité des Assemblages Biologiques) (to M.D.).
Author contributions: J.G. and M.D. designed research; J.G., P.J.L., F.P., and H.L. performed research; J.G. and P.J.L. analyzed data; and M.D. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ITS, initially transcribed sequence; IC, initial complex; EC, elongation complex; RNAP, RNA polymerase.
References
- 1.Yin, Y. W. & Steitz, T. A. (2002) Science 298, 1387-1395. [DOI] [PubMed] [Google Scholar]
- 2.Tahirov, T. H., Temiakov, D., Anikin, M., Patlan, V., McAllister, W. T., Vassylyev, D. G. & Yokoyama, S. (2002) Nature 420, 43-50. [DOI] [PubMed] [Google Scholar]
- 3.Martin, C. T., Muller, D. K. & Coleman, J. E. (1988) Biochemistry 27, 3966-3974. [DOI] [PubMed] [Google Scholar]
- 4.Ikeda, R. A. (1992) J. Biol. Chem. 267, 11322-11328. [PubMed] [Google Scholar]
- 5.Bonner, G., Lafer, E. M. & Sousa, R. (1994) J. Biol. Chem. 269, 25120-25128. [PubMed] [Google Scholar]
- 6.He, B., Rong, M., Durbin, R. K. & McAllister, W. T. (1997) J. Mol. Biol. 265, 275-288. [DOI] [PubMed] [Google Scholar]
- 7.Jia, Y. & Patel, S. S. (1997) Biochemisty 36, 4223-4232. [DOI] [PubMed] [Google Scholar]
- 8.Skinner, G. M., Baumann, C. G., Quinn, D. M., Molloy, J. E. & Hoggett, J. G. (2004) J. Biol. Chem. 279, 3239-3244. [DOI] [PubMed] [Google Scholar]
- 9.Lopez, P. J., Guillerez, J., Sousa, R. & Dreyfus, M. (1997) J. Mol. Biol. 269, 41-51. [DOI] [PubMed] [Google Scholar]
- 10.Milligan, J. F., Groebe, D. R., Witherell, G. W. & Uhlenbeck, O. C. (1987) Nucleic Acids Res. 15, 8783-8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davanloo, P., Rosenberg, A. H., Dunn, J. J. & Studier, F. W. (1984) Proc. Natl. Acad. Sci. USA 81, 2035-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makarova, O. V., Makarov, E. M., Sousa, R. & Dreyfus, M. (1995) Proc. Natl. Acad. Sci. USA 92, 12250-12254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Humphreys, G. O., Willshaw, G. A., Smith, H. R. & Anderson, E. S. (1976) Mol. Gen. Genet. 145, 101-108. [DOI] [PubMed] [Google Scholar]
- 14.He, B., Rong, M., Lyakhov, D., Gartenstein, H., Diaz, G., Castagna, R., McAllister, W. T. & Durbin, R. K. (1997) Protein Expression Purif. 9, 142-151. [DOI] [PubMed] [Google Scholar]
- 15.McAllister, W. T. & Raskin, C. A. (1993) Mol. Microbiol. 10, 1-6. [DOI] [PubMed] [Google Scholar]
- 16.McClure, W. R. & Chow, Y. (1980) Methods Enzymol. 64, 277-297. [DOI] [PubMed] [Google Scholar]
- 17.Gong, P., Esposito, E. A. & Martin, C. T. (2004) J. Biol. Chem. 279, 44277-44285. [DOI] [PubMed] [Google Scholar]
- 18.Guajardo, R., Lopez, P. J., Dreyfus, M. & Sousa, R. (1998) J. Mol. Biol. 281, 777-792. [DOI] [PubMed] [Google Scholar]
- 19.Brieba, L. G. & Sousa, R. (2001) Biochemistry 40, 3882-3890. [DOI] [PubMed] [Google Scholar]
- 20.Villemain, J., Guajardo, R. & Sousa, R. (1997) J. Mol. Biol. 273, 958-977. [DOI] [PubMed] [Google Scholar]
- 21.Ikeda, R. A. & Richardson, C. C. (1986) Proc. Natl. Acad. Sci. USA 83, 3614-3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cazenave, C. & Uhlenbeck, O. C. (1994) Proc. Natl. Acad. Sci. USA 91, 6972-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carpousis, A. J. & Gralla, J. D. (1980) Biochemistry 19, 3245-3253. [DOI] [PubMed] [Google Scholar]
- 24.Holstege, F. C. P., Fiedler, U. & Timmers, H. T. M. (1997) EMBO J. 16, 7468-7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steitz, T. A. (2004) Curr. Opin. Struct. Biol. 14, 4-9. [DOI] [PubMed] [Google Scholar]
- 26.Hsu, L. M. (2002) Biochim. Biophys. Acta 1577, 191-207. [DOI] [PubMed] [Google Scholar]
- 27.Cashel, M., Hsu, L. M. & Hernandez, V. J. (2003) J. Biol. Chem. 278, 5539-5547. [DOI] [PubMed] [Google Scholar]
- 28.Liu, C. & Martin, C. T. (2002) J. Biol. Chem. 277, 2725-2731. [DOI] [PubMed] [Google Scholar]
- 29.Cheetham, G. M. & Steitz, T. A. (1999) Science 286, 2305-2309. [DOI] [PubMed] [Google Scholar]
- 30.Brieba, L. G. & Sousa, R. (2001) EMBO J. 20, 6826-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Theis, K., Gong, P. & Martin, C. T. (2004) Biochemistry 43, 12709-12715. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.