Abstract
Organophosphorus (OP) insecticides and chemical warfare agents act primarily by inhibiting acetylcholinesterase. There are many secondary targets for OP toxicants as observed for example with the major insecticide chlorpyrifos and its bioactivated metabolite chlorpyrifos oxon (CPO). Therefore, it was surprising that the predominant mouse brain protein labeled in vitro by [3H-ethyl]CPO (1 nM) (designated CPO-binding protein or CPO-BP) is not one of these known OP toxicant targets. CPO-BP is a 50-kDa membrane-bound serine hydrolase measured by derivatization with [3H]CPO and SDS/PAGE or filtration binding assay. It appears to undergo rapid diethylphosphorylation by [3H]CPO followed by either dephosphorylation and reactivation or aging on loss of an ethyl group. CPO and several other OP toxicants potently inhibit CPO-BP in vivo (i.p., 2 h) (50% inhibition at 2-25 mg/kg) and in vitro (50% inhibition at 8-68 nM). Using three chemical labeling reagents, i.e., [3H]CPO and the activity-based proteomic probes fluorophosphonate-biotin and fluorophosphonate-rhodamine, mouse brain CPO-BP is identified as serine hydrolase KIAA1363 of unknown function. Brains from KIAA1363-/- mice show greatly reduced levels of CPO labeling and hydrolytic metabolism compared to brains from wild-type mice. KIAA1363 therefore is the principal enzyme for metabolizing low levels of CPO in brain and may play a more general role in detoxification of OP nerve poisons.
Keywords: detoxification, insecticide, phosphorylation, serine hydrolase KIAA1363
Organophosphorus (OP) compounds are among the most studied and best known toxicants because they are a major class of insecticides and the most potent chemical warfare agents. Their primary mechanism of toxicity is inhibition of the serine hydrolase acetylcholinesterase (AChE), thereby prolonging the action of acetylcholine as the cholinergic neurotransmitter. The human genome contains several hundred other predicted serine hydrolases of unknown OP sensitivity and relevance (1, 2). Multiple approaches are required to sort this daunting array of candidate OP toxicant secondary targets (1).
Chlorpyrifos oxon (CPO) is of particular interest as a probe to study OP insecticide secondary targets. It is the bioactivated metabolite of chlorpyrifos, which is one of the most important OP insecticides, so extensively used that its trichloropyridinol metabolite appeared in the urine of 82% of American adults (3). Chlorpyrifos and CPO are the most extensively studied OP insecticides for secondary targets, and CPO has high potency on more OP targets than any other insecticide (1). It has also been prepared as [3H-ethyl]CPO (4). [3H]CPO is used here to recognize sensitive serine hydrolases with the expectation that it will label some of the already known toxicant targets. Surprisingly, when [3H]CPO at 1 nM is incubated with mouse brain membranes, it labels a site, referred to here as CPO-binding protein (CPO-BP), that was not previously considered as an OP insecticide target. The properties observed for this protein radiolabeled with [3H]CPO focused attention on a recently reported serine hydrolase KIAA1363 of known primary structure but unknown function (5). We report here the recognition of the binding protein CPO-BP in mouse brain membranes, the reactions of [3H]CPO and other OP compounds with CPO-BP in vivo and in vitro, its identification as KIAA1363, and its role as an OP toxicant target.
Materials and Methods
Chemicals. The OP inhibitors studied (Table 1) and [3H-ethyl]CPO (67 Ci/mmol; 1 Ci = 37 GBq) (4) were available from previous studies in the Berkeley laboratory. Two activity-based proteomic probes were used: fluorophosphonate (FP)-biotin (6) from Toronto Research Chemicals (Toronto) and FP-rhodamine (7) (Table 1).
Table 1. OP toxicants and activity-based proteomic probes.
| Structure
|
|||
|---|---|---|---|
| No./name or designation | R | R1 | R2 |
| CPO and analogs [(RO)2 P(O)O-2- pyridinyl-3,5,6- Cl3] | |||
| 1 CPO | C2H5 | ||
| 5 CPO-methyl | CH3 | ||
| 3 CPO-pentyl | n-C5C11 | ||
| Other insecticides and analogs | |||
| 2 paraoxon | |||
| 7 diazoxon | |||
| 11 dichlorvos | |||
| 8 profenofos | |||
| Fluorophosphorus compounds [R1R2P(O)F] | |||
| 4 EOPF | n-C8H17 | C2H5O | |
| 6 IDFP | n-C12H25 | i-C3H7O | |
| 10 DFP | i-C3H7O | i-C3H7O | |
| 9 FP-biotin | i-C3H7O | biotin-NH(CH2)6O | |
| FP-rhodamine | C2H5O | tetramethylrhodamine -9-C(O)NH (CH2)5- NHC(O)(OCH2 CH2)4 | |
CPO, paraoxon and diazoxon are the P=O (oxon) activation products of the major P=S (thion) insecticides chlorpyrifos, parathion and diazinon (1).
Mouse Strains and Treatments. Male albino Swiss-Webster mice (25-30 g) were from Harlan Laboratories. Experimental details for the generation of KIAA1363-/- mice are available on request. The animals used in this study were littermates on a mixed genetic background (129S6/SvEv and C57BL/6). Test compounds were administered i.p. by using DMSO as the carrier vehicle (1 μl per g of body weight) or DMSO alone was injected (control). Doses for 50% lethality (LD50) and for 50% inhibition of [3H]CPO labeling (ED50) were determined at 2 h after treatment.
Brain Preparations and Incubations with [3H]CPO. Brains (freshly obtained or from frozen storage at -80°C) were homogenized in 100 mM phosphate buffer (pH 7.4). Three preparations were used for assays: the homogenate after removing debris by centrifugation at 1,000 × g for 20 min and the cytosol (supernatant) and membrane (pellet) fraction from centrifugation of the homogenate at 100,000 × g for 60 min. The membranes were resuspended in phosphate buffer to the original volume and used directly or held at -80°C until analyzed. Protein was determined by the method of Bradford (8). Brain membrane, cytosol, or homogenate proteins (200 μg) in 100 mM phosphate buffer (pH 7.4; 500 μl) were incubated with [3H]CPO (1 nM final concentration unless indicated otherwise) added in ethyl acetate (5 μl) for 15 min (or as specified) at 25°C. Nonspecific binding was determined by 15-min preincubation with unlabeled CPO (100 μM). Potassium fluoride (100 mM, 15 min at 25°C) was used to differentiate diethylphosphorylated (reactivatable) from monoethylphosphorylated (nonreactivatable or aged) proteins based on studies with other serine hydrolases (9, 10). To determine in vitro inhibitor potency, the test OP in a 1, 3, 10, etc., nM final concentration series was introduced in DMSO (5 μl) before adding the [3H]CPO. The inhibitor concentration for 50% inhibition (IC50) in filtration assays was determined by iterative nonlinear least squares regression using the sigmaplot 8.0 program (SPSS, Chicago).
Analysis for [3H]CPO Binding, Hydrolysis, and Metabolism. Protein binding was determined by filtration or electrophoresis. For filtration assay, the binding was terminated by adding cold 0.9% sodium chloride (2 ml) and filtering through GF/B filters presoaked in 0.9% sodium chloride for 2 h. The tubes and filters were washed three times with fresh cold sodium chloride solution (2 ml), and each filter was mixed with scintillation mixture (Safety-Solve, Research Products International) (10 ml) and counted. For SDS/PAGE, a 50-μl aliquot (20 μg of protein) from the incubation mixture was added to loading buffer (62.5 mM Tris, pH 7.4/5% SDS/10% glycerol/5% 2-mercaptoethanol/0.1% wt/vol bromophenol blue) (50 μl). This mixture was loaded directly onto an 8% gel and run at 150 V for 2 h. The gel was stained with Commassie G250 (0.1% wt/vol) in 10% acetic acid and 40% methanol and destained with 10% acetic acid and 40% methanol. Gel sections prepared with a Bio-Rad gel slicer (Bio-Rad) were placed into scintillation vials with 30% hydrogen peroxide (500 μl) and incubated overnight at 80°C. Then, 1 M hydrochloric acid (100 μl) and scintillation mixture were added before counting.
A partitioning method was used to determine [3H]CPO hydrolysis and metabolism, selecting the extracting solvent so that the more accessible upper phase is analyzed for [3H](C2H5O)2P(O)O- and [3H]CPO, respectively. Hydrolysis was terminated by addition of potassium carbonate (200 mM, 0.83 ml) and extraction with chloroform/methanol/hexane (1.25:1.4:1) (2.5 ml). After centrifugation, a 1.5-ml portion of the upper aqueous phase (≈2 ml) was counted to determine the hydrolysis product. For analyzing unmetabolized [3H]CPO, the reactions were terminated by addition of ethyl acetate (2.0 ml), hexane (0.8 ml), and water (0.2 ml), and the upper organic layer (2.5 ml) was counted. In a series of experiments, a selective OP inhibitor (unpublished data) was used to block [3H]CPO reaction in brain homogenate and membrane fractions with 15-min preincubation for inhibition, and 60-min incubation with [3H]CPO for hydrolysis and metabolism.
FP-Biotin and FP-Rhodamine Labeling. Serine hydrolases were analyzed by the FP-biotin (11) and FP-rhodamine (5) procedures. Brain membrane proteins (200 μg) in 100 mM phosphate buffer (pH 7.4; 500 μl) were incubated with FP-biotin (100 μM final concentration, added in 5 μl of DMSO) for 30 min at 25°C. For in vitro inhibition studies, the OP (added in 5 μl of DMSO) was preincubated with brain membranes for 15 min to allow it to react before competing with FP-biotin. An aliquot (20 μl) was then added to SDS/PAGE loading buffer (30 μl). The reaction mixtures were run on SDS/PAGE as above (20 μg of protein per gel lane) and transferred by electroblotting onto nitrocellulose membranes, which were blocked in Tris-buffered saline (TBS) with 1% Tween (TBS-Tween) and 3% BSA for 1 h at 25°C. Blots were then treated with an avidin-horseradish peroxidase conjugate (Bio-Rad) (1:1,000 dilution) in TBS-Tween with 1% BSA for 30 min at 25°C. The blot was washed with TBS-Tween three times (10 min per wash), treated with SuperSignal chemiluminescence reagents (Bio-Rad), and exposed to film for 1 min before development.
FP-rhodamine analyses followed the procedure of Leung et al. (5) with 10-min incubation of OP (0.5-100,000 nM final concentration added in 1 μl of DMSO) and brain membranes (1,000 μg of protein per ml) in 50 mM Tris·HCl (pH 8.0) before FP-rhodamine addition (100 nM final concentration from DMSO stock). Reactions were quenched after 10 min with SDS/PAGE loading buffer, subjected to SDS/PAGE, and visualized in-gel by using a flatbed fluorescence scanner (MiraiBio, Alameda, CA). Labeled proteins were quantified by measuring integrated band intensities (normalized for volume); control samples (DMSO alone) were considered 100% activity, and inhibitor-treated samples were expressed as a percentage of remaining activity. Potent inhibitors (IC50 values < 10 nM) were also tested at 0.5, 1, and 5 nM with proteomes adjusted to 100 μg of protein per ml, so that the estimated concentration of target enzymes was kept at least 5-fold below the inhibitor concentration (enzyme concentrations were estimated by activity-based protein profiling as described in ref. 12). IC50 values were determined from dose-response curves from three trials at each inhibitor concentration by using prism software (Graph-Pad, San Diego).
Cells of Varying KIAA1363 Expression. Two human breast cancer cell lines, with low (MCF-7) and high (MDA-MB-231) KIAA1363 expression (12), were grown to 80% confluence in DMEM containing 10% FBS and then cultured in serum-free medium for 48 h. The adhering cells were harvested and centrifuged at 2,400 × g for 5 min. The pellets were resuspended in 100 mM phosphate buffer (pH 7.4) and sonicated. The 100,000 × g membrane fraction was then prepared and assayed in the same way as with the mouse brain membranes.
Results
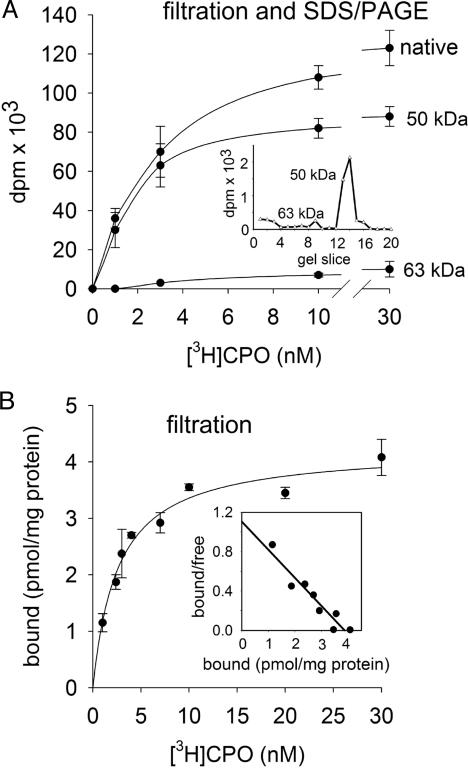
[3H]CPO Labeling. Incubation of [3H]CPO (1-30 nM) with mouse brain membranes for 15 min leads to covalent derivatization based on analysis by the filtration assay of the 100,000 × g pellet (native membranes) and by SDS/PAGE (50- and 63-kDa proteins) (Fig. 1A). There is predominantly a single labeled band of ≈50 kDa tentatively assumed to be one protein and designated CPO-BP (Fig. 1 A Inset). With 1 nM [3H]CPO, there is comparable recovery for the total membrane-bound material by filtration (36,000 ± 5,000 dpm) and the 50-kDa fraction by SDS/PAGE (30,000 ± 5,000 dpm). On this basis, the filtration and SDS/PAGE assays at 1 nM [3H]CPO can be used interchangeably as convenient to quantitate bound material. [3H]CPO binding was also examined at 1-30 nM to determine the possible presence of additional binding sites (Fig. 1). Although CPO-BP is essentially the only labeled protein at 1 nM [3H]CPO, there is increasing labeling of a 63-kDa band with 3, 10, and 30 nM [3H]CPO; this protein may be fatty acid amide hydrolase (FAAH) (1). The filtration assay measures the sum of CPO-BP and the presumed FAAH. The binding isotherm and pseudoScatchard analysis indicate that CPO-BP is a single labeled component with apparent maximal binding (Bmax) of 4.2 pmol per mg of protein and apparent phosphorylation constant of 2.6 nM (Fig. 1B). The minimal CPO-BP amount can be calculated, based on a 50-kDa protein, as 220 μg per g of brain.
Fig. 1.
[3H]CPO labeling of mouse brain membranes analyzed by filtration assay of the 100,000 × g pellet (native membranes) and by SDS/PAGE (50- and 63-kDa proteins). Membranes (200 μg of protein) were incubated for 15 min with [3H]CPO at 1 nM (A Inset shows labeled 50- and 63-kDa proteins) or 1-30 nM. Data in B are shown as binding isotherm (mean ± SD, n = 3) and pseudoScatchard plot (r2 = 0.91) (Inset).
Brain cytosol incubated with 1 nM [3H]CPO under the standard conditions gives a single labeled band at ≈67 kDa (data not shown) that may be albumin (13) because it is not detected in perfused brain.
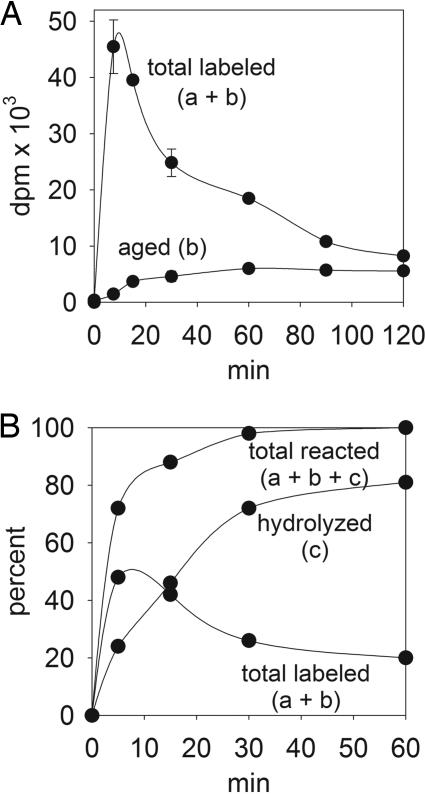
Reaction of [3H]CPO with CPO-BP Involves Diethylphosphorylation, Aging, and Hydrolysis. The reaction of [3H]CPO with mouse brain membranes was monitored in vitro. Membrane preparations were incubated with 1 nM [3H]CPO for up to 120 min before filtration. Labeling is rapid, and then falls to ≈18% of the 7.5-min value by 120 min (Fig. 2A, total labeled). The initial and final binding involve a single 50-kDa band based on SDS/PAGE analysis (data not shown) and probably therefore a single labeled protein during the entire time course examined. The same pattern of labeling and loss of label is repeated upon addition of fresh [3H]CPO after 60 min (data not shown), implying reactivation by dephosphorylation. Total labeling is largely displaced by adding potassium fluoride 15 min after [3H]CPO, i.e., a major portion can be reactivated. The 12% portion of maximum labeling not displaced by fluoride increases for 60 of the 120 min and is designated aged serine hydrolase. Formation of [3H](C2H5O)2P(O)O- corresponds to 81% hydrolyzed at 60 min. The total reacted CPO (bound and hydrolyzed) at 1 nM is 72% at 5 min increasing to 100% at 60 min (Fig. 2B). With excess substrate (10 nM), only 36% is hydrolyzed after 60 min (data not shown).
Fig. 2.
[3H]CPO (1 nM) labeling of mouse brain membranes (200 μg of protein) in vitro showing the proposed diethylphosphorylated (a), aged (b), and hydrolyzed (c) portions. Time course is shown as dpm (A) and percent of total dpm added (B). In the aged curve (b), potassium fluoride was added for 15 min after the incubation times indicated. Data are mean ± SD (n = 3).
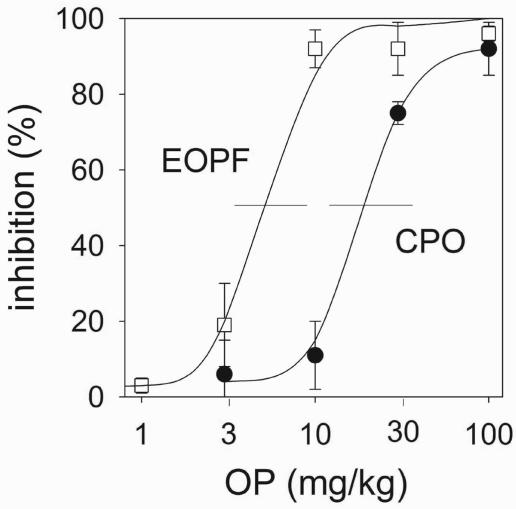
CPO-BP is Inhibited by Several OPs in Vivo. Unlabeled CPO and four other OPs were administered i.p. to mice. After 2 h, brain membranes were prepared to measure [3H]CPO binding by the filtration assay. All five OPs inhibit [3H]CPO binding in a dose-dependent manner. Comparable inhibition data with similar variances are obtained for diisopropyl fluorophosphate (DFP), paraoxon, and isopropyl dodecylfluorophosphonate (IDFP) to those shown for ethyl octylphosphonofluoridate (EOPF) and CPO (Fig. 3). The ED50 values are 2-25 mg/kg for the acute cholinergic toxicants (DFP, CPO, and paraoxon) and 4-7 mg/kg for the delayed toxicants (EOPF and IDFP) (Table 2). The ED50 is below the LD50 for DFP, EOPF, and IDFP, but not for CPO and paraoxon, which are lethal at the ED50 dose.
Fig. 3.
Potency of two OP compounds administered i.p. as in vivo inhibitors of mouse brain CPO-BP at 2 h after treatment. Shown is [3H] CPO labeling of brain membranes from the treated mice with 30-min incubation. In vivo inhibition curves are used to determine ED50 values shown for five compounds in Table 2. Data are mean ± SD (n = 3).
Table 2. Relation for five OP compounds between in vivo inhibition of mouse brain CPO-BP and their toxicity at 2 h after treatment.
| Dose, mg/kg
|
||
|---|---|---|
| OP | ED50 | LD50 |
| Acute cholinergic toxicants | ||
| DFP | 2 | 7 |
| CPO | 21 | 9 |
| paraoxon | 25 | 2 |
| Delayed toxicants | ||
| EOPF | 4 | 50 |
| IDFP | 7 | >100 |
Data for mortality are n = 4 at LD50.
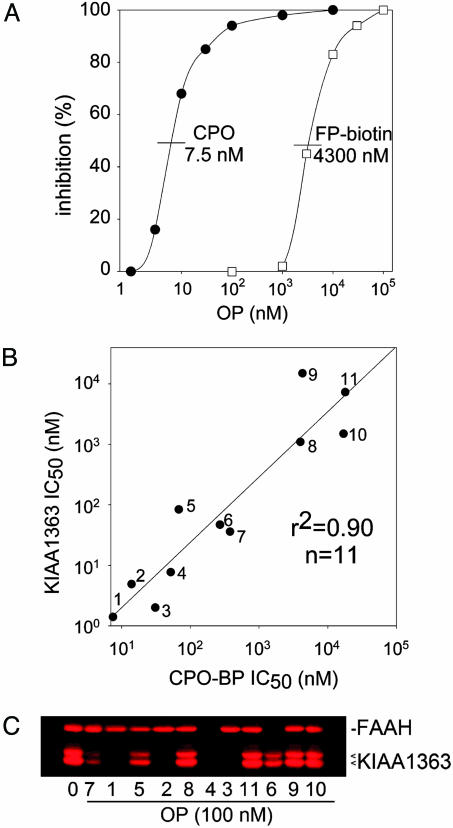
Cross Reactivity of CPO-BP and KIAA1363 with FP-Biotin and CPO. The activity-based proteomic probe FP-biotin was tested for potency in the [3H]CPO filtration-binding assay (Fig. 4A). It is much less potent than CPO (IC50 values of 4,300 and 7.5 nM, respectively), but is still suitable for the biotinylation-SDS/PAGE procedure. It labels at least nine bands, two of which (50 and 63 kDa) are sensitive to CPO at 1,000 and 10,000 nM (data not shown). The 50-kDa doublet and 63-kDa band were previously identified with FP-biotin as serine hydrolase KIAA1363 and FAAH, respectively (5). This cross reactivity experiment suggests the possibility that CPO-BP is KIAA1363.
Fig. 4.
Similar OP inhibitor specificity profiles for inhibition of [3H]CPO binding to CPO-BP and FP-rhodamine labeling of KIAA1363. (A) FP-biotin and CPO inhibit [3H]CPO binding. Mouse brain membranes and CPO or FP-biotin were incubated together for 15 min before addition of 1 nM [3H]CPO and further incubation for 15 min before filtration. (B) Correlation plot for inhibition of [3H]CPO binding to CPO-BP and FP-rhodamine labeling of KIAA1363. Data are given in Table 3. (C) Inhibition of FP-rhodamine labeling for 11 compounds at 100 nM compared with the control (0). Membranes were incubated with inhibitors for 10 min before addition of 100 nM FP-rhodamine for 10 min. KIAA1363 appears as two glycosylated forms at ≈50 kDa. Some OPs also inhibit FAAH (63 kDa) as discussed in the text. The OP compounds (1-11) are identified in Table 1.
Similar OP Inhibitor Specificity Profiles for CPO-BP and KIAA1363. Each serine hydrolase has a unique OP inhibitor specificity profile (1), providing a useful guide in determining whether two different assays measure the same enzyme. CPO-BP was assayed as [3H]CPO binding by the filtration assay, and KIAA1363 was determined by using the FP-rhodamine procedure (Table 3). The best inhibitors in both assays are CPO, paraoxon, CPO-pentyl, and EOPF (compounds 1-4) with IC50 values of 1-52 nM. The inhibitor potency is less for CPO-methyl, IDFP, and diazoxon (compounds 5-7) (IC50 values of 37-379 nM), and lowest for profenofos, FP-biotin, DFP, and dichlorvos (compounds 8-11) (IC50 values of 890-18,000 nM) (Fig. 4B and Table 3) confirmed by comparing potencies at 100 nM OP (Fig. 4C). There is a good correlation between the [3H]CPO binding and FP-rhodamine assays (r2 = 0.90, n = 11; Fig. 4B), suggesting that they recognize the same site, i.e., CPO-BP is KIAA1363.
Table 3. Potency of eleven OP compounds for inhibition of [3H]CPO binding to CPO-BP and FP-rhodamine labeling of KIAA1363 (two glycosylated forms at ≈50 kDa) and FAAH.
| IC50 ± SD, nM
|
|||
|---|---|---|---|
| OP | CPO-BP | KIAA1363 | FAAH |
| 1 | 7.5 ± 2 | 1.1 ± 0.6 | 613 ± 39 |
| 2 | 14 ± 3 | 4.5 ± 1.2 | 5,800 ± 770 |
| 3 | 31 ± 1 | 2.2 ± 0.2 | 3,800 ± 2,200 |
| 4 | 52 ± 9 | 8.3 ± 1.4 | 1.3 ± 0.2 |
| 5 | 68 ± 4 | 99 ± 17 | 220 ± 83 |
| 6 | 271 ± 37 | 37 ± 4 | 2.1 ± 3.3 |
| 7 | 379 ± 138 | 45 ± 6.3 | >100,000 |
| 8 | 4,000 ± 200 | 890 ± 230 | 3,300 ± 500 |
| 9 | 4,300 ± 1,200 | 12,000 ± 1,500 | 12,000 ± 1,500 |
| 10 | 17,000 ± 5,000 | 1,600 ± 140 | >100,000 |
| 11 | 18,000 ± 750 | 7,300 ± 3,400 | 14,000 ± 1,830 |
IC50 values are means (n = 3).
FAAH (63 kDa) is also inhibited [particularly by EOPF (4) and IDFP (6)], but with a different specificity profile than KIAA1363 (Table 3), as determined by a correlation plot for the two serine hydrolases (r2 = 0.28, n = 11; data not shown) and inhibition at 100 nM OP (Fig. 4C). The method of activity-based protein profiling is further validated by comparing the IC50 values obtained for mouse brain FAAH assayed by using the FP-rhodamine procedure with those measured by the conventional [14C]oleamide hydrolysis technique (1); a good correlation (r2 = 0.94) is obtained for assays with seven compounds (DFP, EOPF, IDFP, paraoxon, CPO, profenofos, and dichlorvos).
[3H]CPO Labeling Related to KIAA1363 Expression in Cancer Cell Lines. In a supplemental test, radioligand binding was compared for membranes from MCF-7 cells with low KIAA1363 expression and MDA-MB-231 cells with 7-fold higher KIAA1363 expression (12). [3H]CPO binding is higher in the MDA-MB-231 cells by 5-fold, as determined by filtration assay, and 3-fold, as determined by SDS/PAGE (data not shown).
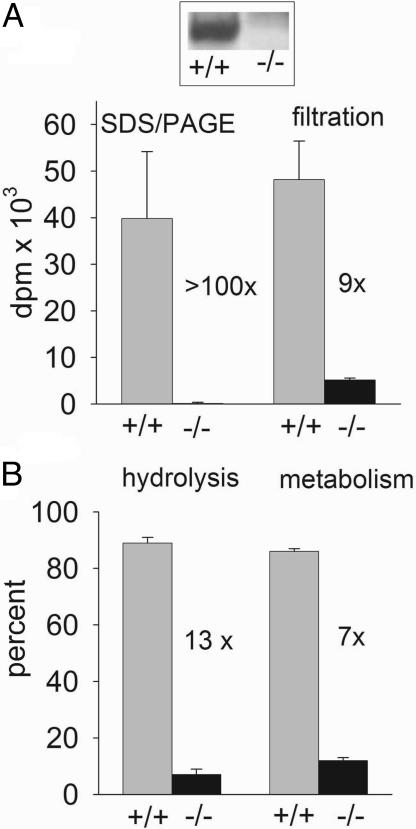
KIAA1363-Deficient Mice Are also CPO-BP Deficient. KIAA1363-/- mice were compared with their +/+ wild-type littermates to determine the possible relationship between CPO-BP and KIAA1363. There is no FP-biotin labeling of KIAA1363 in the -/- mice compared with the +/+ mice (Fig. 5A Inset). Most importantly, using brain membrane preparations, [3H]CPO (1 nM) labeling is >100-fold lower for -/- versus +/+ mice by the specific SDS/PAGE procedure and 9-fold lower by the somewhat less discriminating filtration assay (Fig. 5A). This difference is also evident for hydrolysis and overall metabolism, which are 13- and 7-fold lower, respectively, for -/- versus +/+ animals (Fig. 5B). The studies also considered brain cytosol and whole brain homogenate. There is relatively little metabolism of [3H]CPO (1 nM) by brain cytosol compared with the membranes (Fig. 5). Although not detailed here, the use of a selective inhibitor [n-C13H27O(CH3)P(O)OC6H4-4-NO2, 100 nM] also established that the membrane enzyme accounts for essentially all of the [3H]CPO (1 nM) hydrolysis and metabolism by brain homogenate.
Fig. 5.
KIAA1363-deficient mice are also deficient in CPO-BP based on [3H]CPO binding (A) and hydrolysis or metabolism (B) by mouse brain membranes. Inset in A illustrates FP-biotinylation of 50-kDa band in +/+ and -/- mice. Binding assays with 200 μg of membrane protein and 1 nM [3H]CPO labeling for 15 min with analysis by SDS/PAGE and filtration are shown. Hydrolysis and metabolism assays involve 60-min incubation and analysis for hydrolysis product or loss of parent compound. Comparative results for the same incubations replacing membrane with cytosol protein (200 μg) were as follows: hydrolysis of 6 ± 0% for -/- and 7 ± 1% for +/+; metabolism of 20 ± 4% for -/- and 14 ± 10% for +/+. Data are mean ± SD (n = 4 for binding and n = 3 for hydrolysis and metabolism).
Discussion
CPO-BP Is the Principal High-Affinity [3H]CPO-Binding Protein in Brain. CPO disrupts a large number of brain enzymes and receptors and other biochemical and organismal systems. One way to study these targets is to determine the most sensitive brain proteins undergoing diethylphosphorylation (1). [3H]CPO at 1 nM apparently labels only one major protein of ≈50 kDa, designated CPO-BP, and, surprisingly, it is not a known target of OP toxicants. CPO-BP can be determined equally well by filtration and SDS/PAGE assays with more rapid analysis by the former procedure. At higher radioligand levels, the filtration assay is not satisfactory because there is also another labeled protein, probably FAAH. When calculated on the basis of 50 kDa and 4.2 pmol per mg of protein, CPO-BP makes up 0.02% of the total mouse brain membrane proteins. The CPO-BP concentration in the standard filtration binding assay is therefore at least 1.7 nM, compared with 1 nM [3H]CPO.
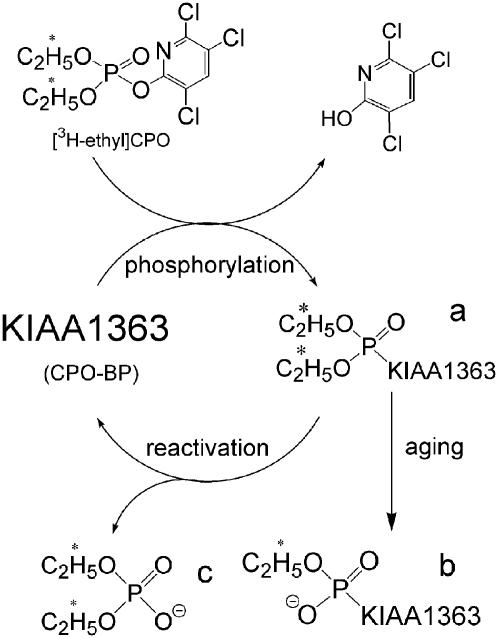
Reactions of CPO-BP with [3H]CPO. CPO-BP in mouse brain membranes is very rapidly labeled by [3H]CPO with subsequent loss of a portion of the label. This pattern can be rationalized as initial formation of the enzyme-inhibitor complex (data not shown) followed by rapid diethylphosphorylation and a somewhat slower dephosphorylation or reactivation, i.e., CPO-BP hydrolyzes [3H]CPO (Fig. 6). Potassium fluoride greatly accelerates the reactivation. A portion of the inhibited hydrolase rapidly changes to a derivative not reactivated by potassium fluoride, presumably the aged enzyme formed on dealkylation. Aging of diethylphosphorylated-CPO-BP is much more rapid (half-time, 10-20 min) than that of the corresponding adducts of AChE and butyrylcholinesterase (half-time, 12-58 h) (14). The aged target with ≈12% of the label only retains one [3H]ethyl group, which is equivalent to half of the original diethylphosphorylated form, i.e., 24% of the target initially labeled is quickly aged.
Fig. 6.
Reaction is shown of KIAA1363 (also known as CPO-BP) with [3H-ethyl]CPO involving phosphorylation (a) followed by aging (b) (loss of one ethyl group and not reactivated) or reactivation (c) (accelerated by potassium fluoride). At 1 nM [3H]CPO, a ratio of 3:1 is observed for reactivation and aging (adjusted for loss of one labeled residue).
CPO-BP Is KIAA1363. Four observations establish that CPO-BP is KIAA1363. First, CPO-BP and KIAA1363 are both present in mouse brain membranes with a molecular mass of ≈50 kDa (or more specifically 45 and 50 kDa for multiple glycosylated forms evident by PNGaseF treatment of FP-rhodamine-labeled proteomes) (5). Second, FP-biotin inhibits [3H]CPO labeling of CPO-BP and, conversely, CPO inhibits FP-biotin phosphorylation of KIAA1363. Third, the same OP inhibitor specificity profile is observed for CPO-BP determined by [3H]CPO binding and KIAA1363 assayed with FP-rhodamine. Furthermore, MDA-MB-231 cells, with 7-fold higher KIAA1363 expression than MCF-7 cells (12), are 3- to 5-fold higher in [3H]CPO labeling (this study). Fourth, and most definitively, KIAA1363-/- mice have little or no CPO-BP compared with their KIAA1363+/+ littermates based on each of several different methods of analysis, i.e., [3H]CPO binding determined by SDS/PAGE and filtration assay and [3H]CPO loss both as hydrolysis and overall metabolism.
KIAA1363 Is an OP-Detoxifying Enzyme in Brain. The findings with KIAA1363-deficient mice establish that this enzyme is the principal high-affinity CPO-metabolizing site in brain serving as an OP hydrolase. Other targets also react with high levels of CPO because multiple serine hydrolases are inhibited and labeled. Inhibition of KIAA1363 in vivo probably consists of diethylphosphorylated and monoethylphosphorylated enzyme because any excess noncovalently bound inhibitor would have been removed during membrane preparation for assay. The form of inhibited KIAA1363 with a large excess of CPO will change with time after treatment from predominantly reactivatable at early stages to mainly aged at later times. Brain KIAA1363 is inhibited by several OPs in vivo and in vitro, and therefore serves as a sequestration protein or hydrolase for many compounds. The apparent affinity, phosphorylation rate, and extent of aging with KIAA1363 will be greatly affected by the OP structure, as is the case also with AChE (9) and neuropathy target esteraselysophospholipase (15-17). AChE is the presumed target for acute cholinergic toxicants and neuropathy target esteraselysophospholipase for the delayed toxicants studied here. The relationship for a series of OPs between the in vivo ED50 for KIAA1363 and the LD50 suggests that this hydrolase actually reacts with acute and delayed OP toxicants in a concentration and time frame of potential toxicological relevance. KIAA1363-deficient mice appear to be phenotypically normal, so the OP reaction with KIAA1363 in brain is not expected to result in a toxic response but instead a detoxification.
There are several mammalian OP hydrolases of greatly different tissue distribution and properties. A very critical site for OP detoxification is in the nerve because it minimizes AChE and neuropathy target esterase-lysophospholipase inhibition. In contrast to other tissues containing paraoxonase and DFPase enzymes, with histidine moieties and calcium at their active sites (18-20), the serine hydrolase KIAA1363 appears to be the most important protein in mouse brain for CPO metabolism.
KIAA1363 Properties and Function. KIAA1363 belongs to the GDXG family of serine hydrolases having lipase homology with the active site-serine at position 191 (http://myhits.isb-sib.ch/cgi-bin/motif_scan); it also has 44% sequence homology with kidney arylacetamide deacetylase (http://us.expasy.org/tools/blast). The molecular mass is 45 and 50 kDa in different glycosylated forms, and the pI is 6.7. It is highly expressed in several invasive cancer cell lines and is therefore a potential marker for cancer invasiveness (12). However, its role in cancer progression, its endogenous substrate, and its possible genetic polymorphism are not defined. The discoveries and KIAA1363-null mice reported here allow for further testing of the function of KIAA1363, including the proposal that it is a significant detoxifying enzyme for OP nerve poisons in brain.
Acknowledgments
The cancer cell lines were supplied by Leonard Bjeldanes (University of California, Berkeley). This work was supported by National Institutes of Health Grants ES08762 (to J.E.C.), DA017259 (to B.F.C.), and CA87660 (to B.F.C.).
Author contributions: D.K.N., B.F.C., and J.E.C. designed research; D.K.N., D.L., and K.P.C. performed research; B.F.C. contributed new reagents/analytic tools; D.K.N., G.B.Q., and J.E.C. analyzed data; and D.K.N., G.B.Q., and J.E.C. wrote the paper.
Abbreviations: OP, organophosphorus; AChE, acetylcholinesterase; CPO, chlorpyrifos oxon; CPO-BP, CPO-binding protein; FAAH, fatty acid amide hydrolase; DFP, diisopropyl fluorophosphate; IDFP, isopropyl dodecylfluorophosphonate; EOPF, ethyl octylphosphonofluoridate; FP, fluorophosphonate.
References
- 1.Casida, J. E. & Quistad, G. B. (2004) Chem. Res. Toxicol. 17, 983-998. [DOI] [PubMed] [Google Scholar]
- 2.Yousef, G. M., Kopolovic, A. D., Elliott, M. B. & Diamandis, E. P. (2003) Biochem. Biophys. Res. Commun. 305, 28-36. [DOI] [PubMed] [Google Scholar]
- 3.Hill, R. H., Jr., Head, S. L., Baker, S., Gregg, M., Shealy, D. B., Bailey, S. L., Williams, C. C., Sampson, E. J. & Needham, L. L. (1995) Environ. Res. 71, 99-108. [DOI] [PubMed] [Google Scholar]
- 4.Zhang, N., Morimoto, H., Williams, P. G. & Casida, J. E. (2000) J. Labelled Cpd. Radiopharm. 43, 1275-1282. [Google Scholar]
- 5.Leung, D., Hardouin, C., Boger, D. L. & Cravatt, B. F. (2003) Nat. Biotechnol. 21, 687-691. [DOI] [PubMed] [Google Scholar]
- 6.Higson, A. P., Ferguson, M. A. J. & Nikolaev, A. V. (1999) Synthesis 1999, 407-409. [Google Scholar]
- 7.Patricelli, M. P, Giang, D. K., Stamp, L. M. & Burbaum, J. J. (2001) Proteomics 1, 1067-1071. [DOI] [PubMed] [Google Scholar]
- 8.Bradford, M. M. (1976) Anal. Biochem. 72, 248-254. [DOI] [PubMed] [Google Scholar]
- 9.Aldridge, W. N. & Reiner, E. (1972) Enzyme Inhibitors as Substrates (Elsevier, New York).
- 10.Clothier, B. & Johnson, M. K. (1980) Biochem. J. 185, 739-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu, Y., Patricelli, M. P. & Cravatt, B. F. (1999) Proc. Natl. Acad. Sci. USA 96, 14694-14699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jessani, N., Liu, Y., Humphrey, M. & Cravatt, B. F. (2002) Proc. Natl. Acad. Sci. USA 99, 10335-10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peeples, E. S., Schopfer, L. M., Duysen, E. G., Spaulding, R., Voelker, T., Thompson, C. M. & Lockridge, O. (2005) Toxicol. Sci. 83, 303-312. [DOI] [PubMed] [Google Scholar]
- 14.Nachon, F., Asojo, O. A., Borgstahl, G. E. O., Masson, P. & Lockridge, O. (2005) Biochemistry 44, 1154-1162. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, M. K. & Glynn, P. (2001) in Handbook of Pesticide Toxicology, ed. Krieger, R. I. (Academic, San Diego), Vol. 2, pp. 953-965. [Google Scholar]
- 16.van Tienhoven, M., Atkins, J., Li, Y. & Glynn, P. (2002) J. Biol. Chem. 277, 20942-20948. [DOI] [PubMed] [Google Scholar]
- 17.Quistad, G. B., Barlow, C., Winrow, C. J., Sparks, S. E. & Casida, J. E. (2003) Proc. Natl. Acad. Sci. USA 100, 7983-7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mounter, L. A. (1963) in Cholinesterases and Anticholinesterase Agents, ed. Koelle, G. B. (Springer, Berlin), pp. 486-504.
- 19.Costa, L. G. & Furlong, C. E., eds. (2002) Paraoxonase (PON1) in Health and Disease (Kluwer Academic, Norwell, MA).
- 20.Kondo, Y., Ishigami, A., Kubo, S., Handa, S., Gomi, K., Hirokawa, K., Kajiyama, N., Chiba, T., Shimokado, K. & Maruyama, N. (2004) FEBS Lett. 570, 57-62. [DOI] [PubMed] [Google Scholar]