Abstract
The function of the virion-associated protein (VAP) of cauliflower mosaic virus (CaMV) has long been only poorly understood. VAP is associated with the virion but is dispensable for virus morphogenesis and replication. It mediates virus transmission by aphids through simultaneous interaction with both the aphid transmission factor and the virion. However, although insect transmission is not fundamental to CaMV survival, VAP is indispensable for spreading the virus infection within the host plant. We used a GST pull-down technique to demonstrate that VAP interacts with the viral movement protein through coiled-coil domains and surface plasmon resonance to measure the interaction kinetics. We mapped the movement protein coiled-coil to the C terminus of the protein and proved that it self-assembles as a trimer. Immunogold labeling/electron microscopy revealed that the VAP and viral movement protein colocalize on CaMV particles within plasmodesmata. These results highlight the multifunctional potential of the VAP protein conferred by its efficient coiled-coil interaction system and show a plant virus possessing a surface-exposed protein (VAP) mediating viral entry into host cells.
Keywords: movement protein, virion-associated protein, Biacore
Cauliflower mosaic virus (CaMV) is the type member of the Caulimoviridae family (1). Its double-stranded DNA genome contains six major ORFs encoding the functional and structural viral proteins (2). Of these ORFs, only the capsid protein (and/or its polyprotein precursor GAG), the polymerase polyprotein, and the transactivator/viroplasmin are essential for virus replication (2, 3), whereas the movement protein (MP) and the virion-associated protein (VAP) are additionally required for induction of plant disease (4-6).
VAP, a rather small protein (15 kDa) with a bipolar functional structure (Fig. 1), associates with capsid protein in virion shells through its C-terminal region (7, 8). Its N-terminal region has a high α-helical content and harbors a coiled-coil motif assembling into a parallel tetramer with a rod-like structure (9). The tetramer is the stable form of VAP found in infected plants (10). VAP also contains a nonsequence-specific nucleic acid-binding domain mapped at the C-terminal proline-rich domain between amino acids 112-126 (11).
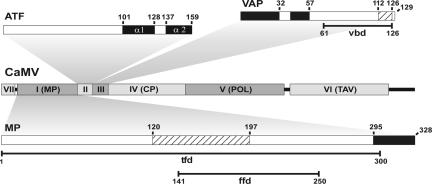
Fig. 1.
Genome organization of CaMV ORFs I-VII, functional maps of MP, ATF, and VAP. CP, capsid precursor polyprotein; POL, polymerase polyprotein (reverse transcriptase/RNase H and aspartic protease); TAV, transactivator/viroplasmin. MP, VAP, and ATF are depicted by boxes with amino acid numbers indicated. Hatched boxes in MP and VAP represent the nucleic acid-binding domains, and the black boxes represent the coiled coils. tfd, tubule-forming domain; ffd, focus-forming domain; vbd, virus-binding domain.
In both plant and animal systems, coiled-coil domains mediate not only protein homo-oligomerization but also interaction between distinct proteins (12). Based on its bipolar structure, we speculated that VAP, anchored by its C-terminal domain in the virus capsid, could act as the virion “arm,” mediating important CaMV functions through interaction with other proteins. Indeed, the VAP coiled-coil cooperates with the aphid transmission factor (ATF), the 18-kDa nonstructural protein encoded by CaMV ORF II, which oligomerizes in a parallel-oriented trimer (Fig. 1 and refs. 13 and 14). The N-terminal region of ATF interacts with the inner lining of the aphid stylet, exposing its C-terminal α-helical domains for the coiled-coil-dependent association with VAP (14), which itself is bound to virions (7). Thus, VAP mediates CaMV transmission by aphids through coiled-coil interactions with ATF.
Transmission by aphids is an important function but is not required for virus infection of a single plant host. The finding that plant infection through mechanical inoculation of a VAP-defective CaMV strain is unsuccessful (6), whereas virus replication in a single cell is independent of this protein (5), suggests that VAP not only mediates aphid transmission but could also be involved in virus movement.
Two different mechanisms of virus cell-to-cell movement have been recognized and studied extensively. In one, MP is associated with viral RNA in a nucleoprotein complex that moves to the neighboring cell (15); in the other, the protein modifies plasmodesmata by insertion of tubular structures, allowing entire virus particles to be transported (16). CaMV MP (encoded by ORF I; Fig. 1 and ref. 4) features both RNA binding activity (17, 18) and formation of tubules containing virus particles that protrude from infected protoplasts (19). However, a number of lines of evidence confirm that CaMV moves as virions through tubules, and the biological relevance of the RNA binding activity remains to be determined. MP accumulates in foci at the periphery of transfected protoplasts, and the tubules it forms extend beyond into the medium (19, 20). The functional domains of MP involved in both these functions have been determined and mapped (Fig. 1 and refs. 21 and 22). CaMV MP is the only component of tubules (23); nevertheless, it colocalizes with the Arabidopsis thaliana MPI7 protein in the cell periphery foci (24), suggesting the possible involvement of host factors in virus movement.
Here, we investigate the involvement of VAP in movement of CaMV. We show that MP, like ATF, oligomerizes as a trimer through a C-terminally located coiled-coil domain and, through the same domain, interacts with the coiled-coil of the VAP located on the virion surface. The MP-VAP interaction occurs within plasmodesmata-containing virus particles.
Materials and Methods
Protein Expression and Purification. CaMV ORF I (encoding MP), ORF III (encoding VAP), and derivatives thereof were amplified by PCR from CaMV clone pCa37 (25) by using gene-specific oligonucleotides containing appropriate restriction endonuclease recognition sequences at the precise extremity of each gene. PCR products were cloned into the BamHI-EcoRI sites of pGEX-2TK (Amersham Pharmacia), which contains a thrombin recognition site, as in-frame fusions with the GST domain, and were overexpressed in Escherichia coli BL21. ORF III was also cloned in the NcoI-BamHI sites of pET-3d (Novagen) and expressed in the same cells. MP and VAP expression were induced by growing the cell cultures for 3 h with 0.4 and 0.2 mM IPTG, respectively.
For Biacore surface plasmon resonance assays, GST-tagged proteins were purified on GSTrap FF columns (Amersham Pharmacia). While bound to the column, proteins were cleaved from GST by a 16-h incubation with thrombin protease (Amersham Pharmacia) at 22°C with gentle rotation. Excess protease was removed, allowing cleaved protein to flow through a HiTrap Benzamidine FF column (Amersham Pharmacia). All purification steps were performed according to the manufacturers' instructions.
GST Pull-Down Assay. The pull-down assay was performed essentially as described in ref. 26 with some modifications. The challenging proteins were produced by overexpression in E. coli BL21. Supernatants obtained after bacterial lysis and centrifugation were incubated for 1 h with proteins fused to GST already bound to beads. Aliquots (5-10 μl) of the bound fraction were separated by SDS/PAGE, and proteins detected by Western blotting by using anti-VAP-coiled-coil antibody (7).
Cross-Linking. Chemical cross-linking of purified MP was performed in 20 mM sodium phosphate buffer/150 mM NaCl (pH 7) at a concentration of 0.3 mg/ml with 1 or 5 mM of Bis 2-sulfosuccinimidooxycarbonyloxyethylsulfone (Pierce) for 2 h at 4°C. The reaction was quenched for 20 min with 50 mM Tris buffer, pH 7.5. Products were analyzed by Western blotting with a polyclonal antibody obtained from a rabbit immunized with MP coiled-coil peptide. Oxidative disulfide cross-linking of the MP coiled-coil peptide was performed as described in ref. 9.
Size Exclusion Chromatography. The MP coiled-coil peptide was separated on SUPERDEX 200 resin prepacked in a PC 3.2/30 column according to the manufacturer's instructions (Amersham Pharmacia), collecting 0.5-ml aliquots of the free-flow rate. The column was calibrated with chymotrypsinogen A (25 kDa), cytochrome c (12.4 kDa), and VAP pep(wt) (4,149 Da; ref. 9).
CaMV Purification. CaMV strain CabbS was propagated in Brassica rapa cv. “Just Right” and virus purified as described by Gardner and Shepherd (27) with an additional centrifugation step through a 10-40% sucrose gradient. Purified virions were isolated by high-speed centrifugation (45,000 × g for 1 h) of the band collected from the gradient diluted 1:1 in water.
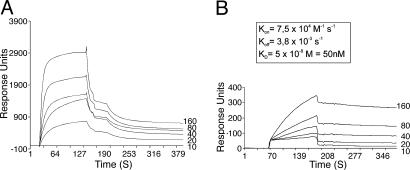
Surface Plasmon Resonance (SPR). SPR is a label-free technology for monitoring biomolecular interactions as they occur. It measures mass changes induced by association or dissociation between an immobilized analyte and a binding partner. A Biacore X optical biosensor system (Biacore, Neuchâtel, Switzerland) was used to measure the kinetics of molecular interactions, expressed in response units. One thousand response units of purified MP, diluted in 10 mM sodium acetate (pH 4.0), were immobilized on the dextran matrix of a CM5 sensor chip by using an amine coupling kit according to the manufacturer's instructions. Real-time interaction analysis was performed by injecting different concentrations of VAP (ranging from 10 to 160 μg/ml) in the absence or presence of virus onto the MP-coated chip. Binding of VAP to an excess of CaMV virions (150 μg/ml) was performed by overnight incubation at 4°C with gentle shaking. All binding experiments were carried out in Hepes-buffered saline (10 mM Hepes/0.15 M NaCl/3 mM EDTA/0.005% surfactant P20), at 25°C and at a flow rate of 20 μg/ml. Ten millimolar HCl pulses were used to regenerate the chip surface. Kinetic rate constants (Kon, Koff, and Kd; see Fig. 6) were calculated on the basis of a single-site model by using Biacore biaevaluation 3.0 software.
Fig. 6.
Biacore SPR analysis of the interaction among MP, VAP, and virions. Sensograms show real-time interaction of immobilized MP with increasing concentrations (10-160 μg/ml, as indicated adjacent to traces) of VAP (A) and VAP associated with CaMV virions (B). A steep upward slope defines the onset of binding partner injection (VAP in A and VAP-virion complex in B). The adjacent ascending regions illustrate association between test partners. The abrupt downward slopes correspond to the end of the injection period and define the start of binding partner dissociation (descending curve). (The steepest parts of the graphs, almost parallel to the y axis, reflect only changes in buffer conditions and do not represent interaction dynamics). Kon, association constant; Koff dissociation constant; Kd = Koff/Kon, equilibrium dissociation constant.
Electron Microscopy. Brassica rapa cv. “Just Right” leaf tissue systemically infected with CaMV was fixed in 4% glutaraldehyde in Pipes (pH 7.4) for 3 h at room temperature under mild vacuum. After extensive washing in Pipes, samples were dehydrated in a graded ethanol series and embedded in LR White resin. Ultra-thin sections of these samples were used for double immunogold labeling as described in ref. 28 with some modifications (see Supporting Methods and Fig. 8, which are published as supporting information on the PNAS web site, for detailed methods). Sections were stained with 2% aqueous uranyl acetate for 15 min and lead citrate for 5 min before viewing with a Zeiss EM 910 transmission electron microscope operated at 80 kV.
Protein Computational Analysis. seqweb, the web-based version of the Wisconsin Package Version 10.2 (Genetics Computer Group, Madison, WI) was used for all protein sequence analyses. Coiled-coils were predicted with coilscan (29, 30).
Results
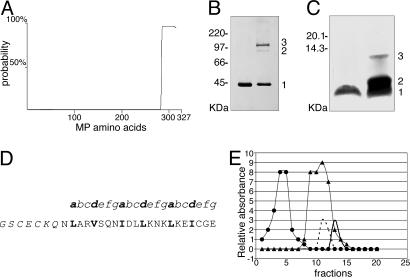
A Coiled-Coil Domain Predicted at the C Terminus of MP Assembles as a Trimer. Inspection of the CaMV MP sequence revealed the presence of a heptad repeat at the C terminus of the protein. In the same region, computer analysis predicts a coiled-coil domain with high probability (Fig. 2A and ref. 29). The ability of MP to form oligomeric structures was investigated by chemical cross-linking of the purified protein. Western blotting analysis with anti-MP antibody showed three forms of MP, corresponding in size to protein monomers, dimers, and trimers (Fig. 2B). Moreover, trimerization of a peptide corresponding to the coiled-coil domain alone demonstrated the latter's involvement in MP oligomerization (Fig. 2C). For this purpose, a peptide of 2,385 Da, encompassing the predicted coiled-coil of MP [coilMP(wt), residues 307-327], was synthesized. A derivative peptide, coilMP, with the addition of the sequence GSCECKQN to the N terminus of coilMP(wt) (Fig. 2D), was constructed to allow us to distinguish whether the peptide assembles to form trimers in parallel or antiparallel orientation. In vitro cysteine oxidative cross-linking would allow formation of S-S bridges between parallel-oriented monomers as the cysteine residues (in bold) would be located in close proximity in the oligomeric state, whereas no cross-linking of three monomers together would occur if the peptides assembled in antiparallel orientation. The highest band detected by SDS/PAGE under denaturing conditions without reducing agent showed that coilMP assembles into a parallelly oriented trimer (Fig. 2C). Chromatography confirmed the trimer as the most stable level of oligomerization of the peptide. CoilMP(wt) peptide loaded on a gel filtration column (Superdex 200) together with the standard molecular mass markers and coilVAP(wt), a synthetic peptide corresponding to the VAP coiled coil that elutes as a tetramer (9), eluted as a single peak with a mass <12 kDa (Fig. 2E), corresponding to the predicted trimer mass (7,155 Da). The absence of MP coiled-coil monomers and dimers suggests that all of the coilMP(wt) exists as a single population of trimers in solution, thus we interpret the presence of monomers and dimers in Fig. 2 B and C to be due to incomplete cross-linking. CoilMP and coilVAP eluted individually, and no heteromers of the two peptides were detected under these conditions.
Fig. 2.
Analysis of CaMV MP structure. (A) Computer prediction of coiled-coil domains within the MP sequence. The entire MP protein is represented on the x axis and coiled-coil probability (%) on the y axis. (B) Chemical cross-linking of MP analyzed by SDS/PAGE and detected by immunoblot analysis with anti-MP coiled-coil antibody. Positions of the monomer and the cross-linked dimers and trimers (1, 2, and 3, respectively) are indicated [CaMV MP (37 kDa) migrates anomalously on SDS/PAGE, ref. 21]. The product in the left lane was not treated with Bis 2-sulfosuccinimidooxycarbonyloxyethylsulfone. (C) Disulfide cross-linking of CoilMP analyzed by oxidative SDS/PAGE and silver staining. Positions of the peptide monomer and cross-linked dimers and trimers (1 = 3,235 Da, 2 = 6,470 Da, and 3 = 9,705 Da) are indicated. The product in the left lane was not treated with oxidized glutathione. (D) Synthetic peptide CoilMP, corresponding to the C-terminal α-helical domain of MP (nonnative amino acids in italics), with the seven amino acid unit (depicted as a-g) of the heptad repeat structure illustrated above; hydrophobic residues of the α-helix occupy positions a and d (highlighted in bold). (E) Separation of reduced CoilMP(wt) by gel filtration on Superdex 200 (Amersham Pharmacia). Standard molecular mass markers are chymotrypsinogen A (25 kDa, •) and cytochrome c (12.4 kDa, ▴). The elution peaks correspond to CoilMP(wt) peptide trimer (7,155 Da, solid black line), and CoilVAP(wt) peptide tetramer (16,596 Da, dashed black line). Protein concentration was determined by monitoring absorbance at 280 nm of each 0.5-ml elution fraction.
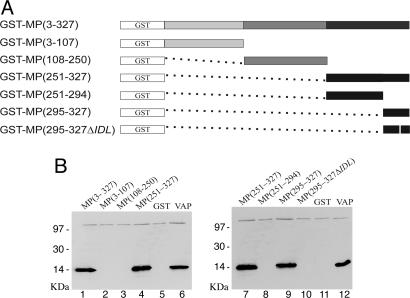
CaMV MP Interacts with VAP Through Its C-Terminal Region. A possible MP-VAP interaction was investigated further in GST pull-down experiments. A GST-MP fusion (Fig. 3A) was overexpressed and purified by using Glutathione-Sepharose 4B beads. The GST-MP-bead complex obtained was incubated with partially purified VAP produced in an E. coli expression system. After extensive washing, proteins bound to the beads were eluted and separated on SDS/PAGE. Western immunoblotting with antibody raised against VAP was used to reveal the presence of the protein in the eluted protein-bead complex. Using this technique, VAP was indeed found to associate with full-length MP (Fig. 3B, lane 1).
Fig. 3.
Mapping of MP domains involved in the interaction with VAP. (A)MP and MP deletion mutants are depicted by boxes of distinct colors with the construct names indicating the corresponding amino acid positions. White boxes represent GST, expressed in fusion with the proteins; black boxes represent the coiled-coil domain. Dotted lines replace deleted amino acids. The white vertical line in construct 295-327ΔIDL represents the three amino acid deletion. (B) Interaction of VAP with MP and its mutants fused to GST and with GST alone (GST, lanes 5 and 11). Proteins were separated by SDS/PAGE and detected by Western blotting with antibodies against VAP. Positions of molecular mass marker proteins are on the left of the gels. Lanes 6 and 12, VAP alone.
To map the MP domains involved in the interaction with VAP, N-terminal, central, and C-terminal fragments of MP were expressed in fusion to the C terminus of GST (Fig. 3A) and used in GST-pull down experiments. VAP associated with the C-terminal region of MP but not with other MP fragments or GST alone (Fig. 3B, lanes 1-5). Analysis of two further mutants of the GST-C-terminal fragment of MP [GST-MP (251-294) and GST-MP (295-327)] revealed the last 32 C-terminal residues of MP to be sufficient to support the interaction with VAP (Fig. 3B, lanes 8 and 9).
Thomas and Maule (31) determined the MP sequences required for virus infectivity by three-amino acid deletion scanning. All mutants unable to mediate systemic infection carried a deletion within the tubule-forming domain except one in which the mutation was located exactly in the region required for VAP binding. When this deletion, ΔIDL (amino acids 314-316), was introduced into GST-MP (295-327), VAP-binding capacity was lost (Fig. 3B, lane 10). This result suggests that the lack of infectivity of the MP(ΔIDL) mutant is due to the inability of MP to interact with VAP and, consequently, to mediate virus cell-to-cell movement.
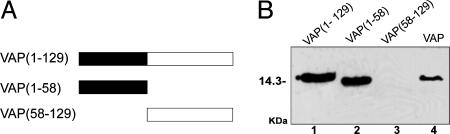
CaMV MP and VAP Interact Through Their Coiled-Coil Domains. The results obtained so far indicate a physical association of VAP with the coiled-coil domain of MP. To determine the region of VAP involved in this interaction, two deletion mutants of VAP, corresponding to the N-terminal coiled-coil (residues 1-58) and the C-terminal region (residues 58-129) were produced and cloned in the pET3a vector (Fig. 4A). GST-MP (295-327) was overexpressed, bound to beads and challenged with both VAP mutants. Only the N-terminal VAP mutant precipitated with the complex, thus confirming that MP and VAP associate through their coiled-coil domains (Fig. 4B).
Fig. 4.
Mapping of VAP domains involved in the interaction with MP. (A) VAP and VAP deletion mutants are depicted by boxes, with the construct names indicating the corresponding amino acid positions. The coiled-coil region is shaded black. (B) Lanes 1-3, interaction of GST-MP (295-327) with VAP and its mutants. Lane 4, VAP alone. Proteins were separated by SDS/PAGE and detected by Western blotting with antibodies against VAP. The position of the 14.3-kDa marker (lysozyme) is on the left of the gel.
Caulimovirus MP and VAP Colocalize in Plasmodesmata. The subcellular localization of MP and VAP, each expressed in fusion with the green fluorescent protein (GFP), has been investigated previously in protoplasts. MP preferentially aggregates in foci at the plasma membrane and in tubular structures emerging from the cell surface (20), whereas VAP forms large aggregates within the cell cytoplasm (L.S., unpublished data).
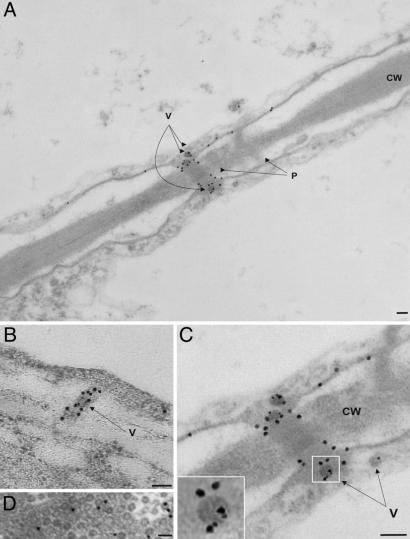
We investigated the localization of these proteins in CaMV-infected Brassica rapa leaf tissue by double immunogold electron microscopy with rabbit antibodies against MP and VAP and, as secondary antibodies, goat anti-rabbit immunoglobulins conjugated to 18- and 6-nm gold particles, respectively. MP was found to be associated with the cell membrane and concentrated within plasmodesmata (Fig. 5 A-C). Notably, MP is more abundant in the inner part of the plasmodesma (Fig. 5B), where the MP-formed tubules are believed to traverse the cell wall (19). No MP aggregates similar to those observed in infected insect cells (21) were found in infected turnip leaf tissue, suggesting that formation of such aggregates could involve other components that are specific to, and/or more abundant in, the insect system.
Fig. 5.
VAP and MP colocalize in modified plasmodesmata. Electron micrographs of ultrathin sections of infected tissue showing two neighboring cells. Immunogold labeling of VAP (6-nm gold particles) and MP (18-nm gold particles) shows the localization of the two proteins. (A) Two plasmodesmata (P) traversing the cell wall (CW) are indicated; one is modified and contains virus particles (V). (B) Longitudinal section of a modified plasmodesma containing CaMV particles (V) showing MP distribution. (C) Magnified image of the modified plasmodesma in A; Inset is enlarged to show MP and VAP localization on a single virion. (D) CaMV inclusion body containing virus particles. Gold particles (6-nm) label VAP. (Scale bars: 100 nm.)
VAP was never observed in isolation, being invariably associated with CaMV virions, located both within and outside the virus inclusion bodies (Fig. 5 and ref. 7). We observed that 18-nm and 6-nm gold particles were closely associated with each other only on virus particles within plasmodesmata (Fig. 5 A and C). This colocalization of MP and VAP was not seen in any other cell compartment, including virus particles close to plasmodesmata (Fig. 5 B and C). This finding suggests that the interaction between the two proteins most probably occurs at the entrance to, or during movement of virions through, the plasmodesmal cytoplasmic sleeve.
Affinity Between MP and VAP Depends on the Presence of CaMV Virions. The electron microscopic observations indicate that interaction between MP and VAP in vivo occurs only in the presence of CaMV virions in plasmodesmata. In addition, MP and VAP were never found associated when overexpressed in fusion with fluorescent proteins or when immunostained after fixation, either in protoplasts or in leaf tissue upon bombardment.
We used SPR to further investigate the affinity and dynamics of the MP-VAP association and the involvement of CaMV virions in the interaction.
Consistent with our in vitro GST pull-down experiments, VAP was trapped by MP immobilized on the sensor chip, albeit in limited amounts and through a rather fast dissociating interaction (Fig. 6A). The anomalous curves illustrating VAP dissociation from MP (Fig. 6A, irregular descending slopes) correlate with the multimeric state of VAP in solution and did not allow calculation of the corresponding kinetic constants.
When a VAP-CaMV virion complex was injected onto immobilized MP, a high-affinity interaction occurred between MP and VAP (Kd ≈ 50 nM, Fig. 6B; CaMV virions alone did not bind measurably to MP, data not shown). Indeed, when trapped in the virion, more VAP bound to MP and subsequent dissociation was much slower [compare ascending (association) and descending (dissociation) parts of the graphs between Fig. 6 A and B]. Thus, when associated with the virion, VAP interacts strongly and stably with MP. This result suggests that, under in vivo conditions, it is the VAP-virion complex, and not VAP alone, that interacts with MP. These results may account for the lack of interaction observed between MP and VAP when overexpressed in vivo.
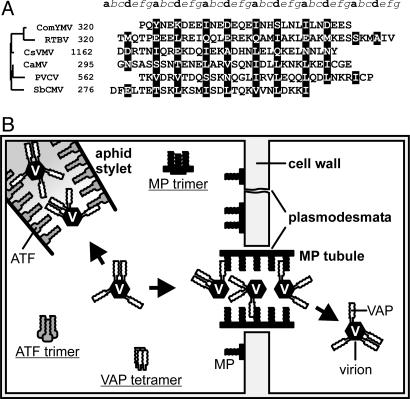
A Coiled-Coil Domain Is Conserved in the MPs of the Caulimoviridae. If, as we propose, the interaction between MP and VAP through coiled coils is essential for CaMV movement and spread of infection in the plant, we would expect this feature to be conserved in other members of the Caulimoviridae, a virus group characterized by well conserved genome architecture and functional strategies (2). Indeed, we previously demonstrated that tetramerization of VAP is a conserved feature of all members of the group (32). Amino acid sequence analysis, supported by specific bioinformatic tools (29), revealed that a coiled-coil motif is indeed conserved, albeit not always perfectly, at the MP C terminus of those caulimoviruses having a defined MP and 3′ of the conserved intercellular transport domain (18) of all other members of the Caulimoviridae family (Fig. 7A), highlighting the importance of both domain and structure in this virus family. The case of petunia vein clearing virus is slightly different in that it encodes a single viral polyprotein containing three closely spaced coiled-coil domains. The presumptive MP, containing only the first coiled-coil domain (Fig. 7A), can trimerize (L.S., unpublished data). This finding suggests that tetramerization of a presumptive petunia vein clearing virus VAP (32) would be directed by the other two coiled coils. However, further studies on petunia vein clearing virus posttranslational regulation are required to clarify whether VAP and MP functions are indeed conserved in this virus.
Fig. 7.
Prediction of Caulimoviridae MP coiled-coil domains and illustration of VAP-mediated CaMV functions. (A) MP C-terminal domains of representative members of the six genera of the Caulimoviridae family are aligned, the numbers representing amino acid position in each MP or polyprotein. The seven positions of the heptad repeat are labeled a-g above the alignment. Positions a and d, occupied by hydrophobic residues that are part of the oligomer core, are highlighted; the other positions are predominantly solvent-exposed polar residues. Sequences are aligned according to their phylogenetic relationships (shown on the left). Phylogeny was inferred from the intercellular transport movement domains located upstream of the coiled coils (corresponding to residues 127-157 in CaMV MP). Cassava vein mosaic virus, CsVMV; commelina yellow mottle virus, ComYMV; rice tungro bacilli-form virus, RTBV; soybean chlorotic mottle virus, SbCMV; petunia vein clearing virus, PVCV. (B) Schematic overview of VAP interactions mediating CaMV functions during the infection cycle. In the aphid stylet, VAP associated with the virion interacts with ATF to mediate aphid transmission. In the MP-modified plasmodesma, the VAP-virion complex interacts with MP, allowing movement to a neighboring cell. Also shown are MP and ATF trimers and VAP tetramers, which are known to occur in planta, although the function of these forms remains unknown. Interactions between the coiled coils of MP and VAP and ATF and VAP are shown only schematically and are not intended to depict the degree of coiled-coil multimerization.
Discussion
CaMV VAP and MP are both essential for the spread of infection in a host plant, but both are dispensable for replication in a single cell. In infected cells, VAP is always associated with the virion but is not an essential component of the viral capsid structure. The fact that VAP binds to virus particles when added externally (14) and is dispensable for genome packaging (5) indicates that VAP is not a scaffolding protein needed for virus assembly but is rather a late addition during the assembly process, suggesting a role in virus spread in the infected host plant.
Our results show that CaMV MP harbors a C-terminal coiled-coil domain, assembled as a parallel trimer, that interacts with the VAP N-terminal coiled coil, supporting the view that VAP is involved in virus movement. Although both proteins contain a nucleic acid binding domain, neither DNA nor RNA mediate the VAP-MP interaction, as shown by the efficient association of VAP and MP deletion mutants containing only the coiled coils and lacking the nucleic acid binding domain.
We have previously speculated that VAP mediates important CaMV functions through its bipolar structure, with one end anchored to the virion and the other, the coiled-coil domain, exposed for interaction with other proteins (32). However, despite the presence of VAP tetramers in planta (10), it remained hard to see how tetramers could fit into an icosahedral virion characterized by 2-, 3-, 5-, and 6-fold, but not 4-fold, symmetry (33). In fact, cryo-electron microscopy experiments have shown that binding to virions triggers a structural shift, changing VAP tetramers into trimeric oligomers emerging from the 3-fold symmetrical axes of the capsid (34). The coiled-coil domains of these parallel trimers become rearranged to form antiparallel dimers with a monomer from a trimer in close proximity, thus creating a network of VAP coiled-coil dimers covering the surface of the virion (34). If this behavior can be extrapolated to ATF and MP, all three of these coiled coil-containing proteins could be reoriented from parallel trimers or tetramers to associate as heterologous, antiparallel coiled-coil complexes that target the virion to different functions of the infectious cycle as depicted in Fig. 7B. In this model, we speculate that the function of the parallel coiled-coil tetrameric form of VAP found in planta could be to impede heterologous interactions until the proper partner is available. In addition, other transport-related functions possibly involving the tetrameric form of VAP, such as vesicular transport (also mediated by coiled-coil interactions; ref. 35), could be envisaged. Such strategies necessarily require strict regulation, the mechanism of which would be fundamental to targeting of different aspects of the infection cycle in CaMV. Thus, investigations are underway to understand how VAP opts between ATF and MP.
The coiled coil we mapped in MP shares three important features with the ATF coiled coil: they are both located in the C-terminal part of the proteins, assemble as parallel trimers, and interact with the amphipathic N-terminal coiled coil of VAP that is exposed on the external surface of the virus particle.
CaMV MP forms tubular structures predicted to cross the cell wall by passing through plasmodesmata (23). Structural analysis, performed by epitope tagging, located the N- and C-termini of MP on the protein surface on both the outer and inner surface of the tubules (31). The model suggested by our results is consistent with this prediction: MP coiled coils exposed inside the tubule are accessible for interaction with virus particles through VAP (Fig. 7B).
Tubules can form on the surface of CaMV-infected protoplasts upon transient expression of MP. Thus, no other virus protein is required for their formation, although host factors might be involved in folding and support of the tubule structure in the host tissue. How tubules are formed is currently unknown. However, the coiled-coil domain is the only part of MP not required for their assembly (20, 31), thus excluding a direct involvement of the MP trimer in tubule structure formation. Nevertheless, a possible role in stabilization or in virion spacing within the tubule cannot be ruled out, especially considering that, as for VAP in the virion shell, a structural shift of the trimer may appropriately reorient the oligomer. In support of such tempting speculations, the coiled-coil region is undoubtedly essential for virus spread within the host (31). Indeed, we demonstrated that a mutation in the MP coiled coil that hinders virus movement (31) also inhibits association with the VAP coiled coil. Interactions between such domains are highly specific as well as highly dependent on proper protein folding (12). Whereas the precise mechanism of virus particle movement through tubules is still unknown, our results indicate an essential role for VAP in mediating the association between virion and tubule through the VAP-MP interaction.
Our EM observations revealed that in infected cells, MP and VAP apparently associate only in the presence of virus particles passing through plasmodesmata. Biacore SPR investigations on the dynamics of the interaction showed that, as for VAP-ATF (36), the affinity of MP for VAP is highly compromised in the absence of virions. In particular, when the two proteins interact alone, faster dissociation is observed. If the MP-VAP association is indeed essential for virion movement through tubules, it is not surprising that the presence of virus particles favors maintenance of this interaction in vivo. Taken together, these observations suggest that the interaction between MP and VAP cannot occur in the cytoplasm, and that the two proteins probably move independently to the plasma membrane, with VAP comigrating with virions. MP can assume different conformations depending on its subcellular localization (31), and these conformations most probably correlate with different MP oligomeric structures that mediate distinct functions, e.g., tubule formation, binding/nonbinding to VAP, etc. Whether the virion-VAP complex interacts with MP before or after the latter forms tubule structures remains to be clarified. However, it is possible that interaction with VAP triggers the MP conformational modification necessary to allow virus movement through the tubules.
CaMV virions invade new cells only by passing through MP-modified plasmodesmata. Through the interaction with MP, VAP protruding from the virion allows virus penetration into neighboring cells. Therefore, like animal viruses, this plant virus has developed a system of cell entry mediated by surface-located viral proteins, albeit adapted to specific conditions in the host organism. Such comparisons justify parallel and complementary research lines also in terms of the evolutionary relationships linking plant and animal viruses (1).
VAP is a multifunctional protein, a tool produced and used by the virus to regulate various functional steps in the viral infection cycle. In a wider context, VAP-ATF/MP is a simple system that can be used to study general regulation of coiled-coil interactions. The α-helical coiled coil is one of the most common oligomerization motifs in proteins (12), and the CaMV model system could be of great benefit in furthering our understanding of a general strategy used by proteins to mediate biological function.
Supplementary Material
Acknowledgments
We thank Helge Rixner for expert technical assistance and advice with EM, Franz Fischer for peptide synthesis, Donato Boscia for producing anti-MP coiled-coil antibody, Mike Rothnie for graphical work, Helen Rothnie and Marcello Russo for critical reading of the manuscript, and Angiola Desiderio, Etienne Herzog, and Kappei Kobayashi for many valuable discussions.
Author contributions: L.S. designed research; L.S., M.E.V., and D.L. performed research; M.E.V. contributed new reagents/analytic tools; L.S., M.E.V., D.L., and T.H. analyzed data; and L.S. and T.H. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ATF, aphid transmission factor; CaMV, cauliflower mosaic virus; MP, movement protein; SPR, surface plasmon resonance; VAP, virion-associated protein.
References
- 1.Rothnie, H. M., Chapdelaine, Y. & Hohn, T. (1994) Adv. Virus Res. 44, 1-67. [DOI] [PubMed] [Google Scholar]
- 2.Hohn, T. & Fütterer, J. (1997) Crit. Rev. Plant Sci. 16, 133-161. [Google Scholar]
- 3.Kobayashi, K. & Hohn, T. (2003) J. Virol. 77, 8577-8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas, C. L., Perbal, C. & Maule, A. J. (1993) Virology 192, 415-421. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi, K., Tsuge, S., Stavolone, L. & Hohn, T. (2002) J. Virol. 76, 9457-9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacquot, E., Geldreich, A., Keller, M. & Yot, P. (1998) Virology 242, 395-402. [DOI] [PubMed] [Google Scholar]
- 7.Leclerc, D., Stavolone, L., Meier, E., Guerra-Peraza, O., Herzog, E. & Hohn, T. (2001) Virus Genes 22, 159-165. [DOI] [PubMed] [Google Scholar]
- 8.Leh, V., Jacquot, E., Geldreich, A., Haas, M., Blanc, S., Keller, M. & Yot, P. (2001) J. Virol. 75, 100-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leclerc, D., Burri, L., Kajava, A. V., Mougeot, J. L., Hess, D., Lustig, A., Kleemann, G. & Hohn, T. (1998) J. Biol. Chem. 273, 29015-29021. [DOI] [PubMed] [Google Scholar]
- 10.Tsuge, S., Kobayashi, K., Nakayashiki, H., Mise, K. & Furusawa, I. (1999) Microbiol. Immunol. 43, 773-780. [DOI] [PubMed] [Google Scholar]
- 11.Mougeot, J. L., Guidasci, T., Wurch, T., Lebeurier, G. & Mesnard, J. M. (1993) Proc. Natl. Acad. Sci. USA 90, 1470-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burkhard, P., Stetefeld, J. & Strelkov, S. V. (2001) Trends Cell Biol. 11, 82-88. [DOI] [PubMed] [Google Scholar]
- 13.Hebrard, E., Drucker, M., Leclerc, D., Hohn, T., Uzest, M., Froissart, R., Strub, J. M., Sanglier, S., van Dorsselaer, A., Padilla, A., et al. (2001) J. Virol. 75, 8538-8546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leh, V., Jacquot, E., Geldreich, A., Hermann, T., Leclerc, D., Cerutti, M., Yot, P., Keller, M. & Blanc, S. (1999) EMBO J. 18, 7077-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujiwara, T., Giesman-Cookmeyer, D., Ding, B., Lommel, S. A. & Lucas, W. J. (1993) Plant Cell 5, 1783-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Lent, J., Storms, M., van der, M. F., Wellink, J. & Goldbach, R. (1991) J. Gen. Virol. 72, 2615-2623. [DOI] [PubMed] [Google Scholar]
- 17.Thomas, C. L. & Maule, A. J. (1995) Virology 206, 1145-1149. [DOI] [PubMed] [Google Scholar]
- 18.Koonin, E. V., Mushegian, A. R., Ryabov, E. V. & Dolja, V. V. (1991) J. Gen. Virol. 72, 2895-2903. [DOI] [PubMed] [Google Scholar]
- 19.Perbal, M. C., Thomas, C. L. & Maule, A. J. (1993) Virology 195, 281-285. [DOI] [PubMed] [Google Scholar]
- 20.Huang, Z., Han, Y. & Howell, S. H. (2000) Virology 271, 58-64. [DOI] [PubMed] [Google Scholar]
- 21.Thomas, C. L. & Maule, A. J. (1999) J. Virol. 73, 7886-7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, Z., Han, Y. & Howell, S. H. (2001) Mol. Plant-Microbe. Interact. 14, 1026-1031. [DOI] [PubMed] [Google Scholar]
- 23.Kasteel, D. T., Perbal, M. C., Boyer, J. C., Wellink, J., Goldbach, R. W., Maule, A. J. & van Lent, J. W. (1996) J. Gen. Virol. 77, 2857-2864. [DOI] [PubMed] [Google Scholar]
- 24.Huang, Z., Andrianov, V. M., Han, Y. & Howell, S. H. (2001) Plant Mol. Biol. 47, 663-675. [DOI] [PubMed] [Google Scholar]
- 25.Lebeurier, G., Hirth, L., Hohn, T. & Hohn, B. (1980) Gene 12, 139-146. [DOI] [PubMed] [Google Scholar]
- 26.Herzog, E., Guerra-Peraza, O. & Hohn, T. (2000) J. Virol. 74, 2073-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gardner, R. C. & Shepherd, R. J. (1980) Virology 106, 159-161. [DOI] [PubMed] [Google Scholar]
- 28.Faoro, F., Tornaghi, G. & Belli, G. (1991) J. Phytopathol. 133, 297-306. [Google Scholar]
- 29.Lupas, A. (1996) Methods Enzymol. 266, 513-525. [DOI] [PubMed] [Google Scholar]
- 30.Lupas, A., Van Dyke, M. & Stock, J. (1991) Science 252, 1162-1164. [DOI] [PubMed] [Google Scholar]
- 31.Thomas, C. L. & Maule, A. J. (1995) Plant Cell 7, 561-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stavolone, L., Herzog, E., Leclerc, D. & Hohn, T. (2001) J. Virol. 75, 7739-7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng, R. H., Olson, N. H. & Baker, T. S. (1992) Virology 186, 655-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plisson, C., Uzest, M., Drucker, M., Froissart, R., Dumas, C., Conway, J., Thomas, D., Blanc, S. & Bron, P. (2005) J. Mol. Biol. 346, 267-277. [DOI] [PubMed] [Google Scholar]
- 35.Clague, M. J. (1999) Curr. Biol. 9, R258-R260. [DOI] [PubMed] [Google Scholar]
- 36.Drucker, M., Froissart, R., Hebrard, E., Uzest, M., Ravallec, M., Esperandieu, P., Mani, J. C., Pugniere, M., Roquet, F., Fereres, A. & Blanc, S. (2002) Proc. Natl. Acad. Sci. USA 99, 2422-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.