Abstract
The central nervous system is fundamentally dependent on guided cell migration, both during development and in adulthood. We report an absolute requirement of the transcription factor serum response factor (SRF) for neuronal migration in the mouse forebrain. Conditional, late-prenatal deletion of Srf causes neurons to accumulate ectopically at the subventricular zone (SVZ), a prime neurogenic region in the brain. SRF-deficient cells of the SVZ exhibit impaired tangential chain migration along the rostral migratory stream into the olfactory bulb. SVZ explants display retarded chain migration in vitro. Regarding target genes, SRF deficiency impairs expression of the β-actin and gelsolin genes, accompanied by reduced cytoskeletal actin fiber density. At the posttranslational level, cofilin, a key regulator of actin dynamics, displays dramatically elevated inhibitory phosphorylation at Ser-3. Our studies indicate that SRF-controlled gene expression directs both the structure and dynamics of the actin microfilament, thereby determining cell-autonomous neuronal migration.
Keywords: actin cytoskeleton, cofilin, transcription
Proper development and functionality of the mammalian brain require migration of postmitotic neurons from their site of origin to their ultimate destination in the brain (1-4). Chain migration of interneuron precursors through the rostral migratory stream (RMS) to the olfactory bulb (5) is a highly informative system for the study of neuronal migration, because it occurs throughout life.
Transcriptional control mechanisms regulating migration-associated gene activities are poorly understood. The transcription factor serum response factor (SRF) (6) regulates the expression of genes encoding cytoskeletal proteins (6-8). Previously, in a solely in vitro study, we observed impaired migration of SRF-deficient murine embryonic stem (ES) cells, accompanied by a disproportionate reduction of the F-actin versus G-actin content (8). To investigate whether SRF was also required for mammalian cell migration in vivo, we now specifically address neuronal migration in the mouse forebrain by using conditional, Cre-mediated, Srf knock-out mutagenesis.
SRF (6-9) is broadly expressed and regulates various target genes in brain, muscle, and other tissues by cooperating with cell type-specific SRF accessory proteins (10-14). Different SRF complexes are controlled by various intracellular signaling pathways, including mitogen-activated protein kinase cascades (15), Rho-dependent signaling (12, 16), and Ca2+ stimulation (17). SRF target genes can be classified into two types, i.e., those representing cellular immediate early genes (IEGs) and those that are activated more slowly and less transiently (18). The latter often encode structural cytoskeletal proteins, such as actins, myosins, tropomyosin, vinculin, and others. SRF is essential in murine embryogenesis, because SRF-deficient embryos do not gastrulate (9) and display enhanced cell death (19).
In Drosophila melanogaster development, SRF, together with its cofactor DMRTF/MAL-D, was found to contribute to mesodermal cell migration and invasive migration of border cells (20, 21). Cell migration requires dynamic remodeling of the actin cytoskeleton (3), with the Rho GTPases Rho, Rac, and Cdc42 as essential regulators of actin polymerization (22). Actin filament turnover is mediated by the severing proteins gelsolin and cofilin, both of which fulfill key regulatory functions in cell migration (23-25). Actin-severing activity of cofilin is regulated by inhibitory phosphorylation at Ser-3 by kinases such as LIM kinase (26, 27) and is reactivated by dephosphorylation through the slingshot and chronophin protein phosphatases (28, 29). SRF activity itself is directly linked to actin turnover by Rho-dependent stimulation of the SRF cofactor MAL (12, 16, 30).
Cre recombinase-mediated conditional deletion of “floxed” murine Srf alleles has been applied previously to studies of the development of the heart (31), skeletal muscle (32), and cardiovascular system (33), the latter study recognizing defects in organizing actin/intermediate filament bundles. Using Cre-mediated Srf deletion in the developing forebrains of Srf(flex1neo/flex1neo)CamKIIα-iCre mice, we demonstrate here that SRF deficiency affects two key regulators of actin filament turnover, gelsolin and cofilin, and causes arrest of neuronal migration. As a result, severe neuroanatomical and pathological abnormalities are generated in mutant mice.
Materials and Methods
Mouse Genetics and Preparation of Brain Sections. Srf(flex1neo/flex1neo) (34) and (CamKIIα-iCre/wt) (35) mice were bred to generate offspring homozygous for the floxed Srf allele and heterozygous for the Cre transgene. These Srf(flex1neo/flex1neo)CamKIIα-iCre mice, abbreviated SrfCamKIIα-iCre or referred to as mutant mice, displayed the conditionally deleted Srf(lx/lx) genotype (34) in Cre-expressing cells of the forebrain (not shown). Mice were of mixed C57BL/6N-129P2/OlaHsd-FVB/N genetic background. Genotyping was according to refs. 34 and 35. To keep mutant mice alive beyond postnatal day 21 (P21), animals were hand-fed with milk substitute (puppy nursing support; Waltham, Melton Mowbray, U.K.) and water-softened food pellets. Experimental animals were 17 days (P17) of age, unless otherwise stated. Deeply anesthetized mice were perfused with 4% phosphate-buffered paraformaldehyde (PFA), and brains were dissected and postfixed for at least 48 h in 4% PFA at 4°C. Before sectioning, brains were embedded in 4% agarose. Coronal or sagittal sections (50 μm) were cut with a Vibratome (Leica VT1000 S) and stored in 0.4% PFA at 4°C. Animal experiments and housing were in accordance with the guidelines of the Federation of European Laboratory Animal Science Associations, approved by the local ethics committee.
Histology and Antibody Staining. Forebrain anatomy was analyzed by using Nissl (cresyl violet) staining. Vibratome sections were mounted and dried on Superfrost plus slides (Langenbrinck, Emmendingen, Germany). Slides were rinsed with water followed by incubation for 10 min in 0.1% cresyl violet. Sections were then washed with water, dehydrated in a graded series of alcohol/water solutions, and mounted under coverslips. All antibody stainings, except for actin staining, were performed on floating Vibratome sections. Primary antibodies were applied overnight at 4°C. Immunofluorescence (Cy2-, Cy3-coupled secondary antibodies; Dianova, Hamburg, Germany) and immunohistochemistry (Vectastain ABC system and peroxidase substrates diaminobenzidine, VectorVip, or NovaRed; Vector Laboratories) applied according to the manufacturers' protocols. Anti-actin staining was done on ultrathin sections (100 nm). Here, the desired brain regions were dissected from 150-μm Vibratome sections (2-mm diameter) and embedded in Lowicryl K11M. After trimming, 100-nm sections were cut and stained with antibody. Primary antibodies used for immunohistochemistry: monoclonal anti-actin (pan-actin) (Cedarlane Laboratories, 1:250), monoclonal anti-BrdUrd (Roche, 1:1,000), polyclonal anti-Cre (Covance Research Products, 1:3,000), polyclonal anti-doublecortin (Santa Cruz Biotechnology, 1:1,000), monoclonal anti-gelsolin (Transduction Laboratories, 1:250), and polyclonal anti-SRF (Santa Cruz Biotechnology, 1:2,500).
Preparative Brain Microdissection and Western Blotting. Forebrain, cerebral cortex, or hippocampus tissue was dissected into PBS and snap-frozen in liquid nitrogen. For microdissection of dentate gyrus, freshly dissected brains were incubated in a 10-fold volume of RNAlater solution (25 mM sodium citrate, pH 5.2/10 mM EDTA and 70 g of ammonium sulfate/100 ml) overnight at 4°C. The brains were then cut into 150-μm sections in RNAlater solution by using a Vibratome. From hippocampal sections, dentate gyri were microdissected into RNAlater solution, washed in PBS, and snap-frozen in liquid nitrogen. For Western blotting of protein extracts (36), tissues were homogenized (Polytron, Kinematica) in lysis buffer [10 mM Tris·HCl, pH 7.2/50 mM NaCl/30 mM Na4P2O7/0.005 mM ZnCl2/0.06 mM 2-mercaptoethanol/1% (vol/vol) Triton X-100] with 1× protease inhibitors (Complete tablets, Roche). Western blotting was done with overnight incubations at 4°C with antisera for actin (2 μg of forebrain or hippocampus extracts, probed with monoclonal anti-actin (Cedarlane Laboratories, 1:1,000) and monoclonal anti-GAPDH (HyTest, 1:20,000)) or gelsolin [5 μg of microdissected dentate gyrus extract, probed with polyclonal anti-gelsolin (1:5,000, generous gift of W. Witke, European Molecular Biology Laboratory, Monterotondo) and monoclonal anti α-tubulin (1:1,000, Sigma)]. Relative actin and gelsolin levels (±SD) were calculated from comparisons of four and three control mice versus four and three mutant mice, respectively. Phosphocofilin was Western blotted by using 50 μg of protein extracts from cerebral cortex (n = 4), hippocampus (n = 6), and liver (n = 2) and a polyclonal anti-phosphocofilin antiserum (1:1,000, Cell Signaling). Total cofilin was quantitated by stripping the same membranes and re-probing with a polyclonal anti-cofilin antiserum (1:500, Cell Signaling Technology), and monoclonal anti-GAPDH antibody.
Real-Time PCR. For total RNA preparations from hippocampus, tissue was dissected and stored in RNAlater solution at 4°C until Polytron disruption and RNA preparation (RNeasy Kit, Qiagen) was performed. Plasmids SRF-VP16 and SRFΔM-VP16 were described in ref. 8. The ES cell line 100 Srf(-/-) was cultured as described in ref. 36. Transient transfection of ES cells was performed with Lipofectamine 2000 (Invitrogen) (8). Total RNA preparation, cDNA synthesis, quantitative PCR using SYBR Green technology (PerkinElmer), and primer sequences for Hprt (hypoxanthine-guanine phosphoribosyltransferase) mRNA expression used for normalization, were described in ref. 36.
BrdUrd Labeling. Eleven-day-old SrfCamKIIα-iCre pups and controls were injected i.p. with a single dose of BrdUrd (50 μg/kg, Roche) and killed at P17. Preparation of brains, fixation, and sectioning were done as described above. Anti-BrdUrd antibody staining visualized BrdUrd-labeled cells.
Matrigel Culture. Culturing of subventricular zone (SVZ) explants was as described in ref. 37.
Results
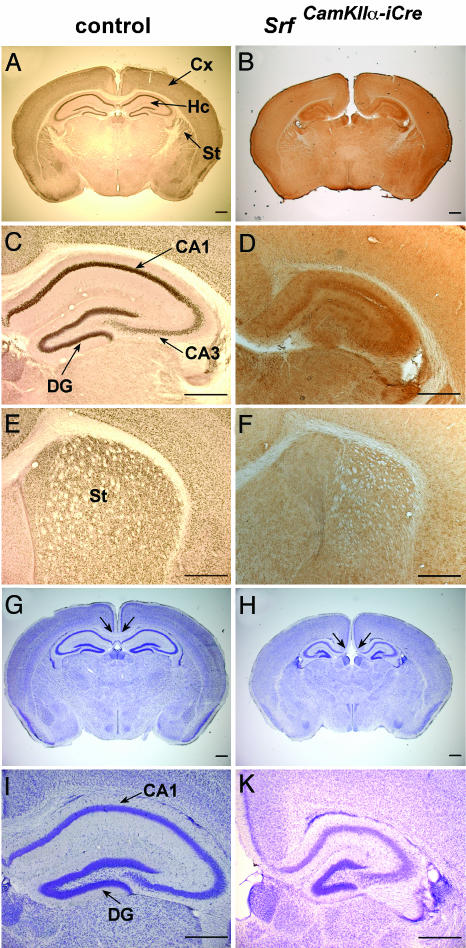
Molecular Characteristics and Neuroanatomical Abnormalities of SrfCamKIIα-iCre Mice. To investigate the role of SRF in murine neuronal migration, we deleted the “floxed” Srf-flex1neo allele (34) by breeding with mice expressing the CamKIIα-iCre transgene (35). The resulting SrfCamKIIα-iCre mutant mice express the Cre recombinase shortly before birth and thereafter in forebrain neurons of the cortex, hippocampus, and striatum (ref. 35 and data not shown), where the SRF protein is also prominently expressed (Fig. 1 A, C, and E). Cre-expressing neuronal cells display the somatic Srf(lx/lx) recombination phenotype (not shown) and the resulting extensive loss of SRF protein (Fig. 1 B, D, and F). Mutant SrfCamKIIα-iCre mice were obtained at Mendelian frequency.
Fig. 1.
Ablation of SRF in forebrains of SrfCamKIIα-iCre mice causes neuroanatomical defects. Coronal forebrain sections of P17 control (A, C, E, G, and I) and SrfCamKIIα-iCre (B, D, F, H, and K) mice stained with an anti-SRF antibody (A-F) or with Nissl stain (G-K). Only control animals display strong nuclear staining of SRF protein in the CA1, CA3, and dentate gyrus (DG) regions of the hippocampus (C versus D) or in the striatum (E versus F). Nissl staining reveals a missing corpus callosum (indicated by arrows) (G versus H) and a deformed hippocampus in SrfCamKIIα-iCre mice (I versus K). (Scale bars, 500 μm.) Cx, cortex; Hc, hippocampus; St, striatum.
Phenotypic abnormalities of neonatal SrfCamKIIα-iCre mice were readily apparent as early as P2 to P5, and they included a pronounced atactic locomotor behavior leading to severe balance impairments, lack of interest in feeding, reduced body size and weight, and, finally, death at ≈P21 (100% penetrance; n > 50). However, assisted feeding permitted the SrfCamKIIα-iCre animals to survive beyond P55, albeit body weight remained stably low at 5-8 g, compared with 20-25 g of weight gained by control littermates at P55 (not shown). Nissl staining of sections containing the hippocampus revealed several morphological defects in mutant mice. The corpus callosum was absent at the shown posterior level of the forebrain but present more anteriorly (compare Fig. 1 G and H with Fig. 2 A and B). As expected from the late prenatal Cre expression, cortical migration appeared unaffected, giving rise to a normally structured neocortex. In normal forebrain development, cortical migration and neocortex development are completed at embryonic day 18, whereas the development of the hippocampus continues at postnatal stages. In contrast with the normal neocortical appearance in mutant mice, the hippocampus was compacted and significantly reduced in size [58 ± 3.6% (SD)], as compared with the hippocampi in control mice (100%) (Fig. 1 G and H), whereby the sizes of respective hippocampi were normalized to the corresponding overall brain sizes. The dentate gyrus was small and the hilus appeared widened and densely filled with ectopic cells (Fig. 1 I and K). Therefore, in SrfCamKIIα-iCre mice, distortion of the hippocampus suggested a disturbed neuronal migration of precursor cells destined to the dentate gyrus but not of the earlier-formed interneurons destined to the cortex.
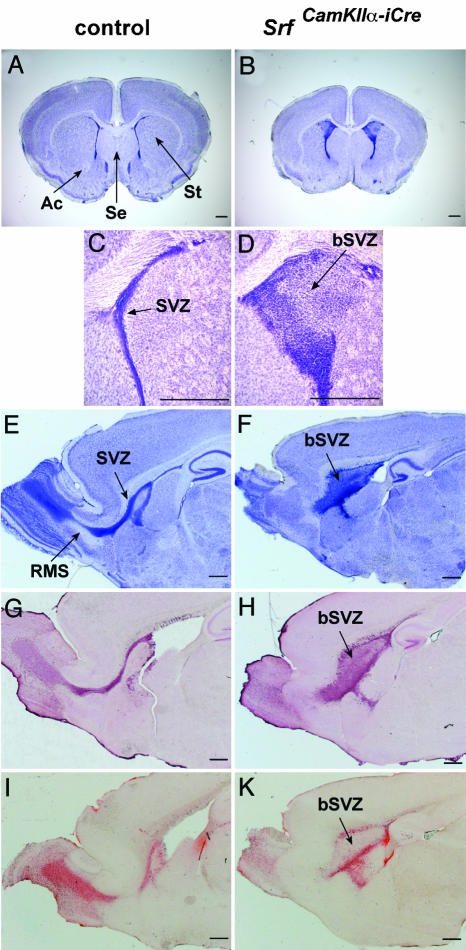
Fig. 2.
SRF-deficient forebrains display cellular accumulation at the SVZ, paralleled by impaired tangential chain migration along the RMS. (A and B) Nissl-stained coronal sections at the striatal plane show abnormal morphology of striatum (St), lateral septum (Se), anterior commissures (Ac), and SVZ in mutant mice. (C and D) Close-up view of the severely broadened SVZ (bSVZ) in brains of SrfCamKIIα-iCre mice compared with the SVZ of control mice. (E and F) Nissl-stained sagittal sections showing the SVZ of control and the bSVZ of mutant mice. The RMS is clearly visible in control mice. (G and H) Staining with an anti-doublecortin antiserum demonstrates a striking retention of neuroblasts in the bSVZ of SrfCamKIIα-iCre mutants. (I and K) BrdUrd labeling reveals an apparent impairment of the ability of neuroblasts born in the SVZ to migrate into the olfactory bulb. (Scale bars, 500 μm.) Ac, anterior commissure; Se, lateral septum; St, striatum.
SRF Deficiency Leads to Accumulation of Cells in the SVZ and Arrests Chain Migration Along the RMS. Nissl staining of anterior sections of P17 brains identified further morphological abnormalities in the SrfCamKIIα-iCre CNS. A relative enlargement of the lateral septum, a size reduction of the striatum, absence of the anterior commissure (Fig. 2 A and B) and, strikingly, a severe broadening of the SVZ (termed bSVZ) (Fig. 2 C and D) (100% penetrance; n > 50) were apparent. During development, the bSVZ was first detectable at P0, and it remained at all postnatal ages investigated, including P57 (Fig. 6 I and K, and Supporting Text, which are published as supporting information on the PNAS web site). The cells deposited inside the bSVZ were of different type, consisting primarily of GABAergic neurons and activated astrocytes (Fig. 6 A-D). Both TUNEL analysis and staining of activated caspase 3 revealed significant levels of apoptotic cells in the bSVZ, whereas neighboring regions did not display obvious changes in the degree of cell death (Fig. 6 E-H). The observed ectopic accumulation of cells in the bSVZ (heterotopia) indicated severely impaired migration of newborn cells out of the SVZ. In postnatal mice, SVZ-derived neuroblasts migrate tangentially through the RMS into the olfactory bulb, where they differentiate into interneurons. Nissl staining of sagittal brain sections of mutant P17 animals revealed abnormal broadening of the caudal segment of the RMS, whereas the rostral part, which links olfactory bulb and SVZ, was missing (Fig. 2 E and F). A similar pattern was observed by doublecortin staining of sagittal sections. Doublecortin is generally expressed in cells with a migratory activation status (38). Here, a string of immature neurons in a thin SVZ and in the olfactory bulb of control animals was labeled (Fig. 2G), whereas mutants showed a striking retention of doublecortin-positive cells in the bSVZ (Fig. 2H). Accumulation of these cells in the mutant bSVZ provides further evidence for impaired neuroblast migration. The retention of neuroblasts correlated directly with an apparent inability of newborn precursor cells to migrate from their site of origin in the SVZ into the olfactory bulb, as revealed by BrdUrd labeling of P17 animals (Fig. 2 I and K).
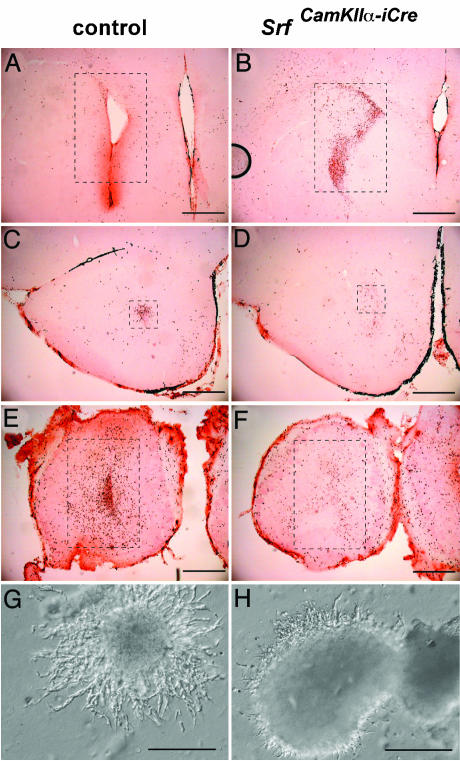
Posterior-to-anterior serial coronal sectioning of BrdUrd-labeled P17 control animals revealed only a few stained cells remaining in the SVZ (12%; Fig. 3A), some migrating cells in the RMS (9.2%; Fig. 3C), but most of the labeled cells accumulated at their destination inside the olfactory bulb (78.8%; Fig. 3E). In contrast, corresponding sections of BrdUrd-labeled mutant mice revealed the majority of labeled cells being retained in the bSVZ (65.8%; Fig. 3B), with few cells in the RMS (4.2%; Fig. 3D) and only one-third of the total cells in the olfactory bulb (30%; Fig. 3F). This result clearly demonstrates an impairment of chain migration along the RMS in SRF mutants and reveals an essential role for SRF-mediated transcriptional activities in neuronal migration.
Fig. 3.
BrdUrd labeling and Matrigel assays demonstrate a defect in neuronal cell migration. (A-F) Three posterior-to-anterior serial coronal sections each of control (A, C, and E) and mutant (B, D, and F) brains were stained with anti-BrdUrd antibody. Labeled cells inside the dashed rectangles were counted (sums of A, C, and E and B, D, and F were each taken as 100%, respectively) and cell numbers in each individual frame are expressed as percentages of the total number of labeled cells per corresponding genotype. Shown are brain sections from the region of the SVZ/bSVZ (12% and 65.8%) (A and B), the region of the RMS (9.2% and 4.2%) (C and D), and the olfactory bulb (78.8% and 30%) (E and F). Note that in brains of mutant mice the majority of labeled cells are still retained in the bSVZ. (G and H) Matrigel assays using cell aggregates dissected from the lateral wall of the lateral ventricles from P6 control (G) and mutant (H) mice, demonstrating impairment in neuronal chain migration in mutant tissue. (Scale bars, 500 μmin A-F and 200 μmin G and H.)
SRF-Deficient bSVZ Cells Exhibit a Cell-Autonomous Migration Defect. To address whether a cell-autonomous defect was responsible for the observed migration defect, we performed Matrigel assays in which chain migration of SVZ explants can be explored in vitro (37). Cell aggregates taken from the bSVZ of P6 SrfCamKIIα-iCre mice displayed a strong impairment in neuronal chain migration, as compared with control aggregates taken from the SVZ area (Fig. 3 G and H). Migratory activities depend on dynamic changes in the actin microfilament network (3). Because actin genes are direct SRF target genes (39, 40), we investigated F-actin microfilament architecture by using actin immunohistochemical staining of hippocampal (Fig. 4 A and B) or striatal (Fig. 4 C and D) ultrathin sections, thereby revealing markedly reduced F-actin fiber densities of mutant microfilaments. Quantitation by Western blotting of β-actin protein levels in mutant forebrains measured a 40% (±5% SD) reduction compared with controls (Fig. 4G). Therefore, the observed neuronal migration impairment in SRF-deficient forebrains is paralleled by significant alterations in the actin cytoskeleton.
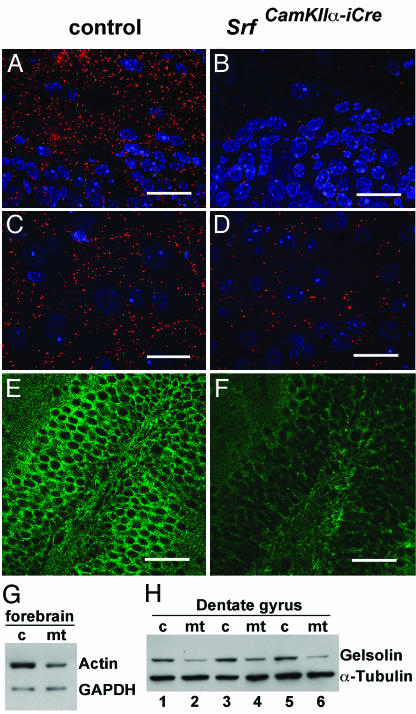
Fig. 4.
Detection of actin and gelsolin proteins by immunohistochemistry and Western blotting. (A-D) Actin stainings (red) were performed on ultrathin Lowicryl sections (100 nm) of dentate gyrus regions (A and B) or the striatum (C and D). Because of the thinness of sections, only F-actin is detected. Nuclei are stained blue. The sections used in C and D are derived from striatal regions also investigated in Fig. 2 (C and D). (E and F) Staining of dentate gyrus regions with an anti-gelsolin antibody. (G) Western blot with 2 μg of forebrain protein extracts of control (c) and mutant (mt) mice probed with an anti-actin antibody. (H) Western blot with 5 μg of protein extracts from microdissected dentate gyrus of three control (c) and three mutant (mt) mice. Comparable loading of proteins in G and H is confirmed by GAPDH or α-tubulin staining. (Scale bars, 20 μmin A-D and 50 μmin E and F.)
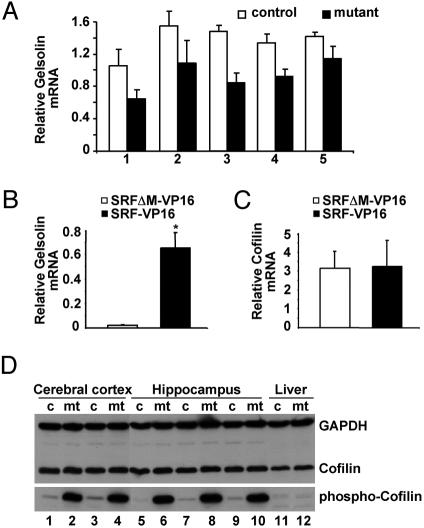
Inhibitory Phosphorylation of Cofilin at Ser-3 Is Dramatically Enhanced in Forebrains of SRF-Deficient Mice. Gelsolin and cofilin are actin-severing proteins that regulate cell migration (23). Both gene expression profiling on SRF-deficient ES cells (41) and preliminary microarray expression profiling performed on SrfCamKIIα-iCre and control mice (T.L., unpublished data) identified gelsolin as a gene down-regulated upon SRF depletion. Gelsolin influences microfilament remodeling by severing and capping actin (42). Gelsolin immunostaining (Fig. 4 E and F), Western blotting (Fig. 4H), and mRNA quantitation by real-time PCR (Fig. 5A) confirmed reduced gelsolin expression in the mutant hippocampus. Anti-gelsolin Western blots revealed a 46% (±17% SD) reduction of gelsolin protein in SRF-deficient dentate gyrus regions compared with controls (Fig. 4H). Supporting this idea of SRF-dependent gelsolin expression, in Srf(-/-) murine ES cells (8), the constitutively active SRF variant SRF-VP16, but not the DNA-binding-deficient SRFΔM-VP16 mutant protein, was able to stimulate gelsolin expression 30-fold (Fig. 5B). We also investigated mRNA expression of additional genes affecting actin turnover, including ADF, Cofilin-1, Profilin-1, and Profilin-2, but we did not detect significant alterations (not shown). Similarly, SRF-VP16 did not stimulate Cofilin-1 mRNA expression in Srf(-/-) murine ES cells (Fig. 5C).
Fig. 5.
SRF deficiency causes reduced expression of gelsolin mRNA and elevated inhibitory phosphorylation of cofilin at Ser-3. (A) Pairwise littermate comparison of relative gelsolin mRNA expression as determined by real-time PCR in the hippocampus of five control and five mutant mice. Each column represents the mean of three different cDNA syntheses, followed by PCR. Error bars correspond to standard deviation. (B and C) Real-time PCR analysis of gelsolin (B) and cofilin (C) mRNA expression in 100 Srf(-/-) ES cells, transiently transfected with SRF-VP16 or SRFΔM-VP16 expression plasmids. Primer sequences are given in Supporting Text. RNA was prepared and analyzed 72 h after transfection (*, P < 0.05, Student's t test). (D) Western blot with 50 μg of protein extracts from control (c) and mutant (mt) animals, derived from cerebral cortex (n = 4), hippocampus (n = 6), and liver (n = 2), probed with antibodies for cofilin and phosphocofilin. Comparable loading of protein is confirmed by GAPDH staining.
At the posttranslational level, phosphorylation of cofilin at Ser-3 represents a key regulatory mechanism to inactivate cofilin and thereby stabilize actin filaments. We wondered whether this mechanism was subject to SRF control and therefore examined the cofilin phosphorylation status in forebrains of wild-type and SRF-deficient mice. Overall cofilin protein levels were unaffected in cortex or hippocampus, as judged by immunohistochemistry (not shown) or Western blotting (Fig. 5D) In striking contrast, however, cofilin phosphorylation at Ser-3 was dramatically increased in mutant brain extracts (Fig. 5D). No difference in cofilin-1 phosphorylation was found in liver tissue of mutant animals (Fig. 5D), where the CamKIIα-Cre transgene is inactive (not shown). This observation demonstrates that the transcription factor SRF is required for posttranslational regulation of cofilin activity.
Together these results reveal that the two F-actin-severing proteins gelsolin and cofilin, which represent two key regulators of cell motility, are down-regulated at the functional level in SRF-deficient neuronal cells. In light of the additionally reduced actin expression levels and the accompanying lowered F-actin density, a key contribution of SRF in determining balanced dynamic changes of the actin filament is revealed.
Discussion
During mouse forebrain development, both radial and tangential modes of cell migration ensure formation of a neuroanatomically defined brain architecture (43-47). We identify here the transcription factor SRF as a major determinant for neuronal migratory activities in the developing mouse forebrain. In our genetic system, chain migration from the SVZ to the olfactory bulb along the RMS is demonstrated here to be defective in the SrfCamKIIα-iCre forebrain, leading to ectopic cell accumulations (bSVZ). Elevated apoptotic activity was observed within the bSVZ but not elsewhere in the forebrain. In addition, we also observed impaired migration of neuroblasts in the hippocampus. Here we noted impaired neuroblast migration out of the subgranular layer, another proliferative zone in the forebrain (data not shown).
In addition to reduced actin expression in SRF-deficient neurons of the hippocampus and the striatum, the down-regulation of hippocampal gelsolin gene expression, together with the strikingly induced inhibitory phosphorylation of cofilin, identify two effectors of cytoskeletal actin turnover to be under SRF control. Gelsolin expression is reduced in SRF-deficient hippocampal neurons at both the mRNA and protein levels and, in support, in SRF-deficient ES cells gelsolin mRNA can be induced by constitutively active SRF-VP16 (Fig. 5B). Cofilin mRNA levels were not affected in SRF-deficient brain regions (S.A., unpublished data), nor could SRF-VP16 induce the gene in SRF-deficient ES cells (Fig. 5C). Importantly, however, at the posttranslational level, cofilin was strongly phosphorylated at Ser-3 in SRF-deficient brain extracts. Phosphorylation of cofilin at Ser-3, known to impair the ability of cofilin both to bind G-actin and to contribute to polymer turnover (48), is mediated by kinases such as LIMK (26, 27, 49, 50).
Collectively, our data demonstrate that in SRF-deficient brain cells reduced levels of expressed and polymerized actin are accompanied by functional down-regulation of two F-actin-severing factors, gelsolin and cofilin. In their capacity to influence the dynamic cycling of actin, both these factors regulate under normal circumstances migratory cell activity (23-25). While this may, in part, explain impaired migration of SRF-deficient cells, we do not yet know the mechanisms by which SRF deficiency results in elevated cofilin phosphorylation. Further work is needed to determine whether the activity of the cofilin kinases is stimulated ectopically by upstream activators at the levels of RhoA/Rac/Cdc42 activity or whether activities of either the slingshot or the chronophin protein phosphatase of cofilin (28, 29) is reduced in the absence of SRF. The reduced density of actin fibers seen in SRF-deficient neurons might impair slingshot activation, which is stimulated under normal circumstances by association with actin filaments (51). It will also be of interest to analyze whether SRF deficiency influences sequestration/stabilization of phosphocofilin by 14-3-3 proteins (51). In the light of our data, a regulatory feedback mechanism is indicated that links SRF function to the balanced activities of cofilin, gelsolin, and G-actin for microfilament turnover during cell migration. Our data highlight the importance of transcriptional control mechanisms, specifically SRF-regulated transcription, to determine cytoskeletal dynamics.
In addition to neuronal migration, other neuronal activities are dependent on actin-based cell motility (52), including neurite extension, branching, and sprouting, dendritic spine formation, growth cone motility, and axon guidance. The absence of both the anterior commissure and the more posteriorly localized parts of the corpus callosum, as observed in our SrfCamKIIα-iCre forebrains, might already indicate a role for SRF in axon guidance. Therefore, given the central involvement of SRF in determining actin dynamics, it is tempting to speculate that SRF might play a widespread role in many motility-based brain functions, in both normal and pathological circumstances (53).
Supplementary Material
Acknowledgments
We greatly appreciate excellent technical support by S. Herberich and C. Krüger and useful comments on the manuscript by B. Knöll. We thank W. Witke (European Molecular Biology Laboratory, Monterotondo) for anti-gelsolin antisera and acknowledge insightful discussions with F. Haiss. A.N. was financed by the Deutsche Forschungsgemeinschaft (120/12-1 and SFB446/B7) and the Fonds der Chemischen Industrie. M.F. and O.K. were supported by the Deutsche Forschungsgemeinschaft (SFB505 and TR-3). U.P. received a Ph.D. scholarship from the Boehringer Ingelheim Fonds.
Author contributions: S.A., S.M.K., O.K., U.P., T.L., H.S., M.F., G.S., and A.N. designed research; S.A., S.M.K., O.K., U.P., T.L., and H.S. performed research; S.A., E.C., and F.F.W. contributed new reagents/analytic tools; S.A., S.M.K., O.K., U.P., T.L., H.S., M.F., G.S., and A.N. analyzed data; and S.A., O.K., M.F., G.S., and A.N. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RMS, rostral migratory stream; SRF, serum response factor; ES, embryonic stem; Pn, postnatal day n; SVZ, subventricular zone; bSVZ, broadened SVZ.
References
- 1.Marin, O. & Rubenstein, J. L. (2003) Annu. Rev. Neurosci. 26, 441-483. [DOI] [PubMed] [Google Scholar]
- 2.Gupta, A., Tsai, L. H. & Wynshaw-Boris, A. (2002) Nat. Rev. Genet. 3, 342-355. [DOI] [PubMed] [Google Scholar]
- 3.Ridley, A. J., Schwartz, M. A., Burridge, K., Firtel, R. A., Ginsberg, M. H., Borisy, G., Parsons, J. T. & Horwitz, A. R. (2003) Science 302, 1704-1709. [DOI] [PubMed] [Google Scholar]
- 4.Hatten, M. E. (2002) Science 297, 1660-1663. [DOI] [PubMed] [Google Scholar]
- 5.Hack, I., Bancila, M., Loulier, K., Carroll, P. & Cremer, H. (2002) Nat. Neurosci. 5, 939-945. [DOI] [PubMed] [Google Scholar]
- 6.Norman, C., Runswick, M., Pollock, R. & Treisman, R. (1988) Cell 55, 989-1003. [DOI] [PubMed] [Google Scholar]
- 7.Miano, J. M. (2003) J. Mol. Cell Cardiol. 35, 577-593. [DOI] [PubMed] [Google Scholar]
- 8.Schratt, G., Philippar, U., Berger, J., Schwarz, H., Heidenreich, O. & Nordheim, A. (2002) J. Cell Biol. 156, 737-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arsenian, S., Weinhold, B., Oelgeschlager, M., Ruther, U. & Nordheim, A. (1998) EMBO J. 17, 6289-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchwalter, G., Gross, C. & Wasylyk, B. (2004) Gene 324, 1-14. [DOI] [PubMed] [Google Scholar]
- 11.Shaw, P. E., Schroter, H. & Nordheim, A. (1989) Cell 56, 563-572. [DOI] [PubMed] [Google Scholar]
- 12.Miralles, F., Posern, G., Zaromytidou, A. I. & Treisman, R. (2003) Cell 113, 329-342. [DOI] [PubMed] [Google Scholar]
- 13.Wang, D., Chang, P. S., Wang, Z., Sutherland, L., Richardson, J. A., Small, E., Krieg, P. A. & Olson, E. N. (2001) Cell 105, 851-862. [DOI] [PubMed] [Google Scholar]
- 14.Chang, D. F., Belaguli, N. S., Iyer, D., Roberts, W. B., Wu, S. P., Dong, X. R., Marx, J. G., Moore, M. S., Beckerle, M. C., Majesky, M. W. & Schwartz, R. J. (2003) Dev. Cell 4, 107-118. [DOI] [PubMed] [Google Scholar]
- 15.Gille, H., Sharrocks, A. D. & Shaw, P. E. (1992) Nature 358, 414-417. [DOI] [PubMed] [Google Scholar]
- 16.Hill, C. S., Wynne, J. & Treisman, R. (1995) Cell 81, 1159-1170. [DOI] [PubMed] [Google Scholar]
- 17.Misra, R. P., Bonni, A., Miranti, C. K., Rivera, V. M., Sheng, M. & Greenberg, M. E. (1994) J. Biol. Chem. 269, 25483-25493. [PubMed] [Google Scholar]
- 18.Gineitis, D. & Treisman, R. (2001) J. Biol. Chem. 276, 24531-24539. [DOI] [PubMed] [Google Scholar]
- 19.Schratt, G., Philippar, U., Hockemeyer, D., Schwarz, H., Alberti, S. & Nordheim, A. (2004) EMBO J. 23, 1834-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Somogyi, K. & Rorth, P. (2004) Dev. Cell 7, 85-93. [DOI] [PubMed] [Google Scholar]
- 21.Han, Z., Li, X., Wu, J. & Olson, E. N. (2004) Proc. Natl. Acad. Sci. USA 101, 12567-12572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raftopoulou, M. & Hall, A. (2004) Dev. Biol. 265, 23-32. [DOI] [PubMed] [Google Scholar]
- 23.Southwick, F. S. (2000) Proc. Natl. Acad. Sci. USA 97, 6936-6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh, M., Song, X., Mouneimne, G., Sidani, M., Lawrence, D. S. & Condeelis, J. S. (2004) Science 304, 743-746. [DOI] [PubMed] [Google Scholar]
- 25.Azuma, T., Witke, W., Stossel, T. P., Hartwig, J. H. & Kwiatkowski, D. J. (1998) EMBO J. 17, 1362-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang, N., Higuchi, O., Ohashi, K., Nagata, K., Wada, A., Kangawa, K., Nishida, E. & Mizuno, K. (1998) Nature 393, 809-812. [DOI] [PubMed] [Google Scholar]
- 27.Arber, S., Barbayannis, F. A., Hanser, H., Schneider, C., Stanyon, C. A., Bernard, O. & Caroni, P. (1998) Nature 393, 805-809. [DOI] [PubMed] [Google Scholar]
- 28.Niwa, R., Nagata-Ohashi, K., Takeichi, M., Mizuno, K. & Uemura, T. (2002) Cell 108, 233-246. [DOI] [PubMed] [Google Scholar]
- 29.Gohla, A., Birkenfeld, J. & Bokoch, G. M. (2005) Nat. Cell Biol. 7, 21-29. [DOI] [PubMed] [Google Scholar]
- 30.Sotiropoulos, A., Gineitis, D., Copeland, J. & Treisman, R. (1999) Cell 98, 159-169. [DOI] [PubMed] [Google Scholar]
- 31.Parlakian, A., Tuil, D., Hamard, G., Tavernier, G., Hentzen, D., Concordet, J. P., Paulin, D., Li, Z. & Daegelen, D. (2004) Mol. Cell. Biol. 24, 5281-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, S., Czubryt, M. P., McAnally, J., Bassel-Duby, R., Richardson, J. A., Wiebel, F. F., Nordheim, A. & Olson, E. N. (2005) Proc. Natl. Acad. Sci. USA 102, 1082-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miano, J. M., Ramanan, N., Georger, M. A., de Mesy Bentley, K. L., Emerson, R. L., Balza, R. O., Jr., Xiao, Q., Weiler, H., Ginty, D. D. & Misra, R. P. (2004) Proc. Natl. Acad. Sci. USA 101, 17132-17137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiebel, F. F., Rennekampff, V., Vintersten, K. & Nordheim, A. (2002) Genesis 32, 124-126. [DOI] [PubMed] [Google Scholar]
- 35.Casanova, E., Fehsenfeld, S., Mantamadiotis, T., Lemberger, T., Greiner, E., Stewart, A. F. & Schutz, G. (2001) Genesis 31, 37-42. [DOI] [PubMed] [Google Scholar]
- 36.Weinhold, B., Schratt, G., Arsenian, S., Berger, J., Kamino, K., Schwarz, H., Ruther, U. & Nordheim, A. (2000) EMBO J. 19, 5835-5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wichterle, H., Garcia-Verdugo, J. M. & Alvarez-Buylla, A. (1997) Neuron 18, 779-791. [DOI] [PubMed] [Google Scholar]
- 38.Brown, J. P., Couillard-Despres, S., Cooper-Kuhn, C. M., Winkler, J., Aigner, L. & Kuhn, H. G. (2003) J. Comp. Neurol. 467, 1-10. [DOI] [PubMed] [Google Scholar]
- 39.Minty, A. & Kedes, L. (1986) Mol. Cell. Biol. 6, 2125-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohun, T., Garrett, N. & Treisman, R. (1987) EMBO J. 6, 667-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Philippar, U., Schratt, G., Dieterich, C., Muller, J. M., Galgoczy, P., Engel, F. B., Keating, M. T., Gertler, F., Schule, R., Vingron, M. & Nordheim, A. (2004) Mol. Cell 16, 867-880. [DOI] [PubMed] [Google Scholar]
- 42.McGough, A. M., Staiger, C. J., Min, J. K. & Simonetti, K. D. (2003) FEBS Lett. 552, 75-81. [DOI] [PubMed] [Google Scholar]
- 43.Lubbers, K., Wolff, J. R. & Frotscher, M. (1985) Neurosci. Lett. 62, 317-322. [DOI] [PubMed] [Google Scholar]
- 44.Altman, J. & Bayer, S. A. (1990) J. Comp. Neurol. 301, 365-381. [DOI] [PubMed] [Google Scholar]
- 45.Bayer, S. A. & Altman, J. (1987) Prog. Neurobiol. 29, 57-106. [DOI] [PubMed] [Google Scholar]
- 46.Brazel, C. Y., Romanko, M. J., Rothstein, R. P. & Levison, S. W. (2003) Prog. Neurobiol. 69, 49-69. [DOI] [PubMed] [Google Scholar]
- 47.Wichterle, H., Turnbull, D. H., Nery, S., Fishell, G. & Alvarez-Buylla, A. (2001) Development (Cambridge, U.K.) 128, 3759-3771. [DOI] [PubMed] [Google Scholar]
- 48.Morgan, T. E., Lockerbie, R. O., Minamide, L. S., Browning, M. D. & Bamburg, J. R. (1993) J. Cell Biol. 122, 623-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng, A. K. & Robertson, E. J. (1995) Mech. Dev. 52, 187-197. [DOI] [PubMed] [Google Scholar]
- 50.Proschel, C., Blouin, M. J., Gutowski, N. J., Ludwig, R. & Noble, M. (1995) Oncogene 11, 1271-1281. [PubMed] [Google Scholar]
- 51.Nagata-Ohashi, K., Ohta, Y., Goto, K., Chiba, S., Mori, R., Nishita, M., Ohashi, K., Kousaka, K., Iwamatsu, A., Niwa, R., et al. (2004) J. Cell Biol. 165, 465-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meyer, G. & Feldman, E. L. (2002) J. Neurochem. 83, 490-503. [DOI] [PubMed] [Google Scholar]
- 53.Feng, Y. & Walsh, C. A. (2004) Nat. Cell Biol. 6, 1034-1038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.