Abstract
We have created a stable transgenic rag2-EGFP-mMyc zebrafish line that develops GFP-labeled T cell acute lymphoblastic leukemia (T-ALL), allowing visualization of the onset and spread of this disease. Here, we show that leukemias from this transgenic line are highly penetrant and render animals moribund by 80.7 ± 17.6 days of life (±1 SD, range = 50-158 days). These T cell leukemias are clonally aneuploid, can be transplanted into irradiated recipient fish, and express the zebrafish orthologues of the human T-ALL oncogenes tal1/scl and lmo2, thus providing an animal model for the most prevalent molecular subgroup of human T-ALL. Because T-ALL develops very rapidly in rag2-EGFP-mMyc transgenic fish (in which “mMyc” represents mouse c-Myc), this line can only be maintained by in vitro fertilization. Thus, we have created a conditional transgene in which the EGFP-mMyc oncogene is preceded by a loxed dsRED2 gene and have generated stable rag2-loxP-dsRED2-loxP-EGFP-mMyc transgenic zebrafish lines, which have red fluorescent thymocytes and do not develop leukemia. Transgenic progeny from one of these lines can be induced to develop T-ALL by injecting Cre RNA into one-cell-stage embryos, demonstrating the utility of the Cre/lox system in the zebrafish and providing an essential step in preparing this model for chemical and genetic screens designed to identify modifiers of Myc-induced T-ALL.
Keywords: lymphoma, tal1/scl, lmo2
T cell lineage acute lymphoblastic leukemia (T-ALL) is an aggressive hematologic malignancy arising when immature T cells acquire mutations that cause differentiation arrest, rapid proliferation, and suppression of apoptosis within developing T lymphocytes (1). Our current understanding of the molecular basis of T cell malignancies has emerged largely from the analysis of recurrent chromosomal translocations, which typically juxtapose T cell oncogenes with strong promoter elements responsible for high expression levels of the T cell receptor (2, 3). These T cell oncogenes encode transcription factors, including (i) basic helix-loop-helix (bHLH) family members such as TAL1/SCL, TAL2, LYL1, and BHLHB1, (ii) LIM-only (LMO) domain genes such as LMO1 and LMO2, and (iii) the orphan homeobox genes HOX11/TLX1 and HOX11L2/TLX3 (3-5). Up-regulation of T-ALL oncogene transcription factors can also occur in the absence of chromosomal translocations (6-8), presumably due to mutations that cause either monoallelic activation, through cis-acting mutations or deletions, or biallelic activation, from the disruption of upstream factors that normally suppress the expression of these genes in developing thymoctyes (8).
In human T-ALL, we have identified five distinct multistep molecular pathways based on the overexpression of (i) TAL1/SCL plus LMO1 or LMO2, (ii) LYL1 plus LMO2, (iii) HOX11, (iv) HOX11L2, and (v) MLL-ENL (1, 9), each of which is characterized by distinct molecular signatures (1, 10, 11). These subgroups are clinically relevant, with event-free survival differing among patient groups (1, 11, 12). For example, patients expressing both TAL1/SCL or LYL1 and a LMO family member (LMO1 or LMO2) have a worse prognosis than those expressing HOX11 (1, 11, 12). Activation of these T cell oncogenes appears to be critical for thymocyte transformation, possibly by causing stage-specific arrest of T cell maturation (1). However, additional mutations are also found in leukemic cells from patients in each of the major subgroups, including those that affect pathways that control apoptosis, proliferation, and genomic instability. Recently, we have identified activating mutations in NOTCH1 that result in increased NOTCH signaling and increased proliferation of developing thymocytes (13). In addition, four of five subgroups of human T-ALL express high levels of either MYC or MYCN (1), suggesting that MYC may be a central regulator of proliferation and/or genomic instability in this malignancy. Finally, most human T-ALLs biallelically delete the CDKN2A locus, which encodes both the p16(INK4A) and p14(ARF) tumor suppressors, thereby disrupting both the RB and p53 pathways and contributing to aberrant control of both cell cycle progression and programmed cell death (1, 14).
The zebrafish has recently emerged as an important vertebrate model of Myc-induced T-ALL (15); however, studies have not been performed to assess how closely zebrafish T-ALL mimics the human disease. In addition, the promise of the zebrafish T-ALL model lies in its utility for chemical (16-18) and genetic modifier screens (19-21), marking the emergence of the zebrafish as a unique vertebrate model with which to identify enhancers that accelerate disease or suppressors that curb tumor growth. Here, we show that transgenic rag2-EGFP-mMyc zebrafish (in which “mMyc” represents mouse c-Myc) develop T-ALLs that faithfully model the most common and most treatment-resistant subtype of human T-ALL, in which SCL and LMO1/2 are coexpressed. However, these rag2-EGFP-mMyc transgenic fish are often severely diseased by the time they reach reproductive maturity, making this line difficult to breed and maintain. Thus, conditional transgenic approaches are needed to establish zebrafish leukemia models that are amenable to forward genetic and small molecule suppressor screens. Our current results indicate that the molecular mechanisms underlying zebrafish T-ALL are remarkably similar to those found in the human disease and establish Cre/lox strategies in transgenic zebrafish that provide a general means to develop conditional models of cancer for genetic analysis in this model organism.
Materials and Methods
Isolation of lmo1 and p16. RNA was obtained from 1- to 5-day-old embryos and made into cDNA, and degenerate PCR primers were used to amplify a fragment of the lmo1 and p16 gene. RACE PCR was used to isolate the full-length lmo1 (GenBank accession no. AF398514). By contrast, the p16 sequence fragment was used to search the zebrafish genome (www.sanger.ac.uk/Projects/D_rerio) and identify a putative full-length ORF for the p16 locus (R. Stewart and A.T.L., unpublished data).
Penetrance of Disease in rag2-EGFP-mMyc Stable Transgenic Zebrafish. Stable transgenic rag2-EGFP-mMyc fish have been generated previously and develop GFP-labeled T-ALL (15). To determine the penetrance of disease in stable transgenic rag2-EGFP-mMyc fish, sperm was harvested from 10- to 20-week-old leukemic male fish and used for in vitro fertilization of AB WT eggs. The resulting progeny were scored for leukemia onset at 30-60 days of life as determined by infiltration of GFP-labeled leukemic cells into sites adjacent to the thymus. At 3 months of age, nonleukemic sibling fish were analyzed for the presence of the mMyc transgene as determined by PCR of genomic DNA isolated from the tail fin as described in ref. 15.
Collection of Leukemias and Determination of DNA Content. Leukemic rag2-EGFP-mMyc fish were killed, and the heads were fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned. The remaining portion of fish was diced over 5 ml of ice-cold 0.9 × PBS plus 5% FBS. The suspension was filtered over a 40-μm filter, washed, and subsequently (i) transplanted into irradiated recipient fish (1 × 106 cells per fish, 2-3 days after receiving 23 Gy of total body irradiation from a 137Cs source), (ii) analyzed for DNA content as determined by DNA flow cytometry, (iii) frozen in 10 million-cell aliquots, and/or (iv) analyzed by FACS to determine the percentage of GFP-labeled leukemic cells contained within each sample. DNA flow cytometric analysis was completed essentially as described in ref. 15. Specifically, tumor cells and WT nucleated red blood cells were stained with propidium iodide in hypotonic sodium-citrate buffer and analyzed for cellular DNA content by flow cytometry alone and as a mixture with normal control cells.
RT-PCR Analysis. RNA was isolated (with TRIzol, GIBCO/BRL) from leukemia cells and control FACS-sorted, GFP-positive thymocytes from rag2-GFP and lck-GFP transgenic fish (15, 22, 23). RNA was treated with DNaseI before reverse transcription, and RT-PCR was performed. (PCR primers and thermocycling conditions are described in detail in the Supporting Text and Table 1, which are published as supporting information on the PNAS web site.)
To confirm that expression of the scl and lmo2 transcripts was confined to the leukemic lymphoblasts, RNA in situ hybridization was completed on paraffin-embedded sections from transgenic rag2-EGFP-mMyc fish essentially as described in refs. 15 and 22. PCR primers used to generate probes for in situ hybridization analysis are described in the Supporting Text.
To determine whether scl and lmo2 expression resulted from transcription of one or both alleles, PCR was used to amplify the 3′ untranslated regions of scl and lmo2 (Table 1). PCR products were purified (QIAquick PCR Purification Kit, Qiagen, Valencia, CA) and sequenced. cDNA was obtained from zebrafish T-ALL samples having polymorphic alleles for either scl or lmo2 and subjected to PCR. PCR fragments were purified and sequenced.
Southern Blot Analysis to Determine Clonality and to Assess Loss of the p16 Locus. Southern blot analysis was used to determine whether leukemia cells have T cell receptor (TCR)-α or IgM receptor rearrangements and whether the p16 genomic locus is lost. Southern blot analysis was performed as described in refs. 15 and 24.
Sequencing of the p53 Locus. Genomic DNA was isolated from zebrafish leukemic samples and subjected to PCR amplification of exons 4-9 of the zebrafish p53 gene. Fragments were purified and sequenced as described in ref. 25 and Table 1.
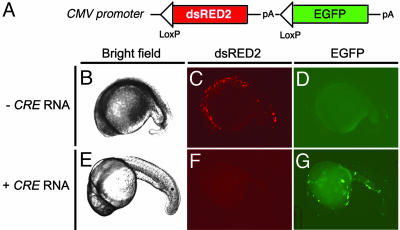
Cre/lox Strategies. To verify that Cre/lox-mediated strategies work in the zebrafish, an expression vector was created that contains loxP sites flanking the dsRED2 transgene and the polyadenylation site contained within the dsRED2-N1 vector (Clontech). The loxed dsRED2 transgene was cloned into the EGFP-N1 expression vector upstream of the EGFP ORF (Fig. 3A). One-cell-stage embryos were injected with either the CMV-loxP-dsRED2-loxP-EGFP plasmid (50 ng/μl) alone or in combination with Cre RNA (25 ng/μl). Cre RNA was made by in vitro transcription by using the pCS2+Cre vector and SP6 RNA polymerase. Transiently injected embryos were analyzed 26 h postfertilization for GFP and dsRED2 expression as determined by fluorescent microscopy.
Fig. 3.
Cre-mediated recombination in transiently injected embryos. (A) Diagram of the CMV-loxP-dsRED2-loxP-EGFP construct. (B-G) One-cell-stage embryos were injected with the CMV-loxP-dsRED2-loxP-EGFP vector in the absence of Cre RNA (- Cre RNA) (B-D) or with Cre RNA (+ Cre RNA, 25 ng/μl) (E-G). Shown are bright-field (B and E), red fluorescence (dsRED2) (C and F), and green fluorescence (EGFP) images (D and G) of embryos at 26 h postfertilization. Anterior is to the left, and dorsal is toward the top.
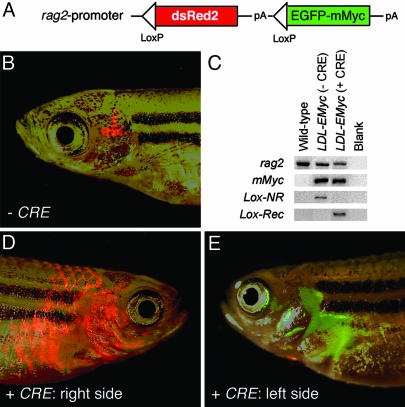
Developing Conditional Transgenic Zebrafish. The loxed dsRED2 coding sequence was cloned into the rag2-EGFP-mMyc plasmid upstream of the EGFP-mMyc transgene by using BamHI restriction enzyme sites (Fig. 4A). The resulting rag2-loxP-dsRED2-loxP-EGFP-mMyc plasmid was linearized with XhoI, phenol/chloroform-extracted, and ethanol-precipitated. Linearized DNA was injected into one-cell-stage AB strain embryos [100 ng/μl DNA in 0.5× TE buffer (10 mM Tris/1 mM EDTA, pH ≈ 7.0) containing 100 mM KCl. Primary injected adult fish were screened for the ability to produce offspring that contained the transgene as determined by detection of dsRED2 fluorescence within developing thymocytes at 6 days postfertilization. Two stable transgenic rag2-loxP-dsRED2-loxP-EGFP-mMyc zebrafish lines were identified (lines G7 and G16, AB strain). F1 and F2 rag2-loxP-dsRED2-loxP-EGFP-mMyc transgenic fish were injected with 25 ng/μl Cre RNA and analyzed for leukemia onset.
Fig. 4.
Cre RNA injection into stable transgenic rag2-loxP-dsRED2-loxP-EGFP-mMyc fish leads to transgene recombination and rapid onset of Myc-induced T-ALL. (A) Diagram of rag2-loxP-dsRED2-loxP-EGFP-mMyc construct. (B) Thymocytes from a 73-day-old rag2-lox-dsRED2-EGFP-mMyc transgenic fish are red-fluorescent-labeled in the absence of Cre expression. (C) PCR of genomic DNA isolated from blood cells of WT control or transgenic rag2-loxP-dsRED2-loxP-EGFP-mMyc (LDL-EMyc) fish. rag2 primers amplify genomic DNA, mMyc primers amplify the mMyc transgene, and Lox primers amplify either a 1.7-kb nonrecombined fragment (Lox-NR) or a 0.4-kb fragment, which results when Cre recombination has occurred (Lox-Rec). (D and E) One-cell-stage rag2-loxP-dsRED2-loxP-EGFP-mMyc embryos were injected with the Cre RNA (25 ng/μl) and grown to 51 days of development, at which time they had both GFP- and dsRED2-labeled (D) or GFP-positive alone (E) leukemias. The same fish is shown, right (D) and left (E) side. Images are composites of dsRED2 and GFP fluorescence and bright-field images.
Analysis of Leukemias from Cre-Injected rag2-loxP-dsRED2-loxP-EGFP-mMyc Fish. Leukemic cells from Cre-injected stable transgenic rag2-loxP-dsRED2-loxP-EGFP-mMyc fish were harvested and (i) transplanted into irradiated adult fish, (ii) analyzed by cytospin and Giemsa/May-Grunwald staining to confirm lymphoblast morphology, (iii) subjected to FACS analysis to assess levels of GFP and dsRED2 expression within lymphoblasts, and/or (iv) extracted for genomic DNA and analyzed for CRE recombination as detected by PCR (forward primer specific to the rag2 promoter region, ATGCTAATTTGAAGCACTAGCA; reverse primer specific to the EGFP coding sequence, GTGCAGATGAACTTCAGGGT).
Results
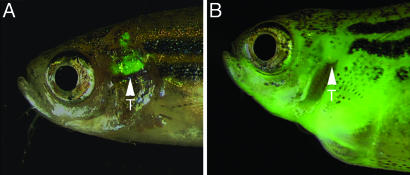
Complete Penetrance of T-ALL in a rag2-EGFP-mMyc Stable Transgenic Line. We have previously described a rag2-EGFP-mMyc stable transgenic zebrafish line in which the onset and progression of T cell malignancy can be monitored by fluorescence microscopy (15). The disease is first detected as an expansion of GFP-labeled T cells in the thymus, and, subsequently, malignant cells infiltrate regions adjacent to the thymus in a phase comparable to human T cell lymphoblastic lymphoma. Then, transformed T cells rapidly spread throughout the skeletal musculature, visceral organs, and kidney marrow, leading to widely disseminated T-ALL (Fig. 1 and Fig. 4 of ref. 15). In a series of 106 stable transgenic rag2-EGFP-mMyc fish, all animals developed T-ALL and succumbed to death by 80.7 ± 17.6 days of life (±1 SD, range = 50-158 days). Sixty-four nonleukemic siblings were raised until 3 months of age, and none harbored the mMyc transgene in somatic DNA. Taken together, these results indicate that Myc-induced leukemias are fully penetrant in our rag2-EGFP-mMyc transgenic line.
Fig. 1.
Stable transgenic rag2-EGFP-mMyc zebrafish develop GFP-labeled thymic lymphoma, which progresses to T-ALL. Fluorescence microscopic analysis at 50 days of life showing the thymus of control rag2-GFP transgenic fish (A) and massive GFP-labeled cellular dissemination of leukemic lymphoblasts in rag2-EGFP-mMyc transgenic fish (B). Fish are oriented with anterior to the left and dorsal to the top. Arrowheads mark location of the thymus (T).
Myc-Induced Leukemias Are of T Cell Origin and Oligoclonal. Because rag2 is expressed in both immature T and B cells (22), we wanted to assess whether leukemias arising in our stable transgenic zebrafish were of T cell or B cell origin. Several lines of evidence indicated that all of the leukemias arising in this transgenic line were of T cell origin. (i) Each malignancy developed as a GFP-labeled lymphoma in the thymus (n = 106). (ii) Southern blot analysis showed that Myc-induced leukemias contained oligoclonal TCR-α gene rearrangements (18 of 36), whereas none of the leukemias contained Ig heavy chain receptor gene (IgM) rearrangements (n = 21; Figs. 6 and 7 and Table 2, which are published as supporting information on the PNAS web site). (iii) RT-PCR analysis showed that leukemic lymphoblasts expressed high levels of the T cell-specific tyrosine kinase gene (lck) (22) (Fig. 2A) and TCR-α (24) (n = 30; Table 3, which is published as supporting information on the PNAS web site).
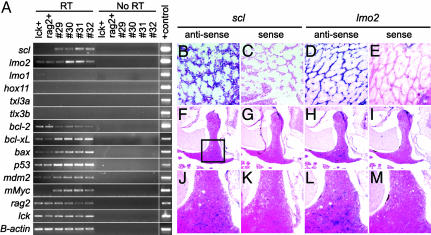
Fig. 2.
Leukemic lymphoblasts express T cell markers and both scl and lmo2.(A) Semiquantitative RT-PCR showing that Myc-induced leukemias are arrested at a stage in which scl and lmo2 are coexpressed. RT-PCR analysis of FACS-sorted, GFP-labeled thymocytes from 70-day-old transgenic lck-GFP (lck+) and rag2-GFP (rag2+) fish or leukemic lymphoblasts isolated from diseased fish (denoted by numbers). RT, reverse transcription reactions; No RT, no reverse transcription controls. (B-M) RNA in situ hybridization of paraffin-embedded sections confirms that scl and lmo2 are coexpressed in lymphoblasts from rag2-EGFP-mMyc transgenic fish (shown here infiltrating the skeletal musculature) and that scl and lmo2 are expressed in a subset of thymocytes in 70-day-old, nonleukemic, lck-GFP transgenic zebrafish. Images are photographed at 400× (B-E), 50× (F-I), and 200× (J-M). Images in J-M are higher-magnification images of the respective regions in F-I (indicated by the boxed region in F).
DNA flow cytometry was also used to determine clonality and to assess whether leukemias acquired chromosomal abnormalities during disease progression (Table 2; see also Fig. 8, which is published as supporting information on the PNAS web site). Ten of 63 leukemias analyzed were hyperdiploid, as indicated by clonal increases in DNA content (range = 1.01-1.45). Although most of these leukemias contained monoclonal populations of cells with increased DNA content (9 of 10), one T-ALL sample contained two distinct populations of hyperdiploid cells but failed to show TCR-α gene rearrangement by Southern analysis (Fig. 8B).
Myc-Induced Leukemias Are Transplantable. Transplantation and propagation of disease into secondary recipients is a hallmark of cancer, so we tested whether leukemic cells from rag2-EGFP-mMyc fish could be transplanted by i.p. injection into irradiated adult recipient fish. Lymphoblasts were isolated from primary leukemic fish, and FACS analysis confirmed that samples were highly enriched for leukemic lymphoblasts, containing 93.8 ± 1.4% (±SD) GFP-labeled leukemic cells (n = 10). GFP-labeled cells multiplied and generated leukemia in irradiated recipient fish within 1 month after injection (n = 11 independently arising leukemias). These results are similar to those reported previously for leukemia cells arising in F0 primary injected fish (15).
Zebrafish Myc-Induced T-ALLs Coexpress both tal1/scl and lmo2. Because cMYC expression is up-regulated in three of five molecular subgroups of human T-ALL (1, 9), including those misexpressing (i) TAL1/SCL plus LMO1 or LMO2, (ii) HOX11, and (iii) HOX11L2/TLX3, we predicted that we might observe several distinct molecular subgroups of zebrafish T-ALL. However, RT-PCR (Fig. 2A and Table 3) and in situ hybridization analyses (Fig. 2 B-E) showed that all Myc-induced leukemias coexpress tal1/scl and lmo2 (n = 20) but not lmo1, hox11, or the zebrafish orthologues of HOX11L2/TLX3, tlx3a, or tlx3b (26). By contrast, RT-PCR analysis revealed that normal thymocytes from lck-GFP and rag2-GFP stable transgenic fish express lower levels of scl and similar levels of lmo2 when compared with Myc-induced T-ALLs (Fig. 2A). Finally, RNA in situ hybridization of paraffin-embedded sections from 70-day-old lck-GFP transgenic animals showed that scl and lmo2 are expressed in only a subset of cortical thymocytes (Fig. 2 F-M).
To investigate the mechanisms by which tal1/scl and lmo2 are overexpressed in zebrafish T-ALL, we asked whether expression was monoallelic or biallelic at the chromosomal level. We identified four zebrafish leukemia sample DNAs that harbored polymorphisms in the 3′ untranslated region of scl and three with polymorphisms in the lmo2 gene (n = 20). RT-PCR analysis of these cDNAs revealed that in each case, scl and lmo2 transcripts were up-regulated equally from both chromosomal alleles (Table 2; see also Fig. 9, which is published as supporting information on the PNAS web site), indicating that expression is biallelic and does not result from chromosomal translocations or other allele-specific deletions or mutations.
Apoptotic Pathways in Zebrafish Myc-Induced T-ALL. Myc-induced transformation in mammals collaborates with mutations that disable components of the cellular apoptotic machinery (1, 14, 27, 28). However, given the rapidity of leukemia onset in our transgenic model, we questioned whether apoptosis was suppressed in zebrafish T-ALL and, if so, whether we could document the mechanisms that deregulate cell death in these tumors. Semiquantitative RT-PCR analysis showed that each of the zebrafish T-ALLs expressed similar levels of p53, mdm2, bcl-xL, bcl-2, and bax RNAs (n = 12; Fig. 2A and Table 3). When compared with thymocyte controls, Myc-induced T-ALLs expressed higher levels of bcl-xL, bax, p53, and mdm2; however, bcl-2 RNA expression was decreased, likely reflecting that T-ALLs are arrested at a stage of development marked by this gene expression profile (8).
Because human T-ALLs have either biallelic deletions of the CDKN2A locus (14) or, less frequently, mutational inactivation of p53 (29, 30), we asked whether zebrafish Myc-induced leukemias harbor abnormalities in orthologues of these loci. The p16 copy number was not decreased in these tumors as detected by Southern blot analysis (n = 21) (Table 2; see also Fig. 10, which is published as supporting information on the PNAS web site). Because >90% of mutations in p53 occur within exons 4-8 (the region that encodes the DNA-binding domain) (31, 32), we analyzed zebrafish T-ALL samples for mutations in p53 in these corresponding exons. We sequenced genomic DNA from zebrafish T-ALL leukemia cells and failed to identify any mutations in exons 4-9 of the p53 genomic locus (n = 12; Table 2).
Cre/lox Conditional Transgenic Strategies in the Zebrafish. T-ALL develops in 100% of stable transgenic rag2-EGFP-mMyc fish and progresses to widespread disease before reproductive maturity, necessitating harvesting sperm from diseased males to maintain the transgenic line by in vitro fertilization (IVF) (15). Because IVF procedures are cumbersome and not amenable to forward genetic approaches, we sought to develop a conditional transgenic approach, allowing identification and maintenance of zebrafish lines that did not develop leukemia until the investigator selectively induced T cell-specific expression of the Myc oncogene. For this purpose, we developed Cre/lox-mediated transgenic approaches, which have been reported in mice (33, 34) and Xenopus (35, 36) but not zebrafish.
We created a vector in which the CMV promoter drives the ubiquitous expression of a dsRED2 transgene that is followed by multiple transcription stop sites and flanked by loxP sites. The EGFP coding sequence was cloned downstream of this cassette (CMV-loxP-dsRED2-loxP-EGFP vector; Fig. 3A). Transient injection of the CMV-loxP-dsRED2-loxP-EGFP construct into embryos without Cre recombinase results in dsRED2 fluorescence and no EGFP expression (50 ng/μl; Fig. 3 C and D). By contrast, coinjection of the CMV-loxP-dsRED2-loxP-EGFP plasmid (50 ng/μl) with Cre RNA (25 ng/μl) resulted in the excision of the dsRED2 allele and juxtaposition of the EGFP transgene next to the CMV promoter, leading to embryos that express EGFP and no red fluorescence (Fig. 3 F and G). Excision was extremely efficient in embryos injected with 25 ng/μl Cre RNA because single cells with red fluorescence were observed in <5% of injected zebrafish embryos (n = 100).
Applying this strategy to our transgenic models of T cell malignancy, the rag2-EGFP-mMyc transgene was modified by inserting the loxP-dsRED2-loxP cassette between the rag2 promoter and the EGFP-mMyc oncogene (Fig. 4A), and two stable transgenic lines were generated (G7 and G16). In the absence of Cre-mediated recombination, rag2-loxP-dsRED2-loxP-EGFP-mMyc transgenic fish exhibited high levels dsRED2 expression in the developing thymocytes but failed to express the EGFP-mMyc transgene or develop lymphoma or leukemia (Fig. 4B). After the injection of Cre RNA into G7 and G16 one-cell embryos, the dsRED2 allele was excised in both lines (Fig. 4C); however, only transgenic zebrafish from the G7 line developed T-ALL. Most leukemias arising in the G7 line expressed both dsRED2 and EGFP-mMyc (Fig. 4D), suggesting that Cre recombination was incomplete in embryos injected with 25 ng/μl Cre RNA. Some of the G7 line leukemias exhibited complete recombination and expressed only the EGFP-mMyc transgene, indicated by green fluorescence without any detectable red fluorescence (Fig. 4E). Heterozygous rag2-loxP-dsRED2-loxP-EGFP-mMyc fish (G7 line) were bred to AB WT fish and injected with Cre RNA at the one-cell stage of development. In total, 12 of 186 CRE-injected progeny developed disease in the G7 line by 151 ± 61 days (range = 52-192 days; n = 5).
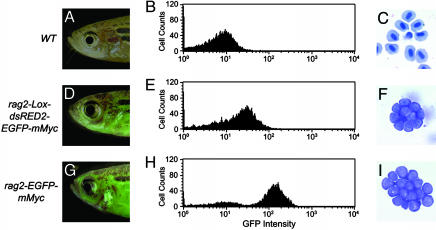
T-ALLs in Cre-Injected Fish Are Similar to Those from rag2-EGFP-mMyc Leukemias. Fluorescent microscopic analysis revealed that rag2-loxP-dsRED2-loxP-EGFP-mMyc transgenic zebrafish injected with Cre RNA at the one-cell stage develop thymic leukemias as adults, indicating that these tumors are of T cell origin. Additionally, fluorescent leukemia cells can be transplanted into irradiated recipients (Fig. 5 D and G; n = 3) and have lymphoblast morphology as determined by Giemsa/May-Grunwald staining (Fig. 5 F and I; n = 2). Taken together, these results indicate that leukemias arising in the conditional rag2-loxP-dsRED2-loxP-EGFP-mMyc transgenic line (G7) are similar to those observed in the rag2-EGFP-mMyc stable transgenic line.
Fig. 5.
Leukemias developing in Cre-injected rag2-loxP-dsRED2-loxP-EGFP-mMyc transgenic fish are transplantable, express low levels of GFP fluorescence, and have typical lymphoblast morphology. WT control (A-C) and irradiated WT fish transplanted with lymphoblasts from rag2-loxP-dsRED2-loxP-EGFP-mMyc (D-F) or rag2-EGFP-mMyc transgenic (G-I) fish. (A, D, and G) Fluorescent microscopic analysis with fish oriented with anterior to the left and dorsal to the top. Images are a composite of GFP fluorescence and bright-field images. (B, E, and H) FACS analysis based on GFP fluorescence. (C, F, and I) Giemsa/May-Grunwald staining of blood cells obtained from cytospin analysis. Original images were photographed at 1,000×.
Discussion
We have previously shown that transgenic zebrafish develop Myc-induced T cell lineage leukemia (15); however, a detailed molecular characterization of these tumors has not been described. Leukemias developing in rag2-EGFP-mMyc transgenic fish are remarkably similar to those found in human patients with T-ALL. For example, zebrafish Myc-induced T-ALLs arise after a defined latency period and have clonal TCR-α gene rearrangements, suggesting that additional mutations are required for malignant transformation of the T cell. Additionally, these zebrafish T-ALLs coexpress both scl and lmo2, resembling the most common and most treatment-resistant molecular subtype of this disease in humans (1, 11). Both scl and lmo2 are biallelically activated in zebrafish Myc-induced T-ALL, in a pattern similar to that of a subset of patients who have overexpression of SCL and LMO2 in leukemic lymphoblasts, indicating that malignant transformation likely results from the disruption of upstream regulatory mechanisms that normally turn off the expression of these transcription factors during double-negative thymocyte development (8).
Although zebrafish Myc-induced T-ALLs are similar to human leukemias, there are also key differences. First, zebrafish Myc-induced T-ALLs resemble only one subclass of human T-ALLs, those that coexpress SCL and LMO2. Remarkably, we never observe expression of hox11/tlx1 or the hox11L2/tlx3 family members in Myc-induced leukemias in the zebrafish (26). It is possible that timing of transgene expression during thymocyte development affects the subtype of T-ALL that is observed. For example, rag2 expression is tightly regulated during T cell development and is only induced in cells undergoing active TCR-α and TCR-β gene rearrangement (37, 38), providing two waves of Myc transgene expression during thymocyte development. Another possibility is that the mechanisms that regulate HOX11- and HOX11L2-induced transformation in humans are not found in the zebrafish. rag2-hox11 and rag2-tlx3 transgenic zebrafish will need to be developed to determine whether overexpression of a hox11 family member can synergize with Myc in the genesis of zebrafish T-ALL.
Human and murine leukemias escape cell death by inactivation of multiple gene products involved in regulating the apoptotic pathways. For example, leukemias developing in Emu-Myc mice harbor mutations that curb apoptosis, including the loss of p19(ARF), mutation of p53, or up-regulation of Mdm2 (27), and most human T-ALLs have biallelic deletion of the CDKN2A locus, which encodes the P14(ARF) gene (14). Because of these findings, we expected to identify abnormalities that down-regulate apoptotic pathways in our zebrafish leukemias; however, we did not find variable expression levels of mdm2, bcl-xL, bcl-2, p53, or bax, nor did we find deletions in the p16 gene locus or mutations in the p53 DNA-binding domain. Thus, we conclude that either suppression of apoptosis is mediated by a currently unidentified mechanism in zebrafish T-ALLs or Myc-induced transformation in zebrafish lymphoid cells does not require an associated mutational inactivation of the apoptotic machinery. Given that teleost fishes apparently lack an ARF gene (39), it is likely that overexpression of myc may not activate the cell death machinery through the p53 pathway in zebrafish, as has been documented in mammalian cells (40). Further experiments using rag2-EGFP-bcl-2 transgenic (41) and p53-deficient fish (25) will likely resolve whether suppression of apoptosis is required for malignant transformation of the T cell in zebrafish.
Leukemias developed in 100% of stable transgenic rag2-EGFP-mMyc fish, which is optimal for performing genetic screens designed to uncover mutations that enhance or suppress leukemogenesis. However, the transgenic rag2-EGFP-mMyc zebrafish line has been difficult to maintain, because these fish develop disease before reaching full reproductive maturity. To resolve this problem, we developed conditional transgenic zebrafish by using Cre/lox technology (33-36). Two transgenic zebrafish lines were identified that contained the rag2-loxP-dsRED2-loxP-EGFP-mMyc transgene. In the absence of Cre RNA expression, both lines had strong expression of dsRED2 within the developing T cells, and no fish developed disease, indicating that the mMyc oncogene was not expressed in the absence of Cre recombination. After injection of Cre RNA into one-cell-stage embryos, both transgenic lines exhibited recombination at the loxP sites; however, only the G7 stable line produced offspring that developed T-ALL. Lack of leukemia onset in the second transgenic line (G16) reinforces the need to develop multiple transgenic lines and suggests that positional effects of integration and/or concatamer orientation may significantly affect Cre-mediated excision and subsequent expression of the second transgene.
Because only 13% of Cre-injected transgenic fish developed disease, it is likely that injection of Cre RNA results in suboptimal recombination and mosaic activation of the EGFP-mMyc transgene after recombination. This interpretation is supported by the fact that most leukemias arising in Cre-injected rag2-loxP-dsRED2-loxP-EGFP-mMyc transgenic fish are both dsRED2- and GFP-labeled, indicating that leukemic clones had partial Cre-mediated recombination at the locus containing transgene concatamers. Similarly, individual fish had a leukemic clone that expressed both dsRED2 and GFP arising in one thymus and a second leukemia clone that expressed only GFP in the other thymus, indicating that Cre recombination occurs in a mosaic fashion within hematopoietic progenitors of individual fish. Similar results have been observed in Xenopus (36). For example, transient injection of Cre RNA into stable transgenic frogs harboring a CMV-loxP-ECFP-loxP-EYFP transgene resulted in mosaic expression of the second ORF. Furthermore, in some animals, neither blue (ECFP) nor yellow (EYFP) fluorescence was detected in transgenic frogs after Cre RNA injection, leading the researchers to conclude that the copy number of the reporter had been reduced by recombination without generating an active EYFP expression cassette (36). By contrast, breeding these CMV-loxP-ECFP-loxP-EYFP transgenic frogs to stable transgenic Cre-expressing animals resulted in ≈100% of doubly transgenic offspring having homogenous expression of the second EYFP ORF. Thus, developing transgenic zebrafish lines that specifically express Cre in the developing T cells will likely aid in the establishment of more penetrant models of disease.
Although zebrafish models of T-ALL exhibit key differences when compared with the human disease, analysis of the conserved mechanisms underlying transformation will likely lead to insights into the pathogenesis of human disease. For example, the molecular mechanisms responsible for regulating biallelic activation of scl and lmo2 are unknown, and the downstream targets of Myc that are responsible for oncogenic transformation and genomic instability have yet to be identified. Because the zebrafish affords the unique opportunity to perform forward genetic screens, it should be possible to dissect the pathways that regulate Myc-induced disease in a genetically tractable vertebrate. The proven feasibility of Cre/lox-mediated strategies in the zebrafish will aid the development of models of leukemia, lymphoma, and other cancers, which will provide opportunities for both genetic and chemical modifier screens designed to identify suppressors and enhancers in carcinogenesis. For example, a dominant modifier genetic screen could be conducted by breeding N-ethyl-N-nitrosourea-mutagenized fish with homozygous rag2-loxP-dsRED2-loxP-EGFP-mMyc fish and then analyzing the progeny for the time of leukemia onset after CRE recombination and expression of the Myc transgene. Mutations that modify the time of leukemia onset could be either enhancers that result in more rapid onset of leukemia due to the inactivation of one allele of a tumor suppressor gene or suppressors that delay or prevent the onset of Myc-induced transformation. Finally, use of the Cre/lox technology in the zebrafish will provide tools for assessing cell lineage commitment and plasticity of stem cells and generating conditional knockouts in developing embryos.
Supplementary Material
Acknowledgments
We thank Y. Yang, N. Campisi, and E. Ronan for expert technical assistance; L. I. Zon, B. Paw, and B. E. H. Langenau for critical review of the manuscript; and J. Vinokur, G. Kourkoulis, and W. Saganic for fish care and husbandry. This work was supported by National Institutes of Health Grants CA-68484 (to A.T.L.) and CA-06516 (to J.L.K.). D.M.L. was a National Science Foundation Predoctoral Fellow and is now the Edmond J. Safra Foundation-Irvington Institute Fellow.
Author contributions: D.M.L. designed research; D.M.L., H.F., and S.B. performed research; S.B. and J.L.K. contributed new reagents/analytic tools; D.M.L., H.F., S.B., J.P.K., and J.L.K. analyzed data; D.M.L. and A.T.L. wrote the paper; J.P.K. and J.L.K. edited the paper; and A.T.L. served as the principal investigator.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: T-ALL, T cell acute lymphoblastic leukemia; mMyc, mouse c-Myc; LMO, LIM-only; TCR, T cell receptor.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF398514).
References
- 1.Ferrando, A. A., Neuberg, D. S., Staunton, J., Loh, M. L., Huard, C., Raimondi, S. C., Behm, F. G., Pui, C. H., Downing, J. R., Gilliland, D. G., et al. (2002) Cancer Cell 1, 75-87. [DOI] [PubMed] [Google Scholar]
- 2.Look, A. T. (1997) Science 278, 1059-1064. [DOI] [PubMed] [Google Scholar]
- 3.Ferrando, A. A. & Look, A. T. (2000) Semin. Hematol. 37, 381-395. [DOI] [PubMed] [Google Scholar]
- 4.Bernard, O. A., Busson-LeConiat, M., Ballerini, P., Mauchauffe, M., Della Valle, V., Monni, R., Nguyen Khac, F., Mercher, T., Penard-Lacronique, V., Pasturaud, P., et al. (2001) Leukemia 15, 1495-1504. [DOI] [PubMed] [Google Scholar]
- 5.Wang, J., Jani-Sait, S. N., Escalon, E. A., Carroll, A. J., de Jong, P. J., Kirsch, I. R. & Aplan, P. D. (2000) Proc. Natl. Acad. Sci. USA 97, 3497-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kees, U. R., Heerema, N. A., Kumar, R., Watt, P. M., Baker, D. L., La, M. K., Uckun, F. M. & Sather, H. N. (2003) Leukemia 17, 887-893. [DOI] [PubMed] [Google Scholar]
- 7.Watt, P. M., Kumar, R. & Kees, U. R. (2000) Genes Chromosomes Cancer 29, 371-377. [DOI] [PubMed] [Google Scholar]
- 8.Ferrando, A. A., Herblot, S., Palomero, T., Hansen, M., Hoang, T., Fox, E. A. & Look, A. T. (2004) Blood 103, 1909-1911. [DOI] [PubMed] [Google Scholar]
- 9.Ferrando, A. A. & Look, A. T. (2003) Semin. Hematol. 40, 274-280. [DOI] [PubMed] [Google Scholar]
- 10.Yeoh, E. J., Ross, M. E., Shurtleff, S. A., Williams, W. K., Patel, D., Mahfouz, R., Behm, F. G., Raimondi, S. C., Relling, M. V., Patel, A., et al. (2002) Cancer Cell 1, 133-143. [DOI] [PubMed] [Google Scholar]
- 11.Ferrando, A. A., Neuberg, D. S., Dodge, R. K., Paietta, E., Larson, R. A., Wiernik, P. H., Rowe, J. M., Caligiuri, M. A., Bloomfield, C. D. & Look, A. T. (2004) Lancet 363, 535-536. [DOI] [PubMed] [Google Scholar]
- 12.Ballerini, P., Blaise, A., Busson-Le Coniat, M., Su, X. Y., Zucman-Rossi, J., Adam, M., van den Akker, J., Perot, C., Pellegrino, B., Landman-Parker, J., et al. (2002) Blood 100, 991-997. [DOI] [PubMed] [Google Scholar]
- 13.Weng, A. P., Ferrando, A. A., Lee, W., Morris, J. P. T., Silverman, L. B., Sanchez-Irizarry, C., Blacklow, S. C., Look, A. T. & Aster, J. C. (2004) Science 306, 269-271. [DOI] [PubMed] [Google Scholar]
- 14.Okuda, T., Shurtleff, S. A., Valentine, M. B., Raimondi, S. C., Head, D. R., Behm, F., Curcio-Brint, A. M., Liu, Q., Pui, C. H. & Sherr, C. J. (1995) Blood 85, 2321-2330. [PubMed] [Google Scholar]
- 15.Langenau, D. M., Traver, D., Ferrando, A. A., Kutok, J. L., Aster, J. C., Kanki, J. P., Lin, S., Prochownik, E., Trede, N. S., Zon, L. I. & Look, A. T. (2003) Science 299, 887-890. [DOI] [PubMed] [Google Scholar]
- 16.Peterson, R. T., Link, B. A., Dowling, J. E. & Schreiber, S. L. (2000) Proc. Natl. Acad. Sci. USA 97, 12965-12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peterson, R. T., Mably, J. D., Chen, J. N. & Fishman, M. C. (2001) Curr. Biol. 11, 1481-1491. [DOI] [PubMed] [Google Scholar]
- 18.Peterson, R. T., Shaw, S. Y., Peterson, T. A., Milan, D. J., Zhong, T. P., Schreiber, S. L., MacRae, C. A. & Fishman, M. C. (2004) Nat. Biotechnol. 22, 595-599. [DOI] [PubMed] [Google Scholar]
- 19.Driever, W., Solnica-Krezel, L., Schier, A. F., Neuhauss, S. C., Malicki, J., Stemple, D. L., Stainier, D. Y., Zwartkruis, F., Abdelilah, S., Rangini, Z., et al. (1996) Development (Cambridge, U.K.) 123, 37-46. [DOI] [PubMed] [Google Scholar]
- 20.Haffter, P., Granato, M., Brand, M., Mullins, M. C., Hammerschmidt, M., Kane, D. A., Odenthal, J., van Eeden, F. J., Jiang, Y. J., Heisenberg, C. P., et al. (1996) Development (Cambridge, U.K.) 123, 1-36. [DOI] [PubMed] [Google Scholar]
- 21.Golling, G., Amsterdam, A., Sun, Z., Antonelli, M., Maldonado, E., Chen, W., Burgess, S., Haldi, M., Artzt, K., Farrington, S., et al. (2002) Nat. Genet. 31, 135-140. [DOI] [PubMed] [Google Scholar]
- 22.Langenau, D. M., Ferrando, A. A., Traver, D., Kutok, J. L., Hezel, J. P., Kanki, J. P., Zon, L. I., Look, A. T. & Trede, N. S. (2004) Proc. Natl. Acad. Sci. USA 101, 7369-7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jessen, J. R., Jessen, T. N., Vogel, S. S. & Lin, S. (2001) Genesis 29, 156-162. [DOI] [PubMed] [Google Scholar]
- 24.Haire, R. N., Rast, J. P., Litman, R. T. & Litman, G. W. (2000) Immunogenetics 51, 915-923. [DOI] [PubMed] [Google Scholar]
- 25.Berghmans, S., Murphey, R. D., Wienholds, E., Neuberg, D., Kutok, J. L., Fletcher, C. D., Morris, J. P., Liu, T. X., Schulte-Merker, S., Kanki, J. P., et al. (2005) Proc. Natl. Acad. Sci. USA 102, 407-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langenau, D. M., Palomero, T., Kanki, J. P., Ferrando, A. A., Zhou, Y., Zon, L. I. & Look, A. T. (2002) Mech. Dev. 117, 243-248. [DOI] [PubMed] [Google Scholar]
- 27.Eischen, C. M., Weber, J. D., Roussel, M. F., Sherr, C. J. & Cleveland, J. L. (1999) Genes Dev. 13, 2658-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yunis, J. J., Frizzera, G., Oken, M. M., McKenna, J., Theologides, A. & Arnesen, M. (1987) N. Engl. J. Med. 316, 79-84. [DOI] [PubMed] [Google Scholar]
- 29.Diccianni, M. B., Yu, J., Hsiao, M., Mukherjee, S., Shao, L. E. & Yu, A. L. (1994) Blood 84, 3105-3112. [PubMed] [Google Scholar]
- 30.Jonveaux, P. & Berger, R. (1991) Leukemia 5, 839-840. [PubMed] [Google Scholar]
- 31.Olivier, M., Eeles, R., Hollstein, M., Khan, M. A., Harris, C. C. & Hainaut, P. (2002) Hum. Mutat. 19, 607-614. [DOI] [PubMed] [Google Scholar]
- 32.Beroud, C. & Soussi, T. (2003) Hum. Mutat. 21, 176-181. [DOI] [PubMed] [Google Scholar]
- 33.Gu, H., Marth, J. D., Orban, P. C., Mossmann, H. & Rajewsky, K. (1994) Science 265, 103-106. [DOI] [PubMed] [Google Scholar]
- 34.Kuhn, R., Schwenk, F., Aguet, M. & Rajewsky, K. (1995) Science 269, 1427-1429. [DOI] [PubMed] [Google Scholar]
- 35.Werdien, D., Peiler, G. & Ryffel, G. U. (2001) Nucleic Acids Res. 29, E53-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryffel, G. U., Werdien, D., Turan, G., Gerhards, A., Goosses, S. & Senkel, S. (2003) Nucleic Acids Res. 31, e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oettinger, M. A., Schatz, D. G., Gorka, C. & Baltimore, D. (1990) Science 248, 1517-1523. [DOI] [PubMed] [Google Scholar]
- 38.Schatz, D. G., Oettinger, M. A. & Baltimore, D. (1989) Cell 59, 1035-1048. [DOI] [PubMed] [Google Scholar]
- 39.Gilley, J. & Fried, M. (2001) Oncogene 20, 7447-7452. [DOI] [PubMed] [Google Scholar]
- 40.Zindy, F., Eischen, C. M., Randle, D. H., Kamijo, T., Cleveland, J. L., Sherr, C. J. & Roussel, M. F. (1998) Genes Dev. 12, 2424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langenau, D. M., Jette, C., Berghmans, S., Palomero, T., Kanki, J. P., Kutok, J. L. & Look, A. T. (2005) Blood 105, 3278-3285. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.