Abstract
Recent studies suggest that lysophosphatidic acid (LPA) and its G protein-coupled receptors (GPCRs) LPA1, LPA2, or LPA3 may play a role in the development of several types of cancers, including colorectal cancer. However, the specific receptor subtype(s) and their signal-transduction pathways responsible for LPA-induced cancer cell proliferation have not been fully elucidated. We show by specific RNA interference (RNAi) that LPA2 and LPA3 but not LPA1 are targets for LPA-induced proliferation of HCT116 and LS174T colon cancer cells. We determined that LPA-induced colon cancer cell proliferation requires the β-catenin signaling pathway, because knockdown of β-catenin by RNAi abolished LPA-induced proliferation of HCT116 cells. Moreover, LPA activates the main signaling events in the β-catenin pathway: phosphorylation of glycogen synthase kinase 3β (GSK3β), nuclear translocation of β-catenin, transcriptional activation of T cell factor (Tcf)/lymphoid-enhancer factor (Lef), and expression of target genes. Inhibition of conventional protein kinase C (cPKC) blocked the effects, suggesting its involvement in LPA-induced activation of the β-catenin pathway. Thus, LPA2 and LPA3 signal the proliferation of colon cancer cells through cPKC-mediated activation of the β-catenin pathway. These results link LPA and its GPCRs to cancer through a major oncogenic signaling pathway.
Keywords: colon cancer cell, G protein-coupled receptor
Lysophosphatidic acid (LPA; 1-acyl-2-hydroxy-sn-glycero-3-phosphate) has diverse cellular functions affecting cell proliferation, morphology, and motility (1, 2). These effects of LPA are implicated in pathophysiological processes such as wound healing, vascular diseases, and cancer progression (3-5). It has previously been demonstrated that cell-surface GPCRs mediate the cellular effects of LPA. Three closely related G protein-coupled receptors (GPCRs), LPA1, LPA2, and LPA3 (formerly EDG2, EDG4, and EDG7), have been identified as specific receptors for LPA (6-9). LPA2 and LPA3 can effectively couple to Gαq, which triggers robust calcium signaling and activates downstream PKC. LPA2 can also couple to Gα12/13. In contrast, LPA1 displays weak coupling to Gαq but robust coupling to Gαi/o and Gα12/13 (9).
Previous studies suggest that LPA and LPA receptors may play a role in the development of ovarian, prostate, breast, and head and neck cancers (5). Recently, Shida et al. (10) reported that stimulation of LPA could induce proliferation of DLD1, WiDR, and HT29 colon cancer cells. However, the signaling mechanism of LPA-induced cell proliferation in these colon cancer cells was not elucidated.
The most important signaling pathway in the etiology of colorectal cancer is the β-catenin pathway (11-13). β-Catenin is a transcriptional coactivator of T cell factor (Tcf)/lymphoid-enhancer factor (Lef) transcription factors. The stability of β-catenin is regulated by a multiprotein complex, which includes adenomatous polyposis coli (APC) and glycogen synthase kinase 3β (GSK3β), and axin. Phosphorylation of β-catenin by GSK3β targets β-catenin to ubiquitination and proteasome degradation. Thus, activation of the pathway represses β-catenin degradation, resulting in nuclear accumulation of β-catenin. In the nucleus, Tcf/Lef/β-catenin activates target genes such as c-myc and cyclin D1, which are involved in oncogenic transformation.
Through a functional screen for LPA-induced cell proliferation, we discovered that LPA promoted cell proliferation of a number of colon cancer cell lines. Notably, HCT116 and LS174T, two colon adenocarcinoma cell lines, yielded substantial responses to LPA in cell proliferation. By using RNA interference, we demonstrated that LPA2 and LPA3, but not LPA1, are responsible for the LPA-induced cell proliferation of HCT116 and LS174T. Interestingly, we observed that the LPA-induced proliferation of HCT116 cells is mediated by the β-catenin pathway.
Materials and Methods
Materials. All cell lines were obtained from American Type Culture Collection. Oligonucleotides of small interfering RNA (siRNA) for LPA receptors were synthesized by Dharmacon (Lafayette, CO). Oligonucleotides of siRNA for β-catenin were purchased from Ambion (Austin, TX). A One Solution Cell Proliferation Assay kit was purchased from Promega. A thymidine uptake 14C Cytostar-T assay kit and cell proliferation ELISA Biotrak System kit were purchased from Amersham Pharmacia. Antibodies against β-catenin and phospho-GSK3β were purchased from Cell Signaling Technology (Beverly, MA). Antibodies against c-myc, cyclin D1, actin, PKCβI, PKCβII, PKCγ, and Sam68 were purchased from Santa Cruz Biotechnology. Antibodies against PKCα and all protein kinase inhibitors were obtained from Calbiochem. LPA (1-oleoyl-2-hydroxy-sn-glycero-3-phosphate) was purchased from Avanti Polar Lipids. Hepatocyte growth factor and fatty-acid-free BSA were purchased from Sigma. TOPglow and FOPglow luciferase reporters were purchased from Upstate Biotechnology (Lake Placid, NY). Athymic nude mice were purchased from Harlan-Sprague-Dawley.
Luciferase Reporter Assay. HeLa cells or HCT116 cells were transfected with expression-vector coding for a 4× Tcf-directed luciferase reporter and β-gal with or without expression vectors for LPA receptors. After serum starvation for 16 h (HeLa cells) or 42 h (HCT116 cells), cells were stimulated by agents for 5 h. Luciferase activity was determined by a chemiluminescence imaging plate reader (CLIPR version 2.3.3, Molecular Devices) with Promega reagents. Relative luciferase activity was normalized against the activity of β-gal, determined with luminescence reagent from Clontech. All luciferase assays were performed with triplicate samples.
Quantitative Real-Time RT-PCR (qPCR). Total RNA was isolated and purified from cells by using an RNAEasy mini kit (Qiagen). In brief, qPCR was done by using 60 ng of total RNA, TaqMan RT-PCR Mastermix (Applied Biosystems), 600 nM each primer, and 200 nM FAM/TAMRA-labeled probe [designed by the program primer express (Applied Biosystems)] at 48°C for 30 min (for reverse transcription) then at 95°C for 15 s and at 60°C for 1 min (for PCR) by using a program of 40 cycles on a Prism 7700 sequence detector (Applied Biosystems). For the sequences of primers and probes, see Supporting Text, which is published as supporting information on the PNAS web site.
Cell Proliferation Assay. Cells in DMEM containing 0.1% fatty-acid-free BSA were seeded in 96-well plates at a density of 5 × 103 cells per well. The cells were then incubated for 3 d in the absence or presence of 1 μM LPA that was renewed daily. CellTiter 96 AQueous (MTS) One Solution reagent (Promega) was added to each well, and absorbance was recorded at 490 nm by using a SpectraMax Plus plate reader (softmaxpro 401, Molecular Devices). Cell numbers were then calculated by using a standard curve correlating the absorbance to the cell number counted under a microscope.
DNA Synthesis Assay. For thymidine incorporation, cells in DMEM with 0.1% fatty-acid-free BSA were seeded in 96-well scintillating microplates at a density of 5 × 103 cells per well. The cells were then treated with LPA for 24 h, and [methyl-14C]thymidine was added thereafter to each well at a final concentration of 0.5 μCi/ml (1 Ci = 37 GBq). Radioactivity was measured by using a MicroBeta 1450 liquid scintillation and luminescence counter (MicroBeta JET, Wallac, Gaithersburg, MD) after 24-h labeling. BrdUrd incorporation was conducted according to the manufacturer's instructions with the Cell Proliferation ELISA Biotrak System kit (Amersham).
Western Blot Analysis. Proteins from whole cell extracts were separated by electrophoresis in a pH 8.7 Tris·glycine/8%, 10%, or 12% polyacrylamide gel and transferred to a nitrocellulose membrane. The nitrocellulose membrane was pretreated with 5% nonfat dry milk in Tween-Tris-buffered saline (TTBS) [20 mM Tris·HCl (pH 7.5)/120 mM NaCl/0.05% Tween-20] for 1 h and incubated with primary antibody in TTBS with 5% BSA for 16 h. The membrane was then washed three times with TTBS and incubated with horseradish peroxidase-conjugated secondary antibody for 1 h. After three washes with TTBS, the bound antibody was detected by enhanced chemiluminescence (Cell Signaling Technology). To detect protein levels in the nucleus, nuclear protein extracts were prepared as described in ref. 14.
siRNA Knockdown Experiments. HCT116 or LS174T cells were transiently transfected with siRNA oligonucleotides by using Lipofectamine 2000 (Invitrogen). At least four different siRNAs against each target were tested for reduction of mRNA level, protein level, or calcium signaling by each LPA receptor. The most effective siRNA oligonucleotide for LPA1 (GAAAUGAGCGCCACCUUUA), LPA2 (GGTCAATGCTGCTGTGTAC), and LPA3 (CAGCAGGAGTTACCTTGTT) or β-catenin (siRNA ID no. 42816, Ambion) were chosen.
Xenograft. HCT116 cells were transiently transfected with siRNA oligonucleotides by using Lipofectamine 2000. The cells were harvested 18 h later, washed twice with Opti-MEM I, resuspended in Opti-MEM I, and mixed 1:1 (vol/vol) with Matrigel (BD Biosciences) at a density of 3.0 × 107 cells per ml. Athymic nude mice were injected s.c. on the right flank with 0.2 ml by using a 1-ml syringe fitted with a 27-gauge needle. Tumor size was documented by direct measurement in three perpendicular directions by using Pro-Max calipers (Fowler Instruments, Newton, MA), and tumor volumes were calculated by multiplication of these three values. The measurements were recorded as tumor volumes (mm3) from groups of 10 mice each. All experimental implantations and measurements were performed blinded by investigators.
Results
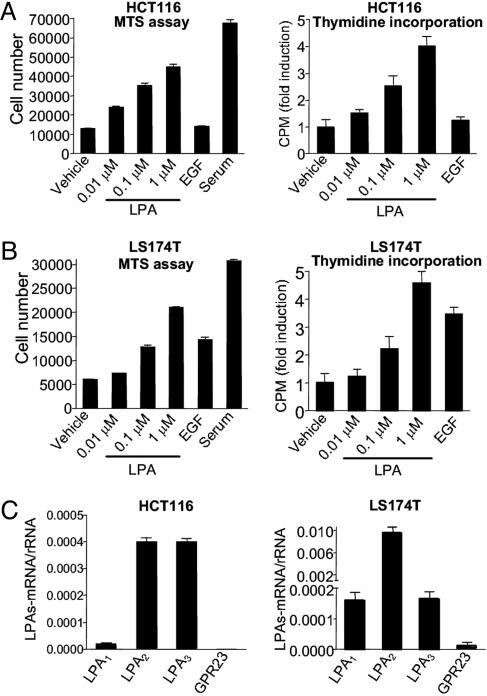
LPA Induces Proliferation of HCT116 and LS174T Colon Cancer Cells. Previous studies have demonstrated that LPA receptors LPA1, LPA2, and LPA3 are overexpressed in several types of tumors and cancer cell lines, including colon cancer cell lines (10). To further study the functional role of LPA in colon cancer cells, we first performed a screen-search for colon cancer cell lines that respond to LPA in cell-proliferation assays. Several cell lines responded to LPA to various extents, with HCT116 and LS174T showing the most robust responses. As shown in Fig. 1A, LPA treatment increased the number of HCT116 cells in a dose-dependent manner. After 3 d of treatment, cell numbers increased >3-fold in 1 μM LPA. LPA also increased DNA synthesis of HCT116 cells as shown by an increase in thymidine incorporation (Fig. 1 A Right) and BrdUrd incorporation (data not shown). Treatment of EGF did not increase the number of HCT116 cells, excluding a role of EGFR transactivation in LPA-induced cell proliferation. Stimulation of LPA also increased the proliferation of LS174T cells in a dose-dependent manner to a similar extent as that seen in HCT116 cells (Fig. 1B).
Fig. 1.
LPA induces proliferation of colon cancer cells. (A) HCT116 cells cultured in a 96-well plate in DMEM containing 0.1% BSA were treated with vehicle, various concentrations of LPA, 50 ng/ml EGF, or 2% FBS. All agents were replenished daily. (Left) Cell number after 3 d. (Right) Thymidine incorporation. (B) Cell proliferation of LS174T measured by both MTS One Solution Assay (Left) and thymidine incorporation (Right). (C) mRNA expression levels (normalized by rRNA) of LPA1, LPA2, LPA3, and GPR23 in HCT116 (Left) or LS174T cells (Right). Results presented are mean ± SEM. All figures shown are representatives from three independent experiments.
Using quantitative real-time RT-PCR (qPCR), we found that HCT116 cells express mRNA for LPA1, LPA2, and LPA3, with LPA2 and LPA3 expressed more abundantly than LPA1 (Fig. 1C Left). LS174T cells also express all three LPA receptors, with the mRNA-expressing level for LPA2 higher than those of LPA1 or LPA3 (Fig. 1C Right). qPCR analysis also shows that neither HCT116 cells nor LS174T cells express a detectable amount of GPR23, a recently described subtype of LPA receptor (15) (Fig. 1C).
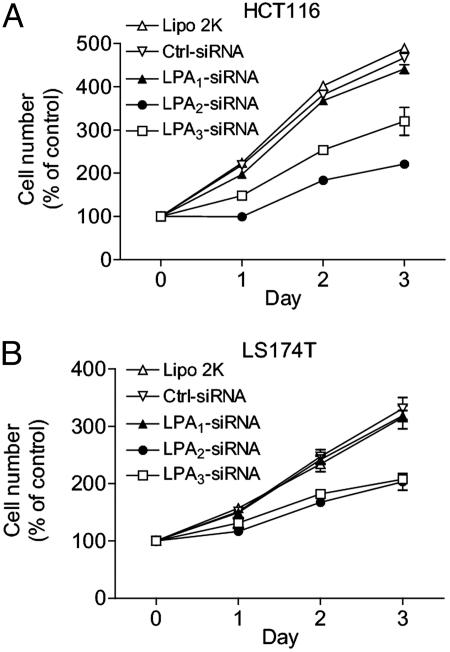
Both LPA2 and LPA3 Contribute to LPA-Induced Proliferation of HCT116 Cells. To assess which of the LPA receptors expressed on HCT116 and LS174T cells is responsible for LPA-induced cell proliferation, we used gene-specific knockdown by siRNA. Transfection of the siRNA directed toward LPA2 or LPA3 markedly diminished LPA-induced proliferation of HCT116 cells (Fig. 2A). Cotransfection of both siRNAs against LPA2 and LPA3 gave an additive effect (data not shown). Similarly, knockdown of LPA2 or LPA3 also diminished LPA-induced proliferation of LS174T cells (Fig. 2B). In both HCT116 and LS174T cells, siRNA against LPA1 did not significantly affect cell proliferation.
Fig. 2.
Effect of siRNA on LPA-induced proliferation of colon cancer cells. Cells of HCT116 (A) or LS174T (B) were transfected with different siRNA oligonucleotides (20 nM each) for 24 h, seeded in 96-well plates, and treated with vehicle or 1 μM LPA. Cell proliferation was measured by the MTS One Solution Assay at various times. Cell numbers are presented as the percentage of cell number in no-LPA-treatment controls. Results presented are mean ± SEM, and representatives of five independent experiments are shown.
The knockdown effect appeared to be target-specific because: (i) Each siRNA specifically reduced the mRNA level of its respective target without an effect on other targets (see Fig. 8A, which is published as supporting information on the PNAS web site). (ii) The knockdown effects are significant with siRNA concentrations as low as 1 nM, a concentration unlikely to generate nonspecific off-target effects (16) (Fig. 8 B and C). (iii) The knockdown effect by LPA2 siRNA can be completely rescued by hepatocyte growth factor, which activates an unrelated receptor (Fig. 8D).
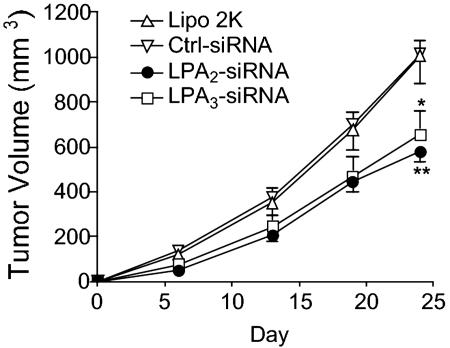
Knockdown of LPA2 or LPA3 Reduces Tumor Formation of HCT116 Cells. To examine the role LPA receptors play in tumorigenicity, we compared the effects of LPA-receptor knockdown on HCT116 cell growth in xenografts. HCT116 cells were transfected with siRNA directed toward LPA2 or LPA3 and implanted s.c. in athymic nude mice. Knockdown of LPA2 or LPA3 inhibited tumor growth (Fig. 3).
Fig. 3.
Effect of siRNA on the xenograft of HCT116 cells. HCT116 cells were transfected with siRNA oligonucleotides (20 nM each) as shown for 18 h. The cells were implanted, and tumors were measured as described in Materials and Methods. Statistical analysis was performed by using one-way ANOVA followed by Dunnett's multiple comparison test (prism 4.0, GraphPad, San Diego). *, P < 0.05 or **, P < 0.01, compared with control (Ctrl-siRNA) mice.
Signaling of LPA-Mediated Cell Proliferation of HCT116: The β-Catenin Pathway. Constitutive activation of the β-catenin pathway plays a central role in colorectal carcinogenesis (17, 18). Mutations of several components along this pathway increase the stability and nuclear translocation of β-catenin, which then acts with Tcf/Lef transcription factors to activate target genes. Many of the target genes control the process of cell proliferation (19, 20).
To investigate whether the β-catenin pathway plays a role in LPA-induced proliferation of colon cancer cells, we tested whether reduction of β-catenin protein by siRNA knockdown would block the LPA-induced proliferation of HCT116 cells.
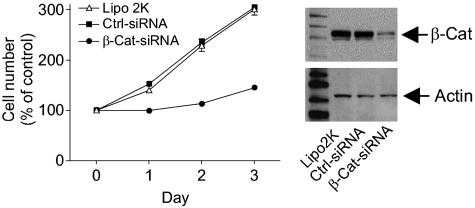
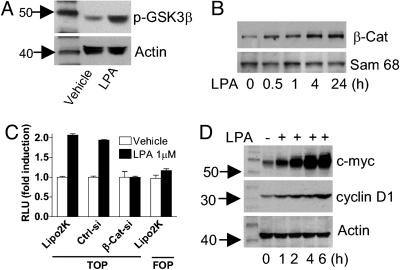
β-Catenin siRNA abolishes LPA-induced cell proliferation. Transfection of β-catenin siRNA almost abolished LPA-stimulated proliferation of HCT116 cells (Fig. 4 Left). This effect was dose-dependent, with the inhibitory effect readily seen at 1 nM β-catenin siRNA (see Fig. 9, which is published as supporting information on the PNAS web site). After 48 h, the cells were collected and analyzed for the expression of β-catenin protein. We observed a concomitant reduction in β-catenin by Western blot, correlating with the decrease of cell proliferation (Fig. 9). Importantly, cell proliferation in response to 10% FBS was unaffected by β-catenin siRNA (data not shown). These data suggest that β-catenin is required for the LPA-activated proliferation of HCT116 cells.
Fig. 4.
Effect of β-catenin siRNA on LPA-induced cell proliferation. HCT116 cells were transfected with Lipofectamine 2000 reagent (Lipo 2K), control siRNA (Ctrl-siRNA, 20 nM), or β-catenin siRNA (β-Cat-siRNA, 20 nM) for 24 h. Cells were then seeded in 96-well plates in DMEM containing 0.1% BSA. Vehicle or 1 μM LPA was added daily. Cell numbers were calculated from absorbance in MTS One Solution Assay. (Left) Effect of β-catenin siRNA on cell number expressed as the percentage of cell number in no-LPA-treatment controls. (Right) aliquots of cells from day 1 were used to detect the protein levels of β-catenin or actin.
LPA activates components of the β-catenin pathway. The observation that knockdown of β-catenin abolished LPA-induced cell proliferation suggested that the β-catenin pathway is activated by LPA. We therefore investigated several key steps along this pathway, namely GSK3β phosphorylation, β-catenin nuclear translocation, and β-catenin-induced transcription.
Because phosphorylation of GSK3β at Ser-9 results in the inhibition of kinase activity (21), we examined whether LPA treatment could induce GSK3β phosphorylation at Ser-9. Indeed, Western blot analysis showed that stimulation with LPA increased phosphorylation of GSK3β (Fig. 5A), indicative of GSK3β inhibition and β-catenin pathway activation.
Fig. 5.
LPA activates β-catenin pathway and downstream target genes. (A) GSK3β phosphorylation. Serum-starved HCT116 cells were treated with vehicle or 1 μM LPA for 20 min. Proteins from whole cell lysates were separated by Tris·glycine/10% polyacrylamide gel. GSK3β was detected by a phosphospecific antibody (phospho-Ser-9) with actin as loading control. (B) Nuclear translocation of β-catenin. Serum-starved HCT116 cells were treated with 1 μM LPA, and nuclear fractions were isolated and used to detect β-catenin protein level by Western blot with nuclear protein Sam68 as a loading control. (C) Tcf reporter gene assay. HCT116 cells were transiently transfected with TOPglow or FOPglow luciferase reporter gene plasmid and expression plasmid for β-gal. Luciferase activity was measured and normalized by β-gal activity. RLU, relative light units. Data shown are fold induction (mean ± SEM) with vehicle control set as unity. For β-catenin knockdown, HCT116 cells were cotransfected with 20 nM β-catenin siRNA (β-Cat-si) or control siRNA (Ctrl-si). (D) Serum-starved HCT116 cells were stimulated by 1 μM LPA. Whole cell lysates were used to detect protein expression levels of c-myc (Top) or cyclin D1 (Middle) by specific antibodies with actin as loading control (Bottom). Results are representatives of two to four independent experiments.
To determine whether LPA can activate β-catenin-mediated transcription in HCT116 cells, we investigated whether LPA could promote the nuclear accumulation of β-catenin. Serum-starved HCT116 cells were stimulated with 1 μM LPA, and the nuclear fractions of cells were harvested for detection of β-catenin protein level at various time points. Western blot analysis revealed that LPA significantly elevated the β-catenin protein level in the nuclear fraction after 30 min of stimulation, peaking at 4 h and remaining at this level for at least 24 h (Fig. 5B).
Subsequently, we assessed whether LPA could induce Tcf/Lef/β-catenin transcriptional activation by using a reporter gene (TOPglow) containing four copies of the Tcf-binding site in HCT116 cells. As shown in Fig. 5C, LPA increased TOPglow activity without affecting the negative control FOPglow. The TOPglow activation by LPA was abolished by β-catenin siRNA (Fig. 5C), suggesting that β-catenin is a direct mediator of the LPA activation of Tcf transcription.
To examine whether LPA could increase the expression of Tcf/Lef target genes such as c-myc and cyclin D1, HCT116 cells were stimulated with LPA for various periods of time, cell lysates were prepared, and c-myc and cyclin D1 proteins were detected by Western blot. Indeed, stimulation with LPA elevated the protein expression of both c-myc and cyclin D1, which reached the highest levels after 4 h (Fig. 5D).
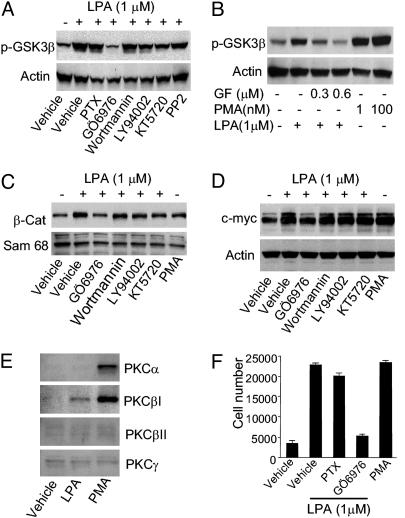
cPKC mediates LPA-induced activation of the β-catenin pathway. To determine the signal transduction between LPA receptors and the β-catenin pathway, we examined the effects of pertussis toxin (PTX) and an array of protein kinase inhibitors on LPA-induced phosphorylation of GSK3β. PTX, a specific inhibitor of Gαi/o, had no effect on LPA-induced GSK3β phosphorylation, thus excluding the involvement of Gαi/o. Inhibitors of phosphatidylinositol 3-kinase (PI3K), wortmannin or LY940024, did not attenuate phosphorylation of GSK3β induced by LPA. Addition of the protein kinase A (PKA) inhibitor KT5720 did not prevent the LPA-induced phosphorylation of GSK3β, excluding the involvement of PKA. Similarly, LPA-induced phosphorylation of GSK3β was not blocked by the tyrosine kinase inhibitor PP2, excluding the involvement of c-Src. By contrast, both a nonspecific PKC inhibitor, GF109203X (22), and a cPKC-specific inhibitor, Gö6976 (23), abolished LPA-induced GSK3β phosphorylation (Fig. 6 A and B), suggesting the involvement of calcium-dependent PKC in activating the β-catenin pathway. Moreover, the PKC activator PMA induced phosphorylation of GSK3β, further supporting the idea that PKC is a mediator leading to GSK3β phosphorylation (Fig. 6B).
Fig. 6.
cPKC is required for LPA-induced activation of the β-catenin pathway. (A) Serum-starved HCT116 cells were pretreated with pertussis toxin (PTX) (500 ng/ml) for 5 h or protein kinase inhibitors for 30 min before treatment with vehicle or 1 μM LPA for 20 min. Proteins from whole cell lysates were separated by Tris·glycine/10% polyacrylamide gel, and phosphorylated GSK3β was detected by anti-phospho-GSK3β antibody (Upper) or actin as loading control (Lower). The concentrations of inhibitors are as follows: 0.5 μMGö6976, 1 μM wortmannin, 10 μM LY94002, 0.5 μM KT5720, and 1 μM PP2. (B) A PKC inhibitor, GF109203X, was used for pretreatment for 30 min. The cells were then stimulated with LPA or phorbol 12-myristate 13-acetate (PMA) for 10 min. (Upper) The phosphorylation of GSK3β.(Lower) Actin loading control. (C) Serum-starved HCT116 cells were pretreated with various inhibitors for 30 min. Nuclear fractions of β-catenin were detected by Western blot (Upper). The nuclear protein Sam68 was used as loading control (Lower). (D) Serum-starved HCT116 cells were pretreated with various inhibitors for 30 min before treatment with vehicle, 1 μM LPA, or 100 nM PMA for 4 h. Whole cell lysates were used to detect the protein level of c-myc by Western blot (Upper) with actin as loading control (Lower). (E) Serum-starved HCT116 cells were treated with vehicle, 1 μM LPA, or 100 nM PMA for 10 min. Membrane fractions were harvested for detection of PKC isoforms by Western blot. (F) HCT116 cells cultured in DMEM containing 0.1% BSA in 96-well plates were pretreated with 100 ng/ml PTX for 12 h or 0.5 μM Gö6976 for 30 min. The cells were then treated with vehicle, 1 μM LPA, or 1 nM PMA, replenished daily. Cell numbers were measured by the One Solution Assay. Results shown are mean ± SEM of representatives of two to four independent experiments.
Independent assessment of β-catenin pathway activation by nuclear translocation resulted in a similar conclusion. As shown in Fig. 6C, cPKC inhibitor Gö6976 strongly inhibited the LPA-induced nuclear accumulation of β-catenin. In contrast, no effect was seen with phosphatidylinositol 3-kinase inhibitors (wortmannin or LY940024) or PKA inhibitor (KT5720). Through examination of β-catenin target genes, we further demonstrated that cPKC is important for LPA-induced up-regulation of key target genes such as c-myc (Fig. 6D). Moreover, the PKC activator PMA significantly increased the expression of c-myc (Fig. 6D).
To further define which isoform of cPKC is activated by LPA stimulation in HCT116 cells, we examined membrane translocation of different cPKC isoforms. HCT116 cells were serum-starved overnight, then stimulated by LPA or PMA for 10 min, membrane fractions were separated, and membrane-bound cPKC isoforms were detected by Western blot. As shown in Fig. 6E, LPA treatment increased the membrane fraction of PKCβI. PMA, on the other hand, increased the membrane fraction of both PKCα and PKCβI. Together, these results suggest that PKCβI mediates the LPA-induced phosphorylation of GSK3β, leading to the activation of the β-catenin pathway.
We then examined the role of cPKC in the LPA-induced proliferation of HCT116 cells. As shown in Fig. 6F, the addition of cPKC inhibitor Gö6976 significantly diminished LPA-induced cell proliferation. Conversely, PKC activator PMA promoted proliferation of HCT116 cells to a similar extent as LPA. These findings suggest that cPKC is a key mediator of the LPA-induced activation of the β-catenin pathway, leading to cell proliferation.
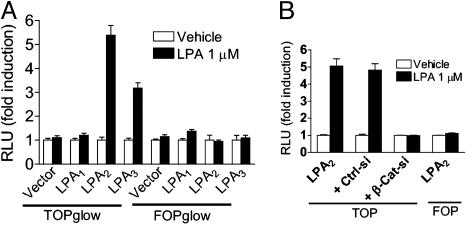
Both LPA2 and LPA3 Are Capable of Activating the β-Catenin Pathway. To determine which of the LPA receptors expressed in HCT116 and LS174T cells are capable of activating the β-catenin pathway, we examined Tcf reporter gene activation in HeLa cells after the transient transfection of each LPA receptor and Tcf reporter gene. When HeLa cells were transiently transfected with either LPA2 or LPA3, but not LPA1, LPA significantly increased Tcf luciferase reporter gene activity (Fig. 7A). Knockdown of β-catenin by siRNA completely blocked LPA2-mediated Tcf reporter gene activation (Fig. 7B). Similar results were obtained for LPA3 (data not shown). These observations indicate that the stimulation of either LPA2 or LPA3 is capable of activating the β-catenin pathway.
Fig. 7.
Both LPA2 and LPA3 can activate β-catenin pathway. (A) HeLa cells were transiently transfected with pcDNA3.1 containing empty vector, LPA1, LPA2, or LPA3. All cells were cotransfected with the TOPglow luciferase reporter gene plasmid and the β-gal expression plasmid. At 24 h, serum-starved cells were treated with vehicle or 1 μM LPA for 5 h. Tcf luciferase activity was measured and normalized against β-gal activity. (B) HeLa cells were transfected as above, some samples were transfected with 20 nM either β-catenin siRNA (β-Cat-si) or control siRNA (Ctrl-si), and luciferase activity was measured after treatment of LPA for 5 h. Data shown are fold induction with vehicle control set as unity. A representative of two independent experiments is shown.
Discussion
Recent studies suggest that LPA and LPA receptors may play a role in the development of several types of cancers (5). For colorectal cancer, Shida et al. (10) reported that LPA receptors are overexpressed in several lines of cultured colon cancer cells, and stimulation by LPA induced the proliferation of these cells. However, it is unclear which particular subtype(s) of LPA receptors mediate(s) the proliferative response of these cells. Moreover, the activated signaling pathway relevant to colon cancer cells has not been elucidated.
We report that LPA2 and LPA3, but not LPA1, are responsible for the LPA-induced proliferation of HCT116 and LS174T colon cancer cells. Furthermore, our study revealed that the LPA-induced proliferation of HCT116 cells is mediated by the β-catenin pathway. Our findings link LPA and its GPCRs to the main tumorigenic pathway in colorectal cancer.
A large body of evidence shows that the β-catenin pathway plays an important role in the development of several types of cancer, particularly colorectal cancer (17-20). In the affected cancers, genetic mutations of APC, axin, or β-catenin invariably cause stabilization and subsequent nuclear accumulation of β-catenin. In the nucleus, β-catenin binds to Tcf/Lef transcription factors, leading to the activation of target genes including c-myc and cyclin D1. In addition to constitutive activation of the β-catenin pathway caused by mutations, extracellular stimuli also activate this pathway. A canonical pathway activated by the secreted Wnt proteins binding to the Frizzled receptors has been well characterized (24, 25). Recently, accumulating evidence indicates that the β-catenin pathway can also be activated by extracellular stimuli other than Wnt. For example, signaling from receptor tyrosine kinases such as EGFR or insulin-like growth factor receptors activate this pathway through phosphatidylinositol 3-kinase (PI3K)-Akt (26, 27). More recently, it has been shown that GPCR signaling also has the potential to activate the β-catenin pathway. Regan and colleagues (28) discovered that stimulation by prostaglandin F (PGF) could activate the β-catenin pathway through PI3K in HEK293 cells overexpressing PGF receptor FP(B). Subsequently, they reported that prostaglandin E2 stimulation of the EP2 or EP4 receptor also activated β-catenin signaling in HEK293 cells stably expressing each receptor (29). Fang and colleagues (30) recently found that LPA treatment resulted in phosphorylation of GSK3 in HEK293 cells stably expressing LPA2 or LPA3. However, because phosphorylation of GSK3 is not always sufficient to activate the downstream β-catenin pathway (31, 32), that study did not draw the conclusion that LPA activates the β-catenin pathway. More importantly, none of the above studies attempted to functionally link these GPCRs and the β-catenin pathway to cancer cell proliferation. We show not only that LPA can activate various components of the β-catenin pathway in colon cancer cells but also that the pathway is required for LPA-induced proliferation of these cells. Our results implicate GPCR activation of the β-catenin pathway in the transformation of colon cancer cells where activation of this pathway plays an essential role in tumorigenesis.
Because the majority of colorectal tumors have mutations in either APC or β-catenin, their basal β-catenin activities are elevated. It remains unknown whether LPA/LPA receptors can further activate the pathway in those colorectal cancer cells that already have APC or β-catenin mutations. We noticed that the two cell lines with the most robust response to LPA, HCT116 and LS174T, have the wild-type allele for APC (33). Both HCT116 and LS174T have mutations in β-catenin at Ser-45 (heterozygous deletion in HCT116 and homozygous missense mutation Ser → Phe in LS174T). It was demonstrated by homologous deletion that this Ser-45 mutant allele was not sufficient to support tumorigenesis of HCT116 cells (34, 35). It is possible that the β-catenin pathway in some cancer cells is not fully activated by mutations and therefore can be stimulated by additional extracellular messengers such as LPA, resulting in further oncogenic transformation. Whether LPA can provide additional input to those cancer cells with mutations that activate the pathway to a higher extent requires further investigation. In addition to colon cancer, the LPA-activated β-catenin pathway may play a role in other tumor types where the β-catenin pathway plays an important role, such as endometrial ovarian cancer and certain forms of liver, prostate, and thyroid cancers (13).
Our study also revealed a signal-transduction mechanism by which GPCRs can signal to the β-catenin pathway. It is known that both LPA2 and LPA3 couple to Gαq, leading to activation of PLC-Ca2+-PKC. In contrast, LPA1 does not effectively couple to Gαq. Indeed, knockdown of either LPA2 or LPA3 but not LPA1 inhibited LPA-induced calcium flux in HCT116 cells (data not shown), supporting the proposition that LPA2 or LPA3 activates PKC in these cells. Previously, PKC has been suggested to be partially involved in a canonical Wnt signaling pathway to β-catenin activation (36, 37). However, direct evidence for Wnt-induced, PKC-mediated activation of the β-catenin pathway in colon cancer cells, particularly for the role of PKC in nuclear translocation of β-catenin and activation of target genes, is still lacking. On the other hand, a noncanonical Wnt signaling pathway, where PKC can be activated by type 2 Frizzled receptors, has not been linked to β-catenin activation (38). Very recently, activation of the Frizzled receptors by the Wnt proteins has been linked to the cAMP pathway in mouse embryos (39), supporting an earlier proposal by Malbon et al. (40) that the Frizzled receptors could function as GPCRs.
Colorectal cancer is one of the most common malignancies, occurring in a significant percentage of the population. More than 80% of sporadic and hereditary colorectal cancers may be caused by aberrations in the β-catenin pathway (17-20). Our findings identify a mechanism by which the β-catenin pathway can be modulated by GPCRs LPA2 and LPA3, resulting in the proliferation of colorectal cancer cells. Together with the observed overexpression of LPA receptors, these results provide the basis for a functional role of LPA and LPA receptors in colorectal cancer cells. Very recently, aberrant expression of LPA receptors, particularly overexpression of LPA2, has been observed in primary colorectal tumors (41). Thus, blockage of LPA2 and/or LPA3 by specific antagonists may be explored as a therapeutic or chemopreventive approach to colorectal cancer.
Supplementary Material
Acknowledgments
We thank Dr. Jeff Reagan for helpful suggestions on the manuscript and Calvin Chang for technical support.
Author contributions: T.H. and S.A. designed research; M.Y., W.W.Z., N.S., A.S., and J.Y. performed research; M.Y., A.S., and S.A. analyzed data; M.Y. and S.A. wrote the paper; and T.H. edited and commented on the manuscript.
Abbreviations: APC, adenomatous polyposis coli; cPKC, conventional protein kinase C; GPCR, G protein-coupled receptor; GSK3β, glycogen synthase kinase 3β; Lef, lymphoid-enhancer factor; LPA, lysophosphatidic acid; PMA, phorbol 12-myristate 13-acetate; siRNA, small interfering RNA; Tcf, T cell factor.
References
- 1.Moolenaar, W. H. (1999) Exp. Cell Res. 253, 230-238. [DOI] [PubMed] [Google Scholar]
- 2.Goetzl, E. J. & An, S. (1998) FASEB J. 12, 1589-1598. [PubMed] [Google Scholar]
- 3.Contos, J. J., Ishii, I. & Chun, J. (2000) Mol. Pharmacol. 58, 1188-1196. [DOI] [PubMed] [Google Scholar]
- 4.Tigyi, G. (2001) Prostaglandins 64, 47-62. [DOI] [PubMed] [Google Scholar]
- 5.Mills, G. B. & Moolenaar, W. H. (2003) Nat. Rev. Cancer 3, 582-591. [DOI] [PubMed] [Google Scholar]
- 6.Hecht, J. H., Weiner, J. A., Post, S. R. & Chun, J. (1996) J. Cell Biol. 135, 1071-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.An, S., Bleu, T., Hallmark, O. G. & Goetzl, E. J. (1998) J. Biol. Chem. 273, 7906-7910. [DOI] [PubMed] [Google Scholar]
- 8.Bandoh, K., Aoki, J., Hosono, H., Kobayashi, S., Kobayashi, T., Murakami-Murofushi, K., Tsujimoto, M., Arai, H. & Inoue, K. (1999) J. Biol. Chem. 274, 27776-27785. [DOI] [PubMed] [Google Scholar]
- 9.Chun, J., Goetzl, E. J., Hla, T., Igarashi, Y., Lynch, K. R., Moolenaar, W., Pyne, S. & Tigyi, G. (2002) Pharmacol. Rev. 54, 265-269. [DOI] [PubMed] [Google Scholar]
- 10.Shida, D., Kitayama, J., Yamaguchi, H., Okaji, Y., Tsuno, N. H., Watanabe, T., Takuwa, Y. & Nagawa, H. (2003) Cancer Res. 63, 1706-1711. [PubMed] [Google Scholar]
- 11.Kinzler, K. W. & Vogelstein, B. (1996) Cell 87, 159-170. [DOI] [PubMed] [Google Scholar]
- 12.Bienz, M. & Clevers, H. (2000) Cell 103, 311-320. [DOI] [PubMed] [Google Scholar]
- 13.Polakis, P. (2000) Genes Dev. 14, 1837-1851. [PubMed] [Google Scholar]
- 14.Yang, M., Sang, H., Rahman, A., Wu, D., Malik, A. B. & Ye, R. D. (2001) J. Immunol. 166, 6885-6892. [DOI] [PubMed] [Google Scholar]
- 15.Noguchi, K., Ishii, S. & Shimizu, T. (2003) J. Biol. Chem. 278, 25600-25606. [DOI] [PubMed] [Google Scholar]
- 16.Persengiev, S. P., Zhu, X. & Green, M. R. (2004) RNA 10, 12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morin, P. J., Sparks, A. B., Korinek, V., Barker, N., Clevers, H., Vogelstein, B. & Kinzler, K. W. (1997) Science 275, 1787-1790. [DOI] [PubMed] [Google Scholar]
- 18.Korinek, V., Barker, N., Morin, P. J., van Wichen, D., de Weger, R., Kinzler, K. W., Vogelstein, B. & Clevers, H. (1997) Science 275, 1784-1787. [DOI] [PubMed] [Google Scholar]
- 19.Giles, R. H., van Es, J. H. & Clevers, H. (2003) Biochim. Biophys. Acta 1653, 1-24. [DOI] [PubMed] [Google Scholar]
- 20.Morin, P. J. (1999) BioEssays 21, 1021-1030. [DOI] [PubMed] [Google Scholar]
- 21.Dajani, R., Fraser, E., Roe, S. M., Young, N., Good, V., Dale, T. C. & Pearl, L. H. (2001) Cell 105, 721-732. [DOI] [PubMed] [Google Scholar]
- 22.Toullec, D., Pianetti, P., Coste, H., Bellevergue, P., Grand-Perret, T., Ajakane, M., Baudet, V., Boissin, P., Boursier, E., Loriolle, F., et al. (1991) J. Biol. Chem. 266, 15771-15781. [PubMed] [Google Scholar]
- 23.Wenzel-Seifert, K., Schachtele, C. & Seifert, R. (1994) Biochem. Biophys. Res. Commun. 200, 1536-1543. [DOI] [PubMed] [Google Scholar]
- 24.Moon, R. T., Bowerman, B., Boutros, M. & Perrimon, N. (2002) Science 296, 1644-1646. [DOI] [PubMed] [Google Scholar]
- 25.Cadigan, K. M. & Nusse, R. (1997) Genes Dev. 11, 3286-3305. [DOI] [PubMed] [Google Scholar]
- 26.Cheon, S. S., Nadesan, P., Poon, R. & Alman, B. A. (2004) Exp. Cell Res. 293, 267-274. [DOI] [PubMed] [Google Scholar]
- 27.Desbois-Mouthon, C., Cadoret, A., Blivet-Van Eggelpoel, M. J., Bertrand, F., Cherqui, G., Perret, C. & Capeau, J. (2001) Oncogene 20, 252-259. [DOI] [PubMed] [Google Scholar]
- 28.Fujino, H. & Regan, J. W. (2001) J. Biol. Chem. 276, 12489-12492. [DOI] [PubMed] [Google Scholar]
- 29.Fujino, H., West, K. A. & Regan, J. W. (2002) J. Biol. Chem. 277, 2614-2619. [DOI] [PubMed] [Google Scholar]
- 30.Fang, X., Yu, S., Tanyi, J. L., Lu, Y., Woodgett, J. R. & Mills, G. B. (2002) Mol. Cell. Biol. 22, 2099-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding, V. W., Chen, R. H. & McCormick, F. (2000) J. Biol. Chem. 275, 32475-32481. [DOI] [PubMed] [Google Scholar]
- 32.Staal, F. J., Burgering, B. M., van de Wetering, M. & Clevers, H. C. (1999) Int. Immunol. 11, 317-323. [DOI] [PubMed] [Google Scholar]
- 33.Ilyas, M., Tomlinson, I. P., Rowan, A., Pignatelli, M. & Bodmer, W. F. (1997) Proc. Natl. Acad. Sci. USA 94, 10330-10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan, T. A., Wang, Z., Dang, L. H., Vogelstein, B. & Kinzler, K. W. (2002) Proc. Natl. Acad. Sci. USA 99, 8265-8270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sekine, S., Shibata, T., Sakamoto, M. & Hirohashi, S. (2002) Oncogene 21, 5906-5911. [DOI] [PubMed] [Google Scholar]
- 36.Chen, R. H., Ding, W. V. & McCormick, F. (2000) J. Biol. Chem. 275, 17894-17899. [DOI] [PubMed] [Google Scholar]
- 37.Cook, D., Fry, M. J., Hughes, K., Sumathipala, R., Woodgett, J. R. & Dale, T. C. (1996) EMBO J. 15, 4526-4536. [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, H. Y. & Malbon, C. C. (2004) Cell. Mol. Life Sci. 61, 69-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen, A. E., Ginty, D. D. & Fan, C. M. (2005) Nature 433, 317-322. [DOI] [PubMed] [Google Scholar]
- 40.Malbon, C. C., Wang, H. & Moon, R. T. (2001) Biochem. Biophys. Res. Commun. 287, 589-593. [DOI] [PubMed] [Google Scholar]
- 41.Shida, D., Watanabe, T., Aoki, J., Hama, K., Kitayama, J., Sonoda, H., Kishi, Y., Yamaguchi, H., Sasaki, S., Sako, A., et al. (2004) Lab. Invest. 84, 1352-1362. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.