Abstract
Stat3 protein has an important role in oncogenesis and is a promising anticancer target. Indirubin, the active component of a traditional Chinese herbal medicine, has been shown previously to inhibit cyclin-dependent kinases, resulting in cell cycle arrest. Here, we show that the indirubin derivatives E564, E728, and E804 potently block constitutive Stat3 signaling in human breast and prostate cancer cells. In addition, E804 directly inhibits Src kinase activity (IC50 = 0.43 μM) in an in vitro kinase assay. Levels of tyrosyl phosphorylation of c-Src are also reduced in cultured cells 30 min after E804 treatment. Tyrosyl phosphorylation of Stat3, which is known to be phosphorylated by c-Src, was decreased, and constitutive Stat3 DNA binding-activity was suppressed in cells 30 min after E804 treatment. The antiapoptotic proteins Mcl-1 and Survivin, which are encoded in target genes of Stat3, were down-regulated by indirubin derivatives, followed by induction of apoptosis. These results demonstrate that E804 directly blocks the Src-Stat3 signaling pathway, suggesting that the antitumor activity of indirubin compounds is at least partially due to inhibition of this pathway.
Keywords: Src, signal transducer and activator of transcription 3
Signal transducer and activator of transcription (STAT) proteins have been shown to play an important role in tumor cell survival and proliferation (1-4). One STAT family member, Stat3, is often constitutively activated in many human cancer cells and tumor tissues and has been shown to induce expression of genes involved in cell proliferation and survival (1-4). Recently, Stat3 has been implicated as a promising target for cancer therapy (1, 5, 6). Tyrosine kinases that phosphorylate Stat3, particularly Jak and Src kinases, also have been investigated as potential targets for cancer treatment (7-10).
The c-Src kinase acts upstream of Stat3 and has a key role in cell proliferation, tumorigenesis, and metastasis (11, 12). Elevated c-Src protein levels and kinase activities have been demonstrated in numerous human cancer cell lines and tumor tissues of patients (13-15). The c-Src kinase phosphorylates tyrosyl residues of critical cellular substrates, which results in activation of oncogenic signal transduction pathways (11, 16). As a substrate of c-Src kinase, tyrosyl phosphorylation and activation of Stat3 modulates cell proliferation, survival, angiogenesis, and immune evasion of cancer cells (1, 2). Many studies have demonstrated that inhibition of Src-Stat3 signaling induces growth arrest and apoptosis of human tumor cells (1, 11).
Indirubin has been shown to be the active component of a traditional Chinese herbal medicine, Danggui Longhui Wan, used for treatment of chronic myelogenous leukemia (17). Indirubin derivatives (IRDs) were found to act as potent inhibitors of cyclin-dependent kinase (CDK)1/cyclin B, CDK2/cyclin A, CDK2/cyclin E, GSK3β, and CDK5/p25, displaying potent growth inhibitory effects in human tumor cells (18-20). The cell cycle of tumor cells treated with indirubins is arrested in the G1/SorG2/M phases, resulting in inhibition of cell proliferation, and ultimately, induction of apoptosis (18, 19).
Indirubins competitively inhibit ATP binding in the catalytic domain of CDK enzymes. X-ray structures of the enzyme-inhibitor complex revealed that three hydrogen bonds formed in the ATP-binding pocket by indirubins NH, CO, and N′H groups are essential for binding to the peptide backbone (18, 21). Molecular modifications in this key region of the molecule abolish binding affinity and ATP-competitive inhibition of CDKs. In contrast, the 5′ and 3′ positions have been found to be amenable to molecular permutations for improved inhibitory potency (21). The molecular mechanism by which indirubins may cause apoptosis of tumor cells, however, has not been demonstrated previously.
In the present study, we report that the IRD E804 directly inhibits c-Src kinase activity in vitro and causes reduction of phosphotyrosyl c-Src levels in cells. We also demonstrate a corresponding reduction of constitutive Stat3 DNA-binding activity upon E804 treatment of human breast and prostate cancer cells. Moreover, inactivation of Stat3 DNA-binding activity results in down-regulation of the antiapoptotic proteins Mcl-1 and Survivin and induction of apoptosis of human breast cancer cells. These results indicate that the IRD E804, may be a novel Src inhibitor that blocks downstream Stat3 signaling, demonstrating the potential of this compound for treatment of epithelial tumors.
Materials and Methods
Cell Lines and Reagents. MDA-MB-468 and MDA-MB-435 human breast cancer cells were cultured in DMEM containing 10% FBS. Human DU145 prostate cancer cells were grown in RPMI medium 1640 supplemented with 10% FBS. Normal NIH 3T3 and transformed NIH 3T3/v-Src mouse fibroblasts were cultured in DMEM with 5% iron-supplemented bovine calf serum. Normal MCF-10A human breast epithelial cells were cultured in DMEM/F-12 supplemented with 20 ng/ml EGF/100 ng/ml cholera toxin/10 μg/ml insulin/500 ng/ml hydrocortisone/5% horse serum.
Polyclonal antibodies to phospho-Tyr-705-Stat3 (p-Stat3), phospho-Tyr-416-Src (p-Src), and phospho-Tyr-1022/1023-Jak1 (p-Jak1) were obtained from Cell Signaling Technologies (Cambridge, MA). Polyclonal antibodies to Stat3, Jak1, Bcl-xL, Mcl-1, and β-actin were from Santa Cruz Biotechnology. Polyclonal antibody to Survivin was obtained from Alpha Diagnostic International (San Antonio, TX). Monoclonal antibody to c-Src was obtained from Upstate Biotechnology (Lake Placid, NY). Polyclonal antibody to poly (ADP-ribose) polymerase (PARP) was purchased from Roche Molecular Biochemicals (Indianapolis). An In situ cell death detection kit was from Roche Applied Science (Indianapolis, IN).
Indirubin Compounds. Indirubin-3′ oximes were prepared by condensation of indirubines with hydroxylamine hydrochloride. Indirubins were produced by the reaction of appropriately substituted isatins with indoxyl acetate as described (18). Indirubin-3′ oxime ethers were synthesized by reaction of indirubin-3′ oximes with substituted, functionalized bromo alkanes according to published methods (22). Structures and purities of all compounds were ascertained by 13C- and 1H-NMR spectroscopy and elemental analyses (S.M., F.H., S.V., K.-H.M., and G.E., unpublished data).
EMSA. Nuclear extracts were prepared as described (8, 23). For mobility shift assay of Stat3-DNA binding activity, 8 μg of nuclear protein was incubated with 32P-radiolabeled oligonucleotide probe containing the high-affinity sis-inducible element, m67 variant (hSIE) derived from the c-fos gene promoter (5′-AGCTTCATTTCCCGTAAATCCCTA-3′). For supershifts, 1 μl of antibody to Stat3 was preincubated with the extract for 30 min before the addition of 32P-labeled hSIE probe. Nuclear extract (5 μg) from NIH 3T3/v-Src-transformed fibroblasts was preincubated with E804 for 10 min before reaction with 32P-labeled hSIE probe for EMSA. Resolution of protein-DNA complexes was performed by 5% nondenaturing PAGE and detected by autoradiography.
Western Blot Analysis. Enhanced chemiluminescence (ECL) Western blot analysis was performed as described (24, 25). Seventy micrograms of total protein was resolved on SDS/PAGE gels and immunoblotted with specific antibodies. Primary phosphospecific antibodies were incubated in TBS (pH 7.5) with 0.1% Tween-20 and 5% BSA with gentle agitation overnight at 4°C as described in the supplier's instructions. Other antibodies were diluted with PBS containing 5% nonfat milk and 0.1% Tween-20 overnight at 4°C. Horseradish peroxidase-conjugated secondary antibodies were incubated in TBS (pH 7.5) with 5% nonfat milk and 0.1% Tween-20, or PBS with 5% nonfat milk and 0.1% Tween-20, at a 1:2,000 dilution for 1 h at room temperature. Positive immunoreactions were detected by using the ECL system (Amersham Pharmacia, Piscataway, NJ).
Src Kinase Assay in Vitro. MDA-MB-468 cells were harvested, washed, and lysed with a RIPA 150 buffer (10 mM Tris·HCl pH 7.5/150 mM NaCl/10% glycerol/5 mM EDTA/100 μM sodium orthovanadate/0.1% SDS/1% Triton X-100/10 μg/ml aprotinin/10 μg/ml leupeptin/1 μg/ml antipain) with gentle rocking for 30 min at 4°C. Src kinase assays in vitro were performed as described with some modifications (26, 27). Briefly, whole-cell lysates were clarified by centrifugation at 13,000 rpm for 30 min (Eppendorf, Westbury, NY, model 5415R microfuge). Then, 800-μg aliquots of cell lysates were incubated with 2 μg of c-Src antibody for 3 h at 4°C. Protein A/G PLUS-agarose (25 μl) was added with gentle rocking for 1 h at 4°C. Immunoprecipitates were washed with RIPA 150 buffer three times, and twice with RIPA 10 buffer (10 mM Tris·HCl, pH 7.5/10 mM NaCl/10% glycerol/5 mM EDTA/100 μM sodium orthovanadate/0.1% SDS/1% Triton X-100/10 μg/ml aprotinin/10 μg/ml leupeptin/1 μg/ml antipain). Immunoprecipitates were then washed once with kinase reaction buffer (50 mM Hepes, pH 7.5/10 mM MgCl2/5 mM MnCl2/10 mM DTT). The pellet was preincubated with DMSO solvent, E804, PD180970, or AG490 in 25 μl of reaction buffer containing 5 μg of denatured exogenous enolase substrate for 3 min before the addition of 4 μl with a mixture of 100 μM ATP and 10 μCi (1 Ci = 37 GBq) of [γ-32P]ATP. Reaction mixtures were incubated with gentle rocking for 20 min at 30°C, and the kinase reaction was stopped with 2× SDS/PAGE loading buffer. Samples were boiled, resolved on SDS/10.5% PAGE gels, and phosphorylated enolase was visualized by autoradiography.
Jak1 Kinase Assay in Vitro. Jak1 autophosphorylation kinase assay in vitro was carried out as described with minor modifications (25, 26). MDA-MB-468 cells were lysed with lysis buffer (25 mM Hepes, pH 7.5/0.5 mM DTT/0.1% Triton X-100/1 mM sodium orthovanadate/1 mM PMSF/10 μg/ml aprotinin/10 μg/ml leupeptin). Cell lysates (800 μg) were incubated with 2 μg of Jak1 antibody with gentle rocking for 3 h at 4°C. Then, 25 μl of protein A/G PLUS-agarose was added to each sample. The pellet was washed with buffer (50 mM Hepes, pH 7.4/0.5 mM DTT/0.1% Triton X-100/150 mM NaCl) three times and once with kinase reaction buffer (50 mM Hepes, pH 7.4/0.5 mM DTT/0.1% Triton X-100/6.25 mM MnCl2/100 mM NaCl). DMSO solvent, E804, PD180970, or AG490 was preincubated in 25 μl of reaction buffer for 3 min before the addition of 4 μl containing 100 μM ATP and 20 μCi of [γ-32P]ATP. The reaction mixture was incubated with gentle rocking for 20 min at 30°C and terminated with 10 mM EDTA. Samples were resolved on SDS/8% PAGE gels, and Jak1 autophosphorylation was visualized by autoradiography.
CDK Kinase Assay in Vitro. Human recombinant CDK/cyclin complexes, expressed in Sf-21 insect cells, were obtained from Upstate Biotechnology (CDK1/cyclin B, CDK2/cyclin A, and CDK2/cyclin E) with specific activities according to the individual lot numbers. The in vitro kinase assays were performed following the manufacturer's protocol, with 0.4 μCi of [γ-32P]ATP per sample vial. Briefly, the respective CDK/cyclin assay mixture was incubated for 10 min at 30°C. There-after, the solution (20 μl) was spotted on P81 phosphocellulose squares. After being washed with 0.75% phosphoric acid three times on a rocking platform, the sheets were rinsed with acetone, dried, and transferred into scintillation vials for β-counting. Each assay was performed in duplicate in at least three independent experiments.
Apoptosis Assays and Cell Cycle Analysis. Whole-cell lysates were immunoblotted with specific antibodies to PARP to detect PARP cleavage as an apoptosis indicator. Positive immunoreactions were detected by using the ECL blotting system. All procedures were performed as described (28). DNA fragmentation assay for detection of DNA strand breaks was carried out by using an Apo-BrdUrd kit as described by the supplier (BD Biosciences PharMingen, San Diego). Cells were analyzed by using a FACScan flow cytometer with cellquest 3.3 software (Becton Dickinson Immunocytometry, San Jose, CA) and modfit lt cell cycle analysis software (Verity Software, Topsham, ME).
Results
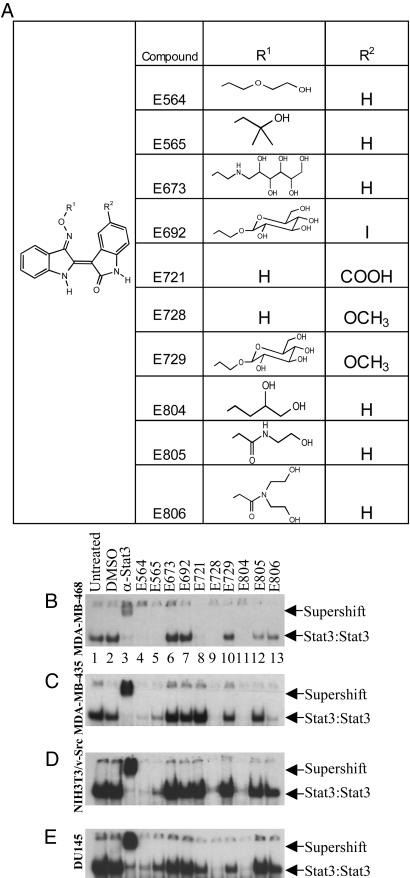
Inhibition of Stat3 DNA-Binding Activity by IRDs. Ten IRDs with a common core structure and different substituents (Fig. 1A) were synthesized and tested to determine their effect on Stat3 activation in MDA-MB-435 and MDA-MB-468 human breast cancer cells, DU145 human prostate cancer cells, and Src-transformed NIH 3T3 mouse fibroblasts. Most IRDs are poorly soluble in water and show limited bioavailability. Therefore, substituents bearing hydrophilic components in the 3′ oxime ether position were synthesized to enhance bioavailability. However, inhibitory potency was impaired or abolished for the compounds with sugar or aminopolyol moieties (E673, E692, and E729) or the substituted glycine amides (E805 and E806). In contrast, unbranched short-chain oxime ethers with one or two hydroxyl groups (as represented by E804) were found to be favorable (Fig. 1).
Fig. 1.
IRDs inhibit Stat3 DNA-binding activity. (A) Structures of IRDs. (B-E) The cell lines MDA-MB-468 (B), MDA-MB-435 (C), NIH 3T3/v-Src (D), and DU145 (E) were treated with or without 10 μM of IRDs for 24 h. Controls are untreated, DMSO vehicle alone, or supershift with antibody to Stat3, as indicated. Nuclear extracts were incubated with radiolabeled hSIE probe and Stat3 DNA-binding activities were determined by EMSA. The electrophoretic mobilities of Stat3:Stat3 homodimers bound to DNA probe and antibody supershifted Stat3 complexes are indicated by arrows.
Stat3 DNA-binding activity measured by EMSA was strongly inhibited by 3 of the 10 IRDs, E564, E728, and E804 (Fig. 1 B-E) in all four cell lines. E721 showed inhibition of Stat3 DNA-binding activity only in MDA-MB 468 cells (Fig. 1B, lane 8). Compounds E728 and E804 displayed the greatest inhibition of Stat3 DNA binding. Because the latter derivative was slightly more soluble, it was selected for further studies on MDA-MB-468 breast cancer cells. This breast cancer cell line was chosen because it has high constitutive Stat3 DNA-binding and Src kinase activities (34).
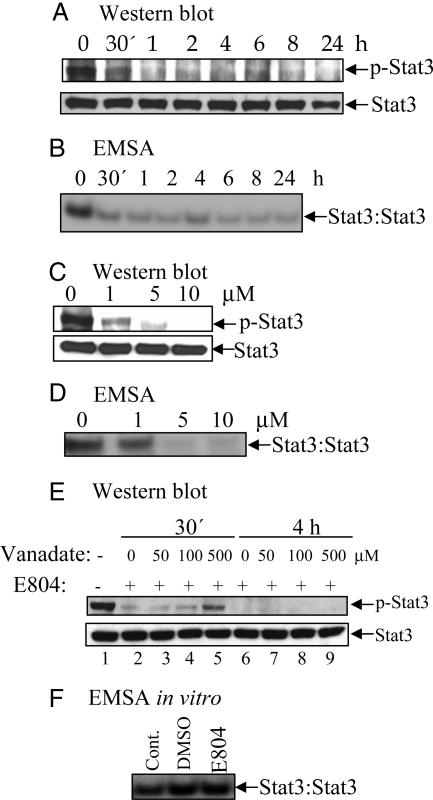
E804 Reduces p-Stat3 Levels in Cells. Western blot analysis with specific antibodies to p-Stat3 revealed that the p-Stat3 level was significantly decreased as early as 30 min after 10 μM E804 treatment (Fig. 2A Upper), whereas total Stat3 protein level was unchanged (Fig. 2 A Lower). A dose-response study using whole lysates of cells treated with E804 for 4 h revealed a significant decrease in the p-Stat3 level at 1-10 μM (Fig. 2C Upper). Consistent with these results, we found a significant decrease in nuclear Stat3 DNA-binding activity as measured by EMSA after 30 min of 10 μM E804 treatment (Fig. 2B). A dose-response study revealed inhibition of constitutive Stat3 DNA-binding activity at 1-10 μM E804 treatment for 4 h (Fig. 2D).
Fig. 2.
E804 blocks Stat3 tyrosyl phosphorylation. MDA-MB-468 cells were treated with 10 μM E804 in a time-dependent manner (A and B) or with E804 for 4 h in a dose-response manner (C and D). DMSO was used as vehicle control. (E) Cells were cotreated with or without 10 μM E804 (lanes 2-9) or the tyrosine phosphatase inhibitor sodium orthovanadate (lanes 3-5 and 7-9) for 30 min or 4 h, as indicated. For Western blot analysis, 70 μg of cell-free extracts was subjected to immunoblotting with specific antibodies to p-Stat3 and Stat3. (A, C, and E Upper) Tyrosyl phosphorylation levels (p-Stat3). (A, C, and E Lower) Total Stat3 protein levels. (F) Nuclear extracts from NIH 3T3/v-Src-transformed fibroblasts were preincubated in vitro with 10 μM E804, or controls, for 10 min before incubation with radiolabeled hSIE probe.
To assess the potential role of tyrosine phosphatase activity, which may account for the decrease in p-Stat3 levels, cells were treated with E804 plus vanadate, an inhibitor of tyrosine phosphatase activities (Fig. 2E). If the reductions of p-Stat3 levels were due to an increase in tyrosine phosphatase activity, vanadate would be expected to reverse the effects of E804 on p-Stat3 levels. At 30 min after treatment, vanadate was found to partially increase the p-Stat3 level only at 500 μM concentration (Fig. 2E). However, at 4 h after treatment, vanadate did not reverse the p-Stat3 level at all concentrations studied.
EMSA was next performed to determine whether E804 directly interacts with p-Stat3 protein in vitro, resulting in disruption of Stat3 DNA binding. E804 at 10 μM did not reduce Stat3 DNA-binding activity when added to nuclear extracts (Fig. 2F), suggesting that E804 does not directly disrupt constitutive Stat3 DNA-binding activity in intact cells.
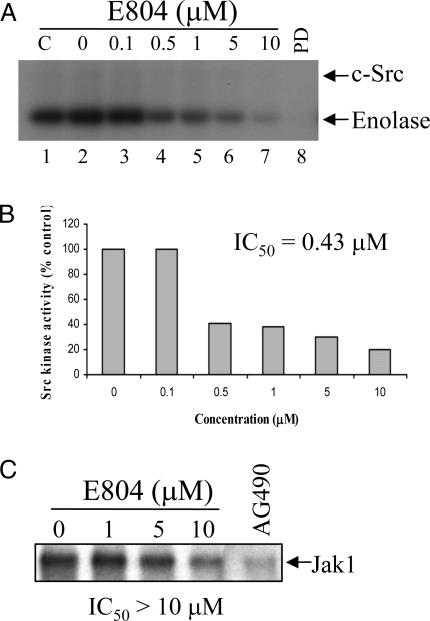
E804 Directly Inhibits Src Kinase Activity in Vitro. To investigate the effect of E804 on Src kinase activity, c-Src was immunoprecipitated with a specific antibody and incubated with E804 and the exogenous substrate enolase in an in vitro kinase assay (Fig. 3A). E804 exhibited significant inhibitory effects on c-Src kinase activity with an IC50 = 0.43 μM (Fig. 3B). As a positive control, the known Src kinase inhibitor PD180970 (1 μM) was used (Fig. 3A, lane 8). E804 also inhibited CDK1/cyclin B, CDK2/cyclin A, and CDK1/cyclin E complexes in an in vitro kinase assay (data not shown), which is consistent with previous results (18). The IC50 values for inhibition of the above CDK/cyclin complexes were 1.65, 0.54, and 0.21 μM, respectively, suggesting that E804 directly inhibits both Src and CDK kinase activities in a similar range of concentrations.
Fig. 3.
E804 directly inhibits Src kinase activity. (A) E804 directly inhibits Src kinase activity in vitro. Equal amounts of cell-free extracts (800 μg) were divided and incubated with 2 μg of antibody to c-Src. Immunocomplexes were preincubated in the presence/absence of various concentrations of E804 for 10 min, and then kinase reactions were performed for 20 min. C, 0, and PD indicate untreated, DMSO, and 1 μM PD180970 (a Src inhibitor) as controls, respectively. (B) Src kinase activities shown in the autoradiograph in A were quantified with imagequant software (Molecular Dynamics) to determine the IC50 of Src inhibition by E804. (C) E804 inhibits Jak1 autophosphorylation activity only at high concentrations. Jak1 immnuocomplexes were preincubated with DMSO or E804 for 10 min, and then kinase reactions were performed for 20 min. AG490 (100 μM), a Jak1 inhibitor, was used as a positive control (C, lane 5). Jak1 autophosphorylation activities were visualized by autoradiography.
To determine the effect of E804 on Jak kinase activity, Jak1 was immunoprecipitated with a specific antibody and incubated with E804 in an in vitro kinase assay (Fig. 3C). E804 was found to only partially inhibit Jak1 autophosphorylation activity at 10 μM. As a positive control, the Jak-selective inhibitor AG490 (100 μM) was used. These results suggest that E804 directly inhibits Src, and to a lesser extent, Jak1, thereby inhibiting tyrosyl phosphorylation and DNA-binding activity of Stat3 in cells.
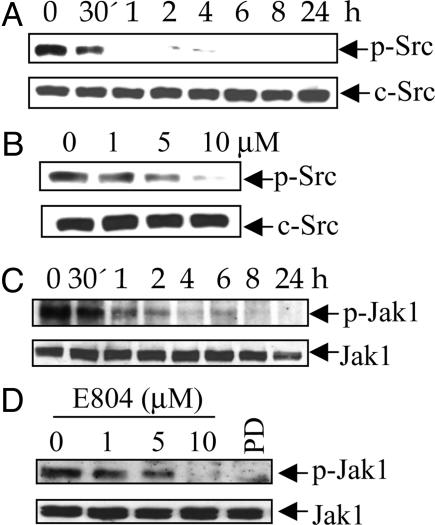
E804 Reduces Levels of p-Src and p-Jak1 in Vivo. Thirty minutes to 1 h of 10 μM E804 treatment resulted in significant reduction of p-Src and p-Jak1 levels in intact cells (Fig. 4 A and C). A dose-response study showed a decrease in p-Src and p-Jak1 levels (Fig. 4 B and D) at 1-10 μM, whereas E804 did not affect total c-Src and Jak1 protein levels. Interestingly, the Src inhibitor PD180970 (1 μM) also inhibited p-Jak1 levels, suggesting that c-Src phosphorylates Jak1 in these cells (Fig. 4 D, lane 5).
Fig. 4.
E804 blocks tyrosyl phosphorylation of Src and Jak1 in cells. MDA-MB-468 cells were treated with 10 μM E804 in a time-dependent manner (A and C) or with E804 for 4 h in a dose-response manner (B and D). DMSO was used as vehicle control. PD180970 (1 μM) was used for comparison (D, lane 5). Seventy micrograms of cell-free extracts was subjected to immunoblotting with specific antibodies to p-Src, c-Src, p-Jak1, or Jak1. Positive immunoreactivity was detected by using ECL.
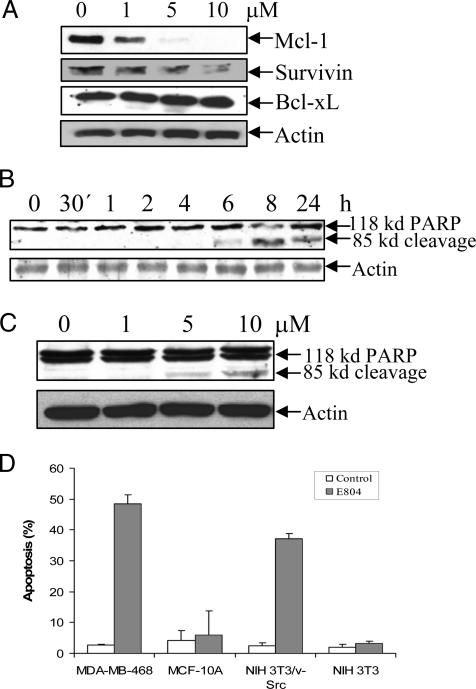
E804 Down-Regulates Mcl-1 and Survivin Proteins and Induces Apoptosis. To determine whether reduction of Stat3 activity by E804 can result in down-regulation of the antiapoptotic proteins, Bcl-xL, Mcl-1, and Survivin, immunoblot analysis of lysates from cells treated with E804 for 24 h was performed. E804 significantly down-regulated Mcl-1 and Survivin expression at 1 to 10 μM, whereas Bcl-xL expression was unchanged (Fig. 5A). Importantly, the reduction of Mcl-1 and Survivin levels correlates well with the inhibition of Stat3 DNA-binding activity shown above (Fig. 2D).
Fig. 5.
E804 down-regulates Mcl-1 and Survivin proteins and induces apoptosis. MDA-MB-468 cells were treated with E804 in a time-dependent manner (B) or with E804 for 24 h in a dose-response manner (A and C). DMSO was used as vehicle control. Seventy micrograms of cell-free extracts was subjected to immunoblotting with specific antibodies to Mcl-1, Survivin, Bcl-xL, PARP, and β-actin. PARP cleavage products were detected by ECL. (D) Apo-BrdUrd labeling and flow cytometry. Cells were treated with DMSO or 10 μM E804 for 24 h were fixed with 1% paraformaldehyde. After being labeled, cells were incubated with the fluorescein-conjugated anti-BrdUrd antibody and analyzed by using a FACScan flow cytometer. The percent of apoptotic cells is shown. Each assay was performed in triplicate.
The effect of E804 on induction of apoptosis was investigated by immunoblotting for PARP cleavage. PARP cleavage was detected at 6 h with 10 μM E804 (Fig. 5B). A dose-response analysis revealed the appearance of PARP cleavage products at 5 μM E804 concentration for 24 h (Fig. 5C). To assess whether E804 could induce apoptosis in both normal and tumor cells, a flow cytometry assay with Apo-BrdUrd was performed on MDA-MB-468 breast cancer cells, MCF-10A normal breast epithelial cells, NIH 3T3/v-Src-transformed cells, and NIH 3T3 normal fibroblasts. E804 induced apoptosis in transformed MDA-MB-468 and NIH 3T3/v-Src cells, which contain high levels of constitutively active Stat3 protein, but did not induce apoptosis in normal MCF-10A and NIH 3T3 cells lacking activated Stat3 (Fig. 5D). An in situ TUNEL assay also revealed apoptosis in MDA-MB-468 cells after treatment with 1-10 μM E804 for 24 h (data not shown).
In contrast to the apoptosis results, E804 induced cell cycle arrest in all of the normal and transformed cell lines shown in Fig. 5D (Fig. 6, which is published as supporting information on the PNAS web site). These results are consistent with the inhibition of CDKs by E804 and with previous reports (18-20). Collectively, these data demonstrate that inhibition of Src-Stat3 signaling by E804 is associated with down-regulation of Mcl-1 and Survivin expression and induction of apoptosis.
Discussion
Indirubin was originally identified as an active compound in the herbal medicine, Danggui Longhui Wan, which has been widely used as a traditional Chinese treatment for chronic myelogenous leukemia. Indirubin was found to display marked antitumor properties and relatively low toxicity in animal model studies (18). Hoessel et al. (18) demonstrated that indirubins inhibit CDKs, resulting in cell cycle arrest and inhibition of tumor cell proliferation. In this study, we demonstrate that IRDs such as E804 and E728 also block Stat3 signaling and subsequently induce apoptosis in solid tumor cells. Furthermore, the indirubin E804 was found to block c-Src kinase activities both in vitro and in vivo. Whereas E804 inhibits tyrosyl phosphorylation and activation of Stat3 in vivo, it has no direct effect on Stat3 DNA-binding activity in vitro. These findings suggest that E804 blocks Stat3 signaling in breast cancer cells by inhibiting upstream c-Src kinase activity. This result is similar to the finding that PD180970, a selective Src kinase inhibitor, blocks Stat3 activation and induces apoptosis of breast cancer cells (34).
Inhibitors of CDKs, including indirubins and roscovitine, contain a flat heterocyclic ring that occupies the purine-binding pocket at the catalytic site (31). Indirubins and roscovitine are ATP-mimics, inhibiting CDK activities through hydrogen bonds in the ATP-binding pocket (18). E804 directly and potently inhibits c-Src kinase activity (IC50 = 0.43 μM) in vitro. Thus, E804 as an ATP-mimicking molecule may interact with the ATP-binding site of Src tyrosine kinase, resulting in inhibitory activity similar to the mechanism by which indirubin inhibits CDKs. Because E804 has a chiral center, it is a racemate. The two enantiomers have been prepared by separate syntheses. Both of them showed similar inhibition of phosphorylation of Src and Stat3, resulting in induction of apoptosis (S.M., F.H., S.V., K.-H.M., G.E., S.N., and R.J., unpublished results).
Stat3 can directly regulate expression of several survival genes (1, 2). The antiapoptotic genes encoding Bcl-xL, Mcl-1, and Survivin proteins are Stat3 target genes (1, 3). Mcl-1 is highly expressed in hematopoeitic cells (30), and Niu et al. (36) reported that inhibition of Src or Stat3 by a Src inhibitor (PD180970) results in down-regulation of expression of the Mcl-1 gene in melanoma cells. In addition, activation of Stat3 signaling induces Survivin gene expression with antiapoptotic activity in human breast cancer cells (T. Gritsko and R.J., unpublished data). In this study, Mcl-1 and Survivin expression is dramatically reduced in response to disruption of Stat3 signaling, and apoptosis is induced in breast cancer cells. These findings suggest that the decrease in Mcl-1 and Survivin expression contributes to E804 mediated-apoptosis.
We observed that E804 induced apoptosis in transformed cells, which harbor high levels of activated Stat3, but not normal cells lacking activated Stat3. Indirubin-3′ monoxime, a CDK inhibitor (IC50 = 0.18-0.44 μM), was known to arrest cells in the G2/M phase in various cell lines (18). We also found that 10 μM E804 arrested the cell cycle in G2/M phase in both normal and tumor cells, which is consistent with inhibition of CDK/cyclin complexes in vitro (Fig. 6). Together with our finding that E804 does not induce apoptosis in cells lacking activated Stat3, these data suggest that induction of apoptosis by E804 is mediated, at least in part, by inhibition of Src-Stat3 signaling in tumor cells.
E804 inhibited Jak1 kinase activity in vitro at 10 μM, whereas p-Jak1 levels were inhibited at 1-5 μM in vivo. Similarly, 1 μM PD180970 down-regulated p-Jak1 levels in vivo (Fig. 4D), but not in vitro (26). This result is consistent with an earlier study showing that PD180970 inhibits Jak1 phosphorylation in NIH 3T3/v-Src cells, thereby reducing Stat3 DNA-binding activity (26). Previous studies indicated that Src and Jak kinase activities cooperate to mediate constitutive activation of Stat3 (34, 35). Our findings suggest that E804 may block cooperation of Src and Jak1 involved in tyrosyl phosphorylation of Stat3.
In summary, numerous studies have established Stat3 signaling as a potential target for cancer therapy (1, 5, 6). Several small molecules have been identified as lead compounds that directly block Stat3 signaling, but they remain to be further developed in terms of potency and specificity (25, 29, 37). We have discovered that E804 blocks Stat3 signaling through inhibition of Src and induces apoptosis of human breast cancer cells. These findings provide evidence of another mechanism for indirubin's antitumor activity in addition to inhibition of CDK activity. Thus, E804 is a potent inhibitor of the Src-Stat3 signaling pathway and has potential as an antitumor therapeutic agent in epithelial malignancies.
Supplementary Material
Acknowledgments
We thank members of our laboratories for stimulating discussions. This work was supported by National Institutes of Health Grants CA55652 and CA82533.
Author contributions: S.N., R.B., J.T., D.K., J.Q.C., S.M., F.H., S.V., K.-H.M., G.E., and R.J. designed research; S.N., R.B., D.K., J.Q.C., S.M., F.H., S.V., K.-H.M., G.E., and R.J. performed research; S.N., R.B., J.T., D.K., J.Q.C., S.M., F.H., S.V., K.-H.M., G.E., and R.J. analyzed data; and S.N., R.B., G.E., and R.J. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: IRD, Indirubin derivative; STAT, signal transducer and activator of transcription; CDK, cyclin-dependent kinase; PARP, poly (ADP-ribose) polymerase; p-Stat3, phospho-Tyr-705-Stat3; p-Src, phospho-Tyr-416-Src; p-Jak1, phospho-Tyr-1022/1023-Jak1; hSIE, high-affinity sis-inducible element, m67 variant; ECL, enhanced chemiluminescence.
References
- 1.Yu, H. & Jove, R. (2004) Nat. Rev. Cancer 4, 97-105. [DOI] [PubMed] [Google Scholar]
- 2.Buettner, R., Mora, L. B. & Jove, R. (2002) Clin. Cancer Res. 8, 945-954. [PubMed] [Google Scholar]
- 3.Bromberg, J. F., Wrzeszczynska, M. H., Devgan, G., Zhao, Y., Pestell, R. G., Albanese, C. & Darnell, J. E., Jr. (1999) Cell 98, 295-303. [DOI] [PubMed] [Google Scholar]
- 4.Bromberg, J. F. & Darnell, J. E., Jr. (2000) Oncogene 19, 2468-2473. [DOI] [PubMed] [Google Scholar]
- 5.Darnell, J. E., Jr. (2002) Nat. Rev. Cancer 2, 740-749. [DOI] [PubMed] [Google Scholar]
- 6.Turkson, J. & Jove, R. (2000) Oncogene 19, 6613-6626. [DOI] [PubMed] [Google Scholar]
- 7.Luo, C. & Laaja, P. (2004) Drug Discov. Today 9, 268-275. [DOI] [PubMed] [Google Scholar]
- 8.Yu, C. L., Meyer, D. J., Campbell, G. S., Larner, A. C., Carter-Su, C., Schwartz, J. & Jove, R. (1995) Science 269, 81-83. [DOI] [PubMed] [Google Scholar]
- 9.Heinrich, P. C., Behrmann, I., Muller-Newen, G., Schaper, F. & Graeve, L. (1998) Biochem. J. 334, 297-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowman, T. L., Garcia, R., Turkson, J. & Jove, R. (2000) Oncogene 19, 2474-2488. [DOI] [PubMed] [Google Scholar]
- 11.Yeatman, T. J. (2004) Nat. Rev. Cancer 4, 470-480. [DOI] [PubMed] [Google Scholar]
- 12.Courtneidge, S. A. & Fumagalli, S. (1994) Trends Cell Biol. 4, 345-347. [DOI] [PubMed] [Google Scholar]
- 13.Moasser, M. M., Srethapakdi, M., Sachar, K. S., Kraker, A. J. & Rosen, N. (1999) Cancer Res. 59, 6145-6152. [PubMed] [Google Scholar]
- 14.Muthuswamy, S. K. & Muller, W. J. (1994) Adv. Cancer Res. 64, 111-123. [DOI] [PubMed] [Google Scholar]
- 15.Irby, R. B. & Yeatman, T. J. (2000) Oncogene 19, 5636-5642. [DOI] [PubMed] [Google Scholar]
- 16.Parsons, S. J. & Parsons, J. T. (2004) Oncogene 23, 7906-7909. [DOI] [PubMed] [Google Scholar]
- 17.Xiao, Z., Hao, Y., Liu, B. & Qian, L. (2002) Leuk. Lymphoma 43, 1763-1768. [DOI] [PubMed] [Google Scholar]
- 18.Hoessel, R., Leclerc, S., Endicott, J. A., Nobel, M. E., Lawrie, A., Tunnah, P., Leost, M., Damiens, E., Marie, D., Marko, D., et al. (1999) Nat. Cell Biol. 1, 60-67. [DOI] [PubMed] [Google Scholar]
- 19.Marko, D., Schätzle, S., Friedel, A., Genzlinger, A., Zankl, H. & Eisenbrand, G. (2001) Br. J. Cancer 84, 283-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leclerc, S., Garnier, M., Hoessel, R., Marko, D., Bibb, J. A., Snyder, G. L., Greengard, P., Biernat, J., Wu, Y. Z., Mandelkow, E. M., et al. (2001) J. Biol. Chem. 276, 251-260. [DOI] [PubMed] [Google Scholar]
- 21.Davies, T. G., Tunnah, P., Meijer, L., Marko, D., Eisenbrand, G., Endicott, J. A. & Noble, M. (2001) Structure (London) 9, 389-397. [DOI] [PubMed] [Google Scholar]
- 22.Eisenbrand, G. (2000) International Patent WO 00/61555.L.O.
- 23.Turkson, J., Bowman, T., Garcia, R., Caldenhoven, E., De Groot, R. P. & Jove, R. (1998) Mol. Cell. Biol. 18, 2545-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nam, S., Smith, D. M. & Dou, Q. P. (2001) J. Biol. Chem. 276, 13322-13330. [DOI] [PubMed] [Google Scholar]
- 25.Blaskovich, M. A., Sun, J., Cantor, A., Turkson, J., Jove, R. & Sebti, S. M. (2003) Cancer Res. 63, 1270-1279. [PubMed] [Google Scholar]
- 26.Zhang, Y., Turkson, J., Carter-Su, C., Smithgall, T., Levitzki, A., Kraker, A., Krolewski, J. J., Medveczky, P. & Jove, R. (2000) J. Biol. Chem. 275, 24935-24944. [DOI] [PubMed] [Google Scholar]
- 27.Cunnick, J. M., Dorsey, J. F., Standley, T., Turkson, J., Kraker, A. J., Fry, D. W., Jove, R. & Wu, J. (1998) J. Biol. Chem. 273, 14468-14475. [DOI] [PubMed] [Google Scholar]
- 28.Nam, S., Smith, D. M. & Dou, Q. P. (2001) Cancer Epidemiol. Biomarkers Prev. 10, 1083-1088. [PubMed] [Google Scholar]
- 29.Turkson J., Kim J. S., Zhang S., Yuan J., Huang M., Glenn M., Haura E., Sebti S., Hamilton A. D. & Jove R. (2004) Mol. Cancer Ther. 3, 261-269. [PubMed] [Google Scholar]
- 30.Epling-Burnette, P. K., Liu, J. H., Catlett-Falcone, R., Turkson J., Oshiro, M., Kothapalli, R., Li, Y., Wang, J.-M., Yang-Yen, H.-F., Karras, J., et al. (2001) J. Clin. Invest. 107, 351-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gray, N., Detivaud, L., Doerig, C., and Meijer, L. (1999) Curr. Med. Chem. 6, 859-875. [PubMed] [Google Scholar]
- 32.Meijer, L., Borgne, A., Mulner, O., Chong, J. P., Blow, J. J., Inagaki, N., Inagaki, M., Delcros, J. G. & Moulinoux, J. P. (1997) Eur. J. Biochem. 243, 527-536. [DOI] [PubMed] [Google Scholar]
- 33.Mohapatra, S., Chu, B., Wei, S., Djeu, J., Epling-Burnette, P. K., Loughran, T., Jove, R. & Pledger, W. J. (2003) Cancer Res. 63, 8523-8530. [PubMed] [Google Scholar]
- 34.Garcia, R., Bowman, T. L., Niu, G., Yu, H., Minton, S., Muro-Cacho, C. A., Cox, C. E., Falcone, R., Fairclough, R., Parsons, S., et al. (2001) Oncogene 20, 2499-2513. [DOI] [PubMed] [Google Scholar]
- 35.Campbell, G. S., Yu, C. L., Jove, R. & Carter-Su, C. (1997) J. Biol. Chem. 272, 2591-2594. [DOI] [PubMed] [Google Scholar]
- 36.Niu, G., Bowman, T., Huang, M., Shivers, S., Reintgen, D., Daud, A., Chang, A., Kraker, A., Jove, R. & Yu, H. (2002) Oncogene 21, 7001-7010. [DOI] [PubMed] [Google Scholar]
- 37.Turkson, J. (2004) Expert Opin. Ther. Targets 8, 409-422. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.