Abstract
RB60 is an atypical protein disulfide isomerase (PDI) that functions as a member of a redox regulatory protein complex controlling translation in the chloroplast of Chlamydomonas reinhardtii, but also contains a C-terminal endoplasmic reticulum (ER) retention signal, -KDEL. Here, we show by fluorescence microscopy that RB60 resides in the chloroplast but also outside of the chloroplast colocalized with BiP, an ER marker protein. RB60 accumulates in microsomes that exhibit a typical ER magnesium-shift, and cotranslationally translocates into ER microsomes. The first 50-aa leader of RB60 is sufficient for both chloroplast and ER targeting. The leader is cleaved upon translocation into the ER, whereas it remains intact after import to the chloroplast. The leader sequence also contains an acidic domain that appears necessary for the protein's association with the thylakoid membranes. Based on these and additional results, we propose that the dual localization of RB60 occurs via the two conserved transport mechanisms, to the chloroplast and to the ER, that the chloroplast RB60 most likely carries an additional function in the ER, and that its mode of transport, including the differential cleavage of its N terminus, plays an important role in its suborganellar localization and organellar-specific function.
Keywords: dual subcellular localization, redox-responsive regulator, membrane association, GFP
RB60, a protein disulfide isomerase (PDI)-like protein, is localized in plant chloroplasts, where it serves as a redox sensor component of an mRNA-binding protein complex. This complex binds to the 5′ untranslated region of the chloroplast psbA mRNA, where it is thought to regulate the translation of the downstream ORF in response to photosynthetic and light signals (1-3). RB60 controls the assembly of the regulatory protein complex on psbA mRNA through two mechanisms (1, 2, 4-6). In the first, complex assembly is stimulated by the reduction, and diminished by the oxidation, of vicinal disulfide groups in RB60 (2, 6). Because the pool of RB60 disulfides in the chloroplast becomes proportionally reduced with increasing light exposure, this regulatory mechanism is thought to modulate psbA mRNA translation in parallel to light intensity (6). In the second mechanism, ADP-dependent phosphorylation of RB60 inactivates protein complex assembly under ADP concentrations that are typically attained in chloroplasts only in the dark; hence, this pathway is thought to inhibit psbA mRNA translation in the dark (1). RB60, a nucleus-encoded protein, is taken up posttranslationally by isolated chloroplasts of both Chlamydomonas reinhardtii and the higher plant pea via a transit peptide-dependent mechanism (7), indicating a conserved import mechanism to chloroplasts. In the chloroplast under steady-state conditions, RB60 is partitioned between a soluble form in the stroma and a form that is tightly bound to the photosynthetic thylakoid membranes. However, the newly imported RB60 is apparently directed first to the thylakoids (7).
The high homology of RB60 to PDI is intriguing, and may suggest an additional function for this protein. PDI is an oxidoreductase that was identified first as a highly abundant, essential protein in the lumen of the endoplasmic reticulum (ER), where it catalyzes the formation, reduction, and isomerization of disulfide bridges of nascent proteins during their folding in the ER (8, 9). However, there is accumulating evidence that, similarly to RB60, additional PDI-like proteins might also carry out other biological functions and that they may reside in additional subcellular compartments. Regulatory roles have been proposed for PDIs on the extracellular face of a number of cell types (10-13), in nuclei of maturing spermatids (14), associated with the nuclear matrix in chicken liver cells (15), and probably also in mitochondria (16). Markedly, RB60 is the sole PDI gene identified thus far in the C. reinhardtii genome (www.ncbi.nlm.nih.gov), thus raising the possibility that it might also function as the essential ER PDI. Furthermore, RB60 contains a C-terminal signal for ER retention (5). Typically, an ER-retention signal, -(K/H)DEL, prevents ER resident proteins from being transported to subsequent locations of the secretory system (17). The basis for the presence of a -KDEL signal at the C terminus of RB60 is, as yet, unknown. It may be cryptic, derived as an evolutionary remnant from an ancestral PDI gene. Alternatively, this sequence may indicate an additional ER-localized function of RB60.
Here, we show that RB60 is located in the chloroplast but also colocalizes in C. reinhardtii cells with the ER marker protein BiP. Import assays demonstrated that RB60 is taken up by ER microsomes via a cotranslational mechanism, consistent with its import through the conserved signal recognition particle (SRP) system to the ER. We found that the 50-aa-long leader of RB60 is sufficient for its correct targeting to chloroplasts and ER. The leader is cleaved upon translocation into the ER, whereas it remains intact after import to the chloroplast. In the chloroplasts, RB60 appears associated with the thylakoid membranes, an association that is likely dependent on an acidic domain in the N terminus of the protein. These data, together with our previous results demonstrating RB60's unique role in the chloroplast as a translational regulator, suggest that RB60 might have multiple functions depending on its subcellular localization, posttranslational processing, and its organelle-specific protein-protein interactions.
Experimental Procedures
Algae Cell Fixation and Immunostaining. A total of 1 × 106 to 2 × 106 cells were washed in PBS before and in between each step, and fixed with 4% (vol/vol) paraformaldehyde in PBS for 20 min at room temperature. Fixed cells were permeabilized with 0.2% (vol/vol) Triton X-100/PBS for 8 min and then blocked in 1% (wt/vol) BSA/PBS for 1 h. RB60 mouse antisera or BiP rabbit antisera (a kind gift from E. Herman, Donald Danforth Plant Science Center, St. Louis) were added for 1 h. AlexaFluor goat anti-mouse (Molecular Probes) or Cy3 goat anti-rabbit (Jackson ImmunoResearch) Abs were added for 1 h.
ER Import and Fractionation. In vitro protein synthesis reactions were performed by using the T3 TNT-coupled reticulocyte lysate system according to the manufacturer's instructions (Promega) with 2 μg of plasmid DNA. Microsomal import reactions were performed posttranslationally by incubating 1 × 105 cpm of translation product in the presence or absence of 1-μl canine pancreatic microsomal membranes. In cotranslational import assays, 1-μl microsomal membranes were included in the T3 TNT-coupled reticulocyte lysate system. Where indicated, import reactions were treated with 2.5 mg/ml proteinase K and/or 1% (vol/vol) Triton X-100 for 45 min at 4°C.
A total of 2 × 109 CW15 C. reinhardtii cells were broken in an ice cold solution of 100 mM Tris·Cl, pH 7.8/10 mM KCl containing 12% (wt/vol) sucrose and either 5 mM MgCl2 or 5 mM EDTA. The homogenate was centrifuged for 10 min at 1,000 × g at 4°C. Next, 600 μl of the supernatant was loaded on a 12-ml linear 16-55% (wt/wt) sucrose gradient made in the same buffer. After centrifugation at 154,400 × g and at 4°C for 2 h, 650-μl fractions were collected from the top and assayed by immunoblotting.
Plant Growth and Protoplast Transformation. C. reinhardtii 2137a cells were grown in TAP medium to a density of ≈1 × 107 cells per ml as in ref. 3. Physcomitrella patens B.S.G. was grown, and protoplasts were isolated and transformed as in ref. 18. We routinely obtained high transformation rates with no selectable marker (30-40% of the protoplasts express GFP). For assays of protein secretion, total protoplast proteins or trichloroacetic acid (TCA)-precipitated proteins of the media were separated on SDS/10% PAGE and immunoblotted by using anti-GFP mouse antibodies (Roche Diagnostics).
Construction of Fusion Proteins. The RB60:GFP fusion gene was assembled by ligating the RB60 ORF in frame and upstream of GFP5 (19). The RB60:GFP was then subcloned between the actin promoter and terminator in the expression vector, pCOR. The RB60:GFPK construct was produced by replacing the GFP5 in the RB60:GFP construct with GFP5 containing the -KDEL signal at its C terminus. The RB60 deletion mutant constructs, Δ28RB60, L28, and L50 were generated by PCR amplification and subcloned in frame and upstream of either GFP5 or GFPK. The LHCII:GFP and SSU:GFP constructs were similarly assembled by fusing the coding region of C. reinhardtii LHCII or the N terminus of C. reinhardtii small subunit (SSU), respectively, with GFP5. Yellow fluorescence variant of ER-targeted protein (Chit:YFPK) was produced by replacing the GFP ORF of psGFP5K (kindly provided by Jean-Marc Neuhaus, University of Neuchātel, Neuchātel, Switzerland; ref. 20) with enhanced yellow fluorescent protein (eYFP).
Fluorescence Microscopy. The fluorescence images were obtained by using a confocal laser scanning microscope Olympus Fluo-view FV500. For imaging expression of GFP constructs, excitation lines of an argon ion laser of 488 nm were used with a 505/520-nm bandpass filter. For imaging colocalization of YFP and GFP constructs, we followed the settings recommended by Brandizzi et al. (21) with slight modifications. Excitation lines of an argon ion laser of 458 nm for GFP and 514 nm for YFP were used alternately with line switching by using the multitrack facility of the microscope. Fluorescence was detected by using a 505/525-nm bandpass filter for GFP and a 560/615-nm bandpass filter for YFP. Images of chlorophyll fluorescence were collected by using a 660IF filter. Where indicated, brefeldin A was included at 100 μg/ml.
In Silico Analysis. Multiple alignment of 250 polypeptides that share the highest homology with RB60 (based on blast, ref. 22), was generated by using clustalw (23). The putative cleavage site for RB60 was identified by using the computer program of Nielsen et al. (24). The secondary structure common to the leader of the aligned proteins was predicted by using tmap (25).
Results
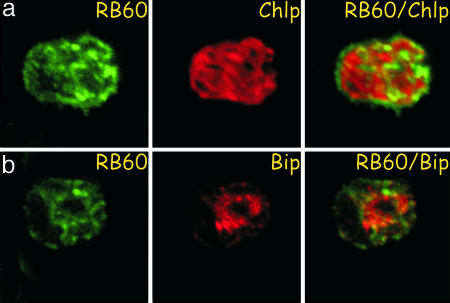
The ER function of PDI is essential and RB60 is the sole PDI gene identified in the genome of C. reinhardtii. Together with the presence of a -KDEL signal at the C terminus of RB60 these data may indicate that, in addition to its function in the chloroplast, RB60 is also localized to the ER. Hence, we assayed the subcellular localization of RB60 in C. reinhardtii cells by immunostaining with anti-RB60 monoclonal antibody by using confocal laser microscopy. The green fluorescence of immunolabeled RB60 and the red autofluorescence of the thylakoid membranes of the chloroplast were acquired separately. The micrographs show that, in addition to its chloroplast localization, RB60 is also found outside of the chloroplast (Fig. 1). Staining the ER with BiP antisera showed that the portion of RB60 that is found outside of the chloroplast is colocalized with BiP, suggesting that RB60 also resides in the ER.
Fig. 1.
RB60 colocalizes with chloroplast and ER markers in C. reinhardtii cells. C. reinhardtii cells were fixed and stained for RB60 by using anti-RB60 monoclonal antibody and imaged by using confocal laser microscopy (green channel, RB60) focusing on the chloroplast (a) and ER (b). The red autofluorescence of the photosynthetic thylakoid membranes of the single cup-shaped chloroplast (red channel, Chlp) only partially overlapped the green fluorescence of RB60 resulting in a yellow color in the combined image (RB60/Chlp), suggesting that RB60 is also localized outside the chloroplast. Staining the ER with BiP antisera (red channel, BiP) showed that RB60 also colocalized with BiP (RB60/BiP), suggesting a second location of RB60 in the ER.
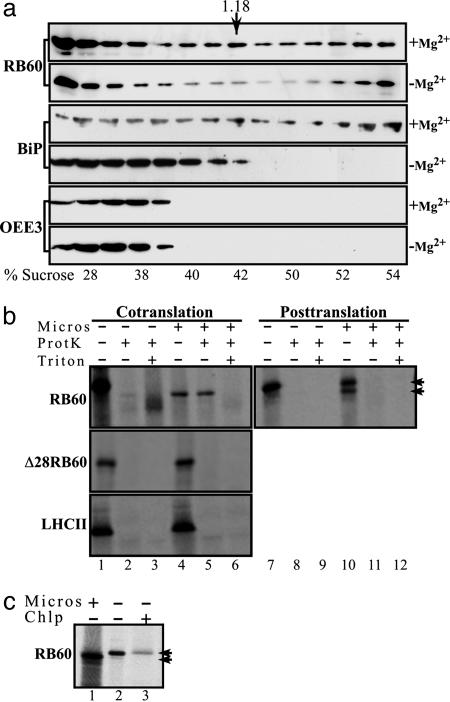
Next, we assayed the accumulation of RB60 in microsomes of C. reinhardtii cells. Cytoplasmic protein extracts were centrifuged on sucrose gradients in the presence or absence of magnesium and analyzed by immunoblot assays to determine whether RB60 cofractionates with ER microsomes (Fig. 2a). Membranes decorated with anti-RB60 monoclonal antibody or with antisera specific to BiP showed that similarly to BiP the peak of RB60 bands at a density of ≈1.18 g·ml-1. Moreover, in a magnesium-shift assay, an established test for microsomes of the rough ER, the peaks of RB60 and BiP displayed a similar shift to the lighter sucrose fractions (Fig. 2a). Parallel immunoblot assays with antisera specific to OEE3, a chloroplast protein, showed that the microsome-containing fractions were not contaminated with chloroplast proteins (Fig. 2a), indicating that these fractions of RB60 are ER localized.
Fig. 2.
Biochemical evidence of the ER localization and independent import of RB60. (a) RB60 is found in microsomes of C. reinhardtii. Sucrose gradient fractionation of cytoplasmic extracts of C. reinhardtii cells was performed in the presence (+Mg2+) or the absence (-Mg2+) of Mg2+, followed by immunoblot analysis using anti-RB60 (RB60) or anti-BiP (BiP) sera or sera raised against the chloroplast OEE3 protein (OEE3). Similarly to BiP, the peak of RB60 bands at a density of ≈1.18 g·ml-1 (the density of peak fraction of rough ER, denoted by an arrow), and in a magnesium shift assay, an established test for microsomes of the rough ER, the peaks of RB60 and BiP displayed a similar shift to the lighter sucrose fractions. The actual sucrose density of each fraction is denoted below each lane (% sucrose). (b) RB60 imports cotranslationally into microsomes. Cotranslational translocation (Cotranslation) into dog pancreas microsomes was performed with in vitro synthesized, 35S-labeled RB60 (RB60), a leaderless RB60 (Δ28RB60), and LHCII (LHCII) recombinant proteins in the presence of microsomes (Micros) as indicated above the lanes. For posttranslational assays (Posttranslation), 35S-labeled proteins were first synthesized in vitro in the absence of microsomes. Protease-protected microsomal translocation was ensured by proteinase K treatment (ProtK). Treatment with Triton X-100 (Triton) verified that the proteinase K-treated proteins were indeed taken up by microsomes. The two arrowheads indicate the migration of the precursor and cleaved forms of RB60. (c) In contrast to the chloroplast, the leader of RB60 is cleaved after uptake by microsomes. In-vitro-synthesized, 35S-labeled RB60 (lane 2) was separated by using SDS/PAGE alongside RB60 imported by dog pancreas microsomes (lane 1) and chloroplast-imported RB60 (lane 3).
The import of proteins to the ER in plants has been shown to be determined by an N terminus signal peptide and to occur only cotranslationally (26). Thus, to gain better insight into the transport mechanism of RB60 into the ER, radiolabeled recombinant RB60 was synthesized in vitro and incubated either posttranslationally or cotranslationally with dog pancreas microsomes. After incubation, protein that had not entered the microsomes was degraded by treatment with proteinase K. When RB60 was incubated cotranslationally, it was completely taken up by microsomes and was protected from protease treatment (Fig. 2b, lanes 4 and 5). The imported RB60 appeared smaller in size than the precursor protein (Fig. 2b, compare lanes 4 and 5 with lane 1), consistent with the typical cleavage of the signal peptide during import to the ER. Disruption of microsome membranes by treatment with nonionic detergent resulted in the degradation of imported RB60 (Fig. 2b, lane 6), verifying the effectiveness of the protease treatment and that the radiolabeled protein was in fact protected from degradation by being taken up by microsomes. When RB60 was incubated posttranslationally with microsomes, a small fraction of cleaved protein was detected (Fig. 2b, lane 10), but the protein did not enter the microsomes, as shown by its sensitivity to the protease treatment (Fig. 2b, lane 11). This finding suggests that the complete uptake of RB60 occurs only cotranslationally, a mechanism that is unique to ER import. RB60 lacking the first 28 amino acids of its N terminus (Δ28RB60) did not import to microsomes cotranslationally (Fig. 2b) or posttranslationally (data not shown), confirming that the N-terminal sequence is required for targeting of RB60 to the ER. The chloroplast protein LHCII was not imported to microsomes (Fig. 2b), showing that dual targeting to both ER and chloroplasts is not common to all chloroplast proteins. A comparison of RB60 imported to microsomes to RB60 that was imported to chloroplasts showed that, unlike the microsomal RB60 (Fig. 2c, lane 1), the leader of the chloroplast imported protein was not cleaved (Fig. 2c, lane 3 and in ref. 7), strengthening our assumption that the dual import of RB60 to the ER and chloroplasts occurs via the two mutually exclusive conserved targeting mechanisms.
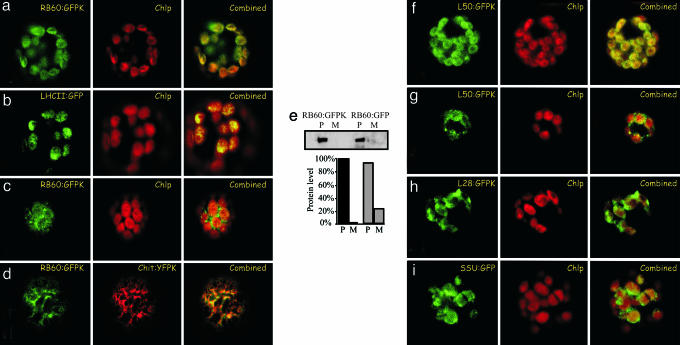
To study the import mechanisms of RB60 in vivo, we compared the accumulation of a transiently expressed RB60 fused with GFP containing -KDEL at its C terminus (RB60:GFPK) to that of GFP fused with the chloroplast-localized light-harvesting complex II protein (LHCII:GFP) and to an ER-localized YFP (Chit:YFPK) in P. patens protoplasts by using confocal laser microscopy. To avoid mislocalization due to overaccumulation of expressed proteins, we imaged only protoplasts displaying the earliest signal of GFP fluorescence. The proper size of the fusion proteins expressed in the transformed protoplasts was verified by immunoblot assays (see Fig. 6, which is published as supporting information on the PNAS web site). When the images were taken from the chloroplasts plane of the cell, the pattern of the green fluorescence of RB60:GFPK (Fig. 3a) was quite similar to that of LHCII:GFP (Fig. 3b), indicating that a portion of RB60:GFPK was targeted to chloroplasts in vivo and suggesting again that RB60 imports to chloroplasts by a conserved mechanism. When fluorescence images of the cortical part of the cell were obtained, the RB60:GFPK signal was also observed outside of chloroplasts (Fig. 3c). Comparing the fluorescence, in the same cell, of RB60:GFPK (Fig. 3d; RB60:GFPK) with that of an ER-localized YFP protein (Fig. 3d; Chit:YFPK) showed that the two fusion proteins are colocalized.
Fig. 3.
RB60:GFPK fusion protein is targeted in vivo to chloroplast and ER. RB60 fused to GFP containing an ER retention signal at its C terminus (RB60:GFPK) was expressed in protoplasts of the moss P. patens. (a) Images of RB60:GFPK fluorescence and the autofluorescence of the chloroplast thylakoids membranes (Chlp) taken from the chloroplasts plane of the cell were overlaid (Combined) for comparison. (b) Comparing RB60:GFP fluorescence, the thylakoids autofluorescence, and that of the chloroplast thylakoid protein LHCII (LHCII:GFP) showed that RB60 is localized within the chloroplast. (c) When the cortical plane of the cell was imaged, an RB60:GFPK signal outside of the chloroplast was also detected. (d) Comparing RB60:GFPK fluorescence and that of an ER-localized YFP marker protein (Chit:YFPK) showed that a portion of RB60:GFPK is localized in the ER. (e) Removal of ER retention signal stimulates secretion of RB60:GFP. Cellular proteins (P) and secreted proteins of the media (M) of protoplasts expressing RB60:GFPK (RB60:GFPK, construct containing -KDEL) and RB60:GFP (RB60:GFP, construct lacking -KDEL) were analyzed by immunoblot assays using anti-GFP mAbs. The bar graph depicts the digital quantification of the level of secreted protein (M) and cellular protein (P) of RB60:GFPK (dark gray) and RB60:GFP (light gray). The leader of RB60 is sufficient for authentic targeting. (f and g) Fluorescence images of protoplasts transiently expressing construct of the first 50 amino acids of the N terminus of RB60 fused to the N terminus of GFPK (L50:GFPK) that were taken from the chloroplast (f) and the cortical (g) planes of the cell showed that the leader of RB60 is sufficient for the dual ER and chloroplast localization of the protein. (b, f, h, and i) The localization of L50:GFPK (f) was similar to that of LHCII:GFP fusion protein (a marker of the thylakoids) (b), and the localization of a construct of the first 28 amino acids of the N terminus of RB60 fused to the N terminus of GFPK (L28:GFPK) (h) was similar to that of SSU:GFP (a marker of chloroplast stroma) (i), suggesting that L50:GFPK is associated with thylakoids and that L28:GFPK is mostly in the stroma.
The C-terminal signal, -(K/H)DEL, retains proteins in the ER and its deletion typically stimulates secretion of ER proteins (17, 27). We found that protoplasts of P. patens expressing the RB60:GFP construct (lacking the -KDEL signal) secreted notably higher amounts of the fusion protein than cells expressing the RB60:GFPK construct (containing KDEL) (Fig. 3e), indicating an ER localization of RB60:GFPK. The stimulated level of secretion of RB60:GFP was similar to that previously observed for the ER-localized calreticulin (27).
Previous findings in Euglena showed that in some species chloroplast proteins are transported via the ER and the Golgi to the chloroplast (28). Our results showing that RB60 imports in vitro directly to isolated chloroplasts of both C. reinhardtii and the higher plant pea (7) do not support a similar targeting mechanism for RB60. However, we monitored the effect of a known inhibitor of ER-Golgi transport, brefeldin A (BFA), on the accumulation of RB60:GFPK over a period of 2 h (according to ref. 29). The BFA treatment did not affect the chloroplast localization of RB60:GFPK (see Fig. 7, which is published as supporting information on the PNAS web site), indicating that RB60 import to the chloroplast is independent of its uptake by the ER. Together, these results indicate that RB60 itself contains the targeting information for dual localization to the ER and to the chloroplast, and moreover that RB60 is likely routed through the two independent evolutionary conserved targeting mechanisms: to the chloroplast via the posttranslational chaperone-assisted import, and to the ER via the SRP-dependent cotranslational uptake.
The N-terminal 50-aa sequence of RB60, preceding the conserved sequence of PDI, is quite different from those of most PDIs in both length and amino acid composition. It contains a potential cleavage site after the 28th amino acid residue followed by a region of 22 aa, 29-50, enriched with negatively charged residues (7). Alignment of the leader sequence of RB60 with leader sequences of 250 PDIs displaying the highest homology to RB60 identified a small group of PDI-like proteins containing, in a similar position, an acidic domain of variable length (Fig. 4). The proteins with an acidic domain similar in size to that of RB60 include VC_PDI, a Volvox carteri PDI-like protein, and a pancreas-specific human PDI-A2. Also in this group are the PDI-A4/ERp72-type proteins from human, mouse, and rat, which contain a larger acidic domain than that of RB60, and Triticum aestivum and Zea mays PDIs and a Caenorhabditis elegans ERp72-like protein, which include a smaller size acidic domain (Fig. 4). Analysis of the aligned N terminus of this group of proteins for transmembrane domains suggests that they could all form a transmembranal α-helical domain in the segment of their N terminus that precedes the acidic domain (Fig. 4). The processing of RB60 during its import to microsomes (Fig. 2 b and c) and its lack of processing after its uptake by chloroplasts (Fig. 2c and in ref. 7) suggest that this domain is cleaved off after RB60 import to the ER and that it is present in the chloroplast-localized form of RB60.
Fig. 4.
PDIs in a unique class contain acidic domains preceded by potential α-helix transmembrane domains at their N termini. Multiple alignment of the polypeptides was generated by using clustlw. Acidic amino acids are shaded. Alignment of the leader sequence of RB60 with that of other PDIs identified a small group of PDIs that contain a similar domain. The putative cleavage site for RB60 (marked by an arrow) was identified by using the computer program of Nielsen et al. (24). The secondary structure common to the leader of the aligned proteins (α-helix is denoted by cylindrical shading) was predicted by using tmap.
To study the role of the leader of RB60 in the subcellular localization of the protein, we fused the first 50 amino acids of RB60 to GFP containing an ER retention signal at its C terminus (L50:GFPK) and imaged the fluorescence of the fusion protein in the chloroplasts plane (Fig. 3f) and in the cortical region of the cell (Fig. 3g). The high similarity of the fluorescence images of L50:GFPK (Fig. 3 f and g) and of RB60:GFPK (Fig. 3 a and c) indicates that the 50-aa-long leader of RB60 is sufficient for the authentic targeting of the protein. Next, we fused only the first 28 amino acids of RB60 to GFPK and imaged the localization of the fusion protein (L28:GFPK) in the chloroplast plane of the protoplasts. The L28:GFPK fusion protein accumulated in the chloroplasts at a low level (Fig. 3h), and its pattern of accumulation differed from that of L50:GFP (Fig. 3f). Because RB60 was found partitioned between a soluble form in the stroma and a form that is tightly bound to the thylakoid membranes (7), we also imaged, for comparison, the localization of LHCII (Fig. 3b) and a stromal protein, the small subunit of RuBisCO (Fig. 3i; SSU:GFP). The pattern of accumulation of L28:GFPK resembled mostly that of SSU:GFP, whereas the accumulation of L50:GFPK showed greater similarity to that of LHCII:GFP, suggesting that the first 28 amino acids of RB60 are sufficient for chloroplast targeting but not for the association with thylakoids, whereas the 50-aa-long leader of RB60 is sufficient for both. These results also imply that amino acids 29-50 of the leader of RB60 are required for its association with thylakoids. Imaging of the cortical part of the cell revealed a similar pattern of accumulation of L50:GFPK and RB60:GFPK (Fig. 3 g and c, respectively), suggesting that L50 is sufficient for ER targeting. We could not detect accumulation of the L28:GFPK fusion protein in the ER by laser microscopy or immunoblot assay using GFP-specific mAb (data not shown). Therefore, at this point, our results do not distinguish between the two most likely scenarios: that the L28:GFP fusion protein is targeted to but does not accumulate in the ER, or that the first 28 amino acids of RB60 are not sufficient for ER targeting.
Discussion
The C. reinhardtii RB60 is a PDI that functions as a regulatory protein in the chloroplast, where it is found in both soluble and membrane-associated forms (7). Here, we show that, in addition to its chloroplast function, RB60 is localized to the ER (Fig. 1), and begin to study the hierarchy of signals that control the intracellular trafficking of RB60 and its association with thylakoids. The majority of chloroplast proteins is encoded by the nuclear genome, synthesized by cytosolic ribosomes, and are then targeted to chloroplasts by an N-terminal sequence, traditionally termed the transit peptide (30, 31). Unlike the cotranslational transport of proteins to the ER, which is typically mediated by the SRP system (32), the import to chloroplasts occurs posttranslationally, possibly assisted by cytosolic protein chaperones (33). Earlier work demonstrated that RB60 is imported in vitro to isolated chloroplasts of both the unicellular green alga C. reinhardtii and the higher plant pea by a posttranslational mechanism (7), indicating that its uptake by the chloroplast is direct and conserved among evolutionary diverse species of algae and higher plants. Here, we show that, similarly to the two nucleus-encoded chloroplast proteins of C. reinhardtii, LHCII and SSU, RB60 is targeted to the chloroplast in vivo in P. patens protoplasts (Fig. 3), further corroborating that RB60 is imported to the chloroplast through the conserved posttranslational mechanism. Likewise, the ER localization of RB60 in C. reinhardtii cells (Fig. 1) and P. patens protoplasts (Fig. 3d), together with the cotranslational and signal peptide-dependent uptake of RB60 by isolated dog pancreas microsomes (Fig. 2b), implies that RB60 is transported to the ER through the conserved SRP-dependent import mechanism (32). Finally, the processing of the leader of RB60 after transport to the ER (Fig. 2 b and c) and the absence of cleavage after import to chloroplasts (Fig. 2c and ref. 7), together with the resistance of RB60 accumulation in chloroplasts to the inhibitor of the secretory system, brefeldin A (Fig. 7), further argue that the two import mechanisms of RB60 to the chloroplast and to the ER are independent of each other.
We demonstrated that the first 50 amino acids of the RB60 leader sequence are sufficient for its correct targeting to chloroplast thylakoids and to the ER (Fig. 3 f and g). Analysis of the leader of RB60 predicts a transmembranal α-helical domain in its N-terminal segment (Fig. 4). The signal peptide containing this hydrophobic domain is cleaved off during RB60 import to microsomes (Fig. 2 b and c) but remains part of RB60 following its uptake by chloroplasts (7), suggesting that the predicted hydrophobic α-helical domain is required for the association of RB60 with thylakoids. Our finding that the GFP fusion protein containing only the first 28 amino acids of RB60 (α-helical domain) is distributed in the chloroplast stroma (Fig. 3h) suggests that this segment is not sufficient for the protein association with thylakoids, a process that likely requires the last 22 amino acids of the leader, which are rich in negatively charged residues. Intriguingly, acidic residues in the N- or C-terminal domains of the chloroplast PsbX, PsbY, and PsbW proteins are also essential for the integration of these proteins into the thylakoids (34). Therefore, future studies are required to determine whether the association of RB60 with thylakoids (7) might also be regulated by the adjacent acidic region by influencing the formation or disruption of the transmembranal α-helical domain.
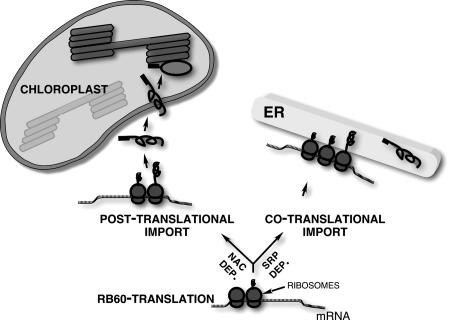
RB60 is targeted to ER and chloroplasts through two alternative mechanisms: the SRP system or cytoplasmic chaperones; notably, both mechanisms are mediated by the N terminus of the protein. How, then, might the dual targeting of RB60 be regulated in the cell? The transport of RB60 to chloroplasts and ER by these two mutually exclusive targeting mechanisms suggests that targeting of RB60 is regulated before its commitment to either path. An ideal candidate for this type of control is the nascent polypeptide-associated complex (NAC), which competes with the SRP for binding to polypeptides as they emerge from the translating ribosomes and is thought to negatively regulate protein transport to the ER (35, 36). Interestingly, NAC has recently been implicated as an enhancer of protein targeting to the mitochondria (37, 38). We suggest a working model (Fig. 5) in which the regulated competition between the SRP and, hypothetically, NAC determines the path of RB60 transport. If SRP binds the emerging signal peptide of RB60, translation is attenuated until the ribosome-bound RB60 associates with ER membranes, where it is then cotranslationally translocated into the ER. However, if the nascent RB60 is bound by the NAC, then RB60 is synthesized in the cytosol and targeted posttranslationally, under chaperone assistance, to the chloroplasts. An example of a similar mechanism is provided by the proapoptotic proteins BAX and BAK, whose subcellular localization to the mitochondria and ER in animal cells is regulated by a death signal (39, 40).
Fig. 5.
Working model of cotranslational and posttranslational targeting of RB60. The targeting path of RB60 is determined during the first steps of its synthesis. The regulated competition of SRP and regulatory proteins, such as NAC, for binding ribosomes translating RB60 mRNA determines whether the protein is directed via the SRP system to the ER or alternatively to the chloroplast. Translation of RB60 mRNA by ribosomes bound to SRP is attenuated until they associate with the SRP receptor in ER membranes. Thereafter, RB60 is cotranslationally translocated into the ER, where it functions as a leaderless form. Translation of RB60 mRNA by ribosomes theoretically bound by NAC or functionally equivalent proteins proceeds uninhibited, and RB60 is synthesized to completion in the cytosol. The leader of the chaperone-associated RB60 then directs and imports the protein into the chloroplast. The leader of RB60 is not cleaved after its import by chloroplasts and directs the protein to the thylakoids.
The dual targeting of RB60 indicates that it bears at least two functions, one as a regulatory protein in the chloroplast and a second in the ER. This finding is not entirely surprising, as only one PDI gene has been identified thus far in the C. reinhardtii genome. Thus, RB60, expressed in the ER, most likely carries out the highly conserved and essential functions of the classical PDI enzyme, i.e., catalyzing and isomerizing disulfide bonds of ER-imported proteins. The ER function of RB60 requires further study.
How does a single polypeptide perform two diverse biological roles? The intrinsic capacity of RB60 for dual targeting to the chloroplast and ER is necessary for this task, but is it sufficient? Our findings indicate that an additional level of regulation is likely required. Notably, the unique regulatory function of RB60 in chloroplasts of C. reinhardtii depends not only on its targeting to the chloroplast but also on its specific association with a unique complex of proteins containing a second regulatory RNA-binding protein, RB47 (2, 41). Likewise, the function of the classical PDI enzyme in the ER was shown in yeast to depend on its interaction with the ER-localized oxidoreductase Ero1p (42, 43). Therefore, depending on its subcellular localization, its specific protein-protein interactions, and unique posttranslational processing, a particular PDI might express several biological roles.
The dual localization of RB60 in chloroplasts and ER bears several implications to the endosymbiotic theory of the chloroplast. The high homology of RB60 to eukaryotic PDIs suggests that it was recruited from the host genome rather than from the cyanobacterial endosymbiont genome. This evolutionary path of organellar proteins might be more prevalent than previously expected, because comparison of proteomes of Arabidopsis chloroplast and of cyanobacterium suggested that only ≈35% of chloroplast proteins originated from the cyanobacterium genome (44). Likewise, studies of the mitochondrial proteome indicate that its evolution included recruitment of α-proteobacterial gene products as well (45). It seems that RB60 gained its chloroplast function in addition to its role in the ER. Determining whether the evolution of RB60 is unique or is shared by additional chloroplast and ER proteins is required for better understanding the evolution of endosymbiotic organelles.
Supplementary Material
Acknowledgments
We thank G. Galili and H. Zehavi-Peled for critical reading of this manuscript. This study was supported by grants from the Israeli Science Foundation and the Minerva Foundation. A.L. is a recipient of a Feinberg Graduate School Fellowship.
Author contributions: A.L., T.T., and A.D. designed research; A.L., T.T., V.K., Y.P., and I.D. performed research; and A.L. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PDI, protein disulfide isomerase; ER, endoplasmic reticulum; SRP, signal recognition particle; YFP, yellow fluorescent protein; SSU, small subunit.
References
- 1.Danon, A. & Mayfield, S. P. (1994) EMBO J. 13, 2227-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Danon, A. & Mayfield, S. P. (1994) Science 266, 1717-1719. [DOI] [PubMed] [Google Scholar]
- 3.Trebitsh, T. & Danon, A. (2001) Proc. Natl. Acad. Sci. USA 98, 12289-12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fedoroff, N. V. (2002) Curr. Opin. Plant Biol. 5, 452-459. [DOI] [PubMed] [Google Scholar]
- 5.Kim, J. & Mayfield, S. P. (1997) Science 278, 1954-1957. [DOI] [PubMed] [Google Scholar]
- 6.Trebitsh, T., Levitan, A., Sofer, A. & Danon, A. (2000) Mol. Cell. Biol. 20, 1116-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trebitsh, T., Meiri, E., Ostersetzer, O., Adam, Z. & Danon, A. (2001) J. Biol. Chem. 276, 4564-4569. [DOI] [PubMed] [Google Scholar]
- 8.Gilbert, H. F. (1998) Methods Enzymol. 290, 26-50. [DOI] [PubMed] [Google Scholar]
- 9.Freedman, R. B., Hawkins, H. C. & McLaughlin, S. H. (1995) Methods Enzymol. 251, 397-406. [DOI] [PubMed] [Google Scholar]
- 10.Pariser, H. P., Zhang, J. & Hausman, R. E. (2000) Exp. Cell Res. 258, 42-52. [DOI] [PubMed] [Google Scholar]
- 11.Bennett, T. A., Edwards, B. S., Sklar, L. A. & Rogelj, S. (2000) J. Immunol. 164, 4120-4129. [DOI] [PubMed] [Google Scholar]
- 12.Couet, J., de Bernard, S., Loosfelt, H., Saunier, B., Milgrom, E. & Misrahi, M. (1996) Biochemistry 35, 14800-14805. [DOI] [PubMed] [Google Scholar]
- 13.Essex, D. W., Li, M., Miller, A. & Feinman, R. D. (2001) Biochemistry 40, 6070-6075. [DOI] [PubMed] [Google Scholar]
- 14.Ohtani, H., Wakui, H., Ishino, T., Komatsuda, A. & Miura, A. B. (1993) Histochemistry 100, 423-429. [DOI] [PubMed] [Google Scholar]
- 15.Ferraro, A., Altieri, F., Coppari, S., Eufemi, M., Chichiarelli, S. & Turano, C. (1999) J. Cell Biochem. 72, 528-539. [DOI] [PubMed] [Google Scholar]
- 16.Rigobello, M. P., Donella-Deana, A., Cesaro, L. & Bindoli, A. (2000) Free Radical Biol. Med. 28, 266-272. [DOI] [PubMed] [Google Scholar]
- 17.Pelham, H. R. (1990) Trends Biochem. Sci. 15, 483-486. [DOI] [PubMed] [Google Scholar]
- 18.Meiri, E., Levitan, A., Guo, F., Christopher, D. A., Schaefer, D., Zryd, J. P. & Danon, A. (2002) Mol. Genet. Genomics 267, 231-240. [DOI] [PubMed] [Google Scholar]
- 19.Haseloff, J., Siemering, K. R., Prasher, D. C. & Hodge, S. (1997) Proc. Natl. Acad. Sci. USA 94, 2122-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fluckiger, R., De Caroli, M., Piro, G., Dalessandro, G., Neuhaus, J. M. & Di Sansebastiano, G. P. (2003) J. Exp. Bot. 54, 1577-1584. [DOI] [PubMed] [Google Scholar]
- 21.Brandizzi, F., Fricker, M. & Hawes, C. (2002) Nat. Rev. Mol. Cell. Biol. 3, 520-530. [DOI] [PubMed] [Google Scholar]
- 22.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403-410. [DOI] [PubMed] [Google Scholar]
- 23.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nielsen, H., Engelbrecht, J., Brunak, S. & von Heijne, G. (1997) Protein Eng. 10, 1-6. [DOI] [PubMed] [Google Scholar]
- 25.Persson, B. & Argos, P. (1997) J. Protein Chem. 16, 453-457. [DOI] [PubMed] [Google Scholar]
- 26.Vitale, A., Ceriotti, A. & Denecke, J. (1993) J. Exp. Bot. 44, 1417-1444. [Google Scholar]
- 27.Sonnichsen, B., Fullekrug, J., Nguyen Van, P., Diekmann, W., Robinson, D. G. & Mieskes, G. (1994) J. Cell Sci. 107, 2705-2717. [DOI] [PubMed] [Google Scholar]
- 28.Sulli, C., Fang, Z., Muchhal, U. & Schwartzbach, S. D. (1999) J. Biol. Chem. 274, 457-463. [DOI] [PubMed] [Google Scholar]
- 29.Steele-King, C. G., Evans, D. E., Satiat-Jeunemaitre, B. & Hawes, G. (1999) J. Exp. Bot. 50, 1465-1469. [Google Scholar]
- 30.Cline, K. (1986) J. Biol. Chem. 261, 14804-14810. [PubMed] [Google Scholar]
- 31.Keegstra, K. & Cline, K. (1999) Plant Cell 11, 557-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rapoport, T. A. (1992) Science 258, 931-936. [DOI] [PubMed] [Google Scholar]
- 33.Schnell, D. J. (2000) Essays Biochem. 36, 47-59. [DOI] [PubMed] [Google Scholar]
- 34.Tissier, C., Woolhead, C. A. & Robinson, C. (2002) Eur. J. Biochem. 269, 3131-3141. [DOI] [PubMed] [Google Scholar]
- 35.Wiedmann, B., Sakai, H., Davis, T. A. & Wiedmann, M. (1994) Nature 370, 434-440. [DOI] [PubMed] [Google Scholar]
- 36.Powers, T. & Walter, P. (1996) Curr. Biol. 6, 331-338. [DOI] [PubMed] [Google Scholar]
- 37.George, R., Beddoe, T., Landl, K. & Lithgow, T. (1998) Proc. Natl. Acad. Sci. USA 95, 2296-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Funfschilling, U. & Rospert, S. (1999) Mol. Biol. Cell 10, 3289-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cartron, P. F., Priault, M., Oliver, L., Meflah, K., Manon, S. & Vallette, F. M. (2003) J. Biol. Chem. 278, 11633-11641. [DOI] [PubMed] [Google Scholar]
- 40.Scorrano, L., Oakes, S. A., Opferman, J. T., Cheng, E. H., Sorcinelli, M. D., Pozzan, T. & Korsmeyer, S. J. (2003) Science 300, 135-139. [DOI] [PubMed] [Google Scholar]
- 41.Yohn, C. B., Cohen, A., Danon, A. & Mayfield, S. P. (1996) Mol. Cell. Biol. 16, 3560-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frand, A. R. & Kaiser, C. A. (1998) Mol. Cell 1, 161-170. [DOI] [PubMed] [Google Scholar]
- 43.Pollard, M. G., Travers, K. J. & Weissman, J. S. (1998) Mol. Cell 1, 171-182. [DOI] [PubMed] [Google Scholar]
- 44.Abdallah, F., Salamini, F. & Leister, D. (2000) Trends Plant Sci. 5, 141-142. [DOI] [PubMed] [Google Scholar]
- 45.Andersson, S. G., Karlberg, O., Canback, B. & Kurland, C. G. (2003) Philos. Trans. R. Soc. London B 358, 165-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.