Abstract
Within its intermediate host, Toxoplasma gondii switches between two forms: a rapidly replicating tachyzoite and an encysted bradyzoite. Bradyzoites persist within the host throughout its life, hidden from antimicrobial agents and the immune system. The signals that mediate switching are poorly understood. A gene trap was employed to isolate genes whose expression is up-regulated early in the switching of bradyzoites via the negative and positive selectable marker hypoxanthine-xanthine-guanine phosphoribosyltransferase (HXGPRT). T. gondii was transfected with promoterless HXGPRT and negatively selected with 6-thioxanthine to inhibit the growth of tachyzoites expressing HXGPRT. The surviving tachyzoites were then induced for in vitro bradyzoite formation and treated with mycophenolic acid and xanthine to positively select for parasites in which the construct had integrated downstream of a bradyzoite-specific gene. Strains were checked for their ability to differentiate by using Dolichos biflorus agglutinin (a bradyzoite-specific lectin) and a monoclonal antibody against P36 (a bradyzoite-specific surface antigen). After differentiation, all gene-trapped clones had Dolichos immunofluorescence and all but one expressed P36. The sequences flanking the insertion site of this P36-negative strain were homologous to the Toxoplasma family of surface antigens, strongly suggesting that P36 is encoded by the disruptive gene. Genetic mapping and complementation of the P36-negative strain further indicated that the disrupted gene is P36. Reverse transcriptase PCR and S1 nuclease digestion were used to compare mRNA levels during the tachyzoite and bradyzoite stages. The presumptive P36 gene does not appear to regulate its mRNA levels between the two stages, indicating a posttranscriptional mechanism of regulation for early bradyzoite-specific genes.
The apicomplexan parasite Toxoplasma gondii causes congenital infections leading to blindness, mental retardation, and hydrocephaly. Recently, attention has been focused on this parasite because of the increased incidence of toxoplasmic encephalitis in patients immunocompromised by AIDS (22). Although T. gondii has a sexual cycle within the feline intestinal epithelium (13), it is usually transmitted asexually by carnivorism and scavenging. The asexual cycle has two developmental stages: a rapidly replicating form called the tachyzoite and a slow-growing, quiescent stage called the bradyzoite. The tachyzoite resides within an intracellular vacuole which is not capable of acidification or fusion with any organelle of the host’s endocytic pathway (18, 35). In response to some as-yet-unknown cellular signal, tachyzoites differentiate into encysted bradyzoites which lie essentially dormant within host tissues for months or years, apparently hidden from the immune system and antimicrobial agents. Among immunocompromised patients, it is thought that toxoplasmic encephalitis is due to recrudescence of a latent infection of bradyzoites as a result of the loss of cellular immune surveillance (22). Determining how this transition occurs is crucial for understanding disease pathology.
The physiological conditions which can trigger the tachyzoite-bradyzoite conversion in vitro have been examined (5, 37), but the molecular signals which induce this transition in vivo have not been identified. This work has been aided by the availability of antibodies against stage-specific markers for bradyzoites (P36, P34, P21, and P18) and tachyzoites (P30) (19, 37, 42). Induction of bradyzoite protein synthesis is thought to be complex since P21 is expressed later than the other bradyzoite-specific proteins during differentiation, and heterogeneous vacuoles containing parasites with tachyzoite and bradyzoite antigens were observed (38). Physiological conditions which can induce bradyzoite formation are alkalizing the culture medium to pH 8, shifting the temperature from 37 to 43°C, and treating infected macrophages with gamma interferon. Use of these conditions has enhanced the study of the bradyzoite stage, including the isolation of three genes which are bradyzoite specific, i.e., the genes encoding lactate dehydrogenase 2 (LDH2 [45]), surface antigen P18 (SAG4 [26]), and a small heat shock protein (BAG1/5 [4, 27]).
Many molecular genetic tools have been developed for T. gondii, including insertional mutagenesis (10, 11). Although insertional mutagenesis appears to be random (31), it is limited to nonessential genes in haploid organisms such as T. gondii. One of the genes that was recently cloned in T. gondii by insertional mutagenesis is that encoding hypoxanthine-xanthine-guanine phosphoribosyltransferase (HXGPRT). T. gondii had been shown to have phosphoribosyltransferase activity for hypoxanthine, xanthine, and guanine presumably within the same protein, since mutants always lack activity for all three nucleotides (29). Mutants defective in HXGPRT were obtained by negative selection with 6-thioxanthine (6-TX) and then back selected for wild-type HXGPRT by positive selection with mycophenolic acid and xanthine (MPA-X [29]). These results were confirmed in the cloning and characterization of the HXGPRT gene (11). In contrast, the Escherichia coli xanthine-guanine phosphoribosyltransferase (GPT) has been a commonly used selectable marker for animal cell transformation (24) and recombinant virus construction (7, 12, 24), but it does not use 6-TX as a substrate. Thus, the negative selections with GPT have been limited to the use of 6-thioguanine or 8-azaguanine in HGPRT-negative cell lines.
The goal of this study was to combine insertional mutagenesis with the strong selectivity of the Toxoplasma HXGPRT system to isolate genes whose expression was up-regulated early in the tachyzoite-to-bradyzoite switch. For this bradyzoite-specific gene trap, promoterless HXGPRT was inserted throughout the genome and then negative selection with 6-TX was performed to inhibit the growth of all recombinants with HXGPRT fused to tachyzoite-specific or constitutive genes. After alkalizing the culture medium to pH 8.1, which causes tachyzoites to convert to bradyzoites, positive selection with MPA-X was used to select recombinants that were expressing HXGPRT in a bradyzoite-specific manner. We obtained eight bradyzoite-specific recombinant (BSR) strains by this technique; the isolation of their genomic flanking sequences and their characterization are described below.
MATERIALS AND METHODS
Strains and plasmids.
The PLK strain was used since it grows well in human foreskin fibroblasts (HFFs) under standard Toxoplasma culture conditions (44) and easily differentiates into bradyzoites in vitro under the appropriate stimuli (6). A PLK strain lacking HXGPRT activity, PLK/HXGPRT− (11), was used in the selection experiments.
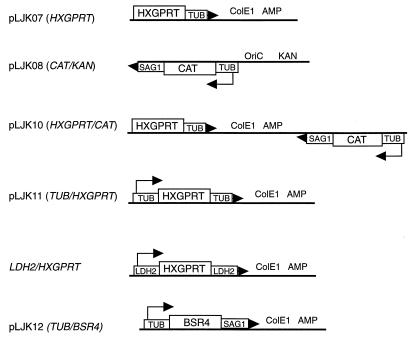
All plasmids described here are pictured in Fig. 1. Promoterless HXGPRT with the α-tubulin 3′ region [including the 3′ untranslated region (UTR) and poly(A) addition site] was constructed when a 325-bp fragment containing the 3′ region from pTAT2 (25a) was ligated onto the HXGPRT cDNA in pBluescript (11) and 164 bp of the HXGPRT promoter was removed by digestion with BamHI and ligation (pLJK07). The chloramphenicol acetyltransferase (CAT) expression plasmid (pLJK08; CAT-KAN) was constructed by ligation of the 1.5-kb XbaI-HindIII fragment (CAT gene with the α-tubulin promoter and p30 3′ UTR) from pT/230 (39) into the 4-kb XbaI- and HindIII-digested pYUB213 (30). Similarly, a 1.5-kb fragment with the CAT gene from pT/230 was ligated into pLJK07, yielding pLJK10 (HXGPRT-CAT). A 500-bp fragment including the α-tubulin promoter from pT/230 was inserted upstream of HXGPRT in pLJK07, yielding pLJK11 (TUB-HXGPRT). The LDH2-HXGPRT expression construct included a 1.0-kb fragment from the LDH2 upstream region (including the promoter) and a 1.7-kb fragment from the LHD2 downstream region (including the 3′UTR), flanking a 708-bp fragment of the coding region of the HXGPRT cDNA in pBluescript SK. To express BSR4 at the tachyzoite stage, a 1.2-kb EcoRI- and PacI-digested PCR fragment containing the BSR4 gene was cloned into EcoRI- and PacI-digested pTUB-βgal (33), yielding pLJK12 (TUB-BSR4).
FIG. 1.
Construction of expression vectors was as described in Materials and Methods. Arrowheads at the end of the 3′ UTR as well as bent arrows within the promoters indicate the direction of transcription.
Transfection and selection.
Transfections and selections for the gene trap clones were performed as follows. One microgram of pLJK07 (HXGPRT), linearized at the SacI site 47 bp upstream of the HXGPRT start codon, was cotransfected with 20 μg of NotI-linearized pLJK08 (CAT-KAN) or 1 μg of pLJK10 (HXGPRT-CAT), linearized at the XhoI site 41 bp upstream of the HXGPRT start codon. Stable integration into the genome was enhanced by restriction enzyme-mediated integration (REMI) (3, 32) using either NotI or DpnII (100 U per transfection). Transformants were selected with 20 μM chloramphenicol and between 40 and 400 μg of 6-TX (Sigma) per ml for 4 to 21 days under tachyzoite conditions (Dulbecco’s modified Eagle medium plus 10% NuSerum, pH 7.2; 37°C in 5% CO2). A total of 106 selected tachyzoites were allowed to invade a new HFF monolayer for 4 h and then changed to bradyzoite-inducing conditions (RPMI with 5% fetal bovine serum and 50 mM HEPES, pH 8.1; 37°C in air). After 18 h, either 100 or 200 μg of mycophenolic acid (Sigma) and 50 μg of xanthine (Sigma) were added for 3 days. Monolayers with induced bradyzoites were either trypsinized, scraped, and syringed with a 27-gauge needle or scraped, syringed, digested in 170 mM NaCl–pepsin (0.1 mg/ml)–60 mM HCl for 1 min at 37°C, and then neutralized with 94 mM Na2CO3. These bradyzoites were allowed to grow in fresh HFF monolayers for 4 to 7 days. The entire selection procedure was repeated once before the selected parasites were cloned to single isolates by limiting dilution in 96-well plates.
Viability assay.
Each of the BSR strains was initially characterized by measuring [3H]uracil incorporation with and without 6-TX under tachyzoite conditions or MPA-X under bradyzoite conditions as follows. A total of 105 tachyzoites were added to each well of a 24-well plate of HFFs and allowed to invade for 4 h. For tachyzoites, the original medium was replaced with fresh medium with and without 100 μg of 6-TX per ml, and after an additional 40 h of growth, 1 μCi of [3H]uracil was added per well for an 8-h incubation. For experiments under bradyzoite conditions, the parasites were under inducing conditions for 18 h and then the medium was replaced with fresh inducing medium with or without 100 μg of MPA and 50 μg of xanthine per ml. After an additional 40 hr of growth, 10 μCi of [3H]uracil was added per well for a 14-h incubation. The protein in each well was trichloroacetic acid precipitated, washed, and counted as previously described (31). Each experiment had duplicate wells, and growth assays were repeated at least four times.
Immunofluorescence microscopy.
Toxoplasma cells were prepared for indirect immunofluorescence by growing the strains in HFFs on glass coverslips in 24-well plates for 36 h as tachyzoites or for 3 days under bradyzoite-inducing conditions. Cells were fixed in 3% formaldehyde for 20 min and then permeabilized with 0.2% Triton X-100 in phosphate-buffered saline for 20 min. Primary antibody Incubations with primary antibodies were for 1 h with a monoclonal antibody against P36 and biotinylated Dolichos biflorus agglutinin (Vector); secondary antibody incubations were 30 min with fluorescein isothiocyanate (FITC)-conjugated Fab (Cappel) and streptavidin-Texas red conjugate (Gibco BRL). Coverslips were embedded with Vectashield (Vector), and epifluorescence was detected with an Olympus BX60 microscope.
Recovery of flanking DNA sequences.
Genomic DNA was prepared from each BSR strain in Tris-EDTA-LiCl-Triton lysis (TELT) buffer (23). This DNA was digested as follows: (i) with EcoRV, StuI, PmlI, and SmaI (blunt-end cutters that do not cut within the plasmid construct); (ii) with HindIII, NcoI, or SacI (rescue of the 5′ flanking sequence); or (iii) with KpnI (rescue of the 3′ flanking sequence). The resulting fragments were then ligated, transformed into E. coli, and selected for ampicillin resistance.
Measurement of mRNA levels.
Total RNA was prepared from tachyzoites by using Ultraspec according to the manufacturer’s instructions (BIOTECX Laboratories Inc.). Total RNA was prepared from bradyzoites under switching conditions for 3 days; then cells were removed from the flask by scraping, centrifuged at 1,200 × g, resuspended in phosphate-buffered saline, and pulsed twice for 1 s each in a Waring blender to disrupt the HFFs. Bradyzoites were isolated by pepsin digestion as described above, and bradyzoite total RNA was prepared by using Ultraspec.
For reverse transcriptase PCR (RT-PCR), total RNA was treated with amplification-grade DNase I according to the manufacturer’s instructions (Gibco BRL). RNA (1 μg) was reverse transcribed with 150 ng of oligo(dT)18 primer and 50 U of SuperScript II RT (Gibco BRL) at 42°C for 1 h in a solution containing 25 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, and 500 μM each deoxynucleoside triphosphate. The reaction was heat inactivated at 70°C for 15 min. The mRNA was removed by incubation at 37°C for 20 min with 10 U of RNase H and purified on GlassMAX DNA isolation spin cartridges (Gibco BRL).
Tenfold serial dilutions of the cDNA products were amplified with 10 pmol of each primer and 2.5 U of AmpliTaq DNA polymerase (Perkin-Elmer) in 100-μl reaction volumes containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.001% gelatin, and 250 μM each deoxynucleoside triphosphate. Thermal cycling conditions were 94°C for 30 s, 1 min at 50 to 60°C (the precise annealing temperature depended on the primer pair), and 1 min at 72°C for 27 cycles (linear range, 12 to 30 cycles). PCR products were electrophoresed on 6 to 15% polyacylamide gels (the polyacylamide concentration depended on the PCR product size), stained for 20 min with SYBR Green I (Molecular Probes), visualized with the Storm 860 PhosphorImager system (Molecular Dynamics), and quantified with ImageQuaNT data analysis software (Molecular Dynamics). Four different negative controls were used: (i) no RT was added in the reverse transcription reaction to detect possible DNA contamination in the RNA sample; (ii) RNA was digested with RNase A before the reverse transcription reaction to ensure that the PCR product originated from the RNA; (iii) cDNA was omitted from the amplification reaction; and (iv) one primer was omitted to ensure that the reaction produced no product. At least two different sets of primers specific for the genomic sequences flanking the insertion site for each of the BSR strains were used. Primers for the α-tubulin gene were 5′-CCCGACCTACACCAACCT-3′ and 5′-CCCTCCTCTTCACCTTCA-3′, for a fragment with a size of 680 bp. For LDH2, primers 5′-GCTCGGCATTCGTACTTCA-3′ and 5′-CTCTACGATCTCTGCCAGT-3′ produced a product of 697 bp. A 72-bp product of the BSR2 gene, with primers 5′-CGATCATTTAGCTCCGAA-3′ and 5′-CGAGAGGACCGAGGAAGA-3′, and a 127 bp product of the BSR3 gene with primers 5′-ACCTTGAGACCTTCCAACA-3′ and 5′-GCAATTATCTTATAGTCCA-3′, were produced. For the BSR4 gene, PCR with primers 5′-CGAGGCGGACCAGCAGTT-3′ and 5′-CCGCATGAACTTACCACA-3′ produced a fragment with a size of 369 bp. The product from BSR5 with primers 5′-GTCTAATTTGATGAAGGA-3′ and 5′-CCAGCGAGATAGTGTTGT-3′ was 504 bp. For BSR6, primers 5′-GTCTCACCGAACGGCTTCA-3′ and 5′-GGGCTCTGTGTTGATTGT-3′ produced a 253-bp fragment. For BSR7, primers 5′-GGCTAGCGTGATCTTATT-3′ and 5′-CACACACAGATCGTCGCA-3′ produced a 429 bp fragment, while 5′-CGGACTCTGGGATACACT-3′ and 5′-CAGGCTCCCACCATGTCT-3′ produced a 102-bp product for BSR8.
For the S1 nuclease digestions, 10 μg of DNase I-treated total RNA from either tachyzoites or bradyzoites was hybridized to completion with a 100-fold excess of the appropriate 32P-end-labeled oligonucleotide and treated with S1 nuclease as described elsewhere (8). The oligonucleotides all contain 6 residues at their 3′ ends that are not complementary to the mRNA, thereby permitting an easy distinction between bands attributable to appropriate RNA-DNA hybrids and undigested probe: sequences used were 5′-GGTAAGTGCCGGTGCGAACCTCATCGACGACGGTGGGCTCCAAATCCAAGAAGAGTGTAG-3′ for the α-tubulin gene and 5′-GTCCGATGCAGGAGTCCTTCAAGAAGATTATCAGCAACAGCCTGTCCGCTAGAGTTGTTG-3′ for BSR4. Reaction mixtures were slightly overdigested to ensure that the signal in the digested lanes was from the protected RNA-DNA hybrids (i.e., there is no detectable undigested probe in these lanes).
Nucleotide sequence accession numbers.
The flanking genomic DNA sequences for BSR2 to BSR8 were submitted to GenBank with accession numbers AF015289, AF015288, AF015290, AF015291, AF015292, AF015293, and AF015294, respectively.
RESULTS
Selections for BSR.
Eight BSR strains (BSR1 to BSR8) were obtained by using the selection strategy detailed above. Briefly, the first three BSR strains were created by cotransfection of a 20-fold excess of the promoterless HXGPRT construct compared to the CAT-expressing construct. (It should be noted that since T. gondii has a low transfection efficiency, the gene trap protocol did not successfully produce any BSR strains without a prior CAT selection.) Differential rescue of the DNA flanking the HXGPRT gene was achieved because the promoterless HXGPRT construct contains the ColE1 origin of replication and an ampicillin resistance gene whereas the CAT construct contains the oriC origin and a kanamycin resistance gene. However, linear DNA has a propensity to integrate into the genome of T. gondii in tandem arrays, especially with REMI (3); this made rescue of the genomic DNA flanking the insertion site difficult. Several unsuccessful attempts were made to isolate the flanking sequences of BSR1, and since it appeared to have significant tachyzoite expression of HXGPRT (Fig. 2) (see below), further investigations were not done. For BSR2 and BSR3, the CAT construct integrated downstream of the HXGPRT construct in a tandem array, and therefore we were only able to rescue the 5′-end insertion site sequences.
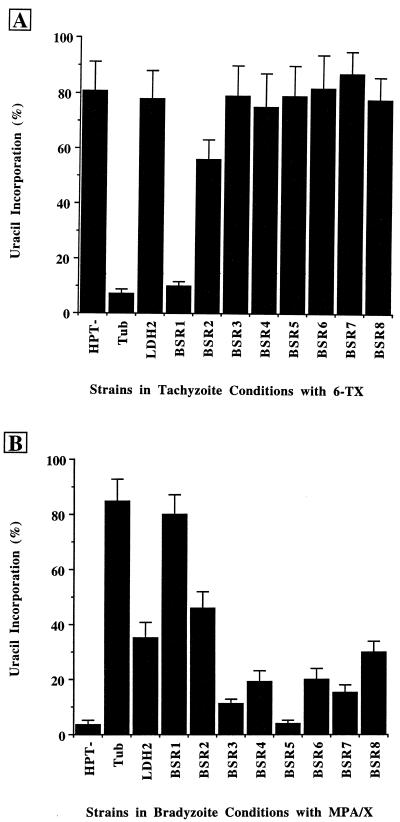
FIG. 2.
Measurement of BSR viability with 6-TX under tachyzoite conditions and with MPA-X under bradyzoite conditions by [3H]uracil incorporation. PLK/HXGPRT− (HPT−), TUB/HXGPRT (Tub), LDH2/HXGPRT (LDH2), and all of the BSR strains were labeled with [3H]uracil with and without 6-TX under tachyzoite conditions (A) and with and without MPA-X under bradyzoite conditions (B). TUB/HXGPRT and LDH2/HXGPRT are cloned strains from stable transfections. Results are expressed as a percentage: total amount of incorporation with drug divided by the total amount of incorporation without drug + standard deviation.
To reduce tandem arrays, we changed to a single-plasmid transfection system. The CAT gene cassette was spliced into the promoterless HXGPRT plasmid such that when the construct was linearized, HXGPRT and CAT were in a tail-to-tail arrangement with the E. coli ori and ampicillin resistance gene in between. With this HXGPRT-CAT construct, we were able to use 20-fold less DNA per transfection. This virtually eliminated tandem arrays (as seen by Southern blot analyses [data not shown]), and we were able to rescue both the 5′ and 3′ ends of all of the strains (BSR4 to BSR8) made this way.
Since BSR1 had significant tachyzoite expression of HXGPRT (Fig. 2), we increased the stringency of the 6-TX selection for the experiment that yielded the second set of mutants (BSR4 to BSR8). 6-TX is a static inhibitor, not a cidal inhibitor, and the “leaky” BSR1 was selected with only 40 μg of 6-TX per ml for 4 days. Thus, the later transfections were split after an initial pass and grown as tachyzoites for a further 15 to 21 days (five to seven passages) in either 40 or 200 μg of 6-TX per ml. The result was five additional BSR clones, with four from the selection with 40-μg/ml 6-TX (BSR4, BSR5, BSR6, and BSR8) and two from the selection with 200-μg/ml 6-TX (BSR5 and BSR7; note that BSR5 was selected at both 6-TX concentrations).
Characterization of BSR strains.
To begin to characterize the BSR strains, we used [3H]uracil incorporation as a measure of their viability with and without 6-TX under tachyzoite conditions and MPA-X under bradyzoite-inducing conditions (Fig. 2). We attempted to measure Toxoplasma HXGPRT activity directly via [3H]xanthine incorporation; however, [3H]xanthine incorporation is 60-fold lower in bradyzoites (MPA-X selection is also significantly less effective in bradyzoites than in tachyzoites), and thus the weak bradyzoite-specific genes were barely above background (data not shown). When looking at [3H]uracil incorporation with and without 6-TX under tachyzoite conditions, we saw that like the parental PLK/HXGPRT− strain, the LDH2/HXGPRT and BSR3-8 strains were inhibited by 6-TX by approximately 20%; thus, they probably do not express significant levels of HXGPRT at the tachyzoite stage. Conversely, the TUB/HXGPRT and BSR1 strains must express HXGPRT in tachyzoites because they are strongly inhibited by 6-TX (7 and 10% uracil incorporation with 6-TX compared to without 6-TX, respectively [Fig. 2A]). Similarly, BSR2 seems to have a minimal amount of tachyzoite HXGPRT expression, since its uracil incorporation was reduced to 56% with 6-TX compared to the PLK/HXGPRT− strain at 80%. Under bradyzoite conditions with and without MPA-X, the parental PLK/HXGPRT− strain was strongly inhibited (4% uracil incorporation with MPA-X compared to without [Fig. 2B]). In contrast, the TUB/HXGPRT strain was only slightly inhibited by MPA-X (85% with compared to without). LDH2/HXGPRT and all of the BSR strains were less inhibited by MPA-X than was PLK/HXGPRT− (range, 11% for BSR3 to 80% for BSR1), except for BSR5, which clearly does not confer MPA-X resistance at the bradyzoite stage (4% incorporation with MPA/X [Fig. 2B]). For the selection strategy, the cultures were passed in either 40 or 200 μg of 6-TX per ml for tachyzoites and in either 100 or 200 μg of MPA per ml for bradyzoites. Based on Southern analyses, the BSR5 strain was selected under all four of these sets of conditions; thus, we did not expect its uracil incorporation to be severely affected by MPA-X at the bradyzoite stage. Perhaps the knockout of the BSR5 gene by the insertion construct enabled the BSR5 strain to survive the selection (see below).
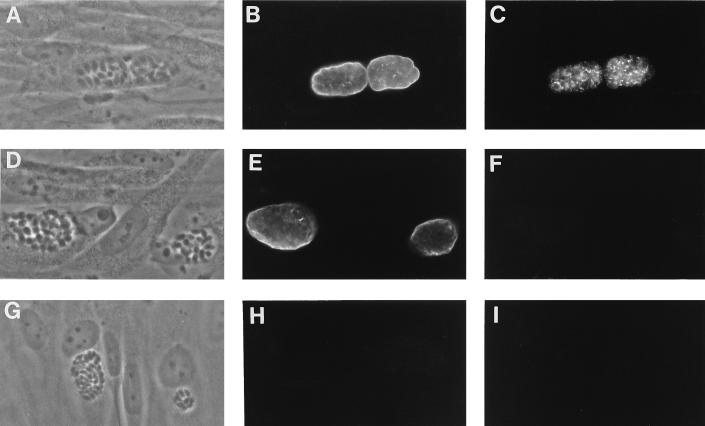
Since the insertion of the promoterless HXGPRT construct would be apt to functionally knock out the gene into which it had inserted, some of which might be required for differentiation, we tested the BSR strains’ abilities to differentiate into bradyzoites. We placed all of the BSR strains under bradyzoite switching conditions for 3 days and then subjected them to pepsin digestion, which is lethal for tachyzoites; all of the strains were resistant to pepsin (data not shown). In addition, after 36 h under tachyzoite conditions or 3 days under switching conditions, BSR strains were fixed, permeabilized, and incubated with the biotinylated bradyzoite-specific lectin D. biflorus agglutinin (6, 26a) and a monoclonal antibody against the known bradyzoite-specific surface antigen P36 (42). Immunofluorescence with a Texas red-streptavidin conjugate showed that the cyst wall staining of all of the BSR strains with the D. biflorus agglutinin was characteristic of 3-day in vitro bradyzoites (Fig. 3B and E). Surprisingly, BSR4 had a negative P36 signal, while all of the other BSR strains had the expected strong P36 immunofluorescence (Fig. 3; compare panels C and F). Under tachyzoite conditions, all strains lacked an immunofluorescence signal for both D. biflorus and P36 (Fig. 3 panels G to I). Western blots confirmed the absence of P36 protein in BSR4 bradyzoites (data not shown). These results strongly suggest that BSR4 encodes the P36 gene.
FIG. 3.
Immunofluorescence microscopy of the wild type (WT) and the BSR4 mutant strain. (A, B, and C) WT in vitro bradyzoite cysts; (D, E, and F) the BSR4 in vitro bradyzoite cysts; (G, H, and I) WT grown under standard tachyzoite conditions. Strains were grown for either 3 days under bradyzoite-inducing conditions or 36 h under tachyzoite conditions and then incubated with biotinylated D. biflorus agglutinin and a monoclonal antibody against P36. This was followed by incubations with streptavidin-Texas red and FITC-conjugated goat anti-mouse antiserum and photography with the Olympus BX60 microscope system using either phase-contrast (A, D, and G), the Texas red filter (specific for D. biflorus agglutinin) B, E, and H), or the FITC filter (specific for the P36 monoclonal antibody) (C, F, and I).
Analysis of the genomic DNA sequences flanking the insertion site.
The genomic DNA adjacent to the insertion sites of the various BSRs was retrieved by restriction enzyme digestion, ligation, and transformation of E. coli. The Toxoplasma genomic DNA fragments were sequenced with antisense primers from HXGPRT and sense primers from the 3′ end of the vector. When comparing the genomic DNA-vector junction, we saw that 0 to 13 bp of vector sequence had been “chewed in”, which is within the range previously seen (31). Upon further examination of the genomic DNA-vector junction, we saw that of the BSR strains whose transfection was enhanced with NotI REMI, three of four genomic sequences had a partial NotI site. No evidence of a DpnII site was seen in the rescued genomic sequences (although DpnII has only a 4-bp recognition site). BSR4 was created with NotI REMI; an RT-PCR product for BSR4 that spanned the insertion site was sequenced, revealing an intact NotI site. It was reported previously that in T. gondii, REMI does not leave an intact NotI site at the genomic DNA-vector junction (3). The results presented here provide an explanation for that observation: the NotI site in the genomic DNA is indeed targeted by the enzyme, but insertion of a chewed-back vector results in a failure to recreate the NotI site. This is unlike the situation in yeast, in which insertion of an intact plasmid results in recreated sites flanking the insert (32).
The genomic DNA sequences of the BSR genes were compared to the entire nucleic acid and protein databases by using the BLASTN and BLASTX homology search programs (1, 15) to identify homologous sequences. A 161-bp portion of sequence at the 3′ end of BSR5 had exact identity with an expressed sequence tag (EST) of unknown function from the Toxoplasma in vitro bradyzoite cDNA library (gb:AA012249; TgESTzz18d06.r1). However, the AA012249 EST predicts that the orientation of the BSR5 gene is opposite that predicted by the HXGPRT insertion, assuming the latter to be transcribed and active as some sort of gene fusion. A 3′ rapid amplification of cDNA ends with BSR5 primers supported the 5′-to-3′ orientation predicted by the EST; thus, insertion of the gene trap construct into the BSR5 gene probably disrupted its function. Perhaps this loss caused the BSR5 strain to survive the selection, since, as already mentioned above, no direct evidence for HXGPRT expression could be obtained for this strain.
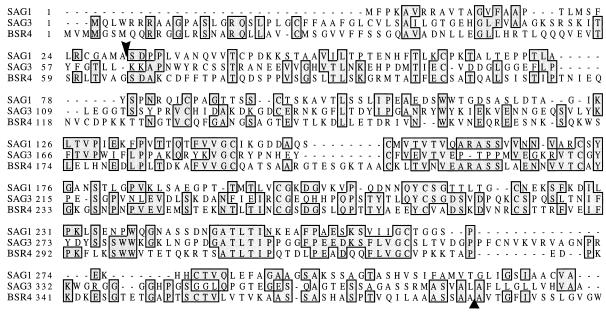
In a BLASTX search, the predicted BSR4 protein product showed the highest degree of homology to Toxoplasma surface antigens SAG3 (score, 200; probability, 3 × 10−18), SAG5 (score, 116; probability, 6.6 × 10−11), and SAG1 (score, 74; probability, 8 × 10−7). The SAG3 and SAG1 protein sequences were aligned with that of the BSR4 protein product (Fig. 4). Based on its predicted size, the presence of a likely signal peptide and a glycosylphosphatidylinositol (GPI) addition signal, and homology to other known surface antigens, the sequence data strongly support the conclusion reached above that BSR4 encodes the P36 surface antigen. None of the other BSR genes showed significant similarity to sequences in the databases.
FIG. 4.
Sequence alignment of SAG1, SAG3, and BSR4. Areas of sequence identity are boxed and shaded. Protein sequences for SAG1, SAG3, and BSR4 were compared by using the ClustalW multiple sequence alignment program (41). The arrowhead indicates the known signal peptide cleavage site for SAG1. The triangle indicates the site of HXGPRT insertion into BSR4.
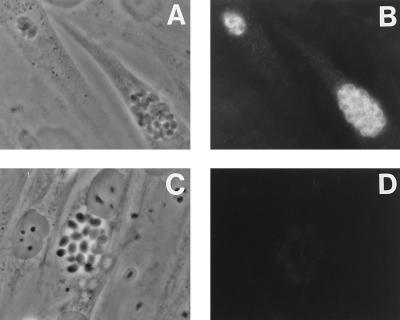
To test the reactivity of the BSR4 protein with the monoclonal antibody against P36, the BSR4 gene was constitutively expressed from the α-tubulin promoter (Fig. 5). The BSR4 strain transiently transfected with the TUB-BSR4 expression construct showed a strong P36 immunofluorescence signal (Fig. 5A and B), whereas the mock-transfected strain had a negative P36 signal (Fig. 5C and D). Thus, the BSR4 gene, under the control of the α-tubulin promoter, complements the P36-negative phenotype in the BSR4 strain and gives a strong P36 signal in the normally P36-negative tachyzoite stage, further evidence that P36 is encoded by the BSR4 gene.
FIG. 5.
Complementation of the BSR4 strain with a TUB-BSR4 construct. The BSR4 strain was transiently transfected with either pLJK12 (TUB/BSR4) (A and B) or water (C and D), grown in tachyzoite conditions for 2 days, and used in immunofluorescence experiments with the monoclonal antibody against P36.
To confirm the identity of the BSR4 gene, genetic mapping was performed. We have previously mapped polymorphic genes in the progeny of a cross of strains ME49 and CEP (34). These two strains have a restriction fragment length polymorphism for BSR4 and are dimorphic for reactivity with the anti-P36 monoclonal antibody (ME49 does and the CEP strain does not react with the monoclonal antibody [6]). The results show that the BSR4 sequence polymorphism cosegregates with the reactivity of the P36 monoclonal antibody (17a). These genetic data, together with the immunofluorescence (Fig. 3 and 5) and sequence (Fig. 4) data, further strengthen the conclusion that P36 is encoded by the BSR4 gene.
Analysis of mRNA levels of the BSR genes.
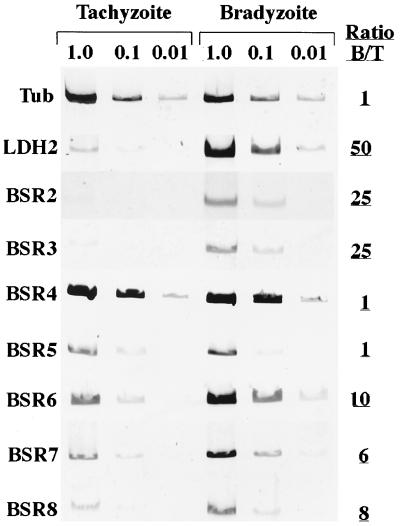
Semiquantitative RT-PCR was used to compare the mRNA levels of the BSR genes in tachyzoites and bradyzoites. Even though equal amounts of tachyzoite and bradyzoite total RNA were used in the RT reaction, α-tubulin primers were used as a control to ensure that equal amounts of cDNA from each stage were being compared (Fig. 6). We used the monomeric fluorescent dye SYBR green I as our nucleic acid gel stain because of the ease of detection with the PhosphorImager system, and the fluorescence intensity versus DNA concentration had been shown to be linear over more than 3 orders of magnitude (2, 36). When we examined two previously described bradyzoite-specific genes, LDH2 (Fig. 6) and SAG4 (data not shown), we saw, as expected, a greater than 50-fold increase in message abundance in bradyzoites compared to tachyzoites.
FIG. 6.
RT-PCR amplification of tachyzoite and bradyzoite mRNA. The dilutions of cDNA used in the α-tubulin (Tub) reactions were 1:1,000, 1:10,000, and 1:100,000. All of the other primer sets used cDNA diluted to 1:1, 1:10, and 1:100 for their reactions. The control that omitted the cDNA from the amplification reaction for each primer set (all of which gave no detectable signal) was used as the background for subtraction from the values obtained for the other bands. Ratios listed are the values within the linear range for the bradyzoite stage (B) divided by the linear-range values obtained for the tachyzoite stage (T).
Figure 5 shows the wide range of relative abundances for the BSR transcripts. BSR2 was approximately 20-fold more abundant in the bradyzoite stage (14- to 25-fold depending on the experiment and the primer set). Similarly, the mRNA for BSR3 was consistently increased at least 25-fold in the bradyzoite stage. Surprisingly, there was little or no change in abundance of the BSR4 (P36) gene in the bradyzoite stage (0- to 3-fold depending on the experiment). Even though the BSR4 transcript appears to be constitutively expressed, the evidence clearly indicates that its protein product is the bradyzoite-specific surface antigen P36 (38, 42) (Fig. 3C, F, and I). We also sequenced an RT-PCR fragment produced from BSR4-specific primers and tachyzoite cDNA; we saw that it contained a single sequence that corresponded exactly to the BSR4 gene (data not shown).
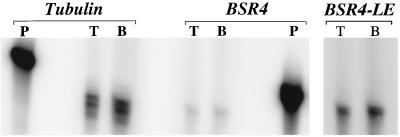
To confirm that the BSR4 transcript is equally abundant in tachyzoites and bradyzoites, S1 nuclease digestions were performed on tachyzoite and bradyzoite RNA hybridized to primers specific for either the α-tubulin or the BSR4 gene. The S1 nuclease experiments confirmed the results of the semiquantitative RT-PCR, showing that the abundances of both α-tubulin and BSR4 mRNAs are not significantly different in the tachyzoite and bradyzoite stages (Fig. 7) and that the α-tubulin mRNA appears to be massively more abundant than the BSR4 mRNA. Note that the RT-PCR and S1 nuclease primers were tested from both the 5′ and 3′ ends of the BSR4 gene, where it is most divergent from the other SAG family members (Fig. 4).
FIG. 7.
S1 nuclease digestion of tachyzoite and bradyzoite RNA protected with oligonucleotides against either the α-tubulin gene or BSR4. The levels of tachyzoite (T) and bradyzoite (B) RNAs for the α-tubulin gene and BSR4 were compared. Undigested probes (P) were diluted 1:1,000 and included to show the completeness of the reaction. The right panel is a long exposure of BSR4.
There also was no difference in abundances of the BSR5 transcript between the tachyzoite and bradyzoite stages (Fig. 6), but this was not so surprising, since, as already discussed, the data argue against bradyzoite-specific expression of the BSR5-HXGPRT fusion as an explanation for its selection. The last three BSR genes showed minimal increases in bradyzoite stage expression versus tachyzoite levels; BSR6 was increased 5- to 10-fold, BSR7 was increased 3- to 6-fold, and BSR8 was increased 5- to 9-fold (Fig. 6).
DISCUSSION
The essence of the gene trap protocol is selection for the activity of a promoterless reporter gene after it is randomly integrated into an organism’s genome (16). For this study, promoterless HXGPRT was nonhomologously inserted into the Toxoplasma genome; negative selection with 6-TX was performed to inhibit the growth of all recombinants fused to tachyzoite-specific or constitutive genes, and then positive selection with MPA-X was used to select recombinants that were expressing HXGPRT in a bradyzoite-specific manner. This approach could isolate genes which are essential for tachyzoite-to-bradyzoite conversion, because a switch gene is most likely not essential for tachyzoite growth and the trypsin treatment (as opposed to pepsin) does not kill nonswitched tachyzoites.
Unlike the E. coli GPT marker, Toxoplasma HXGPRT uses 6-TX as a substrate, and this inhibition works in a dose- and time-dependent fashion. It should be possible to use Toxoplasma HXGPRT as a marker in other eukaryotes (e.g., vertebrate cell lines, yeast, etc.) that have only hypoxanthine and guanine phosphoribosyltransferase activity, since 6-TX is not a substrate for HGPRT. Even though strains BSR1 and, to a lesser extent, BSR2 express HXGPRT under tachyzoite conditions, they presumably survived the 6-TX selection because it was present at only 40 μg/ml for 4 days. Consistent with this, BSR3 was selected with the more stringent concentration of 6-TX, 400 μg/ml, for 4 days, and it showed no evidence of HXGPRT expression at the tachyzoite stage. Along the same line, we did not isolate any obviously strong bradyzoite promoters in the later selections, perhaps because the 6-TX selection was lengthened from 4 days to 15 or 21 days and, thus, any leakiness would be lethal (e.g., BSR1 and BSR2 have the most MPA resistance at the bradyzoite stage and were selected for only 4 days at 40 μg/ml [Fig. 2]). Most likely, no bradyzoite-specific gene is 100% repressed under tachyzoite tissue culture conditions. Thus, during a long 6-TX selection, the growth of the strong bradyzoite expressers may be partially inhibited, and they may be removed from the tachyzoite population. This hypothesis is also supported by the small amount of transcripts of the LDH2 and SAG4 genes detected in tissue culture tachyzoites; in the original reports, no LDH2 or SAG4 transcripts were seen in tachyzoites grown intraperitoneally in mice (26, 45). However, this phenomenon cannot explain the BSR4 mRNA expression in tachyzoites, since similar amounts of BSR4 transcript were detected in tachyzoites grown in tissue culture or in the mouse intraperitoneal cavity, as well as in tachyzoites from the RH strain of T. gondii (data not shown), which is known to be inefficient in switching to bradyzoites in vitro (31, 37). Consistent with these results, a BLASTN search showed that the only exact match to the BSR4 gene sequence was an EST from the RH tachyzoite cDNA library (N82097; TgESTzy41g02.r1).
Normally, Toxoplasma SAG proteins are transported to the cell surface through the secretory pathway, and there a GPI anchor is added to their carboxyl termini (25, 42). Hence, a priori, it would be surprising for a BSR4-HXGPRT fusion protein to allow survival in the selection used here; i.e., it would be expected that such a fusion would be either secreted or trapped within the secretory pathway, thereby preventing productive phosphoribosylation of xanthine to overcome MPA inhibition of IMP dehydrogenase. The GPI anchor addition signal has been shown to be a stretch of about 20 hydrophobic residues at the extreme carboxyl-terminal end of a primary translation product (9). The promoterless HXGPRT gene was fused with BSR4 in the middle of the presumptive GPI anchor signal; this may have caused the hydrophobic residues to function as a transmembrane (stop transfer) domain instead of a GPI anchor signal. This transmembrane domain would place the BSR4 portion of the fusion within the lumen of the endoplasmic reticulum but locate the HXGPRT protein in the cytoplasm, where it would be available to metabolize xanthine to XMP. We know that the HXGPRT of this BSR4-HXGPRT fusion protein is functional, since the transient transfection of the plasmid rescued from BSR4 does confer MPA resistance to bradyzoites (data not shown). The BSR4 portion of this fusion protein is no longer recognized by the monoclonal antibody produced against it, probably because the P36 portion is misfolded or, alternatively, the epitope for the monoclonal antibody includes the GPI anchor.
Using this gene trap scheme, not only have we isolated several bradyzoite-specific genes, apparently including the P36 gene, but we have also uncovered a possible posttranscriptional mechanism for developmental regulation in T. gondii. This protocol was designed to capture genes whose expression is upregulated early in the switch by addition of MPA-X after only 18 h of bradyzoite induction, compared with the ≥3 days needed under in vitro switching conditions for cysts to fully develop (even then they may not be fully mature, as indicated by the low level of P21 expression [37, 38]). Perhaps the genes that are expressed early in the bradyzoite conversion process are already transcribed in tachyzoites, so they can be expressed faster. Although posttranscriptional regulation has not yet been seen for a bradyzoite-specific protein, the tachyzoite-specific form of lactate dehydrogenase (LDH1) has been shown to be posttranscriptionally controlled (46).
The signal for posttranscriptional control of BSR4 is most likely encoded in the 5′ UTR and/or the coding region, since HXGPRT (complete with the α-tubulin 3′ UTR and polyadenylation site) was fused to the carboxyl terminus of the BSR4 gene and expression of this fusion protein was apparently still bradyzoite specific. Kinetoplastid protozoan parasites, like T. brucei, use an unknown posttranscriptional mechanism to regulate many of their stage-specific proteins (28). Translation is regulated by the 5′ UTR sequences in eukaryotes as diverse as humans, yeasts, and plants (14, 20, 40). The classic example of translational control by the 5′ UTR is the iron response element (IRE)/iron regulatory protein (IRP) paradigm. The IRP is an RNA-binding protein that mediates translational regulation of the mammalian ferritin protein by binding a stem-loop structure (i.e., the IRE) in the 5′ UTR of the ferritin gene (43). Similarly, the 5′ UTR of HSP70 is both necessary and sufficient for translational initiation at higher temperatures (21). It will be exciting to study the 5′ UTR of the Toxoplasma BSR4 gene and test its ability to regulate translation.
The positive and negative selection capabilities of the Toxoplasma HXGPRT gene have allowed us to develop a stage-specific gene trap, and while this work was in progress, we learned of similar success with this approach by Roos and colleagues (30a). Typically, vector traps have used as the reporter gene β-galactosidase, which we found to be too labor-intensive for isolating bradyzoite-specific genes of T. gondii (20a). To look for glucocorticoid-regulated genes in a mouse pituitary tumor cell line (17), a fusion gene coding for hygromycin phosphotransferase and herpes simplex thymidine kinase was created, and its expression conferred hygromycin resistance and gangciclovir sensitivity. They were able to isolate genes that were both up-regulated and down-regulated by glucocorticoids, highlighting the usefulness of positive and negative selectable markers. The HXGPRT system adds to the repertoire of such tools and should facilitate studies of developmentally regulated gene expression in T. gondii and other systems.
ACKNOWLEDGMENTS
We thank D. Roos for providing the HXGPRT gene and PLK/HXGPRT− strain (via the NIH AIDS repository); A. Hehl, E. Ortega, and S. Bonnefoy for provision of unpublished data; A. Hehl, R. Striker, and K. Wilson for critical readings of the manuscript; and B. Cormack, A. Hehl, I. Manger, K. Wilson, and other members of the Boothroyd lab for helpful suggestions.
This work was supported by the National Institutes of Health (grants A121423 and A141014). L.J.K. is supported by the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation Fellowship (grant DRG-1341).
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Becker A, Reith A, Napiwotzki J, Kadenbach B. A quantitative method of determining initial amounts of DNA by polymerase chain reaction cycle titration using digital imaging and a novel DNA stain. Anal Biochem. 1996;237:204–207. doi: 10.1006/abio.1996.0230. [DOI] [PubMed] [Google Scholar]
- 3.Black M, Seeber F, Soldati D, Kim K, Boothroyd J C. Restriction enzyme-mediated integration elevates transformation frequency and enables co-transfection of Toxoplasma gondii. Mol Biochem Parasitol. 1995;74:55–63. doi: 10.1016/0166-6851(95)02483-2. [DOI] [PubMed] [Google Scholar]
- 4.Bohne W, Gross U, Ferguson D J P, Heesemann J. Cloning and characterization of a bradyzoite-specifically expressed gene (hsp30/bag1) of Toxoplasma gondii, related to genes encoding small heat-shock proteins of plants. Mol Microbiol. 1995;16:1221–1230. doi: 10.1111/j.1365-2958.1995.tb02344.x. [DOI] [PubMed] [Google Scholar]
- 5.Bohne W, Heesemann J, Gross U. Induction of bradyzoite-specific Toxoplasma gondii antigens in gamma interferon-treated mouse macrophages. Infect Immun. 1993;61:1141–1145. doi: 10.1128/iai.61.3.1141-1145.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boothroyd J C, Black M, Bonnefoy S, Hehl A, Knoll L J, Manger I D, Ortega-Barria E, Tomavo S. Molecular genetic analysis of development in Toxoplasma gondii. Philos Trans R Soc Lond B. 1997;352:1347–1352. doi: 10.1098/rstb.1997.0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyle D B, Coupar B E H. A dominant selectable marker for the construction of recombinant poxviruses. Gene. 1988;65:123–128. doi: 10.1016/0378-1119(88)90424-6. [DOI] [PubMed] [Google Scholar]
- 8.Cormack B P, Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992;69:685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- 9.Cross G A M. Glycolipid anchoring of plasma membrane proteins. Annu Rev Cell Biol. 1990;6:1–39. doi: 10.1146/annurev.cb.06.110190.000245. [DOI] [PubMed] [Google Scholar]
- 10.Donald R G, Roos D S. Insertional mutagenesis and marker rescue in a protozoan parasite: cloning of the uracil phosphoribosyltransferase locus from Toxoplasma gondii. Proc Natl Acad Sci USA. 1995;92:5749–5753. doi: 10.1073/pnas.92.12.5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donald R G K, Carter D, Ullman B, Roos D S. Insertional tagging, cloning, and expression of the Toxoplasma gondii hypoxanthine-xanthine-guanine phosphoribosyltransferase gene. Use as a selectable marker for stable transformation. J Biol Chem. 1996;271:14010–14019. doi: 10.1074/jbc.271.24.14010. [DOI] [PubMed] [Google Scholar]
- 12.Falkner F G, Moss B. Escherichia coli gpt gene provides dominant selection for vaccinia virus open reading frame expression vectors. J Virol. 1988;62:1849–1854. doi: 10.1128/jvi.62.6.1849-1854.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frenkel J K. Toxoplasmosis. Pediatr Clin N Am. 1985;32:343–410. doi: 10.1016/s0031-3955(16)34862-3. [DOI] [PubMed] [Google Scholar]
- 14.Geballe A P, Morris D R. Initiation codons within 5′-leaders of mRNA as regulators of translation. Trends Biochem Sci. 1994;19:159–164. doi: 10.1016/0968-0004(94)90277-1. [DOI] [PubMed] [Google Scholar]
- 15.Gish W, States D J. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 16.Gossler A, Joyner A L, Rossant J, Skarnes W C. Mouse embryonic stem cells and reporter constructs to detect developmentally regulated genes. Science. 1989;244:463–465. doi: 10.1126/science.2497519. [DOI] [PubMed] [Google Scholar]
- 17.Harrison R W, Miller J C. Functional identification of gene up- and down-regulated by glucocorticoids in AtT-20 pituitary cells using an enhancer trap. Endocrinology. 1996;137:2758–2763. doi: 10.1210/endo.137.7.8770895. [DOI] [PubMed] [Google Scholar]
- 17a.Hehl, A., S. Bonnefoy, L. J. Knoll, and J. C. Boothroyd. Unpublished data.
- 18.Joiner K A, Fuhrman S A, Miettinen H M, Kasper L H, Mellman I. Toxoplasma gondii: fusion competence of parasitophorous vacuoles in Fc receptor-transfected fibroblasts. Science. 1990;249:641–646. doi: 10.1126/science.2200126. [DOI] [PubMed] [Google Scholar]
- 19.Kasper L H, Crabb J H, Pfefferkorn E R. Purification of a major membrane protein of Toxoplasma gondii by immunoabsorption with a monoclonal antibody. J Immunol. 1983;130:2407–2412. [PubMed] [Google Scholar]
- 20.Klaff P, Riesner D, Steger G. RNA structure and the regulation of gene expression. Plant Mol Biol. 1996;32:89–106. doi: 10.1007/BF00039379. [DOI] [PubMed] [Google Scholar]
- 20a.Knoll, L. J., and J. C. Boothroyd. Unpublished data.
- 21.Lindquist S. Autoregulation of heat-shock response. In: Ilan J, editor. Translational regulation of gene expression 2. New York, N.Y: Plenum Press; 1993. pp. 279–320. [Google Scholar]
- 22.Luft B J, Remington J S. Toxoplasmic encephalitis in AIDS. Clin Infect Dis. 1992;15:211–222. doi: 10.1093/clinids/15.2.211. [DOI] [PubMed] [Google Scholar]
- 23.Medina-Acosta E, Cross G. Rapid isolation of DNA from trypanosomatid protozoa using a simple ‘mini-prep’ procedure. Mol Biochem Parasitol. 1993;59:327–330. doi: 10.1016/0166-6851(93)90231-l. [DOI] [PubMed] [Google Scholar]
- 24.Mulligan R C, Berg P. Selection for animal cells that express the Escherichia coli gene coding for xanthine-guanine phosphoribosyltransferase. Proc Natl Acad Sci USA. 1981;78:2072–2076. doi: 10.1073/pnas.78.4.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagel S D, Boothroyd J C. The major surface antigen, P30, of Toxoplasma gondii is anchored by a glycolipid. J Biol Chem. 1989;264:5569–5574. [PubMed] [Google Scholar]
- 25a.Nagel S D, Boothroyd J C. The alpha- and beta-tubulins of Toxoplasma gondii are encoded by single copy genes containing multiple introns. Mol Biochem Parasitol. 1988;29:261–273. doi: 10.1016/0166-6851(88)90081-3. [DOI] [PubMed] [Google Scholar]
- 26.Odberg-Ferragut C, Soete M, Engels A, Samyn B, Loyens A, Van Beeumen J, Camus D, Dubremetz J F. Molecular cloning of the Toxoplasma gondii sag4 gene encoding an 18 kDa specific surface protein. Mol Biochem Parasitol. 1996;82:237–244. doi: 10.1016/0166-6851(96)02740-5. [DOI] [PubMed] [Google Scholar]
- 26a.Ortega, E., A. Hehl, and J. C. Boothroyd. Unpublished data.
- 27.Parmley S F, Weiss L M, Yang S. Cloning of a bradyzoite-specific gene of Toxoplasma gondii encoding a cytoplasmic antigen. Mol Biochem Parasitol. 1995;73:253–257. doi: 10.1016/0166-6851(95)00100-f. [DOI] [PubMed] [Google Scholar]
- 28.Pays E, Vanhamme L, Berberof M. Genetic controls for the expression of surface antigens in African trypanosomes. Annu Rev Microbiol. 1994;48:25–52. doi: 10.1146/annurev.mi.48.100194.000325. [DOI] [PubMed] [Google Scholar]
- 29.Pfefferkorn E R, Borotz S E. Toxoplasma gondii: characterization of a mutant resistant to 6-thioxanthine. Exp Parasitol. 1994;79:374–382. doi: 10.1006/expr.1994.1099. [DOI] [PubMed] [Google Scholar]
- 30.Ramakrishnan L, Tran H T, Federspiel N A, Falkow S. A crtB homolog essential for photochromogenicity in Mycobacterium marinum: isolation, characterization, and gene disruption via homologous recombination. J Bacteriol. 1997;179:5862–5868. doi: 10.1128/jb.179.18.5862-5868.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.Roos, D. Personal communication.
- 31.Roos D S, Donald R G K, Morrissette N S, Moulton A L C. Molecular tools for genetic dissection of the protozoan parasite Toxoplasma gondii. Methods Cell Biol. 1994;45:27–63. doi: 10.1016/s0091-679x(08)61845-2. [DOI] [PubMed] [Google Scholar]
- 32.Schiestl R H, Petes T D. Integration of DNA fragments by illegitimate recombination in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991;88:7585–7589. doi: 10.1073/pnas.88.17.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seeber F, Boothroyd J C. Escherichia coli beta-galactosidase as an in vitro and in vivo reporter enzyme and stable transfection marker in the intracellular protozoan parasite Toxoplasma gondii. Gene. 1996;169:39–45. doi: 10.1016/0378-1119(95)00786-5. [DOI] [PubMed] [Google Scholar]
- 34.Sibley L D, LeBlanc A J, Pfefferkorn E R, Boothroyd J C. Generation of restriction fragment length polymorphism linkage map for Toxoplasma gondii. Genetics. 1992;132:1003–1015. doi: 10.1093/genetics/132.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sibley L D, Weidner E, Krahenbuhl J L. Phagosome acidification blocked by intracellular Toxoplasma gondii. Nature. 1985;315:416–419. doi: 10.1038/315416a0. [DOI] [PubMed] [Google Scholar]
- 36.Skeidsvoll J, Ueland P M. Analysis of double-stranded DNA by capillary electrophoresis with laser-induced fluorescence detection using the monomeric dye SYBR green I. Anal Biochem. 1995;231:359–365. doi: 10.1006/abio.1995.9986. [DOI] [PubMed] [Google Scholar]
- 37.Soete M, Camus D, Dubremetz J F. Experimental induction of bradyzoite-specific antigen expression and cyst formation by the RH strain of Toxoplasma gondii in vitro. Exp Parasitol. 1994;78:361–370. doi: 10.1006/expr.1994.1039. [DOI] [PubMed] [Google Scholar]
- 38.Soete M, Fortier B, Camus D, Dubremetz J F. Toxoplasma gondii: kinetics of bradyzoite-tachyzoite interconversion in vitro. Exp Parasitol. 1993;76:259–264. doi: 10.1006/expr.1993.1031. [DOI] [PubMed] [Google Scholar]
- 39.Soldati D, Boothroyd J C. A selector of transcription initiation in the protozoan parasite Toxoplasma gondii. Mol Cell Biol. 1995;15:87–93. doi: 10.1128/mcb.15.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stripecke R, Oliveira C A, McCarthy J E G, Hentze M W. Proteins binding to 5′ untranslated region sites: a general mechanism for translational regulation of mRNAs in human and yeast cells. Mol Cell Biol. 1994;14:5898–5909. doi: 10.1128/mcb.14.9.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomavo S, Fortier B, Soete M, Ansel C, Camus D, Dubremetz J. Characterization of bradyzoite-specific antigens of Toxoplasma gondii. Exp Parasitol. 1991;59:3750–3753. doi: 10.1128/iai.59.10.3750-3753.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walden W E. Repressor-mediated translational control: the regulation of ferritin synthesis by iron. In: Ilan J, editor. Translational regulation of gene expression 2. New York, N.Y: Plenum Press; 1993. pp. 321–334. [Google Scholar]
- 44.Ware P L, Kasper L H. Strain-specific antigens of Toxoplasma gondii. Infect Immun. 1987;55:778–783. doi: 10.1128/iai.55.3.778-783.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang S, Parmley S F. A bradyzoite-specific expressed gene of Toxoplasma gondii encodes a polypeptide homologous to lactate dehydrogenase. Mol Biochem Parasitol. 1995;73:291–294. doi: 10.1016/0166-6851(95)00124-j. [DOI] [PubMed] [Google Scholar]
- 46.Yang S, Parmley S F. Toxoplasma gondii expresses two distinct lactate dehydrogenase homologous genes during its life cycle in intermediate hosts. Gene. 1997;184:1–12. doi: 10.1016/s0378-1119(96)00566-5. [DOI] [PubMed] [Google Scholar]