Abstract
Olfaction, which is an important physiological function for the survival of mammals, is controlled by a large multigene family of olfactory receptor (OR) genes. Fishes also have this gene family, but the number of genes is known to be substantially smaller than in mammals. To understand the evolutionary dynamics of OR genes, we conducted a phylogenetic analysis of all functional genes identified from the genome sequences of zebrafish, pufferfish, frogs, chickens, humans, and mice. The results suggested that the most recent common ancestor between fishes and tetrapods had at least nine ancestral OR genes, and all OR genes identified were classified into nine groups, each of which originated from one ancestral gene. Eight of the nine group genes are still observed in current fish species, whereas only two group genes were found from mammalian genomes, showing that the OR gene family in fishes is much more diverse than in mammals. In mammals, however, one group of genes, γ, expanded enormously, containing ≈90% of the entire gene family. Interestingly, the gene groups observed in mammals or birds are nearly absent in fishes. The OR gene repertoire in frogs is as diverse as that in fishes, but the expansion of group γ genes also occurred, indicating that the frog OR gene family has both mammal- and fish-like characters. All of these observations can be explained by the environmental change that organisms have experienced from the time of the common ancestor of all vertebrates to the present.
Keywords: birth-and-death evolution, multigene family, vertebrate evolution
Vertebrates can discriminate among thousands of different odor molecules in the environment. Odor molecules are detected by olfactory receptors (ORs) that are expressed in sensory neurons of olfactory epithelia in nasal cavities (see refs. 1-3 for review). ORs are encoded by a large multigene family, which consists of ≈1,000 different genes in mammalian species. They are G protein-coupled receptors (GPCRs) that contain seven α-helical transmembrane regions. OR genes are ≈310 codons long on average, do not have any introns in their coding regions, and form genomic clusters that are scattered on many chromosomes. The OR gene family in each species is considered to reflect the ability of olfaction of the species, because different OR genes are thought to bind to different sets of odor molecules.
Analysis of whole-genome sequences has shown that in humans, there are ≈800 OR genes, but ≈50% of them are pseudogenes (4, 5), whereas in mice, there are ≈1,400 OR genes, and the fraction of pseudogenes is ≈25% (6-9). OR genes were first identified from rats (10). In nonmammalian vertebrates, OR genes have been identified from lampreys (11), catfish (12), zebrafish (13, 14), goldfish (15), medaka fish (16, 17), Japanese loaches (18), Xenopus laevis (19), and chickens (20), although the numbers of genes identified are relatively small.
Currently, it is widely accepted that vertebrate OR genes can be classified into two different classes, class I and II genes (21). All of the fish genes are believed to belong to class I genes, and for this reason, they are referred to as “fish-like” genes. By contrast, class II genes are called “mammalian-like” genes, because the majority of mammalian OR genes belong to class II genes. X. laevis is known to have both types of genes. Class I genes are exclusively expressed in the water-filled lateral diverticulum of the nasal cavities and are similar to known fish genes, whereas class II genes are expressed in the air-filled medial diverticulum and are similar to known mammalian genes (19). From this observation and others (22), it has been proposed that class I genes are specialized for detecting water-soluble odorants, and class II genes are for recognizing airborne odorants (19). A small number of class I genes were found in humans, but they were considered to be evolutionary relics (23). Later, however, an extensive search of OR genes from the human and mouse genome sequences revealed that mammals have a substantial number of class I genes (4, 6). Therefore, the functional difference between class I and II genes is now unclear (6, 24).
The purpose of this paper is to study the evolutionary dynamics of this huge multigene family in vertebrates. For this purpose, we first identified OR genes from the draft genome sequences of zebrafish, pufferfish, Xenopus tropicalis, and chickens. We then conducted large-scale phylogenetic analyses by using these genes and compared OR gene families among various vertebrate species.
Materials and Methods
Database Search for Known Fish, Amphibian, and Avian OR Genes. To collect full-length sequences of functional OR genes in nonmammalian vertebrates, we conducted blastp searches (25) against the DAD database (all of the amino acid sequences) on the DNA Databank of Japan (DDBJ) web site (www.ddbj.nig.ac.jp) with the E value below 1e-05. A human class I OR gene (HsOR11.3.2, OR4F16) and a class II gene (HsOR1.1.4, OR52B4) were used as queries. For the names and sequences of human OR genes, see ref. 5. Here the nomenclature adopted by the HUGO Gene Nomenclature Committee is also presented (www.gene.ucl.ac.uk/nomenclature).] From the hits obtained by homology search, mammalian genes and sequences <250 aa long were excluded. The remaining sequences were used for further analysis. We constructed a neighbor-joining (NJ) phylogenetic tree (26) by using the remaining sequences and 388 functional OR genes identified from the complete human genome sequences (5). OR genes formed a monophyletic clade supported with a high bootstrap value (>90%) and were therefore easily distinguishable from non-OR GPCR genes. We excluded these non-OR genes and obtained 106 nonredundant full-length functional OR genes.
Identification of OR Genes from Draft Genome Sequences. The procedure of the identification of functional and nonfunctional OR genes is drawn as a flow chart in Fig. 3, which is published as supporting information on the PNAS web site. The draft genome sequences of zebrafish (Danio rerio), pufferfish (Fugu rubripes), western clawed frogs (X. tropicalis), and chickens (Gallus gallus) were downloaded from the following web sites: www.ensembl.org (assembly Zv3, released in November 2003), genome.jgi-psf.org (assembly v3.0, released in August 2002; ref. 27), genome.jgi-psf.org (assembly v1.0, released in February 2004), and genome.ucsc.edu (galGal2, released in February 2004; ref. 28), respectively. We performed tblastn searches (25) with the E value below 1e-10 against these genome sequences by using 106 fish/amphibian/avian OR genes obtained (see above) and 388 human functional OR genes (5) as queries.
We identified functional OR genes from the blast hits obtained above in the following way, which is modified from the method used in our previous study (5). We first excluded the sequences <250 aa (criterion 1 in Fig. 3). When a sequence contained interrupting stop codons or frameshifts, the longest sequence between a stop codon/frameshift and another stop codon/frameshift was considered, and if the longest sequence was <250 aa, it was excluded. Some of the remaining sequences (sequences of ≥250 aa) were also excluded by additional filtering processes (criteria 2 and 3). Because a blast hit may not cover the entire coding region of a gene, we extended the DNA sequence of each of the blast hits to both 3′ and 5′ directions along the chromosome and extracted the longest sequence that starts with the initiation codon ATG and ends with the stop codon. All of the sequences were translated into amino acid sequences. A multiple alignment for these amino acid sequences and the sequences of 388 human functional OR genes (5) was constructed by using the program fft-ns-i (29), and appropriate positions of the start codon were chosen by visual inspection. We then assigned seven transmembrane regions according to Man et al. (30). The sequences containing long deletions or insertions within transmembrane regions were excluded by visual inspection (criterion 2). The remaining sequences may include non-OR GPCR genes. To exclude non-OR GPCR genes from the remaining sequences, we constructed a phylogenetic tree (see below) by using all of these sequences and several known non-OR GPCR genes collected from the DDBJ database. The non-OR GPCR genes formed a monophyletic clade with some of the remaining sequences in the phylogenetic tree. We regarded these sequences as non-OR genes and excluded them from our data set (criterion 3). The distinction between OR genes and non-OR genes was clear, and there were no gray-area genes. The remaining sequences were regarded as functional OR genes.
As a result, we obtained 608 functional OR genes from the draft genome sequences of the four species. We conducted a tblastn search against these draft genome sequences once again by using the 608 OR genes as queries. All of the processes described above were repeated for the additional blast hits newly obtained by the second-round tblastn search. We then obtained 18 more functional OR genes, most of which were type 2 genes (see Fig. 1B). We therefore identified 626 functional OR genes in total from the four species.
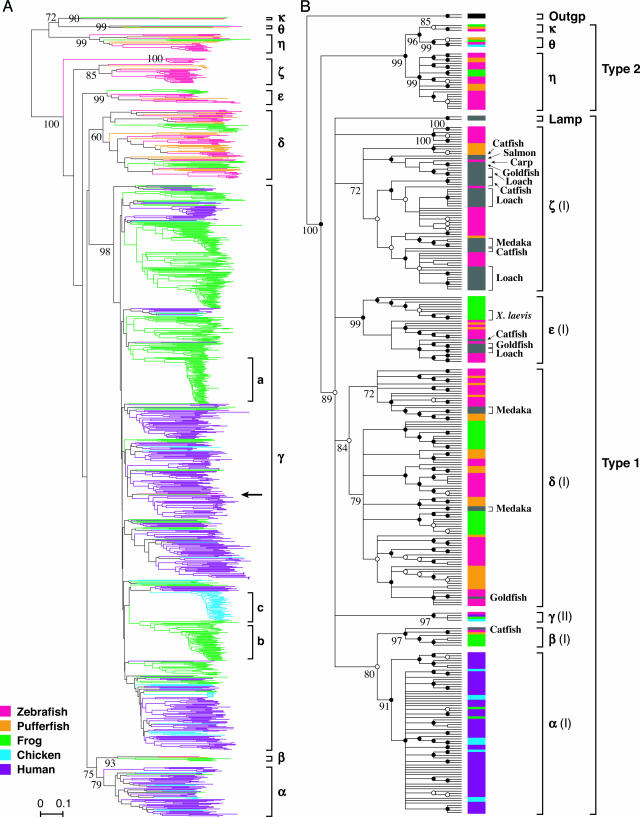
Fig. 1.
Phylogenetic trees of vertebrate OR genes. (A) NJ tree for 1,026 functional OR genes. This tree contains 102 zebrafish, 44 pufferfish, 410 X. tropicalis, 82 chicken, and 388 human (5) OR genes. The number of amino acid sites used was 173 after deletion of all alignment gaps. A branch specific to each species is colored according to the color code at the bottom left. “Frog” indicates X. tropicalis. The scale bar indicates the estimated number of amino acid substitutions per site. The name of each OR gene group, defined in B, is shown. The bootstrap value obtained from 1,000 replications is shown for the clade determining each group. The arrow indicates the zebrafish group γ gene (Dr3OR5.4). Clade a and b and clade c indicate the rapid expansion of group γ genes in the X. tropicalis and chicken lineages, respectively. The GenBank accession numbers of the genes not assigned to the genome sequences are U42392, U42394, and AF283560-AF283561 for zebrafish genes; AB031380 and AB031383-AB031385 for pufferfish genes; and Z79585 and Z79587-Z79589 for chicken genes. (B) Condensed phylogenetic tree (31) at the 70% bootstrap value level for 310 functional OR genes and two outgroup non-OR GPCR genes. This condensed tree was produced from the NJ tree shown in Fig. 4 by assuming that all of the interior branches showing <70% bootstrap values had branch length 0. Bootstrap values obtained from 1,000 replications are shown for major clades. Black and white dots at nodes indicate the branches supported by >90% and >80% bootstrap values, respectively. (I) or (II) after the name of an OR gene group (α-ζ) indicates class I or II in the currently accepted classification of vertebrate OR genes. As the outgroup (Outgp), the bovine adenosine A1 receptor (GenBank accession no. X63592) and rat α2B-adrenergic receptor (AF366899) were used. As for group γ genes, four representative genes from zebrafish (Dr3OR5.4), X. tropicalis (Xt1OR1178.1), chickens (Gg2OR5.8), and humans (HsOR11.18.9, OR10G6) were used. Fish and amphibian OR genes found in the DDBJ database were also included. The color code for each bar showing the species is the same as in A. Four X. laevis genes (AJ249404 and AJ250750-AJ250752) are indicated by the same color (green) as X. tropicalis. Fish genes from the species other than zebrafish and pufferfish are shown by gray bars with their names: Carp, common carp (Cyprinus carpio, AB038166); Catfish, channel catfish (Ictalurus punctatus, L09217-L09225); Goldfish, goldfish (Carassius auratus, AF083076-AF083079); Lamp, European river lamprey (Lampetra fluviatilis, AJ012708-AJ012709); Loach, Japanese loach (Misgurnus anguillicaudatus, AB115055-AB115078); Medaka, Japanese medaka (Oryzias latipes, AB022646-AB022647 and AB029474-AB029480); Salmon, Atlantic salmon (Salmo salar, AY007188).
We next detected OR pseudogenes and partial sequences of OR genes from the blast hits obtained by conducting tblastn search against the draft genome sequences. We excluded blast hits <50 aa long (criterion 4 in Fig. 3), because it is difficult to distinguish between OR and non-OR genes for short sequences. Each of the blast hit sequences was translated into an amino acid sequence, and blastp (25) search was conducted against the DAD database on the DDBJ web site (www.ddbj.nig.ac.jp) by using the translated amino acid sequences as queries. A blastp search was also conducted against the 626 functional OR genes from the four species and 388 functional OR genes from humans. When the best hit of a given query was an OR gene, we regarded the query sequence as an OR pseudogene or a partial sequence of an OR gene. Otherwise, we regarded the query sequence as a non-OR gene and excluded the sequence from our data set (criterion 5). OR pseudogenes were clearly distinguishable from non-OR genes by using this procedure.
Phylogenetic Analysis. Phylogenetic trees were constructed in the following way. Translated amino acid sequences were aligned by the program fft-ns-i (29). Poisson correction distances (31) were calculated after all alignment gaps were eliminated. A phylogenetic tree was constructed from these distances by using the NJ method given in the program lintree (32).
Classification of Functional and Nonfunctional OR Genes. The functional OR genes from zebrafish, pufferfish, X. tropicalis, and chickens were classified into groups α-κ on the basis of the phylogenetic tree in Fig. 1. The classification of OR pseudogenes and partial sequences into the nine groups was done in the following way. In this analysis, we used 1,088 OR genes containing all of the functional OR genes identified from the four species and humans and all of the nonredundant functional OR genes in fishes, amphibians, and birds collected from the public database. We conducted a blastp search against the 1,088 functional OR genes by using the translated amino acid sequence of each of the OR pseudogenes and OR partial sequences as a query. The query sequence was assigned to the group to which the best hit of the query belonged.
Results
OR Genes in Zebrafish, Pufferfish, X. tropicalis, and Chickens. We identified OR genes from the draft genome sequences of zebrafish, pufferfish, X. tropicalis, and chickens by conducting a homology search (see Materials and Methods). Table 1 shows the numbers of functional OR genes identified for these four species. Note that these numbers are the minimal estimates, because the genome sequencing of the four species has not been completed. Therefore, an OR gene may be split into two different contigs, or a part of a gene sequence may be missing from the draft genome sequence. In Table 1, therefore, the sequences identified as pseudogenes or partial sequences may contain a considerable number of functional genes.
Table 1. Functional and nonfunctional OR genes in six vertebrate species.
| Zebrafish | Pufferfish | X. tropicalis | Chicken | Mouse* | Human* | |
|---|---|---|---|---|---|---|
| Total | 133 | 94 | 888 | 554 | 1,391 | 802 |
| Functional genes | 98 | 40 | 410 | 78 | 1,037 | 388 |
| Pseudogenes or partial sequences† | 35 | 54 | 478 | 476 | 354‡ | 414‡ |
| Functional genes in database§ | 4 | 4 | 0 | 4 | —¶ | —¶ |
Total number of OR genes minus the number of functional genes. A partial sequence indicates a part of a functional gene or of a pseudogene that was generated because of incomplete sequencing (see text).
Number of pseudogenes.
Number of full-length functional OR genes found in the DDBJ database but not assigned to the genome.
Not examined.
As shown in Table 1, the numbers of OR genes found from the zebrafish and pufferfish genomes are close to the previous estimate from catfish (≈100; ref. 12). We found that X. tropicalis has a larger number of functional OR genes than humans. We identified only 78 functional genes of >500 OR gene sequences in the chicken genome. However, the actual number of functional OR genes in the chicken genome appears to be much larger than 78, because the chicken draft genome sequence includes numerous short contigs that are a few kilobases long. (Approximately one-half of the contigs are shorter than 2 kb.) The lists and the nucleotide and amino acid sequences for the OR genes identified in this study are available as Tables 3-6 and Data Sets 1-8, which are published as supporting information on the PNAS web site. Several functional OR genes in the DDBJ database were not assigned to the genomes but are used for the phylogenetic analysis in addition to the genes identified from the genome sequences.
Phylogenetic Analysis. To examine the evolutionary relationships among the functional OR genes obtained above, we constructed a NJ tree for 1,026 amino acid sequences of all functional genes identified from the zebrafish, pufferfish, X. tropicalis, chicken, and human genome sequences (Fig. 1A). Here we did not use mouse OR genes, because they are evolutionarily close to human genes, as shown in previous studies (6, 9). Fig. 1A shows that human class II OR genes form a monophyletic clade with most of the OR genes from X. tropicalis and chickens, and this clade is supported with a 98% bootstrap value. We refer to the genes included in this clade as group γ genes. Group γ genes are equivalent to class II genes for humans or mice. (OR gene groups are determined on the basis of the phylogenetic tree in Fig. 1B; see below.) Approximately 90% of the functional genes in X. tropicalis or chickens are group γ genes, and this fraction is similar to that in humans or mice (Table 2). Fig. 1A also indicates that group γ OR genes rapidly expanded during a short period in the X. tropicalis lineage (clades a and b) and chicken lineage (clade c; ref. 28). We found that one zebrafish gene (indicated by the arrow sign in Fig. 1A) is included in the group γ gene clade, although it had been thought that fishes do not have any class II genes.
Table 2. Numbers of functional and nonfunctional OR genes belonging to different groups in six species.
| Zebrafish
|
Pufferfish
|
X. tropicalis
|
Chicken
|
Mouse†
|
Human†
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group* | Funct | Total | Funct | Total | Funct | Total | Funct | Total | Funct | Total | Funct | Total |
| α (I) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (0.5) | 6 (0.7) | 9 (11.0) | 14 (2.5) | 115 (11.1) | 163 (11.7) | 57 (14.7) | 102 (12.7) |
| β (I) | 1 (1.0) | 2 (1.5) | 1 (2.3) | 1 (1.0) | 5 (1.2) | 19 (2.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| γ (II) | 1 (1.0) | 1 (0.7) | 0 (0.0) | 0 (0.0) | 370 (90.2) | 802 (90.3) | 72 (87.8) | 543 (97.3) | 922 (88.9) | 1,228 (88.3) | 331 (85.3) | 700 (87.3) |
| δ (I) | 44 (43.1) | 55 (40.1) | 28 (63.6) | 61 (62.2) | 22 (5.4) | 36 (4.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| ε (I) | 11 (10.8) | 14 (10.2) | 2 (4.5) | 2 (2.0) | 6 (1.5) | 17 (1.9) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| ζ (I) | 27 (26.5) | 40 (29.2) | 6 (13.6) | 8 (8.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| η | 16 (15.7) | 23 (16.8) | 5 (11.4) | 24 (24.5) | 3 (0.7) | 6 (0.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Θ | 1 (1.0) | 1 (0.7) | 1 (2.3) | 1 (1.0) | 1 (0.2) | 1 (0.1) | 1 (1.2) | 1 (0.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| κ | 1 (1.0) | 1 (0.7) | 1 (2.3) | 1 (1.0) | 1 (0.2) | 1 (0.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Total | 102 (100.0) | 137 (100.0) | 44 (100.0) | 98 (100.0) | 410 (100.0) | 888 (100.0) | 82 (100.0) | 558 (100.0) | 1,037 (100.0) | 1,391 (100.0) | 388 (100.0) | 802 (100.0) |
The numbers in parentheses represent the percentage of genes for each group. Funct, functional genes; Total, total number of genes including functional genes, pseudogenes, and partial sequences.
(I) and (II) indicate class I and II, respectively, in the currently accepted classification of vertebrate OR genes. Groups η, Θ, and κ were newly identified in this study and therefore are neither class I nor class II.
The data for humans and mice were obtained from refs. 5 and 9, respectively.
To examine the evolutionary relationships among non-group-γ OR genes, we constructed another NJ tree after excluding all group γ genes except four representative genes from zebrafish, X. tropicalis, chickens, and humans. We also used fish and amphibian functional OR genes found in the DDBJ database. As the outgroup, two divergent non-OR GPCR genes were used. In Fig. 1B, we showed a condensed tree at the 70% bootstrap value level to understand the relationships of different clades (31). Note that this tree presents the topology only, and the branch lengths do not reflect evolutionary distances. The original tree used for constructing the condensed tree is shown in Fig. 4, which is published as supporting information on the PNAS web site.
Fig. 1B shows that all of the OR genes identified in this study formed a monophyletic clade with a 100% bootstrap support. This OR gene clade was divided into two phylogenetic clades supported with 89% and 99% bootstrap values. We named the genes contained in these clades type 1 and 2 genes. All of the known OR genes found in the public database were type 1 genes, whereas type 2 genes were previously undescribed. Because the function of type 2 genes is unknown, type 2 genes may be non-OR genes (see Discussion). However, for simplicity, here we call them OR genes, because type 2 genes are more similar to known (type 1) OR genes than to any other non-OR GPCR genes. This is supported by the following facts: (i) type 2 genes formed a monophyletic clade together with known OR genes in a phylogenetic tree when all non-OR GPCR genes obtained by blastp search were included, and (ii) the best hit of a type 2 gene was always an OR gene rather than a non-OR GPCR gene when a blastp search was conducted against all known genes in the DDBJ database. The type 1 gene clade included lamprey genes, suggesting that the separation between type 1 and 2 OR genes occurred before the divergence between jawed and jawless vertebrates (see Discussion).
As shown in Fig. 1B, type 1 and 2 gene clades are subdivided into several phylogenetic clades supported with high bootstrap values. Here we define an OR gene group for jawed vertebrates by taking a well supported phylogenetic clade that diverged before the divergence between fishes and tetrapods. When such a clade was nested in another larger clade, we adopted the smaller one. In other words, each group corresponds to at least one ancestral OR gene in the most recent common ancestor (MRCA) between fishes and tetrapods. For example, group ε clade was supported with a 99% bootstrap value, and this clade contained a tetrapod- and a fish-specific clade, suggesting that the MRCA between fishes and tetrapods had at least one ancestral group ε gene. Similarly, the clades determining groups β, γ, δ, η, θ, and κ were supported with high (>80%) bootstrap values, and each of them contained both fish and tetrapod genes. Groups α and ζ are tetrapod- and fish-specific, respectively. However, Fig. 1B indicates that both group α and ζ genes diverged from other group genes before the fish-tetrapod divergence, because all other groups (β, γ, δ, and ε) in type 1 clade were well supported and contained both fish and tetrapod genes. Moreover, the monophyly of group α and ζ genes was not supported, because group α genes formed a monophyletic clade with group β genes in Fig. 1B, suggesting that group α and ζ genes originated from different ancestral genes in the MRCA of fishes and tetrapods. Group ζ was supported only with a 67% bootstrap value (see Fig. 4), and therefore this group is not represented as a monophyletic clade in Fig. 1B. However, for simplicity, we regarded that group ζ is a monophyletic group. In this way, we classified type 1 and 2 genes into six (α-ζ) and three groups (η, θ, and κ), respectively. Note that one group may correspond to two or more ancestral genes in the fish-tetrapod MRCA. Therefore, this classification gives a minimal estimate of the number of ancestral OR genes in the MRCA between fishes and tetrapods. All of the human class I genes belonged to group α. However, known fish OR genes found in the public database, which are also called class I genes, belonged to four different groups, β, δ, ε, and ζ. Therefore, class I genes correspond to genes belonging to five groups (α, β, δ, ε, and ζ) in our new classification, whereas class II genes correspond to group γ genes.
We also classified OR pseudogenes and partial sequences into the nine groups defined above. Table 2 shows the numbers of functional and nonfunctional OR genes belonging to different groups in six species. The distribution of OR genes in the nine groups is quite different among species. Zebrafish have genes belonging to all groups except group α. By contrast, humans and mice have only group α and γ genes. (We conducted a tblastn search against human and mouse genome sequences by using all of the functional OR genes identified in this study as queries. However, we did not find any OR functional genes or pseudogenes belonging to the groups other than α or γ.) Almost all chicken OR genes belong to either group α or γ, but one putatively functional gene belonging to group θ was also found. The OR genes in X. tropicalis belong to all of the different groups except group ζ. Most X. tropicalis genes are group γ genes, which are very rare in fishes but are abundant in mammals or birds. However, X. tropicalis also have a considerable number of group δ genes, which are common in fishes.
Discussion
Evolution of Vertebrate OR Genes. The results in Table 2 can be summarized as follows. (i) Although the total number of OR genes is much smaller in fishes than in mammals or birds, the diversity of an OR gene family is much larger in fishes. (ii) OR gene groups present in mammals or birds (groups α and γ) are absent or very rare in fishes and vice versa. (iii) The OR gene family in amphibians is as diverse as that in fishes, but the majority of OR genes in amphibians and mammals or birds belong to the same group (group γ). In other words, the OR gene repertoire in amphibians is similar to that in fishes in one aspect and to that in mammals or birds in another aspect. As we have mentioned, it is possible that type 2 genes are actually non-OR genes. Note, however, that observations i-iii do not essentially change even if we confine our argument only to type 1 OR genes. The presence or absence of OR gene groups in the organisms living in the aquatic environment (fishes and amphibians) and those in the terrestrial environment (amphibians, birds, and mammals) suggests that group α and γ OR genes are specialized for detecting airborne odorants, and other group genes are for detecting water-soluble odorants.
Observations i-iii can be well understood by considering the long-term evolution of vertebrate OR genes (Fig. 2). As we mentioned earlier, the divergence between type 1 and 2 genes appears to have occurred before the divergence between jawed and jawless vertebrates. Therefore, the MRCA between jawed and jawless vertebrates is thought to have had at least two OR genes. Here we should note that another family of OR genes in lampreys (L1OR) has been reported (1, 33). However, we did not use L1OR genes, because L1OR genes are more similar to non-OR GPCRs such as histamine receptors than to any OR genes used in this study (data not shown), and thus the evolutionary origin of L1OR genes is different from that of the OR genes.
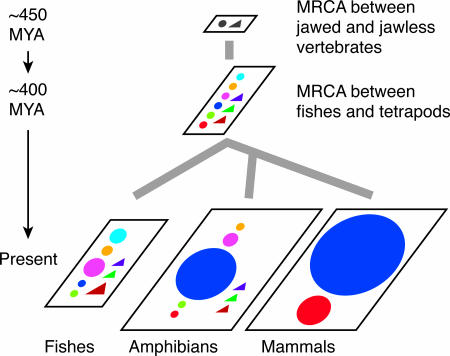
Fig. 2.
Evolutionary dynamics of vertebrate OR genes. The MRCA between jawed and jawless vertebrates had at least two ancestral OR genes, type 1 (gray circle) and type 2 (gray triangle). In the jawed vertebrate lineage, the ancestral type 1 genes had diverged to group α-ζ genes (colored circles), whereas ancestral type 2 genes became group η, θ, and κ genes (colored triangles). The MRCA between fishes and tetrapods had at least nine ancestral genes corresponding to the nine groups. At present, some extant fish species retain OR genes belonging to eight different groups except group α, which is illustrated by five colored circles and three triangles. In the mammalian lineage, seven groups of genes have been lost, but the number of group α (a red circle) and γ genes (a blue circle) have greatly increased to generate an enormous OR gene family. Expansion of group γ genes is also observed in amphibians, but they have genes belonging to eight different groups. The OR gene repertoire in birds (not shown) is similar to that in mammals. The divergence times between jawed and jawless vertebrates and between fishes and tetrapods were obtained from ref. 35.
As indicated in Fig. 1B, the MRCA between fishes and tetrapods was estimated to have had at least six type 1 and three type 2 OR genes. Group α genes are specific to tetrapods, suggesting either that this group of genes have been lost in the fish lineage or that fish group α genes have not yet been found. Fish-specific group ζ genes can be explained in a similar manner. The MRCA between fishes and tetrapods is believed to have lived in the sea, and the OR genes in the MRCA therefore should have detected water-soluble odorants. Fishes retain OR genes belonging to almost all of the nine groups at present, probably because their environment has not changed substantially compared with that of the MRCA. In the tetrapod lineage, group α and γ genes apparently acquired the ability to detect airborne odorants at the time of terrestrial adaptation. After that, the OR genes specific to water-soluble odorants, i.e., genes other than group α or γ genes, appear to have been lost in the mammalian and avian lineages. Although the number of different OR gene groups substantially decreased in mammals and birds, the number of different OR genes, especially those belonging to group γ, have enormously increased. In the amphibian lineage, both the OR gene groups for detecting water-soluble odorants and those for airborne odorants are still present, reflecting that amphibians have adapted to both aquatic and terrestrial environments. Group γ gene expansion was observed in every tetrapod species examined, including amphibians. The reason for the expansion of group γ genes is unclear, but it is possible that more OR genes would have been required in the terrestrial than in the aquatic environment.
As we have seen above, the evolutionary dynamics of an OR gene family is characterized by processes in which some gene lineages have produced many descendants, whereas other gene lineages have been completely lost. Therefore, the OR multigene family is an excellent example of birth-and-death evolution (5, 9, 34).
Classification of Vertebrate OR Genes. As we have mentioned, it is currently accepted that all vertebrate OR genes can be grouped into fish-like class I genes and mammalian-like class II genes. However, our phylogenetic analysis (Fig. 1B) suggested that vertebrate OR genes reported so far (i.e., type 1 OR genes) are classified into at least six different groups. This difference can be explained in the following way. The classification of OR genes into class I and II genes is primarily based on Freitag et al.'s study (19), which proposed that X. laevis have two different classes of OR genes, one (class I) being similar to known fish genes and the other (class II) being close to known mammalian genes. Our phylogenetic analysis supported Freitag et al.'s study, because X. laevis genes used in their study were assigned to group γ and ε in our classification, which contained mammalian and fish genes, respectively. (These X. laevis genes are not included in Fig. 1B, because they are partial, but we confirmed they are closest to group ε genes by homology search. Four X. laevis group ε genes in Fig. 1B are from ref. 22.) Moreover, many studies supported the view that mammalian OR genes are grouped into class I and II genes (4-6, 9). Our current analysis is also consistent with this view, because mammalian class I (group α) and II genes (group γ) were clearly separated in Fig. 1B. Therefore, this portion of Fig. 1B is not inconsistent with previous studies. However, classification of all vertebrate OR genes into two classes would not be appropriate, because Fig. 1B indicated that class I genes have originated from at least five ancestral genes in the MRCA between fishes and tetrapods, whereas class II genes are likely to have originated from one ancestral gene. Moreover, as we have mentioned, the function of class I genes was unclear, because both fishes and mammals have a substantial number of class I genes. According to our new classification, however, OR genes belonging to each group are likely to be specific to either water-soluble or airborne odorants. Therefore, our classification of OR genes appears to be more reasonable than the previous one from both evolutionary and functional aspects. Our hypothesis of the functional specificity of each group of genes, however, should be tested experimentally. One way to test this hypothesis might be to examine the difference in gene expression pattern at the whole-genome level between larval and adult amphibians.
Supplementary Material
Acknowledgments
We thank Shozo Yokoyama, Alex Rooney, Jianzhi Zhang, Jan Klein, and Blair Hedges for helpful comments and discussion. This work was supported by National Institutes of Health Grant GM20293 (to M.N.). The sequence data for the zebrafish genome were produced by the Zebrafish Sequencing Group at the Sanger Institute and can be obtained from www.ensembl.org/Danio_rerio. The data for the pufferfish genome have been provided freely by the Fugu Genome Consortium for use in this publication only. The data for the X. tropicalis genome were provided by the Department of Energy's Joint Genome Institute. The data for the chicken genome were provided by the Genome Sequencing Center, Washington University School of Medicine (St. Louis).
Author contributions: Y.N. and M.N. designed research; Y.N. performed research; Y.N. analyzed data; and Y.N. and M.N. wrote the paper.
Abbreviations: OR, olfactory receptor; NJ, neighbor-joining; MRCA, most recent common ancestor; GPCR, G protein-coupled receptor; DDBJ, DNA Databank of Japan.
References
- 1.Dryer, L. (2000) BioEssays 22, 803-810. [DOI] [PubMed] [Google Scholar]
- 2.Firestein, S. (2001) Nature 413, 211-218. [DOI] [PubMed] [Google Scholar]
- 3.Mombaerts, P. (2004) Nat. Rev. Neurosci. 5, 263-278. [DOI] [PubMed] [Google Scholar]
- 4.Glusman, G., Yanai, I., Rubin, I. & Lancet, D. (2001) Genome Res. 11, 685-702. [DOI] [PubMed] [Google Scholar]
- 5.Niimura, Y. & Nei, M. (2003) Proc. Natl. Acad. Sci. USA 100, 12235-12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang, X. & Firestein, S. (2002) Nat. Neurosci. 5, 124-133. [DOI] [PubMed] [Google Scholar]
- 7.Young, J. M., Friedman, C., Williams, E. M., Ross, J. A., Tonnes-Priddy, L. & Trask, B. J. (2002) Hum. Mol. Genet. 11, 535-546. [DOI] [PubMed] [Google Scholar]
- 8.Zhang, X., Rodriguez, I., Mombaerts, P. & Firestein, S. (2004) Genomics 83, 802-811. [DOI] [PubMed] [Google Scholar]
- 9.Niimura, Y. & Nei, M. (2005) Gene 246, 13-21. [DOI] [PubMed] [Google Scholar]
- 10.Buck, L. & Axel, R. (1991) Cell 65, 175-187. [DOI] [PubMed] [Google Scholar]
- 11.Freitag, J., Beck, A., Ludwig, G., von Buchholtz, L. & Breer, H. (1999) Gene 226, 165-174. [DOI] [PubMed] [Google Scholar]
- 12.Ngai, J., Dowling, M. M., Buck, L., Axel, R. & Chess, A. (1993) Cell 72, 657-666. [DOI] [PubMed] [Google Scholar]
- 13.Barth, A. L., Dugas, J. C. & Ngai, J. (1997) Neuron 19, 359-369. [DOI] [PubMed] [Google Scholar]
- 14.Dugas, J. C. & Ngai, J. (2001) Genomics 71, 53-65. [DOI] [PubMed] [Google Scholar]
- 15.Cao, Y., Oh, B. C. & Stryer, L. (1998) Proc. Natl. Acad. Sci. USA 95, 11987-11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun, H., Kondo, R., Shima, A., Naruse, K., Hori, H. & Chigusa, S. I. (1999) Gene 231, 137-145. [DOI] [PubMed] [Google Scholar]
- 17.Kondo, R., Kaneko, S., Sun, H., Sakaizumi, M. & Chigusa, S. I. (2002) Gene 282, 113-120. [DOI] [PubMed] [Google Scholar]
- 18.Irie-Kushiyama, S., Asano-Miyoshi, M., Suda, T., Abe, K. & Emori, Y. (2004) Gene 325, 123-135. [DOI] [PubMed] [Google Scholar]
- 19.Freitag, J., Krieger, J., Strotmann, J. & Breer, H. (1995) Neuron 15, 1383-1392. [DOI] [PubMed] [Google Scholar]
- 20.Nef, S., Allaman, I., Fiumelli, H., De Castro, E. & Nef, P. (1996) Mech. Dev. 55, 65-77. [DOI] [PubMed] [Google Scholar]
- 21.Glusman, G., Bahar, A., Sharon, D., Pilpel, Y., White, J. & Lancet, D. (2000) Mamm. Genome 11, 1016-1023. [DOI] [PubMed] [Google Scholar]
- 22.Mezler, M., Fleischer, J. & Breer, H. (2001) J. Exp. Biol. 204, 2987-2997. [DOI] [PubMed] [Google Scholar]
- 23.Fuchs, T., Glusman, G., Horn-Saban, S., Lancet, D. & Pilpel, Y. (2001) Hum. Genet. 108, 1-13. [DOI] [PubMed] [Google Scholar]
- 24.Malnic, B., Godfrey, P. A. & Buck, L. B. (2004) Proc. Natl. Acad. Sci. USA 101, 2584-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saitou, N. & Nei, M. (1987) Mol. Biol. Evol. 4, 406-425. [DOI] [PubMed] [Google Scholar]
- 27.Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J. M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. (2002) Science 23, 1301-1310. [DOI] [PubMed] [Google Scholar]
- 28.International Chicken Genome Sequencing Consortium (2004) Nature 432, 695-716. [DOI] [PubMed] [Google Scholar]
- 29.Katoh, K., Misawa, K., Kuma, K. & Miyata, T. (2002) Nucleic Acids Res. 30, 3059-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Man, O., Gilad, Y. & Lancet, D. (2004) Protein Sci. 13, 240-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nei, M. & Kumar, S. (2000) in Molecular Evolution and Phylogenetics (Oxford Univ. Press, New York).
- 32.Takezaki, N., Rzhetsky, A. & Nei, M. (1995) Mol. Biol. Evol. 12, 823-833. [DOI] [PubMed] [Google Scholar]
- 33.Berghard, A. & Dryer, L. (1998) J. Neurobiol. 15, 383-392. [PubMed] [Google Scholar]
- 34.Nei, M., Gu, X. & Sitnikova, T. (1997) Proc. Natl. Acad. Sci. USA 94, 7799-7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nei, M. (1987) in Molecular Evolutionary Genetics (Columbia Univ. Press, New York), p. 10.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.