Abstract
Many aspects of the genetic control of mammalian embryogenesis cannot be extrapolated from other animals. Taking a forward genetic approach, we have induced recessive mutations by treatment of mice with ethylnitrosourea and have identified 43 mutations that affect early morphogenesis and patterning, including 38 genes that have not been studied previously. The molecular lesions responsible for 14 mutations were identified, including mutations in nine genes that had not been characterized previously. Some mutations affect vertebrate-specific components of conserved signaling pathways; for example, at least five mutations affect previously uncharacterized regulators of the Sonic hedgehog (Shh) pathway. Approximately half of all of the mutations affect the initial establishment of the body plan, and several of these produce phenotypes that have not been described previously. A large fraction of the genes identified affect cell migration, cellular organization, and cell structure. The findings indicate that phenotype-based genetic screens provide a direct and unbiased method to identify essential regulators of mammalian development.
Keywords: ethylnitrosourea, hedgehog, intraflagellar transport, mesoderm, neural tube closure
Mammalian embryogenesis differs fundamentally from the development of other animal models. Localized maternal components are not essential for axis specification in mammals (1), whereas the establishment of the body axes in Drosophila, Caenorhabditis elegans, and zebrafish relies on localization of maternal determinants (2-4). Mammalian development has a unique requirement for complex interactions between embryonic and uterine tissues, and the first cell fate decision in the mouse embryo is the choice between embryonic and extraembryonic lineages (5). After implantation, germ layer organization and tissue specification in the mouse embryo depend on coupled morphogenetic movements and intercellular signals, processes not paralleled in invertebrate embryos. A standard approach to study the genetic control of mouse embryogenesis has been to inactivate evolutionarily conserved genes by targeted mutagenesis; this approach may overlook components that are of particular importance in mammals. In contrast, phenotype-based screens in the mouse have the potential to identify the molecules that control mammalian-specific events.
Phenotype-based screens depend on the ability to induce a large number of random mutations in germ cells and on rapid identification of mutants of interest. It has been known for 25 years that ethylnitrosourea (ENU) is an extremely potent mutagen in the mouse (6, 7); and a screen of the progeny of only 700 F1 progeny of ENU-treated animals should yield an average of one allele of each gene in the genome. We previously described a pilot phenotype-based screen to identify recessive mutations that produce easily visible disruptions in the morphology of the midgestation mouse embryo (8). Similar phenotype-based approaches in other laboratories that focused on later stages of embryonic and fetal development in the mouse have also identified novel mutations successfully (9-11). With the availability of the mouse genome sequence, it has become straight-forward to identify the genes responsible for the mutant phenotypes.
Here, we describe the identification of 43 recessive mutations that map to single loci and cause clear morphological abnormalities in the midgestation mouse embryo. Most of the mutations affect previously uncharacterized genes that control a variety of processes, including the morphogenetic movements of gastrulation and early neural patterning. Developmental mechanisms are conserved among mammals, and several mutations described here will help define the developmental basis of inherited human syndromes.
Materials and Methods
Identification of Mutants with Abnormal Morphology. We used the ENU mutagenesis protocol and crossing scheme defined previously, in which C57BL/6J (B6) males were treated with three doses of 100 μg of ENU per gram of body weight, and all outcrosses were to C3HeB/FeJ (C3H) females (8). For each F1 founder male, five to eight G3 litters of embryos potentially homozygous for newly induced mutations were analyzed. Candidate mutants were identified when more than two embryos of similar, distinctive abnormal morphology were identified in a line. Additional G2 × F1 or G2 × G2 crosses were analyzed to confirm the presence of a genetic variant. We examined the morphology of embryonic day 9.5 (e 9.5) G3 embryos from 380 lines. Based on the specific locus test data (7), there should be ≥30 newly induced gene-inactivating mutations in each line. By using this estimate, the lines studied here would represent ≈10,000 gene-inactivating mutations, affecting ≈25-30% of the genes in the genome.
Linkage Mapping. All lines that produced reliable heritable phenotypes were inherited as simple recessive Mendelian traits with complete or nearly complete penetrance. To map the mutation responsible for the phenotype, an initial genome scan panel used DNA from adult parents of mutant litters identified in test crosses (obligate carriers). A panel of 40 MIT simple sequence length polymorphism (SSLP) markers (http://mouse.ski.mskcc.org) analyzed in 20 carriers defined linkage of the mutation to a single segment of the C57BL/6J genome for a majority of mutations. In the remaining cases, the initial genome scan ruled out most genomic segments, and linkage was established with analysis of additional markers and/or additional carriers and mutant embryos. Forty-three mutations were mapped to single genomic regions. In addition, four lines that produced interesting phenotypes did not map to single loci; these and other lines were preserved as frozen sperm.
For most genomic segments, mutations could be mapped to <5 cM by using MIT markers and a small number of additional carriers and mutant embryos. Higher resolution mapping required the generation of additional polymorphic markers. Potentially polymorphic simple sequence repeats in the interval of interest were identified in the genome sequence by using the etandem program from the European Molecular Biology Open Software Suite (emboss) (http://athena.bioc.uvic.ca/pbr/pise). Primers flanking repeats of at least 60 base pairs, identified by using primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi), were tested for polymorphism between C3HeB/FeJ and C57BL/6J; 10-30% of primer pairs identified a useful polymorphism. More than 250 previously uncharacterized SSLP markers have been identified (http://mouse.ski.mskcc.org). Lines were maintained by identification of potential carriers based on flanking SSLP markers.
If initial genome scan did not reveal linkage, we tested markers at chromosome ends, where linkage can be difficult to detect. Some genomic intervals are not highly polymorphic between C57BL/6J and C3HeB/FeJ. For example, no useful MIT markers existed in the centromere-proximal 29 Mb of chromosome 9. After testing a large number of simple sequence repeat-containing segments, we identified several useful SSLPs in this region (http://mouse.ski.mskcc.org).
There were five cases in which two mutations were identified in a single line. Because >10% of lines contained at least one mutation of interest, this was approximately the number expected in a sample of 43 mutations. In four cases, the phenotypes were different and segregated independently so that it was clear that more than one mutation was present in the line. In one case, two different mutations affecting neural patterning arose on the same chromosome but could be separated by recombination.
Map-Based Gene Identification. For lines with interesting phenotypes, we bred animals to generate embryos for phenotypic analysis. These additional carriers and embryos provided enough opportunities for recombination so that the gene could be mapped to a small genomic interval. The MIT SSLP markers could be used to map most genes to 1- to 2-cM intervals, but those intervals generally included too many candidate genes to evaluate. Generation of previously uncharacterized SSLP markers and analysis of ≈200 litters of embryos and their parents made it possible to map most genes to 1 Mb or less. In most regions of the genome, a 1-Mb region would include ≈12 genes, and it was feasible to evaluate all of the genes in such an interval. Candidate genes were evaluated by sequencing RT-PCR products and, if necessary, genomic PCR products.
Pattern of Expression of Molecular Markers. In situ hybridization on whole-mount embryos and immunofluorescent detection on frozen sections were carried out as described (12, 13).
Results and Discussion
Overview of Mutations Identified. Forty-two recessive lethal mutations that disrupted the morphology of the e9.5 embryo were identified and mapped to single loci (Tables 1, 2, 3, 4). Mutations of interest were identified in a large fraction of the lines examined (more than one mutation per 10 lines screened), undoubtedly because many different genetic pathways affect the form of the embryo at this stage. In addition, 150 lines were screened at both e9.5 and e12.5 in collaboration with L. A. Niswander, which nearly doubled the number of mutations identified in those lines (L. A. Niswander, personal communication).
Table 1. Alleles of previously characterized genes.
| Mutant | Phenotype | Gene | Mutation | Nature of allele |
|---|---|---|---|---|
| bent body (bnb) | Dorsalized spinal cord, left-right randomization, die at e9.0 | Smo (19) | Splice site | Null |
| 2A | Dorsalized spinal cord, left-right randomization, die at e9.0 | Kif3a | Not determined | Null |
| flexo (fxo) | Abnormal diencephalon, no floor plate in caudal spinal cord, left-right randomization, polydactyly, die at e13-14 | Polaris (13) | Splice site | Hypomorph |
| M1un | Arrested unturned, variable cardia bifida; arrest at e9.0 | Pten | Missense (M134K) | Hypomorph |
| DD1 | Caudal paraxial mesoderm replaced by neural tubes | Tbx6 | Splice site | Null |
Table 2. Mutations in previously uncharacterized genes that alter dorsal-ventral patterning of the spinal cord.
| Mutant | Phenotype | Map position (Chr; cM) | Gene (ref.) |
|---|---|---|---|
| open brain (opb2) | Exencephaly, ventralized caudal spinal cord, polydactyly, die at e13-14 | 1: 18.5 | Rab 23 (23) |
| wimple (wim) | Exencephaly, dorsalized spinal cord, left-right randomization; die at e10-11 | 5: 16-18 | IFT172 (13) |
| hennin (hnn) | No floor plate in caudal spinal cord, left-right randomization; die at e13-14 | 16: 36-42 | Novel* |
| icbins (icb) | Dorsalized spinal cord, left-right randomization; die at e9.0 | 1: 102 | Disp1 (19) |
| ling-ling (lln) | Dorsalized spinal cord, left-right randomization; die at e10-11 | 9: 0-7.5 | Dnchc2† |
| sister of open brain (sopb) | Exencephaly, ventralized caudal spinal cord, polydactyly; die at e13-14 | 6: 50.5-60.5 |
T.C. and K.V.A., unpublished results.
D.H. and K.V.A., unpublished results.
Table 3. Mutations in previously uncharacterized genes that cause neural tube closure defects.
| Mutant | Phenotype | Map position (Chr: cM) |
|---|---|---|
| open mind (opm) | Exencephaly; survive to the end of gestation | 12: 17-18 |
| F10 | Exencephaly, incompletely penetrant dominant and penetrant recessive; survive to the end of gestation | 8: 21-50* |
| F11 | Exencephaly; survive to the end of gestation, rare escapers in outcrosses are small, with craniofacial defects | 3: 22-35 |
| 97C2 | Exencephaly and open caudal spinal cord; survive to the end of gestation | 7: 2.5-8 |
| G2-E | Exencephaly and open caudal spinal cord; survive to the end of gestation | 4: 60-79 |
| 12A | Exencephaly; survive to the end of gestation | 4: 49-57 |
| 31B | Exencephaly, small forebrain; die at e10 | 2: 77-82 |
| 7A5 | Exencephaly, small forebrain; die at e10 | 5: 36 |
| 7A6 | Exencephaly, small forebrain; die at e10 | 6: 38-60 |
| lilR3 | Exencephaly; die at e10 | 16: 27-32 |
| 33C | Exencephaly, abnormal brain morphology, die at e10-12 | 19: 24-26 |
Line lost due to incompletely penetrant dominant lethality.
Table 4. Mutations in previously uncharacterized genes that affect early morphogenesis.
| Mutant | Phenotype | Map position Chr:cM | Gene |
|---|---|---|---|
| Gastrulation arrest | |||
| F8 | Arrest at e8.0, small embryonic region | 7: 53-57.5 | |
| W3 | Arrest at e8.0, small embryonic region | 11: 49-54 | |
| dimple (dmpl) | Arrest at e8.0, abnormal primitive streak | 6: 37-43 | |
| S2 | Arrest at e8.0, dead cells in amniotic cavity | 15: 39-44 | |
| T6 | Arrest at e8.0, small embryonic region | 5: 18-19 | |
| M1ec | Arrest at e7.0 | 3: 39-41 | |
| lazy mesoderm (lzme) | Arrest at e8.0; streak bulges into amniotic cavity | 5: 38 | Ugdh (12) |
| Early somite stage arrest wing-shaped neural plate (wsnp) | Deficit in paraxial and axial mesoderm, arrest at e8.5 | 16: 27-28 | |
| limulus (lulu) | Deficit in paraxial and axial mesoderm; arrest at e8.5 | 1: 63 | Epb4.115* |
| chato | Arrest at e8.5 outside of yolk sac | 7: 10-16 | |
| mermaid | Deficit in paraxial mesoderm; arrest at e8.5 | 8: 59-72 | |
| kahloista (khlo) | Arrest unturned, small somites | 2: 45-47.8 | Nap1† |
| 99J1 | Arrest unturned | 5: 64-80 | |
| 15A | Deficit in paraxial mesoderm | 17: 30-42 | |
| 1A | Arrest unturned, abnormal heart | 11: 52-70 | |
| nero | Arrested unturned, no left-right asymmetry; arrest at e9.5 | 3: 25-35 | dolichyl-phosphate β-glucosyltransferase‡ |
| M2 | Attest at e9.0, unturned | 4: 56.5 | |
| e9.5 arrest | |||
| 19C | Abnormal heart, arrest at e9.5 | 16: 28-57 | |
| 38D/p | Swollen lateral plate, abnormal somites | 2: 41-48 | |
| 8A | Small, twisty tail | 13: 30-43 | |
| Left-right abnormal | |||
| 3B | Randomized polarity of heart looping, survives past midembryogenesis | 7: 10-46.5 |
J.D.L. and K.V.A., unpublished results.
A.S.R. and K.V.A., unpublished results.
E.H.F., A.S.R. and K.V.A., unpublished results.
Unlike screens that focus on specific genomic regions (14-17), the phenotype-based approach is designed to identify interesting mutations in genes located on any mouse autosome. The advantage of this strategy is that mutants are selected for phenotypes that affect specific aspects of development, but the approach relies on efficient mapping of mutations to chromosomal regions. By using DNA polymorphisms as genetic markers, it was possible to map each mutation to a defined genomic interval, once a cohort of carriers was identified (see Materials and Methods).
Using map-based cloning, we identified the genes responsible for 14 of the mutant phenotypes. In each case, a single nucleotide change was identified in a gene in the interval where the gene mapped, and independent evidence confirmed that the gene was responsible for the phenotype (Tables 1, 2, 3, 4). For several other mutations, nucleotide substitutions likely to be the cause of the phenotype have been identified, but additional experiments will be necessary to confirm that the correct gene has been identified (data not shown). All mutations changed the sequence of the mature RNA. Forty percent of the mutations affected splice sites and were detected initially based on the altered length of an RT-PCR product. The remaining mutations were divided approximately equally between missense and nonsense mutations.
Information about all of the mutations and new polymorphic DNA mapping markers is available at http://mouse.ski.mskcc.org.
Null and Hypomorphic Alleles of Previously Characterized Genes. Based on their map positions and phenotypes, most of the mutations affected genes that had not been previously characterized. However, alleles of five genes were identified that had been previously shown to play essential roles in mouse development. (Table 1 and Fig. 1).
Fig. 1.
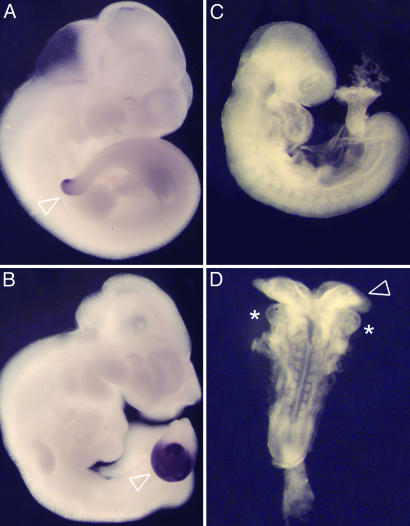
New ENU induced alleles of previously characterized genes. (A and B) DD1 is an allele of Tbx6. e10.5 wild-type (A) and DD1 homozygous (B) embryos, assayed by in situ hybridization of the primitive streak marker Brachyury. Mesenchymal cells accumulate in the tail bud of DD1 mutants (B, arrowhead). This phenotype is identical to that observed in Tbx6 mutants (18), and in DD1/Tbx6 heterozygotes (not shown). The Tbx6DD1 mutation was associated with an A to C substitution in the splice acceptor of Tbx6 exon 3 that led to an abnormally spliced product lacking the DNA binding domain. (C and D) M1un is an allele of Pten. In contrast with e9.5 wild-type littermates (C, lateral view), PtenM1un/Ptennull embryos (D, dorsal view) fail to undergo embryonic turning, have broad open headfolds (arrowhead), and cardia bifida (asterisks indicate the two laterally positioned heart fields).
Three mutations seemed to represent null alleles of characterized genes: Tbx6, Smoothened (Smo), and Kif3a. The Tbx6DD1 mutation caused a phenotype like that of the targeted null allele of Tbx6 (18), failed to complement the null allele, and was associated with an abnormally spliced product that lacked the DNA binding domain. The bent body (bnb) mutation (8) was associated with defective splicing of Smo, produced the same phenotype as the null allele, and failed to complement the null allele (19). The 2A mutation mapped to the same interval as Kif3a, produced the same e9.0 arrest with neural patterning defects as the null Kif3a allele (13), and failed to complement the Kif3a-null allele.
Two mutations were partial loss-of-function alleles of genes, Polaris and Pten, that had been inactivated by targeted mutagenesis. The flexo mutation, which affected neural patterning and causes preaxial polydactyly, was associated with an in-frame deletion caused by a splice site mutation in the Polaris gene (13). The Polarisflexo allele was intermediate in strength between the targeted null allele and a viable insertional mutation, and each allele revealed different aspects of the function of the gene (13, 20). The M1un mutation, mapped to the same interval as Pten, caused arrest at e9.0 without completion of embryonic turning and produced a phenotype similar to that of the null allele of Pten (21). The M1un allele failed to complement the null allele and was associated with a missense mutation (M134K) in a conserved residue adjacent to the phosphatase domain. A mutation at the same position in the human Pten gene (M134R) is a cause of the dominant Bannayan-Riley-Ruvalcaba syndrome (BRRS) (22). PtenM1un homozygotes survived longer than null embryos, suggesting that the PtenM1un allele is a hypomorphic mutation. Analysis of the developmental defects in the PtenM1un embryos should help define the developmental basis of the defects seen in the different human syndromes associated with mutations in Pten.
Mutations in Previously Uncharacterized Genes. The remaining 90% of the genes identified (38 mutations) seemed to affect genes not previously shown to affect embryonic development. Based on the stage of embryonic lethality and the similarity of the phenotypes, we classified the mutations based on the primary defect observed (Tables 2, 3, 4). The findings for each category are described in the sections below.
Proteins Required for Sonic Hedgehog (Shh) Signaling. Nine mutants that showed striking morphological abnormalities in the brain were found to affect the specification of cell types along the dorsal-ventral axis of the spinal cord (Fig. 2 and Tables 1 and 2). Specification of ventral cell types in the spinal cord depends on the activity of the Shh signaling pathway, and phenotypic and epistasis analysis demonstrated that all of the mutations influence Shh signaling (refs. 13, 19, and 23 and J.T.E, T.C., D.H., and K.V.A, unpublished results). As described above, one mutation, Smobnb, was an allele of Smo, an evolutionarily conserved component of the Hedgehog (Hh) signaling pathway. A second mutation, icbins, also affected a homologue of a Drosophila Hh pathway gene, dispatched, and was identified at the same time as targeted null alleles of the gene (19, 24, 25).
Fig. 2.
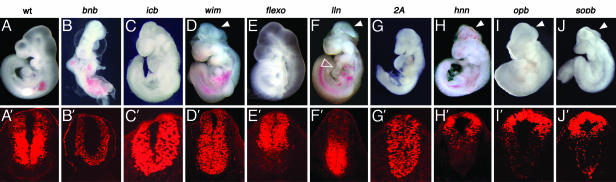
Mutants with altered dorsal-ventral patterning of the neural tube. Morphological phenotype of whole wild-type (A), Smobnb (B), Disp1icb (C), wim (D), Polarisflexo (E), lln (F), Kif3a2A (G), hnn (H), open brain (I) and sopb (J) embryos. B, C, and G are e9.5 and A, D, E, F, H, I, and J are e10.5 embryos. All these mutants have abnormal brain morphology and show abnormal laterality of heart looping (visible in lln; empty arrowhead in F), and several show exencephaly and spina bifida (arrowheads in D, F, H, I, and J). The effects on dorsal-ventral neural tube patterning are illustrated in the corresponding lower panels (A′-J′), which show the pattern of expression Pax6 in transverse sections of the neural tube. Pax6 is a marker of lateral neural cell types in wild-type embryos (A). Pax6 is expressed throughout the neural tube of bnb and icb mutants (B′ and C′), is shifted ventrally in wim and lln mutants (D′ and F′), is nearly normal in flexo mutants (E′), and is restricted to the dorsal neural tube in hnn, open brain, and sopb mutants (H′, I′, and J′).
The other seven mutations mapped to regions that did not include homologues of characterized Hh pathway genes. At least five of these genes seem to regulate intracellular trafficking or localization of proteins that mediate Hh signal transduction.
The first of these genes to be identified was open brain (opb). Both the ENU-induced allele and a spontaneous allele were associated with early stop codons in the ORF of Rab23, which encodes a vesicle transport protein (23). Double mutant analysis placed Rab23 downstream of Smo and upstream of the Gli transcription factors that implement Hh signals (J.T.E. and K.V.A., unpublished results). These findings were the first indication of an essential role for vesicle transport in the Sonic hedgehog signal transduction pathway.
Four of the mutations affected proteins required for intraflagellar transport (IFT) (ref. 13 and D.H. and K.V.A., unpublished results), a process that is required for the structure and maintenance of cilia and flagella (26). Based on double mutant analysis, the mouse IFT proteins are also required at a step in the Hh signal transduction pathway downstream of the membrane receptors and upstream of the Gli transcription factors (ref. 13 and D.H. and K.V.A., unpublished results). The mechanism of action of these cilia-associated proteins in Hh signaling will require further biochemical and cell biological studies.
Rab23 and the IFT genes have orthologues in Drosophila, where Hh signaling has been studied extensively. The IFT proteins are required for the production of sensory cilia in Drosophila (27) and in C. elegans (28), but no role for these genes in Drosophila Hh signaling has been reported. Other approaches have identified additional vertebrate-specific components required for Hh signaling (29-32), suggesting that there are fundamental differences between Hh signaling in Drosophila and the mouse. Some forms of the human developmental disorder Bardet-Biedl syndrome (BBS) affect IFT proteins (33). Some individuals with BBS show reversal of left-right polarity and polydactyly (34), both features of the inappropriate Hh signaling caused by lack of IFT function. It is therefore likely that Hh signaling in humans, as in mice, depends on IFT proteins.
Mutations That Prevent Neural Tube Closure. Neural tube closure in mammals depends on normal proliferation, apoptosis, morphogenesis, and patterning of the neural tube and underlying mesenchyme (35). Defects in neural tube closure, such as anencephaly and spina bifida, are the second most common class of birth defect, perhaps because of the large number of genes that are required for this process. As expected, the screen identified a large number of mutations of this class. Four mutations associated with altered dorsal-ventral neural patterning caused neural tube closure defects (Table 2 and Fig. 2), consistent with the finding that proper neural cell type specification is required for formation of the hinge points, in the neural plate, that allow closure (36). Eleven additional mutations also showed exencephaly (Table 3). Six of these mutations allowed survival to the end of gestation and seemed to be specifically required for neural tube closure. Five mutations caused defects outside the neural tube that led to death during embryogenesis; based on their external morphology, some of these mutations seem to affect anterior-posterior or dorsal neural patterning.
Mutations That Affect Early Morphogenesis. Approximately half of the mutations identified (21 of 43) caused arrest at or before midgestation (e9.5), associated with early abnormalities in tissue organization (Table 4).
Seven mutants arrested during gastrulation, at approximately e7.5. Although embryos at this stage have a simple structure, a few mutants showed distinctive morphology as early as e7.5 (Fig. 3). For example, cells from the primitive streak accumulate in a ball in the amniotic cavity of lazy mesoderm (lzme) embryos, similar to the phenotype seen in Fgf8 mutants. Additional analysis demonstrated that lzme specifically disrupted Fgf signaling (12). In situ analysis of the expression of genes expressed in the streak, node, and early mesoderm made it possible to identify additional mutations that affect patterning rather than survival. For example, the primitive streak of dimple (dmpl) mutants was expanded laterally, as revealed by the pattern of expression of Brachyury (Fig. 3C). In the future, incorporation of reporter genes expressed in these cell types into the screen (11) could aid in distinguishing those mutations that affect embryonic patterning and morphogenesis from those that affect more generalized cellular functions.
Fig. 3.
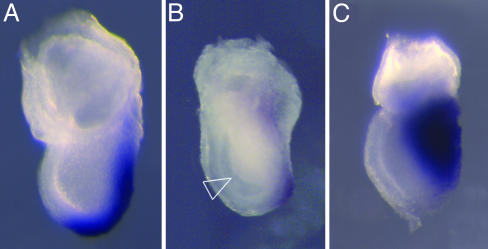
Early morphogenesis mutants that arrest during gastrulation. The morphology of the primitive streak was assayed by in situ hybridization with Brachyury. In contrast to wild-type e7.5 embryos (A), lzme embryos (B) have ectopic cells in the amniotic cavity (arrowhead) and express Brachyury weakly. The morphology of dimple mutant embryos (C) appears normal at e7.5, but they have a laterally expanded domain of Brachyury expression.
Ten mutants arrested at e8.5-9.0, with dramatic disruptions in tissue organization that suggested primary defects in morphogenesis. For example, khlo mutant embryos arrested before embryonic turning, with an open neural plate, a few small abnormal somites, an accumulation of cells in the primitive streak, and cardia bifida (Fig. 4 and A.S.R, unpublished results). Another two mutants, wsnp and lulu also arrested unturned with an open neural plate and cardia bifida (ref. 8 and Fig. 4), but cell-type-specific markers revealed clear differences from the khlo phenotype. For example, the small amount of paraxial mesoderm present in wsnp and lulu embryos never segmented into somites (Fig. 4 and J.D.L., unpublished results). Also in contrast to khlo mutants, which had a normal axial midline, wsnp and lulu mutants had discontinuous axial mesoderm. Candidate genes identified for khlo and lulu suggest that they regulate the cytoskeleton and epithelial organization (A.S.R., J.D.L., and K.V.A, unpublished results). Further analysis of this class of mutants should help define the relationships between morphogenetic movements and the establishment of the mammalian body plan.
Fig. 4.
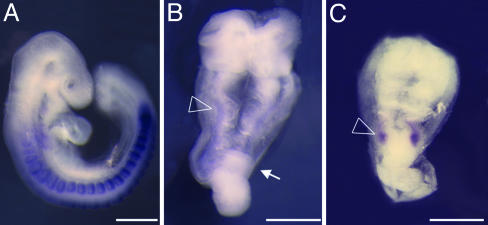
Early morphogenesis mutants that arrest before embryonic turning. Shown are wild-type (A), khlo (B), and lulu (C) embryos assayed for expression of Mox1, a paraxial mesoderm marker. Both khlo and lulu embryos arrest at e9.0, showing abnormal morphogenesis. khlo mutants express Mox1 along the A-P axis (arrowhead in B), although only a few somites can be seen morphologically. In contrast, lulu mutants never differentiate somites, and expression of Mox1 is limited to a small patch of cells (arrowhead in C). The arrow in B points to the accumulation of cells at the primitive streak characteristic of khlo mutants. A is a lateral view of a e9.5 wild-type embryo; B and C are dorsal views of the mutants that have been magnified ×1.3 with respect to A.
Forward Genetic Analysis as a Tool to Analyze Mammalian Development. A relatively large fraction of the mutations identified affect a single process, early dorsal-ventral patterning of the spinal cord. The dramatic morphology of these mutant embryos helped single out these mutations as being of particular interest and led to the identification of nine genes that affect the specification of Hh-dependent cell types in the neural tube, only two of which had been identified as components of the Hh signaling pathway in Drosophila. Approximately 1 of 40 lines tested had a mutation that affected dorsal-ventral neural patterning. If it is assumed that 30 new mutations were induced per line, then ≈1 of 1,200 induced mutations disrupted a gene required for dorsal-ventral patterning in the neural tube. This finding predicts that there are a total of ≈20-40 uncharacterized mouse genes that affect early dorsal-ventral neural patterning that could be detected in this screen. The results demonstrate that modest scale phenotype-based screens in the mouse can successfully identify sets of novel genes, if the mutant phenotypes can be recognized easily.
Many of the genes identified in this screen control aspects of cell biology that are crucial for morphogenesis and patterning of the mouse embryo, which probably reflects the tight coupling between morphogenesis and cell fate determination in mouse embryogenesis. Several mutations, like lzme and dimple, affect the structure of the primitive streak, and should help define the mammalian-specific events of axis specification. The findings presented here suggest that the unbiased approach provided by forward genetics is a particularly useful method to identify the regulators of cellular organization and behavior that are crucial for mammalian development.
A striking finding is that the lack of substantial maternal contribution provides a significant advantage for genetic screens focused on early events in mouse development. Because transcription from the zygotic genome of the mouse begins 1 day after fertilization and patterning of the embryonic body begins 4-5 days later, patterning of the mouse embryo depends almost exclusively on the zygotic genome. In contrast, other model vertebrates rely on stores of maternally transcribed gene products for many aspects of early patterning. The impact of the differences in maternal contribution to embryonic development can be seen by comparing the phenotypes identified in this screen and results from zebrafish. Mouse Smo homozygotes die before midgestation and lack all ventral neural cell types (Fig. 2 and ref. 37). In zebrafish, where Smo is expressed both maternally and zygotically, zygotic Smo mutants make some ventral neural cell types (38). Similarly, lzme mutants lack the enzyme Ugdh, which is required for the production of GAG-modified proteoglycans, and arrest at gastrulation (12), whereas zebrafish homozygous for null Ugdh mutations survive to the end of embryogenesis, when they show heart valve defects (39). Mutations in some IFT genes have been identified in zebrafish, but there are sufficient maternal IFT products to allow the formation of cilia in mutant embryos (40). Morpholino knockdown of zebrafish Smo, Ugdh and IFT RNAs failed to produce stronger phenotypes, perhaps because of maternally contributed protein (38-40). Screens for maternal effect mutants in other vertebrates are difficult, but the genes controlling early developmental events are accessible in a zygotic-lethal screen in the mouse, like the one described here.
Acknowledgments
We thank Joshua Bloomekatz, Aaron Goldberg, Tae-Hee Kim, Karel Liem, Floria Lupu, Lihui Qian, Xin Zhou, Irene Zohn, and Lee Niswander for help with mapping and for generation of new SSLP markers. We thank Aimin Liu for discussions on IFT function, Tim Bestor and Joshua Bloomekatz for comments on the manuscript, Lee Niswander and Elizabeth Lacy for collaborations to screen embryos at later developmental stages, and Virginia Papaioannou (Columbia University, New York), Pier Paolo Pandolfi (Sloan-Kettering Institute, New York), and Robert Benezra (Sloan-Kettering Institute, New York) for mutant mice. All mice carrying ENU-induced mutations are available to the mouse academic community. This work was supported by National Institutional of Health Grants HD35455 and HD043478.
Author contributions: M.J.G.-G. and K.V.A. designed research; M.J.G.-G., J.T.E., T.C., H.L.A., M.R.W., D.H., A.S.R., J.D.L., E.H.F., J.R.T., and K.V.A. performed research; M.J.G.-G., J.T.E., and K.V.A. analyzed data; J.R.T. contributed new reagents/analytic tools; and M.J.G.-G. and K.V.A. wrote the paper.
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on April 20, 2004.
Abbreviations: ENU, ethylnitrosourea; SSLP, simple sequence length polymorphism; en, embryonic day n; Smo, Smoothened; bnb, bent body; Hh, Hedgehog; IFT, intraflagellar transport; lzme, lazy mesoderm.
See accompanying Biography on page 5910.
References
- 1.Rossant, J. & Tam, P. P. (2004) Dev. Cell 7, 155-164. [DOI] [PubMed] [Google Scholar]
- 2.Nüsslein-Volhard, C. (1991) Dev. Suppl. 1, 1-10. [PubMed] [Google Scholar]
- 3.Bowerman, B. (1998) Curr. Top. Dev. Biol. 39, 73-117. [DOI] [PubMed] [Google Scholar]
- 4.Jesuthasan, S. & Strähle, U. (1997) Curr. Biol. 7, 31-42. [DOI] [PubMed] [Google Scholar]
- 5.Rossant, J. & Cross, J. C. (2001) Nat. Rev. Genet. 2, 538-548. [DOI] [PubMed] [Google Scholar]
- 6.Russell, W. L., Kelly, E. M., Hunsicker, P. R., Bangham, J. W., Maddux, S. C. & Phipps, E. L. (1979) Proc. Natl. Acad. Sci. USA 76, 5818-5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hitotsumachi, S., Carpenter, D. A. & Russell, W. L. (1985) Proc. Natl. Acad. Sci. USA 82, 6619-6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kasarskis, A., Manova, K. & Anderson, K. V. (1998) Proc. Natl. Acad. Sci. USA 95, 7485-7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hentges, K., Thompson, K. & Peterson, A. (1999) Development (Cambridge, U.K.) 126, 1601-1609. [DOI] [PubMed] [Google Scholar]
- 10.Herron, B. J., Lu, W., Rao, C., Liu, S., Peters, H., Bronson, R. T., Justice, M. J., McDonald, J. D. & Beier, D. R. (2002) Nat. Genet. 30, 185-189. [DOI] [PubMed] [Google Scholar]
- 11.Zarbalis, K., May, S. R., Shen, Y., Ekker, M., Rubenstein, J. L. & Peterson, A. S. (2004) PLoS Biol. 2, E219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.García-García, M. J. & Anderson, K. V. (2003) Cell 114, 727-737. [DOI] [PubMed] [Google Scholar]
- 13.Huangfu, D., Liu, A., Rakeman, A. S., Murcia, N. S., Niswander, L. & Anderson, K. V. (2003) Nature 426, 83-87. [DOI] [PubMed] [Google Scholar]
- 14.Bode, V. C. (1984) Genetics 108, 457-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shedlovsky, A., King, T. R. & Dove, W. F. (1988) Proc. Natl. Acad. Sci. USA 85, 180-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rinchik, E. M. & Carpenter, D. A. (1999) Genetics 152, 373-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kile, B. T., Hentges, K. E., Clark, A. T., Nakamura, H., Salinger, A. P., Liu, B., Box, N., Stockton, D. W., Johnson, R. L., Behringer, R. et al. (2003) Nature 425, 81-86. [DOI] [PubMed] [Google Scholar]
- 18.Chapman, D. L. & Papaioannou, V. E. (1998) Nature 391, 695-697. [DOI] [PubMed] [Google Scholar]
- 19.Caspary, T., García-García, M. J., Huangfu, D., Eggenschwiler, J. T., Wyler, M. R., Rakeman, A. S., Alcorn, H. L. & Anderson, K. (2002) Curr. Biol. 12, 1628-1632. [DOI] [PubMed] [Google Scholar]
- 20.Murcia, N. S., Richards, W. G., Yoder, B. K., Mucenski, M. L., Dunlap, J. R. & Woychik, R. P. (2000) Development (Cambridge, U.K.) 127, 2347-2355. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki, A., de la Pompa, J. L., Stambolic, V., Elia, A. J., Sasaki, T., del Barco Barrantes, I., Ho, A., Wakeham, A., Itie, A., et al. (1998) Curr. Biol. 8, 1169-1178. [DOI] [PubMed] [Google Scholar]
- 22.Figer, A., Kaplan, A., Frydman, M., Lev, D., Paswell, J., Papa, M. Z., Goldman, B. & Friedman, E. (2002) Clin. Genet. 62, 298-302. [DOI] [PubMed] [Google Scholar]
- 23.Eggenschwiler, J. T., Espinoza, E. & Anderson, K. V. (2001) Nature 412, 194-198. [DOI] [PubMed] [Google Scholar]
- 24.Ma, Y., Erkner, A., Gong, R., Yao, S., Taipale, J., Basler, K. & Beachy, P. A. (2002) Cell 111, 63-75. [DOI] [PubMed] [Google Scholar]
- 25.Kawakami. T., Kawcak, T., Li, Y. J., Zhang, W., Hu, Y. & Chuang, P.-T. (2002) Development (Cambridge, U.K.) 129, 5753-5765. [DOI] [PubMed] [Google Scholar]
- 26.Rosenbaum, J. L. & Witman, G. B. (2002) Nat. Rev. Mol. Cell Biol. 3, 813-825. [DOI] [PubMed] [Google Scholar]
- 27.Avidor-Reiss, T., Maer, A. M., Koundakjian, E., Polyanovsky, A., Keil, T., Subramaniam, S. & Zuker, C. S. (2004) Cell 117, 527-539. [DOI] [PubMed] [Google Scholar]
- 28.Cole, D. G., Diener, D. R., Himelblau, A. L., Beech, P. L., Fuster, J. C. & Rosenbaum, J. L. (1998) J. Cell Biol. 141, 993-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chuang, P.-T. & McMahon, A. P. (1999) Nature 397, 617-621. [DOI] [PubMed] [Google Scholar]
- 30.Sekimizu, K., Nishioka, N., Sasaki, H., Takeda, H., Karlstrom, R. O. & Kawakami, A. (2004) Development (Cambridge, U.K.) 131, 2521-2532. [DOI] [PubMed] [Google Scholar]
- 31.Wolff, C., Roy, S., Lewis, K. E., Schauerte, H., Joerg-Rauch, G., Kirn, A., Weiler, C., Geisler, R., Haffter, P. & Ingham, P. W. (2004) Genes Dev. 18, 1565-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bulgakov, O. V., Eggenschwiler, J. T., Hong, D. H., Anderson, K. V. & Li, T. (2004) Development (Cambridge, U.K.) 131, 2149-2159. [DOI] [PubMed] [Google Scholar]
- 33.Blacque, O. E., Reardon, M. J., Li, C., McCarthy, J., Mahjoub, M. R., Ansley, S. J., Badano, J. L., Mah, A. K., Beales, P. L., Davidson, W. S., et al. (2004) Genes Dev. 18, 1630-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katsanis, N., Lupski, J. R. & Beales, P. L. (2001) Hum. Mol. Genet. 10, 2293-2299. [DOI] [PubMed] [Google Scholar]
- 35.Copp, A. J., Greene, N. D. & Murdoch, J. N. (2003) Nat. Rev. Genet. 4, 784-793. [DOI] [PubMed] [Google Scholar]
- 36.Ybot-Gonzalez, P., Cogram, P. Gerrelli, D. & Copp, A. J. (2002) Development (Cambridge, U.K.) 129, 2507-2517. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, X. M., Ramalho-Santos, M. & McMahon, A. P. (2001) Cell 106, 781-792. [PubMed] [Google Scholar]
- 38.Varga, Z. M., Amores, A., Lewis, K. E., Yan, Y. L., Postlethwait, J. H., Eisen, J. S. & Westerfield M. (2001) Development (Cambridge, U.K.) 128, 3497-3509. [DOI] [PubMed] [Google Scholar]
- 39.Walsh, E.C. & Stainier, D. Y. (2001) Science 293, 1670-1673. [DOI] [PubMed] [Google Scholar]
- 40.Sun, Z., Amsterdam, A., Pazour, G. J., Cole, D. G., Miller, M. S. & Hopkins, N. (2004) Development (Cambridge, U.K.) 131, 4085-4093. [DOI] [PubMed] [Google Scholar]