Abstract
The existence of aneuploid cells within the mammalian brain has suggested the influence of genetic mosaicism on normal neural circuitry. However, aneuploid cells might instead be glia, nonneural, or dying cells, which are irrelevant to direct neuronal signaling. Combining retrograde labeling with FISH for chromosome-specific loci, distantly labeled aneuploid neurons were observed in expected anatomical projection areas. Coincident labeling for immediate early gene expression indicated that these aneuploid neurons were functionally active. These results demonstrate that functioning neurons with aneuploid genomes form genetically mosaic neural circuitries as part of the normal organization of the mammalian brain.
Keywords: cerebral cortex, chromosome paint, mosacism
Aneuploidy, the loss and/or gain of chromosomes producing numerical deviation from haploid multiples, has documented effects on cellular physiology, particularly in pathophysiological settings such as cancer and Down's syndrome (1, 2). Whereas even the quantitative change of a single-gene product can produce major changes in cellular signaling (3), the effects associated with loss or gain of ≈1,000 gene copies on an average chromosome should have even more pronounced changes on a cell's physiology. Approximately 33% of neural progenitor cells display genetic variability, manifested as chromosomal aneuploidy that encompasses both loss and gain of chromosomes (4-7). In the mature brain, aneuploid cells that express neuronal markers have been observed (4, 6). However, an unanswered question is whether these neurons represent functional rather than dying cells, with death being a common fate for aneuploid cells in other systems (8, 9). Toward determining their relevance for adult brain function, we examined aneuploid cells in the normal brain for anatomical connectivity and functional activity.
Methods
Procedures involving live animals were conducted at the University of California at San Diego (UCSD), approved by the Animal Subjects Committee at UCSD, and conform to National Institutes of Health guidelines and public law.
Activation Paradigm for Immediate Early Gene (IEG) Induction. All animals were transferred from the home cage to a clean cage 1 h before killing. To maximize the activation of cortical cells, two mice received additional stimulation that included exposure to a novel male (10) and to various odors (11). Specifically, a novel male was placed in the same cage as the subject male, and the males were allowed to interact for 1-3 min. After the removal of the novel male, subject males were then exposed sequentially to peppermint and banana odors for 15 min each while remaining in the clean cage. Animals were killed 1 h after the beginning of the exposure paradigm, perfused with 0.9% saline, followed by 4% paraformaldehyde in 0.1 M PBS (pH 7.4), and brain tissue was processed as described below.
Retrograde Tracer Injections. Ten male 8-week-old mice from three different strains (BalbC/CR, Swiss Webster, and c129SvJ) received tracer injections. Mice were anesthetized with a 1.2% avertin solution (0.2 ml per 10 g of body weight or 240 mg/g of body weight). A solution of 4% FluoroGold (Fluorochrome, Denver) was injected into the olfactory bulb(s), left motor cortex, and/or somatosensory cortex by using a Hamilton syringe. Alternatively, a small piece of gelfoam soaked in 4% FluoroGold and air-dried was implanted into the areas mentioned above. We chose two injection sites per animal to maximize the number of retrogradely labeled neurons. After 48 h, mice were killed and perfused with 0.9% saline followed by 4% paraformaldehyde in 0.1 M PBS or periodate-lysine-paraformaldehyde fixative (4% paraformaldehyde, 0.1 M l-lysine, and 0.01 M sodium metaperiodate) (12). Brains were removed, postfixed for 2 h, and cryoprotected in sucrose. Brains were then embedded in Tissue-Tek (Sakura, Torrance, CA) and rapidly frozen on dry ice. Tissue was cut coronally at 10 μm on a cryostat, mounted onto Superfrost Plus slides (Fisher Scientific, Pittsburgh), and stored at -20°C.
FluoroGold Immunohistochemistry. To stabilize FluoroGold in back-filled cells, a primary antibody against FluoroGold (Fluorochrome) was used in conjunction with a biotinylated secondary antibody (Vector Laboratories), an avidin:biotinylated enzyme complex (Vectastain ABC elite kit, Vector Laboratories), and 3,3′-diaminobenzidine substrate kit (Vector Laboratories). Tissue sections were incubated in blocking solution (2.5% BSA, 0.3% Triton X-100, and 0.02% sodium azide) for 1 h and rabbit α-FluoroGold (1:50,000; Fluorochrome) for 48 h. Tissue was then washed in PBS and incubated with biotinylated α-rabbit IgG (1:500) for 24 h. Tissue was washed, incubated with ABC for 1 h, washed again, and reacted with 3,3′-diaminobenzidine. After additional rinses, tissue sections were air-dried overnight.
FISH. A modified version of the FISH protocol from Cambio (Cambridge, U.K.) was used here. To aid penetration of chromosome paints, air-dried sections underwent antigen retrieval: slides were microwaved in 10 mM sodium citrate buffer (pH 6.0) for 10 min, slowly cooled, and rinsed with 2× SSC for 5 min. Sections then underwent pepsin treatment: slides were incubated in 0.005% pepsin solution at 37°C for 1-10 min (time varied, depending on the strength of pepsin stock). Slides were washed in 2× SSC for 1 min, rinsed briefly in distilled water, dehydrated in ethanols, and air-dried. Slides were then denatured at 65°C for 2 min in 70% formamide in 2× SSC, quenched in ice-cold ethanols, and air-dried. FITC-labeled whole-mouse chromosome Y paint and Cy3-labeled whole-mouse chromosome X paint (Cambio) were denatured at 65°C for 10 min and held at 37°C for 60 min. Paints were applied to sections and slides were coverslipped, sealed with rubber cement, and incubated in a dark humidified chamber overnight at 37°C. After overnight hybridization, slides underwent stringency washes at 45°C: two washes in 2× SSC for 5 min each, followed by two washes in 50% formamide in 1× SSC for 5 min each, and two washes in 1× SSC for 5 min each. DAPI (0.3 μg/ml; Sigma) was added to the first 1× SSC wash to stain nuclei. Slides underwent a final wash in 4× SSC/0.5% Tween-20 at 45°C for 4 min, were partially air-dried, and coverslipped with Vectashield (Vector Laboratories).
IEG Immunolabeling. Tissue sections previously processed for FluoroGold immunohistochemistry and/or FISH were rinsed in PBS, followed by incubation in 1% BSA blocking solution for 1 h and rabbit α-Egr-1 (1:5,000; Santa Cruz Biotechnology), rabbit α-c-Fos (1:500; Santa Cruz Biotechnology), or sheep α-c-Fos (1:500; Chemicon, Temecula, CA) overnight. Tissue was then washed in PBS and incubated with a cy3-conjugated donkey α-rabbit IgG (1:500; Jackson ImmunoResearch), an Alexa Fluor 546 donkey α-sheep IgG (1:500; Molecular Probes), or an Alexa Fluor 488 donkey α-sheep IgG (1:300; Molecular Probes) for 1 h. Tissue was washed again in PBS and coverslipped with Vectashield.
Analysis of Aneuploid Neurons by Using Deconvolution, Nomarski, and Brightfield Microscopy. To ensure that chromosome paints were confined to a single nucleus, Z stacks of aneuploid cells were captured with a fluorescent deconvolution microscopy setup that included a DeltaVision imaging station (Applied Precision, Issaquah, WA), an Olympus (Melville, NY) IX70 inverted fluorescence microscope, a ×60 oil objective lens, and a CH350 camera (Photometrics, Tucson, AZ). Exposure settings were in the linear range for each fluorophore. Approximately 50 optical sections at a step size of 0.2 μm were collected and compiled into Z stacks. Nomarski and brightfield images of FluoroGold label in aneuploid neurons were taken on a Zeiss Axioskop with Zeiss AxioCam digital camera. Fluorescent images of Egr-1 and c-Fos label were taken on either the deconvolution scope or the Zeiss Axioskop. Images were prepared in Adobe Photoshop (Adobe Systems, Mountain View, CA). Egr-1 staining was pseudocolored green. In some cases, c-Fos staining was pseudocolored red. The nomenclature for brain areas containing back-filled aneuploid neurons was based on that presented in ref. 13.
Results
Neurons Are Colabeled with FluoroGold and FISH Paint. To identify aneuploid neurons with axonal connections, adult mice were injected with the retrograde neuronal tracer FluoroGold, whose labeling is characterized as gold-colored granules and vesicles that are present in the cell cytoplasm but not the nucleus (14). Tissue from these mice was then processed sequentially for FluoroGold immunohistochemistry and interphase FISH by using whole-chromosome paints for the mouse X and/or Y chromosome (Fig. 1). Males were used to take advantage of their single copies of X and Y chromosomes, thus reducing hybridization artifacts associated with paired autosomes, as discussed (6). Back-filled aneuploid neurons were identified by using illumination from both brightfield and fluorescent light sources to detect f luorescent chromosome paints and brightfield FluoroGold label simultaneously within individual cells. As a most stringent criterion, only hyperploid cells (i.e., cells that had gained a sex chromosome) were documented because the identification of hypoploid cells, vastly outnumbering hyperploid cells (6), might be contaminated by sectioning artifacts.
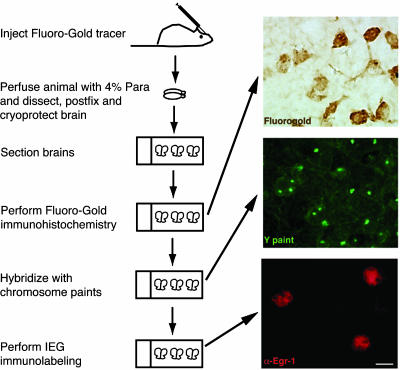
Fig. 1.
Schematic of experimental protocol. Adult male mice were injected with FluoroGold tracer and killed 48 h later. Brains from mice were sectioned and processed sequentially for FluoroGold immunohistochemistry, FISH, and IEG immunolabeling. (Scale bar, 10 μm.)
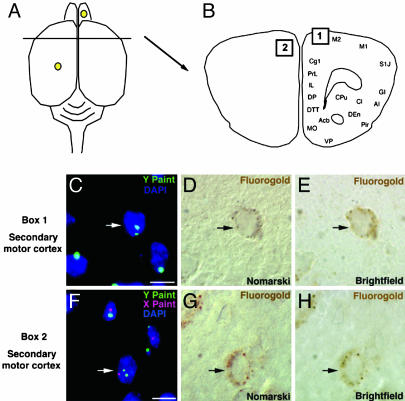
Aneuploid Neurons Are Retrogradely Labeled with FluoroGold. In adult male mice that had received injections of FluoroGold into the right olfactory bulb and left primary motor cortex, retrogradely labeled neurons that presented an extra X chromosome were identified in lateral orbital cortex, primary motor cortex, and secondary motor cortex (Table 1). Deconvolution microscopy was used to obtain high-magnification Z stacks of two of these aneuploid cells (Fig. 2 C and F). The same fields of view were also captured by using Nomarski (Fig. 2 D and G) and brightfield (Fig. 2 E and H) microscopy to illustrate FluoroGold labeling in the cells presenting the extra X chromosome. An adult male that received injections of FluoroGold into both olfactory bulbs displayed retrogradely labeled hyperploid neurons in the olfactory cortical regions that included the anterior olfactory nucleus, dorsal tenia tecta, and dorsal endopiriform nucleus, whereas males that received injections into the right olfactory bulb and left somatosensory cortex displayed back-filled aneuploid neurons in primary and secondary motor cortex, as well as the piriform cortex (Table 1). Examples of back-filled neurons in secondary motor cortex that presented an extra Y chromosome were similarly identified by using deconvolution (Fig. 3 C and F), Nomarski (Fig. 3 D and G), and brightfield (Fig. 3 E and H) microscopy. In total, ≈10,000 neurons were examined for both hyperploidy and positive FluoroGold label. Retrogradely labeled neurons presenting an extra Y or X chromosome were present at approximately equal rates (0.1%).
Table 1. Percent of retrogradely labeled aneuploid neurons in various cortical areas.
| Type of aneuploidy | Location | Percent |
|---|---|---|
| Two Y chromosomes | Dorsal tenia tecta | 22 |
| Two Y chromosomes | Anterior olfactory nucleus, ventral | 22 |
| Two Y chromosomes | Dorsal endopiriform nucleus | 11 |
| Two Y chromosomes | Piriform cortex | 11 |
| Two Y chromosomes | Primary motor cortex | 11 |
| Two Y chromosomes | Secondary motor cortex | 22 |
| Two X chromosomes | Lateral orbital cortex | 25 |
| Two X chromosomes | Primary motor cortex | 25 |
| Two X chromosomes | Secondary motor cortex | 50 |
Fig. 2.
Neurons from adult male mice containing an extra X chromosome were retrogradely labeled with FluoroGold. (A) Dorsal view schematic showing FluoroGold injections in the right olfactory bulb and left motor cortex. (B) Cross section from A showing location of back-filled aneuploid neurons (box 1 corresponds to cell in C and box 2 corresponds to the cell in F). (C and F) Z stacks from a deconvolution microscope showing hyperploid cells (white arrows) in the adult secondary motor cortex (C) and lateral orbital cortex (F) that contain an extra X (red) chromosome. Neighboring cells are euploid for the sex chromosomes. Nuclei are stained blue with DAPI. (D, E, G, and H) Cytoplasmic localization of FluoroGold label (black arrows) in the aneuploid neurons from C and F by using Nomarski (D and G) and brightfield (E and H) microscopy. (Scale bars, 10 μm.) AI, agranular insular cortex; AOM, anterior olfactory nucleus, medial; AOV, anterior olfactory nucleus, ventral; Cg1, cingulate cortex, area 1; DEn, dorsal endopiriform nucleus; IL, infralimbic cortex; LO, lateral orbital cortex; M1, primary motor cortex; M2, secondary motor cortex; MO, medial orbital cortex; Pir, piriform cortex; PrL, prelimbic cortex; VO, ventral orbital cortex.
Fig. 3.
Neurons from adult male mice containing an extra Y chromosome are retrogradely labeled with FluoroGold. (A) Dorsal view schematic showing Fluoro-Gold injections in the right olfactory bulb and left somatosensory cortex. (B) Cross section from A showing location of back-filled aneuploid neurons (Box 1 corresponds to cell in C and Box 2 corresponds to cell in F. (C and F) Z stacks from a deconvolution microscope showing hyperploid cells (white arrows) in secondary motor cortex that contain an extra Y (green) chromosome. Nuclei in C and F are stained blue with DAPI. (D, E, G, and H) Cytoplasmic localization of Fluoro-Gold label (black arrows) in the aneuploid neurons from C and F by using Nomarski (D and G) and brightfield (E and H) microscopy. (Scale bars, 10 μm.) Acb, accumbens nucleus; AI, agranular insular cortex; Cl, claustrum; Cg1, cingulate cortex, area 1; CPu, caudate putamen; DEn, dorsal endopiriform nucleus; DP, dorsal peduncular cortex; DTT, dorsal tenia tecta; GI, granular insular cortex; IL, infralimbic cortex; M1, primary motor cortex; M2, secondary motor cortex; MO, medial orbital cortex; Pir, piriform cortex; PrL, prelimbic cortex; S1J, somatosensory 1, jaw region; VP, ventral pallidum.
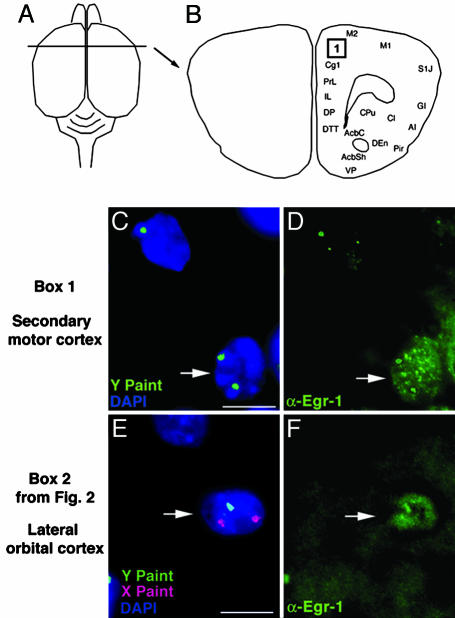
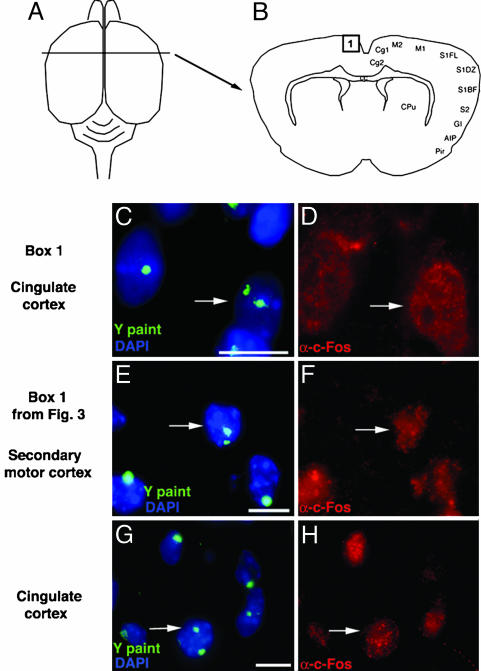
Aneuploid Neurons Are Immunoreactive for Egr-1 and c-Fos IEG Expression. IEGs have been extensively used as markers for functionally active neurons (15), and antiserums against two IEG products, Egr-1 and c-Fos, were therefore used on tissue sections containing FluoroGold back-filled and/or FISH-labeled neurons (Fig. 1). Both Egr-1 and c-Fos produce nuclear immunolabeling, and immunolabeled hyperploid neurons, including triple-labeled neurons (back-filled, FISH-labeled, and IEG-immunolabeled) could be identified throughout the brain. For example, a neuron in the secondary motor cortex (Fig. 4B) containing two Y chromosomes (Fig. 4C) was positive for Egr-1 labeling (Fig. 4D). Likewise, a triple-labeled neuron from the lateral orbital cortex was back-filled with FluoroGold (see Fig. 2 G and H for FluoroGold label in the neuron) contained two X chromosomes (Fig. 4E) and was positive for Egr-1 (Fig. 4F). Similar results were obtained with the IEG, c-fos. For example, a neuron in the cingulate cortex (Fig. 5B) containing two Y chromosomes (Fig. 5C) was positive for c-Fos labeling (Fig. 5D), whereas a triple-labeled neuron from the secondary motor cortex was back-filled with FluoroGold (see Fig. 3 D and E for FluoroGold label in the neuron) contained two Y chromosomes (Fig. 5E) and was positive for c-Fos (Fig. 5F). In a male mouse subjected to a specific activation paradigm (i.e., exposure to a novel male as well as various odors; see Methods), a cell in the cingulate cortex containing two Y chromosomes (Fig. 5G) presented c-Fos label (Fig. 5H).
Fig. 4.
Neurons from male mice containing an extra X or Y chromosome are immunoreactive for Egr-1. (A and B) Dorsal view (A) and cross section (B), indicating location of aneuploid cell in C (box 1). The location of aneuploid cell in E is shown in Fig. 2B (box 2). (C and E) Z stacks from a deconvolution microscope showing hyperploid cells in adult cortex. A cell in the secondary motor cortex contains an extra Y (green) chromosome (white arrow) (C). A retrogradely labeled cell in lateral orbital cortex (see Fig. 2 G and H for FluoroGold label) contains an extra X (red) chromosome (white arrow) (E). Nuclei in C and E are stained blue with DAPI. (D and F) Egr-1 immunoreactivity (white arrows) in aneuploid neurons from C and E. (Scale bars, 10 μm.) AcbC, accumbens nucleus, core; AcbSh, accumbens nucleus, shell; AI, agranular insular cortex; Cl, claustrum; Cg1, cingulate cortex, area 1; CPu, caudate putamen; DEn, dorsal endopiriform nucleus; DP, dorsal peduncular cortex; DTT, dorsal tenia tecta; GI, granular insular cortex; IL, infralimbic cortex; M1, primary motor cortex; M2, secondary motor cortex; Pir, piriform cortex; PrL, prelimbic cortex; S1J, somatosensory 1, jaw region; VP, ventral pallidum.
Fig. 5.
Neurons from male mice containing an extra Y chromosome are immunoreactive for c-Fos. (A and B) Dorsal view (A) and cross section (B), indicating location of the aneuploid cell in C (box 1). The location of the aneuploid cell in E is shown in Fig. 3B (box 1). The location of the aneuploid cell in G is also from the cingulate cortex (see box 1) (C, E, and G) Z stacks from a deconvolution microscope showing hyperploid cells in the adult cortex. A cell in the cingulate cortex contains an extra Y (green) chromosome (white arrow) (C). A retrogradely labeled cell in secondary motor cortex (see Fig. 3 D and E for FluoroGold label) contains an extra Y (green) chromosome (white arrow) (E). A cell in the cingulate cortex from an animal subjected to an activation paradigm (see Methods) contains an extra Y (green) chromosome (white arrow) (G). Nuclei in C, E, and G are stained blue with DAPI. (D, F, and H) c-Fos immunoreactivity (white arrows) in aneuploid neurons from C, E, and G. (Scale bars, 10 μm.) AIP, agranular insular cortex, posterior; cc, corpus callosum; Cg1, cingulate cortex, area 1; Cg2, cingulate cortex, area 2; CPu, caudate putamen; GI, granular insular cortex; M1, primary motor cortex; M2, secondary motor cortex; Pir, piriform cortex; S1FL, somatosensory 1, forelimb region; S1DZ, somatosensory 1, dysgranular zone; S1BF, somatosensory 1, barrel field; S2, secondary somatosensory cortex.
Discussion
Our results demonstrate that aneuploid neurons in the adult mammalian brain can have distant axonal connections and show functional activity. To our knowledge, the combined approaches used in this study have not been previously reported, in part reflecting the challenges of preparing tissues that allow simultaneous preservation of the disparate molecular targets identified by FISH, retrograde immunohistochemistry, and immunolabeling. Back-filled and functional aneuploid neurons are not limited to one type of cortex or neural circuit, being present in all assayed areas spanning the paleocortex through neocortex, and likely, all neuronal populations of the brain.
For multiple reasons, the sampling of aneuploid neurons identified here represents a very conservative estimate of the total percentage of aneuploid neurons integrated within the brain. First, combining FISH with retrograde labeling limited analyses to those neurons that could be both back-filled and accessible by FISH, the latter requiring intact neurons located near the surface of a tissue section. Second, only hyperploid cells were examined to eliminate possible sectioning artifacts that might have led to false-positive identification of hypoploidy (e.g., produced by transection of a single nucleus); importantly, however, hypoploid cells are 7-fold more common than hyperploid cells (6). Third, only sex chromosomes in adult male mice were examined to control for ambiguous signals produced by using a single FISH fluorochrome to identify an autosomal pair within a tissue section; aneuploidy produced by at least some of the remaining 19 autosomes pairs is certain, but was not assessed in this study. It is notable that sampling estimates by using nuclear transfer of cortical neuronal nuclei into oocytes indicated that 64% of nuclei contained deviations from a euploid karyotype (16). Despite these considerations, the rate of sex chromosome aneuploidy documented here, ≈0.2% for combined X or Y hyperploidy, is substantially greater than other neurobiologically important phenomena, such as adult neurogenesis, where S-phase cells have a prevalence of ≈0.03% to 0.05% (17, 18). The overall percentage and complete chromosomal complement of aneuploid neurons; hypoploidy, hyperploidy, and combinations of the two; remain to be determined, and it is important to consider that a cell euploid for sex chromosomes, as assayed here, could well be aneuploid for autosomes. It is therefore probable that the overall extent of neuronal aneuploidy is much greater than the significant levels reported here.
Of what consequence are functionally active aneuploid neurons that are integrated into brain circuitry? One likely outcome is the incorporation of chromosome-limited signaling differences produced by altered gene expression, as documented from yeast through mammalian neoplasms and neural cells (4, 19, 20). Thus, a network composed of intermixed euploid and aneuploid neurons might produce unique signaling properties distinct from a network composed purely of euploid cells. It is also notable that gene expression mapping strategies from different brain regions detect an average or pooled representation of expressed genes within a brain region (21, 22), likely masking heterogeneity of gene expression produced by the mosaicism of intermixed aneuploid and euploid cells. Another function of aneuploid neurons may be to provide brain circuits with selective advantages, analogous to models of aneuploid tumor cell growth (23). Moreover, variation in gene dosage through loss of heterozygosity (24) produced by chromosomal loss, gene duplication through chromosome gain, or possible contributions to large-scale copy number polymorphisms through chromosomal gain and/or loss could influence both the healthy and diseased brain (25-28). Future studies will define the range and functional consequences of aneuplodies in different species.
Acknowledgments
We thank M. Fontanoz and G. Kennedy for excellent technical assistance and K. Spencer for assistance with deconvolution microscopy. This work was supported by the National Institute of Mental Health (J.C.), a postdoctoral fellowship from the Human Frontiers Science Program Grant (to M.A.K.), a National Science Foundation POWER grant (to B.F.), a National Institute of General Medical Sciences Pharmacology Training grant (to M.J.M. and A.H.Y.), a Postdoctoral Fellowship from the PEW Latin American Fellows in the Biomedical Sciences (to S.K.R.), and a Predoctoral Fellowship from the Pharmaceutical Research and Manufacturers of America Foundation (to D.K.).
Author contributions: M.A.K., B.F., and J.C. designed research; M.A.K., B.F., and S.K.R. performed research; M.A.K., B.F., M.J.M., A.H.Y., and D.K. contributed new reagents/analytic tools; M.A.K. analyzed data; M.A.K. and J.C. wrote the paper; and M.J.M., S.K.R., A.H.Y., and D.K. provided feedback to help guide experiments.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: IEG, immediate early gene.
References
- 1.Lengauer, C., Kinzler, K. W. & Vogelstein, B. (1998) Nature 396, 643-649. [DOI] [PubMed] [Google Scholar]
- 2.Schupf, N. & Sergievsky, G. H. (2002) Br. J. Psychiatry 180, 405-410. [DOI] [PubMed] [Google Scholar]
- 3.Gaudet, J. & Mango, S. E. (2002) Science 295, 821-825. [DOI] [PubMed] [Google Scholar]
- 4.Kaushal, D., Contos, J. J., Treuner, K., Yang, A. H., Kingsbury, M. A., Rehen, S. K., McConnell, M. J., Okabe, M., Barlow, C. & Chun, J. (2003) J. Neurosci. 23, 5599-5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McConnell, M. J., Kaushal, D., Yang, A. H., Kingsbury, M. A., Rehen, S. K., Treuner, K., Helton, R., Annas, E., Chun, J. & Barlow, C. (2004) J. Neurosci., 24, 8090-8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rehen, S. K., McConnell, M. J., Kaushal, D., Kingsbury, M. A., Yang, A. H. & Chun, J. (2001) Proc. Natl. Acad. Sci. USA 98, 13361-13366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang, A. H., Kaushal, D., Rehen, S. K., Kriedt, K., Kingsbury, M. A., McConnell, M. J. & Chun, J. (2003) J. Neurosci. 23, 10454-10462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogasawara, M., Aoki, K., Okada, S. & Suzumori, K. (2000) Fertil. Steril. 73, 300-304. [DOI] [PubMed] [Google Scholar]
- 9.Voullaire, L., Slater, H., Williamson, R. & Wilton, L. (2000) Hum. Genet. 106, 210-217. [DOI] [PubMed] [Google Scholar]
- 10.Lai, W. S., Chen, A. & Johnston, R. E. (2004) Horm. Behav. 46, 319-329. [DOI] [PubMed] [Google Scholar]
- 11.Montag-Sallaz, M. & Buonviso, N. (2002) J. Neurobiol. 52, 61-72. [DOI] [PubMed] [Google Scholar]
- 12.McLean, I. W. & Nakane, P. K. (1974) J. Histochem. Cytochem. 22, 1077-1083. [DOI] [PubMed] [Google Scholar]
- 13.Franklin, K. B. J. & Paxinos, G. (1997) The Mouse Brain in Stereotaxic Coordinates (Academic, San Diego).
- 14.Schmued, L. C. & Fallon, J. H. (1986) Brain Res. 377, 147-154. [DOI] [PubMed] [Google Scholar]
- 15.Kaczmarek, L. & Chaudhuri, A. (1997) Brain Res. Rev. 23, 237-256. [DOI] [PubMed] [Google Scholar]
- 16.Osada, T., Kusakabe, H., Akutsu, H., Yagi, T. & Yanagimachi, R. (2002) Cytogenet. Genome Res. 97, 7-12. [DOI] [PubMed] [Google Scholar]
- 17.Kaplan, M. S. & Hinds, J. W. (1977) Science 197, 1092-1094. [DOI] [PubMed] [Google Scholar]
- 18.Kempermann, G., Kuhn, H. G. & Gage, F. H. (1997) Proc. Natl. Acad. Sci. USA 94, 10409-10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phillips, J. L., Hayward, S. W., Wang, Y., Vasselli, J., Pavlovich, C., Padilla-Nash, H., Pezullo, J. R., Ghadimi, B. M., Grossfeld, G. D., Rivera, A., et al. (2001) Cancer Res. 61, 8143-8149. [PubMed] [Google Scholar]
- 20.Hughes, T. R., Roberts, C. J., Dai, H., Jones, A. R., Meyer, M. R., Slade, D., Burchard, J., Dow, S., Ward, T. R., Kidd, M. J., et al. (2000) Nat. Genet. 25, 333-337. [DOI] [PubMed] [Google Scholar]
- 21.Caceres, M., Lachuer, J., Zapala, M. A., Redmond, J. C., Kudo, L., Geschwind, D. H., Lockhart, D. J., Preuss, T. M. & Barlow, C. (2003) Proc. Natl. Acad. Sci. USA 100, 13030-13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gong, S., Zheng, C., Doughty, M. L., Losos, K., Didkovsky, N., Schambra, U. B., Nowak, N. J., Joyner, A., Leblanc, G., Hatten, M. E. & Heintz, N. (2003) Nature 425, 917-925. [DOI] [PubMed] [Google Scholar]
- 23.Cahill, D. P., Kinzler, K. W., Vogelstein, B. & Lengauer, C. (1999) Trends Cell Biol. 9, M57-M60. [PubMed] [Google Scholar]
- 24.Thiagalingam, S., Foy, R. L., Cheng, K. H., Lee, H. J., Thiagalingam, A. & Ponte, J. F. (2002) Curr. Opin. Oncol. 14, 65-72. [DOI] [PubMed] [Google Scholar]
- 25.Lemere, C. A., Blusztajn, J. K., Yamaguchi, H., Wisniewski, T., Saido, T. C. & Selkoe, D. J. (1996) Neurobiol. Dis. 3, 16-32. [DOI] [PubMed] [Google Scholar]
- 26.Sebat, J., Lakshmi, B., Troge, J., Alexander, J., Young, J., Lundin, P., Maner, S., Massa, H., Walker, M., Chi, M., et al. (2004) Science 305, 525-528. [DOI] [PubMed] [Google Scholar]
- 27.Singleton, A. B., Farrer, M., Johnson, J., Singleton, A., Hague, S., Kachergus, J., Hulihan, M., Peuralinna, T., Dutra, A., Nussbaum, R., et al. (2003) Science 302, 841. [DOI] [PubMed] [Google Scholar]
- 28.Yang, Y., Geldmacher, D. S. & Herrup, K. (2001) J. Neurosci. 21, 2661-2668. [DOI] [PMC free article] [PubMed] [Google Scholar]