Abstract
Because of the paucity of available tissue, little information has previously been available regarding the gene expression profiles of primary melanomas. To understand the molecular basis of melanoma progression, we compared the gene expression profiles of a series of nevi, primary melanomas, and melanoma metastases. We found that metastatic melanomas exhibit two dichotomous patterns of gene expression, which unexpectedly reflect gene expression differences already apparent in comparing laser-capture microdissected radial and vertical phases of a large primary melanoma. Unsupervised hierarchical clustering accurately separated nevi and primary melanomas. Multiclass significance analysis of microarrays comparing normal skin, nevi, primary melanomas, and the two types of metastatic melanoma identified 2,602 transcripts that significantly correlated with sample class. These results suggest that melanoma pathogenesis can be understood as a series of distinct molecular events. The gene expression signatures identified here provide the basis for developing new diagnostics and targeting therapies for patients with malignant melanoma.
Keywords: bioinformatics, human, microarray, metastasis, laser capture
In the current staging system for cutaneous melanoma, vertical thickness of the primary tumor is the dominant prognostic factor, belying the fact that a subset of thin tumors metastasize, whereas some thick tumors do not undergo metastasis (1). The original melanoma tumor progression model is characterized by an initial radial growth phase, encompassing in situ and minimally invasive tumors (2). This phase is followed by the development of vertical growth phase, which has been postulated to be the first point at which the tumor gains metastatic capacity. However, metastasis occurs, although with decreased frequency, in patients whose primary melanoma pathology exhibits only a radial growth pattern (3). Previous transcriptome analysis in melanoma defined a cluster of genes expressed in a majority of metastatic melanomas (4); however, this cluster was not related to radial or vertical growth, and precursor nevi (moles) and primary melanomas were not examined. Likewise, mutations in B-RAF occur commonly in both nevi (5) and melanoma (6), and, thus, do not distinguish progressive stages in melanoma progression. In this study, we used cDNA expression array profiling to characterize the global patterns of transcript modulation that underlie the various phases in the known tumor progression pathway of melanoma.
Methods
Study Subjects. Samples from melanoma patients and nevus volunteers presenting to the Melanoma Center were obtained with informed consent under a protocol approved by the UCSF Institutional Review Board. After biopsy, all samples were frozen in OCT freezing medium over dry ice. Subsequently, samples were processed for hematoxylin/eosin staining and confirmed by pathologic review. Only samples comprised of >95% tumor cells were analyzed.
Isolation and Purification of Total RNA from Biopsy Specimens. Depending on the sample size, 5-12 20-μm sections were cryotomed, homogenized [Polytron 1200C, Brinkmann, West-bury, NY, at setting 4 for 30 sec] with 600 μl of RNA lysis buffer plus 1% 2-mercaptoethanol, and RNA was isolated by using RNeasy columns (Qiagen, Valencia, CA). Samples from a large primary melanoma, PM09, were subjected to laser-capture microdissection (Arcturus Instruments, Mountain View, CA) before RNA preparation.
RNA Amplification and Labeling. One microgram of total target RNA, side by side with 1 μg of universal human reference RNA (Stratagene), was linearly amplified through two rounds of modified in vitro transcription (7). Amplified RNAs were converted to aminoallyl-modified cDNA and coupled to N- hydroxysuccinimidyl esters of Cy3 or Cy5 (Amersham-Pharmacia, Piscataway, NJ) (8), and then hybridized to a microarray slide at 65°C for 12-16 h (9); www.microarrays.org). Slides were then washed and immediately scanned with Axon-imager 4000b (Axon Instruments, Foster City, CA), by using GenePixPro3 software.
Microarrays. The 20,862 cDNAs used in these studies were from Research Genetics (Huntsville, AL). On the basis of Unigene build 166, these clones represent 19,740 independent loci.
Microarray Data Analysis: Hierarchical Clustering. Gene expression was analyzed with cluster (10) by using the average linkage metric and displayed using javatreeview (which can be accessed at http://genetics.stanford.edu/~alok/TreeView). genepix (Axon Instruments) median of ratio values from the experiment were subjected to linear normalization in nomad (which can be accessed at http://derisilab.ucsf.edu), log-transformed (base 2), and filtered for genes where data were present in 80% of experiments, and where the absolute value of at least one measurement was >1.
significance analysis of microarrays (sam). After linear normalization, log (base 2) transformation, and hierarchical clustering, the resulting cluster data table was imported into the sam software package. Groups were defined based on the comparison performed; for example, group 1 = radial growth phase, and group 2 = vertical growth phase in Fig. 1. Data were censored if more than one data value was flagged in each group to eliminate poor quality array data. Delta was chosen to limit the output gene list so that fewer than 1% predicted false-positives would be included.
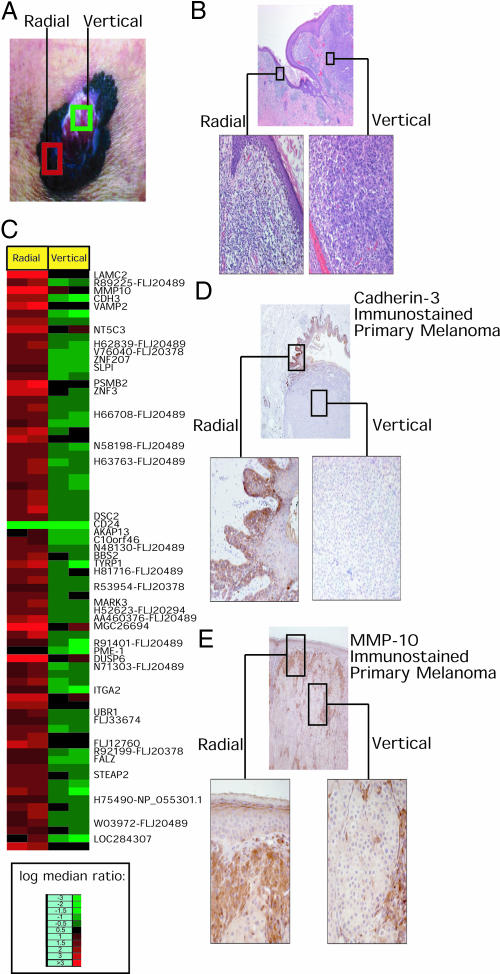
Fig. 1.
Gene expression analysis of the radial and vertical growth phases of primary melanoma. Photograph (A) and photomicrograph (B) of PM09, depicting areas of radial and vertical growth used for laser capture microdissection. (C) List of sam genes shows that many cell adhesion receptors and ESTs are lost in the vertical phase (see Table 1, which is published as supporting information on the PNAS web site, for full list of gene names). Photomicrographs of CDH3 (D) and MMP10 (E) immunostaining in radial growth phase versus vertical growth phase melanoma, with high-power views of the radial growth phase (Left) and vertical growth phase (Right).
Immunohistochemistry. Immunohistochemical staining was performed on 5-μm-thick formalin-fixed paraffin sections by using the avidin-biotin complex method. Tissue sections were deparaffinized with xylene and then rehydrated through graded alcohol. Antigen retrieval was carried out by microwave treatment of the sections in sodium citrate buffer, and endogenous peroxidase and biotin activities were inactivated. The sections were then incubated with a polyclonal rabbit anti-matrix metalloproteinase (MMP) 10 antibody [Lab Vision (Fremont, CA) model no. RB-9235; 1:50 dilution] for 1 h at room temperature or with a polyclonal goat anti-cadherin 3 antibody (Santa Cruz Biotechnology; 1:50 dilution) overnight at 4°C. After the primary antibody, the sections were incubated with the secondary biotinylated goat anti-rabbit antibody (Vector Laboratories catalog no. BA1000; 1:200 dilution) for 30 min and then the avidin-biotin complex (Vector Laboratories catalog no. PK6100 Vectastain ABC kit; 1:100 dilution) for 30 min. All of the stained slides were quantitated by a single pathologist (R.W.S.), with a separate 0-3+ score for intensity of stain in radial and vertical growth phases.
Statistics. Associations between clinical/in vitro parameters and melanoma gene expression were evaluated by using the r software package. For the immunohistochemical analysis, the significance of the difference in immunostaining of cadherin 3 (CDH3) or MMP10 between radial and vertical growth phase was assessed by using the binomial sign test.
B-RAF Mutation Detection. RT-PCR was performed on amplified RNA by using B-RAF exon 15-specific primers, inserts were cloned into TA vector (Invitrogen Life Sciences), and three alleles per patient were sequenced.
Supporting Information. Further information on datasets, software, and analysis can be seen in Data Sets 1-11 and Tables 1-3, which are published as supporting information on the PNAS web site.
Results
To explore whether gene expression profiling could define distinct molecular phenotypes for the radial and vertical growth phase of a large primary melanoma, PM09, we used laser capture (11) to microdissect the tumor (Fig. 1 A and B). Total RNA from the dissected material was isolated, amplified by using a linear T7 method, fluorescently labeled, and hybridized to a microarray comprised of 20,862 cDNA targets (representing 19,740 unique genes). We competitively hybridized duplicate arrays by using the radial and vertical growth phase tumor PM09 RNAs with control universal human reference RNA, which was amplified and fluorescently labeled in parallel with tumor RNA, to facilitate data normalization and sample comparison. sam software was used in two-class mode to determine the list of genes best able to distinguish PM09 radial and vertical growth phases.
Surprisingly, we found no genes activated in the transition to vertical growth, which was only accompanied by loss of expression of a set of genes. Inspection of PM09 gene set 1 (Fig. 1C and Table 1, which is published as supporting information on the PNAS web site) showed predominant loss of cell adhesion and extracellular matrix molecules, such as CDH3, MMP10, integrin α2, and laminin γ2. Strikingly, 10 cDNAs in the list represent different clones from the same transcript, EST FLJ20489.
To confirm that genes differentially expressed in the radial growth phase by expression profiling were also expressed in the radial growth phase at the protein level, immunohistochemical analysis of CDH3 and MMP10 was carried out on an independent panel of 25 primary cutaneous melanoma specimens. For CDH3, 19 cases had evaluable radial and vertical growth phases. In 12 of the 19 cases, CDH3 tumor immunostaining was stronger in the radial growth phase than in the vertical growth phase (Fig. 1D). In the remaining seven cases, the expression was equal between the radial and vertical growth phases. In no case was the CDH3 immunostaining stronger in the vertical growth phase than the radial growth phase. The differences in CDH3 staining between radial growth and vertical growth were statistically significant (P < 0.0005). Intriguingly, in the cases with only the vertical growth phase present (no radial growth identified in the stained sections), only minimal staining of CDH3 was observed.
For MMP10, 22 of 25 cases had evaluable radial and vertical growth phases. In 11 cases, MMP10 tumor immunostaining was stronger in the radial growth phase than in the vertical growth phase (Fig. 1E). In the remaining 11 cases, the expression was equal between the radial and vertical growth phases. Once again, in no case was the MMP10 immunostaining stronger in the vertical growth phase compared with radial growth phase. Differences in MMP10 staining between radial and vertical growth were statistically significant (P = 0.001).
To relate PM09 gene set 1 to the pathogenesis of metastatic melanoma, we carried out gene expression profiling of 19 metastatic melanomas from 17 patients. Fig. 2A shows two separate subtypes of metastatic melanoma identified by unsupervised hierarchical clustering. Intriguingly, when PM09 gene set 1 from Fig. 1 was analyzed across these samples (Fig. 2B), ≈75% of its transcripts were highly expressed only in metastatic melanoma subtype I. Our analysis included two metastases from a supraclavicular lymph node and forearm skin of a single patient, MM-07. The two arrays from this individual are the most closely related in the data set (Pearson correlation of log red/green ratios = 0.95) and are placed side by side (Fig. 2 A and B).
Fig. 2.
Molecular division of metastatic melanoma into two subtypes. (A) Unsupervised hierarchical clustering of metastatic melanoma defines molecular subtypes I and II. (B) Genes identified by sam as lost in the PM09 gene set assessed in the metastatic melanoma dataset. (C) Expression of the PM09 gene set comparing the primary tumor and metastasis within a single patient, MM14. See Data Sets 1-6.
Type II metastatic melanomas express several well known melanoma-associated proteins such as Melan-A (MLANA), tyrosinase, and micropthalmia-associated transcription factor (Fig. 4). Expression of Melan-A and tyrosinase in type II metastatic melanomas was verified by immunoperoxidase studies (data not shown). However, type I metastatic melanomas predominantly express ESTs, suggesting that their growth is promoted by a distinct set of molecular changes that have not yet been well characterized.
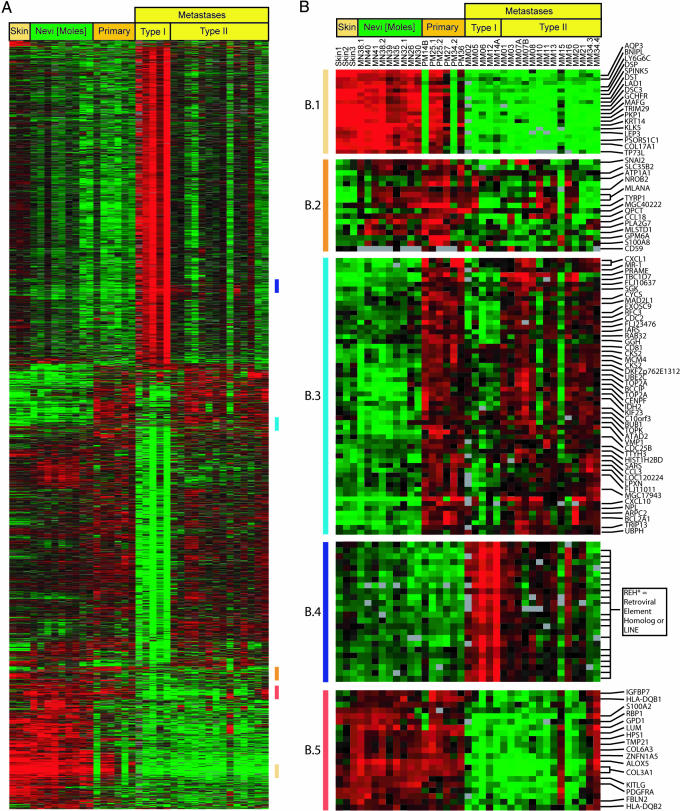
Fig. 4.
Multiclass sam applied to skin, nevi, and primary and metastatic melanoma subtypes predicts 2,602 genes with only 1.6 predicted false-positives able to distinguish among these tissue types. The data table using the 2,602 genes from multiclass analysis was used for supervised hierarchical clustering to facilitate data display. (A) Cluster map of 2,602 genes able to distinguish tissue types. (B) Gene expression nodes particular to each tissue type are highlighted. (B.1) Skin gene expression (B.2) Melanocytic gene expression. (B.3) Melanoma gene expression. (B.4) Metastatic melanoma gene expression. (B.5) Expression lost in metastatic melanoma. See Data Sets 10 and 11.
In addition, we were afforded the rare opportunity to examine melanoma progression within a single patient, MM14, who presented with a nodular primary melanoma and synchronous lymph node metastasis. Fig. 2C shows the PM09 gene set 1 expression pattern of duplicate arrays comparing the MM14 primary tumor and metastasis. Intriguingly, PM09 gene set 1 readily distinguished the primary and metastatic tissue of this patient, suggesting that type I metastasis can arise in patients whose primary tumor exhibits type II gene expression.
Multivariate analysis of age, site of biopsy, and Breslow thickness showed no correlation with either type I or type II metastases (Table 2). In addition, there was no significant difference in frequency of the V599E mutation in the B-RAF gene between type I and type II metastases (Table 2). An exploratory analysis of survival revealed that, whereas all 5 patients with the type I gene expression pattern had died of metastatic melanoma, 4 of 12 patients with the type II gene expression pattern are still alive without evidence of disease.
Subsequently, to complete the comparative molecular evaluation of distinct phases of melanoma tumor progression, we compared signatures of nevi, primary melanomas, and melanoma metastases. As shown in Fig. 3A, unsupervised hierarchical cluster analysis was able to distinguish between benign nevi and primary malignant melanomas. There were both significant gains and losses in gene expression in the transition from nevus to melanoma (Fig. 3B). Two-class sam analysis identified 1,076 genes whose expression distinguishes nevus from primary melanoma with a predicted median false-discovery rate of 2.3 (Table 3). Up-regulated in this gene set were genes known to play an important role in melanoma progression, including SPP1 (osteopontin) (12, 13), and CXCL1 (melanoma growth-stimulating activity) (14), and RAB32 (15). Conversely, genes with potential tumor suppressor activity such as WIF1 (16), ECM2 (17), and SLIT3 (18, 19) were down-regulated. Interestingly, examination of PM09 gene set 1 did not distinguish between nevi and primary melanomas (data not shown).
Fig. 3.
Unsupervised hierarchical cluster analysis applied to nevus and primary melanoma data set. (A) Overall gene expression sample cluster tree shows that nevi and primary melanomas are readily distinguished. (B) Genes more highly expressed in primary melanoma (nodes showing CXCL1 and PRAME) are contrasted with markers of melanocytic differentiation (nodes showing S100B and MLANA). See Data Sets 7-9.
Finally, we performed a multiclass sam comparison of skin, nevi, and primary and metastatic melanoma. The algorithm identified 2,602 genes (multiclass gene set) with a predicted median false-significant rate of 1.6 that significantly distinguish the sample classes (Fig. 4). sam results were visualized by using cluster and treeview of the multiclass gene set.
This analysis demonstrates that unique gene expression patterns characterize each stage of melanoma progression. Fig. 4B.2 shows that melanocytic lineage markers S100B and MLANA (Melan-A) (Fig. 4B.2) are unable to distinguish between any of the multiclass sample groups. The biomarkers identified in Fig. 3 (e.g., CXCL1) not only distinguish primary tumors from moles but also are useful in identifying metastases, and are not expressed in nevi or normal skin (Fig. 4B.3). Biomarkers identified here are also able to distinguish metastatic tumors from all other tissue classes examined.
Among the genes overexpressed in metastatic lesions were several clones representing a reverse transcriptase homolog of an endogenous retrovirus (Fig. 4B.4). In addition, up-regulation of candidate oncogenes such as nuclear receptor coactivator receptor protein 3 (20) and PHIP, a pleckstrin homology domain-containing protein (21) was observed (data not shown). By contrast, genes lost in the transition to metastatic growth (Fig. 4B.5) are implicated in maintenance of normal melanocyte differentiation, including ZNFN1A5 (22) and HPS1 (23).
Discussion
In this study, we examined the gene expression signatures of known points in the tumor progression model of malignant melanoma, from nevus to primary melanoma to metastatic melanoma. Our studies indicate that metastatic melanoma is characterized by two different gene expression signatures, with common features compared with the signatures of radial and vertical growth phases of primary melanoma. Furthermore, gene expression profiling was used to assign specific gene expression signatures to distinct points in the melanoma tumor progression pathway (Fig. 4).
An unexpected finding of this study was that the gene expression signature of the radial growth phase of PM09 was recapitulated in some metastases. Radial growth phase melanomas are considered by some investigators to have little or no propensity for metastasis. These results challenge this notion, suggesting that a small but clinically significant proportion of melanoma metastases may arise from the radial growth phase of primary melanomas. Our hypothesis is supported by the analysis of patient MM14, whose metastasis displayed the type I (radial) signature at the molecular level, whereas the nodular primary displayed the type II (vertical) signature. Nodular melanomas characteristically lack the radial growth phase, and some investigators have postulated that the radial growth phase may be destroyed by the expansile growth of the vertical growth portion. Further analysis is required to determine the specific molecular events that promote tumor metastasis from radial growth phase melanomas.
In addition, the profiling results from PM09 are supported by the analysis of CDH3 and MMP10 immunostaining in a panel of 25 independent primary melanomas. For both markers, protein levels were significantly higher in radial growth compared with vertical growth.
Our study sheds light on the current debate regarding the genesis of the metastasis signature in primary tumors. Recent reports demonstrated the presence of a common metastasis signature in a subset of primary tumors whose origin was different from the metastases studied (24), and that the gene expression signature correlated with metastases was already present in primary breast cancers (25-27). Our study extends these observations by showing that gene expression signatures present in metastases are observed at the in situ or minimally invasive stages that constitute the radial growth phase of primary melanoma (2).
Moreover, the losses in gene expression noted in vertical growth could suggest that the radial growth-dissected samples consisted of genes predominantly expressed in keratinocytes and not melanoma cells. However, our immunohistochemical analyses (Fig. 1), and several previously published studies, demonstrate expression of many PM09 gene set 1 biomarkers in melanomas, including integrin α2 (28), laminin γ2 (29), MMP10 (30), and CDH3 (31). Intriguingly, in an independent study, decreased CDH3 immunostaining correlated with melanoma tumor progression (31), which is similar to our results. Furthermore, the presence of expression of a large proportion of this gene signature in the second subtype of metastatic melanomas, obtained from fine needle aspirates of melanoma metastases, which are predominantly (95%) composed of melanoma cells and devoid of keratinocytes, argues against keratinocyte-specific gene expression alone. Taken together, these results strongly suggest the relevance of the PM09 gene set 1 to the melanocytic lineage.
These results have potentially important implications for the development of melanoma therapy, because many vaccine therapies are currently directed at genes expressed only in type II metastatic melanoma, although we demonstrate that type I metastatic melanoma is also frequent. Further studies are indicated to identify target antigens in the therapy of patients with type I metastatic melanoma.
Examination of the gene expression signatures of nevi versus primary melanoma, suggests the potential utility of these biomarkers as an adjunct to the problematic pathologic diagnosis of melanoma (32). In addition, distinct signatures specific to the metastatic stage were identified (Fig. 4), including activation of several clones of endogenous retrovirus polymerases, which is consistent with reports of human endogenous retrovirus (HERV) retrovirus reactivation in melanoma (33). In this study, whereas primary melanocytes did not shed retroviral particles and were resistant to infection with retrovirus, cell lines derived from melanoma metastases tested produced HERV-K type viruses. We find that long interspersed nuclear element (LINE)-1 and HERV-K type polymerase transcripts are activated in clinical metastatic samples. LINE-1, but not HERV-K, activation permits retrotransposition, which could be one molecular mechanism for genomic damage in advanced melanoma.
Our work indicates that distinct gene expression signatures exist for the known points in the tumor progression pathways of malignant melanoma. In fact, our study implicates different gene sets in the transition from nevus to primary melanoma, and from primary to metastatic melanoma. In addition, a gene expression signature derived from the radial growth phase but absent in the vertical growth phase of a primary melanoma is present in a subset of metastases. Finally, our results provide a basis for developing new molecular diagnostics and targeted therapies for melanoma patients.
Supplementary Material
Acknowledgments
We thank the patients for their participation, Dr. J. DeRisi for helpful advice, and Dr. Alex McMillan for statistical support. We thank Karen Chew of the UCSF Cancer Center Immunohistochemistry Core for assistance with immunohistochemical staining. This work was supported in part by the Herschel and Diana Zackheim Endowment Fund and American Cancer Society Research Scholar Grant RSG-03-247-01-MGO and R03 AR049378 (to M.K.-S.), and National Cancer Institute Grant R01 CA101042-01 and a gift from the Bank of the West (to C.H.).
Abbreviations: sam, significance analysis of microarrays; MMP, matrix metalloproteinase; CDH3, cadherin 3.
References
- 1.Balch, C. M., Buzaid, A. C., Soong, S. J., Atkins, M. B., Cascinelli, N., Coit, D. G., Fleming, I. D., Gershenwald, J. E., Houghton, A., Jr., Kirkwood, J. M., et al. (2001) J. Clin. Oncol. 19, 3635-3648. [DOI] [PubMed] [Google Scholar]
- 2.Clark, W. H., Jr., Elder, D. E., Guerry, D., IV, Epstein, M. N., Greene, M. H. & Van Horn, M. (1984) Hum. Pathol. 15, 1147-1165. [DOI] [PubMed] [Google Scholar]
- 3.Abramova, L., Slingluff, C. L., Jr., & Patterson, J. W. (2002) J. Cutan. Pathol. 29, 407-414. [DOI] [PubMed] [Google Scholar]
- 4.Bittner, M., Meltzer, P., Chen, Y., Jiang, Y., Seftor, E., Hendrix, M., Radmacher, M., Simon, R., Yakhini, Z., Ben-Dor, A., et al. (2000) Nature 406, 536-540. [DOI] [PubMed] [Google Scholar]
- 5.Pollock, P. M., Harper, U. L., Hansen, K. S., Yudt, L. M., Stark, M., Robbins, C. M., Moses, T. Y., Hostetter, G., Wagner, U., Kakareka, J., et al. (2003) Nat. Genet. 33, 19-20. [DOI] [PubMed] [Google Scholar]
- 6.Davies, H., Bignell, G. R., Cox, C., Stephens, P., Edkins, S., Clegg, S., Teague, J., Woffendin, H., Garnett, M. J., Bottomley, W., et al. (2002) Nature 417, 949-954. [DOI] [PubMed] [Google Scholar]
- 7.Baugh, L. R., Hill, A. A., Brown, E. L. & Hunter, C. P. (2001) Nucleic Acids Res. 29, E29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hughes, T. R., Mao, M., Jones, A. R., Burchard, J., Marton, M. J., Shannon, K. W., Lefkowitz, S. M., Ziman, M., Schelter, J. M., Meyer, M. R., et al. (2001) Nat. Biotechnol. 19, 342-347. [DOI] [PubMed] [Google Scholar]
- 9.DeRisi, J., Penland, L., Brown, P. O., Bittner, M. L., Meltzer, P. S., Ray, M., Chen, Y., Su, Y. A. & Trent, J. M. (1996) Nat. Genet. 14, 457-460. [DOI] [PubMed] [Google Scholar]
- 10.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonner, R. F., Emmert-Buck, M., Cole, K., Pohida, T., Chuaqui, R., Goldstein, S. & Liotta, L. A. (1997) Science 278, 1481-1483. [DOI] [PubMed] [Google Scholar]
- 12.Philip, S., Bulbule, A. & Kundu, G. C. (2001) J. Biol. Chem. 276, 44926-44935. [DOI] [PubMed] [Google Scholar]
- 13.Philip, S. & Kundu, G. C. (2003) J. Biol. Chem. 278, 14487-14497. [DOI] [PubMed] [Google Scholar]
- 14.Middleman, B. R., Friedman, M., Lawson, D. H., DeRose, P. B. & Cohen, C. (2002) Mod. Pathol. 15, 532-537. [DOI] [PubMed] [Google Scholar]
- 15.Alto, N. M., Soderling, J. & Scott, J. D. (2002) J. Cell Biol. 158, 659-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wissmann, C., Wild, P. J., Kaiser, S., Roepcke, S., Stoehr, R., Woenckhaus, M., Kristiansen, G., Hsieh, J. C., Hofstaedter, F., Hartmann, A., et al. (2003) J. Pathol. 201, 204-212. [DOI] [PubMed] [Google Scholar]
- 17.Oritani, K. & Kincade, P. W. (1998) Leuk. Lymphoma 32, 1-7. [DOI] [PubMed] [Google Scholar]
- 18.Liu, J., Zhang, L., Wang, D., Shen, H., Jiang, M., Mei, P., Hayden, P. S., Sedor, J. R. & Hu, H. (2003) Mech. Dev. 120, 1059-1070. [DOI] [PubMed] [Google Scholar]
- 19.Yuan, W., Rao, Y., Babiuk, R. P., Greer, J. J., Wu, J. Y. & Ornitz, D. M. (2003) Proc. Natl. Acad. Sci. USA 100, 5217-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu, R. C., Qin, J., Hashimoto, Y., Wong, J., Xu, J., Tsai, S. Y., Tsai, M. J. & O'Malley, B. W. (2002) Mol. Cell. Biol. 22, 3549-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farhang-Fallah, J., Randhawa, V. K., Nimnual, A., Klip, A., Bar-Sagi, D. & Rozakis-Adcock, M. (2002) Mol. Cell. Biol. 22, 7325-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perdomo, J., Holmes, M., Chong, B. & Crossley, M. (2000) J. Biol. Chem. 275, 38347-38354. [DOI] [PubMed] [Google Scholar]
- 23.Feng, L., Novak, E. K., Hartnell, L. M., Bonifacino, J. S., Collinson, L. M. & Swank, R. T. (2002) Blood 99, 1651-1658. [PubMed] [Google Scholar]
- 24.Ramaswamy, S., Ross, K. N., Lander, E. S. & Golub, T. R. (2003) Nat. Genet. 33, 49-54. [DOI] [PubMed] [Google Scholar]
- 25.Huang, E., Cheng, S. H., Dressman, H., Pittman, J., Tsou, M. H., Horng, C. F., Bild, A., Iversen, E. S., Liao, M., Chen, C. M., et al. (2003) Lancet 361, 1590-1596. [DOI] [PubMed] [Google Scholar]
- 26.van de Vijver, M. J., He, Y. D., van't Veer, L. J., Dai, H., Hart, A. A., Voskuil, D. W., Schreiber, G. J., Peterse, J. L., Roberts, C., Marton, M. J., et al. (2002) N. Engl. J. Med. 347, 1999-2009. [DOI] [PubMed] [Google Scholar]
- 27.Sorlie, T., Perou, C. M., Tibshirani, R., Aas, T., Geisler, S., Johnsen, H., Hastie, T., Eisen, M. B., van de Rijn, M., Jeffrey, S. S., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 10869-10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baronas-Lowell, D., Lauer-Fields, J. L., Borgia, J. A., Sferrazza, G. F., Al-Ghoul, M., Minond, D. & Fields, G. B. (2004) J. Biol. Chem. 279, 43503-43513. [DOI] [PubMed] [Google Scholar]
- 29.Pyke, C., Romer, J., Kallunki, P., Lund, L. R., Ralfkiaer, E., Dano, K. & Tryggvason, K. (1994) Am. J. Pathol. 145, 782-791. [PMC free article] [PubMed] [Google Scholar]
- 30.Bodey, B., Bodey, B., Jr., Siegel, S. E. & Kaiser, H. E. (2001) In Vivo 15, 57-64. [PubMed] [Google Scholar]
- 31.Seline, P. C., Norris, D. A., Horikawa, T., Fujita, M., Middleton, M. H. & Morelli, J. G. (1996) J. Invest. Dermatol. 106, 1320-1324. [DOI] [PubMed] [Google Scholar]
- 32.Grant-Kels, J. M., Bason, E. T. & Grin, C. M. (1999) J. Am. Acad. Dermatol. 40, 539-548. [DOI] [PubMed] [Google Scholar]
- 33.Muster, T., Waltenberger, A., Grassauer, A., Hirschl, S., Caucig, P., Romirer, I., Fodinger, D., Seppele, H., Schanab, O., Magin-Lachmann, C., et al. (2003) Cancer Res. 63, 8735-8741. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.