Abstract
The restriction of influenza A virus replication to mouse respiratory epithelium means that this host response is anatomically compartmentalized, on the one hand, to sites of T cell stimulation and proliferation in the secondary lymphoid tissue and, on the other hand, to the site of effector T cell function and pathology in the pneumonic lung. Thus, it is hardly surprising that virus-specific CD8+ T cells recovered by bronchoalveolar lavage (BAL) from the infected respiratory tract seem more “activated” in terms of both cytolytic activity and cytokine production than those cells isolated from the spleen. The present analysis uses Affymetrix microarray technology to compare profiles of gene expression in these two lineage-related, yet anatomically separate, lymphocyte populations. Ninety differentially expressed genes were identified for influenza-specific CD8+DbNP366+ T cells obtained directly ex vivo by BAL or spleen disruption, with nine genes being further analyzed by quantitative, real-time PCR at the population level. Integrin αE, for example, was shown by Affymetrix and real-time mRNA analyses and then by single-cell PCR and protein staining to be present at a much higher prevalence on the BAL CD8+DbNP366+ set. The unpredicted finding, however, was that mRNA expression for 75% of the 90 genes was lower in T cells from the BAL than from the spleen. Apparently, the localization of virus-specific CD8+ T cells to the site of virus-induced pathology is associated with a narrowing, or “focusing,” of gene expression that favors enhanced effector function in the damaged, “high-antigen load” environment of the pneumonic lung.
Keywords: Affymetrix gene array, cytotoxic T cell, influenza A virus, viral immunology
The murine influenza model provides an optimal experimental system for analyzing the compartmentalization of immunity in a localized infectious process (1-5). After an intranasal (i.n.) challenge to C57BL/6J (B6) mice, there is no productive infection in the lymphoid tissue, and virus replication is essentially limited to the respiratory epithelium (1). The differentiation of influenza A virus-specific CD8+ T cells from naïve precursors to effector cytotoxic T lymphocytes (CTLs) is characterized by an initial phase of clonal expansion in the regional lymph nodes and spleen, followed by emigration, relocation, and the development of potent CTL activity in the “high-antigen” environment of the infected lung (1, 4, 5). Once the virus is cleared from the respiratory tract, the T cell numbers contract to form a diminished but stable memory population (5, 6).
The response of B6 (H-2b) mice to the influenza A virus is directed to a total of seven different influenza epitopes (7-10), with one of the most prominent being the viral nucleoprotein (NP366-374) presented by H-2Db (7). A range of phenotypic and functional studies with CD8+DbNP366+ T cells have established that CTLs isolated from the airways of influenza virus-infected mice by bronchoalveolar lavage (BAL) exhibit what seems to be an increased activation status (3-5, 9, 11, 12). This status includes higher levels of the “early activation marker” CD69 (5) and the TNF receptor 2 (12) relative to the comparable populations recovered from the spleen. These findings are in accord with other indications that the molecular profiles of the BAL set reflect a level of differentiation beyond that found for the spleen or, indeed, in the T cells recovered by collagenase digestion of the perfused, lavaged, pneumonic lung (3, 11, 13, 14). The proportion of CD8+DbNP366+ T cells capable of producing multiple cytokines is, for instance, significantly greater for the BAL population (9, 11).
The genetic program that drives this sequential maturation process is complex and, even at our present level of understanding, can be thought to involve a spectrum of signaling pathways and the transcription of multiple genes (15). Comparative profiles of mRNA expression in naïve, activated, and memory CTLs have been analyzed for the systemic infection caused by lymphocytic choriomeningitis virus (LCMV) with use of Affymetrix gene array technology. The experiments focused on T cells from the lymphoid tissue that supports LCMV replication (15) and did not, therefore, target the linked issues of anatomical compartmentalization and various antigen load that are readily addressed with the influenza model.
The present analysis compares gene expression profiles for the immunodominant CD8+DbNP366 set recovered from either the BAL or spleen of mice secondarily challenged with the influenza A HKx31 virus (H3N2). The naïve expectation was that the spectrum of gene “readout” would be much higher for the more “activated” T cells in the infected respiratory tract. The results indicate, however, that there is a substantial narrowing (or focusing) of the gene program in effector CTLs recovered from a site of virus-induced pathology.
Materials and Methods
Mice and Viruses. Conventional B6 female mice were purchased from The Jackson Laboratory. Memory mice for secondary challenge were generated by injecting 107.9 EID50 of the influenza A virus A/Puerto Rico/34/8 (PR8; H1N1) i.p. These mice were held for at least 4-6 weeks, then anesthetized with avertin and infected with 106.8 EID50 of the influenza A HKx31 (H3N2) virus via the i.n. route. Whereas the hemagglutinin (H) and neuraminidase (N) proteins of PR8 and HKx31 are not recognized by cross-reactive neutralizing Abs, these two viruses share the immunodominant NP366-374 (ASNENMETM) peptide. Virus-specific CD8+ CTLs were identified by using tetrameric complexes of the H-2Db glycoprotein and the NP366-374 determinant. The generation of the DbNP366 tetramer has been described (4).
Tissue Sampling, Cell Sorting, and Intracellular Cytokine Staining. Single-cell lymphocyte suspensions were prepared from the pooled spleens of secondary challenged B6 mice, erythrocytes were lysed, and B cells were removed by panning for 1 h at 37°C in an atmosphere of 5% CO2 in T175 cm2 flasks coated overnight with 100 μg/ml goat anti-mouse IgG/IgM (Jackson ImmunoResearch, San Francisco). The nonadherent cells were transferred and incubated with mAbs to CD4 (GK1.5) and MHC class II (TIB120) followed by sheep anti-mouse Ig- and sheep anti-rat Ig-coated beads (Dynal, Oslo) then enriched for the CD8+ subset by magnetic depletion. Lymphocytes were obtained from pooled (n = 50) inflammatory cell populations recovered by BAL of the infected lung and incubated on plastic for 1 h at 37°C to remove adherent cells. For sorting, the predominantly CD8+ population was resuspended at 20 × 106 cells per ml in PBS/0.1% BSA (sort buffer) and stained with the phycoerythrin-conjugated DbNP366 tetramer for 1 h at room temperature, followed by anti-CD8-FITC (Pharmingen) for 30 min at 4°C. After being washed again, the CD8+DbNP366+ set was sorted by using a MoFlo high-speed cell sorter (DakoCytomation, Fort Collins, CO). For intracellular cytokine staining, enriched CD8+ lymphocyte populations from the spleen and BAL were stimulated with the cognate peptide and stained and analyzed as described (11).
RNA Extraction and Affymetrix Analysis. RNA was extracted from the sorted cells by using TRIzol reagent (Invitrogen). The quality of the poly(A)+ mRNA was determined by using Affymetrix quality control methods. The cRNA was then prepared by standard Affymetrix methods, and the quality was evaluated by control hybridization with probes matching the 5′, middle, and 3′ sequences of β-actin, GAPDH, and lack of 18S RNA hybridization as a negative control. Affymetrix MG-U74A and MG-U74Av2 GeneChips were used to analyze gene expression. Signal extraction and analyses of the scanned arrays were performed by using the statistical algorithm of Affymetrix gcos software version 1.1. Signal values were scaled to a 2% trimmed mean target value of 500. For signal extraction and expression comparison, 2,511 of the 12,488 probe sets were excluded because of their incorrect sense orientation on the MG-U74A GeneChip array. Three replicate experiments were performed. Two experiments were analyzed by using the MG-U74A GeneChip and the third was analyzed with use of the MG-U74Av2 GeneChip. Intra- and interexperiment expression comparisons were performed for the first two experiments, whereas only intraexperiment comparison was possible for the third. Stringent filtering criteria were applied to identify differentially expressed genes. First, probe sets identified as “increased” or “decreased” in every experiment by gcos software (P < 0.003) were selected. From this pool, only those probe sets with a >1.5-fold change in every experiment were retained.
Real-Time PCR Analysis. After extracting the RNA (see above), the cDNA was synthesized by using an Omniscript kit (Qiagen, Germantown, MD) according to the manufacturer's instructions. Primers for real-time analysis of CD7, G protein-coupled receptor 56, integrin β7, EDG receptor 6, cathepsin S, CLTA2-α, CTLA2-β, and integrin αE were designed by using primer express software (Applied Biosystems) (the sequences are available upon request). Real-time PCR was performed by using SYBR green chemistry (Applied Biosystems) with GAPDH as the normalizer. The PCR conditions were 48°C for 30 min, 95°C for 10 min, then 40 cycles of 95°C for 15 sec and 60°C for 15 sec; the results were run on an Applied Biosystems ABI7700 RT PCR machine. Results were analyzed by using sequence detector software, and relative fold differences were determined with use of the ΔCt method as described by the manufacturer.
Single-Cell RT-PCR. Lymphocytes derived from the BAL and spleen were stained with anti-CD8α and the DbNP366 tetramer (see above) then sorted by using a MoFlo (DakoCytomation) fitted with a Cyclone single-cell deposition unit. Single CD8+ DbNP366+ T cells were sorted directly into a 96-well PCR plate (Eppendorf, Hamburg, Germany) containing 5 μl of cDNA reaction mix (17, 18). The plates were then incubated at 37°C for 90 min for cDNA synthesis, followed by 5 min at 95°C to stop reverse transcription activity, and stored at -80°C. The CD8α and integrin αE transcripts were amplified by nested PCR with use of 2 μl of cDNA for a 25-μl amplification reaction. The first-round PCR was performed with 1.5 units of Taq polymerase (Invitrogen)/1.5 mM MgCl2/0.2 mM dNTP (Invitrogen)/10 pmol of both the external sense and external antisense primers. A 2-μl aliquot of the first-round PCR was used as a template for the nested PCR with the internal sense and antisense primers. The primer sequences are available on request. The PCR conditions were 95°C for 5 min, followed by 33 cycles of 95°C for 30 sec, 57°C for 30 sec, and 72°C for 1 min, followed by 1 cycle of 95°C for 1 min, 57°C for 1 min, and 72°C for 7 min. The PCR products were resolved on a 2% agarose gel, and the frequency of integrin αE+ cells as a proportion of CD8+ cells was determined.
Results
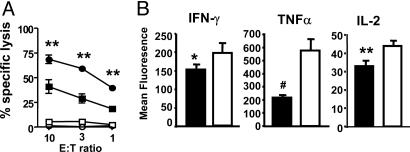
Functional Characteristics of the CD8+DbNP366+ Recall Response. Substantial numbers of tetramer+ T cells are needed to provide sufficient mRNA for the Affymetrix arrays, so the experiments focused on the larger, secondary CD8+DbNP366+ response (4). Earlier analysis of CTL activity in naïve mice infected i.n. with the HKx31 virus showed minimal, if any, lytic activity for the spleen cells, whereas the BAL population contained potent CTL effectors (1). Lymphocyte populations recovered directly ex vivo from secondarily challenged (HKx31 → PR8) mice were assayed for the capacity to induce specific 51Cr release from peptide-pulsed targets. The CD8+DbNP366+ T cell numbers were first adjusted before tetramer staining so that the same effector:target ratio was used for the BAL and spleen populations. The spleen cells showed evidence of CTL activity, but the levels of 51Cr release recorded for the BAL set were significantly (P < 0.001) higher (Fig. 1A) on a per cell basis. Similar differentials were observed when T cells were analyzed for cytokine production (9, 11) by measuring mean fluorescence intensity after a 5-h peptide pulse (Fig. 1B). The CD8+DbNP366+ T cells recovered by BAL of the infected lung demonstrated significantly higher levels of IFN-γ (P < 0.01), TNF-α (P < 0.0007), and IL-2 (P < 0.0005) production when compared with the spleen set (Fig. 1B). Virus-specific CD8+ T cells that have emigrated and localized to the high-antigen environment of the infected respiratory tract are clearly more activated than those that remain in the secondary lymphoid tissue.
Fig. 1.
Differences in ex vivo CTL activity and peptide-induced cytokine production for CD8+DbNP366+ T cells recovered from the respiratory tract by BAL or disaggregation of the spleen. (A) PR8-primed B6 mice (n = 5) were infected i.n. with 104 plaque-forming units of the HKx31 influenza A virus and sampled 8 days later. CD8+ lymphocyte populations were enriched from either the spleen (▪ and □) or BAL (• and ○) populations, and the levels of cytotoxicity measured by specific 51Cr release were determined in vitro after the numbers of CD8+DbNP366+ effectors were adjusted to give the same starting effector:target ratio. Targets were EL4 cells that were either pulsed with 1 μM NP366 peptide (• and ▪) or left unpulsed (○ and □). (B) Other T cells from spleen (▪) and BAL (□) were stimulated for 5 h with 1 μMNP366 peptide in the presence of brefeldin A (8, 11). The CD8α+ sets were analyzed for levels of IFN-γ, TNF-α, and IL-2 expression (mean fluorescence intensity). An unpaired Student t test was used to compare values for spleen and BAL. Results shown are representative of three individual experiments.
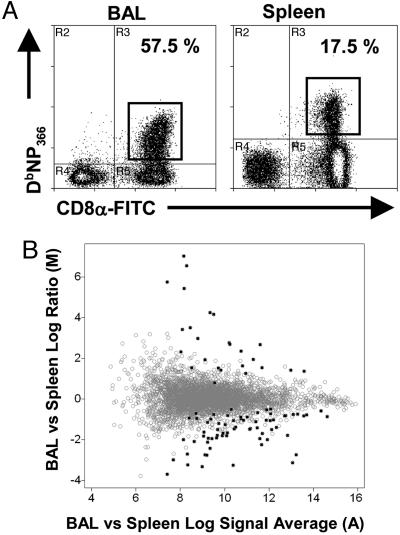
Profiles of Differential Gene Expression Determined by Array Analysis. Spleen and BAL populations were recovered from secondarily challenged mice at 9 d after respiratory exposure to the HKx31 virus. The CD8+DbNP366+ T cells were sorted in the flow cytometer (Fig. 2A), and total RNA was harvested and used to probe U74 Affymetrix GeneChips. The collected gene array data were subjected to gcos analysis in a pairwise fashion. The output from this analysis was identification of genes that had at least a 1.5-fold difference in expression (P < 0.003). The data are presented as a dot plot with the fold change (expressed as a log2 test/control ratio, y axis) versus the signal average (expressed as log2sqrt(test signal × control signal, x axis) (Fig. 2B). Genes expressed at a similar level in the BAL and spleen fall along the “0” line, whereas those detected at a higher level in BAL or spleen fall in the positive or negative directions, respectively. In all, 93 genes were identified as showing at least a 1.5-fold difference (Fig. 2B). Interestingly, only 25 genes were found to have higher expression in BAL than in the spleen, whereas the remaining 65 had the opposite tendency. The general trend was for genes that were more highly expressed in BAL to have a greater average difference in signal strength (Fig. 2B, y axis). While more numerous, those that were prominent in the spleen had lower mean differences in signal strength (Fig. 2B, y axis).
Fig. 2.
Gene array analysis of CD8+DbNP366+ T cells isolated from either the BAL or spleen of PR8 immune mice sampled 9 days after an i.n. challenge with the HKx31 virus. (A) Flow cytometry profiles for the CD8+DbNP366+ populations used as a source of mRNA for the Affymetrix array analysis. (B) Analysis of gene expression in BAL versus spleen after gcos analysis. The y axis shows the magnitude of change between BAL and spleen expression (log2[average BAL signal/average spleen signal]), and the x axis shows the average signal intensity across arrays (log2[average signal]). Differentially expressed probe sets are indicated by filled circles.
A partial list of the genes found to be differentially expressed is provided in Table 1. The complete data set can be obtained from S.J.T upon request. Genes that demonstrated significant expression in the BAL and were not detected above background in the spleen were Chitinase-like 3 (19.1-fold), Isg20 (6.8-fold), integrin αE (17.9-fold), G protein-coupled receptor 56 (10.8-fold), and DNA-damage-induced transcript-4 (3.8-fold). Conversely, six genes were identified as expressed in the spleen population (Table 1) but not detected above background in the BAL set. These genes were Myb (also known as c-myb, -12.9-fold), Lung Krüppel-like factor (LKLF, -10-fold), Deltex 1 homologue (-6.6-fold), Txk tyrosine kinase (-5.3-fold), Rel-B (-3.4-fold), and Kinesin 23 (-3.1-fold).
Table 1. Differential gene expression levels of DbNP366-specific CD8 T cells comparing cells isolated from BAL to spleen.
| GenBank accession no. | Gene name | Fold difference |
|---|---|---|
| Host defense | ||
| M94584 | Chitinase 3-like 3 | 19.1* |
| AW122677 | IFN-stimulated protein 20 | 6.8* |
| X15592 | CTLA-2 β | 5.1 |
| AJ223208 | Cathepsin S | 2.9 |
| X15591 | CTLA-2 α | 2.6 |
| M12302 | Granzyme B | −1.8 |
| AF077347 | IL-18R accessory protein | −2.3 |
| U49513 | CCL9 | −3.5 |
| M13226 | Granzyme A | −4.8 |
| Signal transduction, immune modulation | ||
| U12236 | Integrin αE | 17.9* |
| Al841654 | G protein-coupled receptor 56 | 10.8* |
| X04653 | Ly6a | 2.7 |
| M68903 | Integrin β-7 | −1.7 |
| D86232 | Ly6c | −1.8 |
| AJ237939 | Stat5A | −1.8 |
| AW046181 | Serum/glucocorticoid-regulated kinase | −1.8 |
| AJ006074 | EDG receptor 6 | −2.8 |
| U78171 | RASP-2 | −3.1 |
| AA764261 | Pim 1 | −3.4 |
| D31956 | CD7 antigen | −3.5 |
| AF097357 | KLRG1 | −3.8 |
| U20238 | RAS p21-3 | −3.9 |
| D43963 | TXK kinase | −5.3† |
| U38252 | Deltex 1-homologue (Drosophila) | −6.6† |
| Apoptosis, cell cycle, replication | ||
| NM_029083 | DNA-damage-induced 4 | 3.8* |
| Al642048 | NF-κBi-α | −2 |
| X57800 | Pcna | −2.1 |
| X75316 | RNAP-1 | −2.2 |
| U70494 | H2A-Z | −2.2 |
| AA681998 | CDC28 | −2.6 |
| Al591702 | Kif23 | −3.1† |
| U19799 | NF-κBi-β | −3.2 |
| M83380 | Rel-B | −3.4† |
| Transcription factors | ||
| Al853875 | Znfr 1 | 5 |
| Al835041 | Zfp265 | 2.9 |
| X17459 | RBPS | 2.7 |
| Af053367 | Elfin 1 | −2 |
| X89749 | TG-interacting factor | −2.6 |
| D78382 | Tob | −2.6 |
| Al019193 | TCF-7 | −3.2 |
| M74516 | GA-binding protein | −4.3 |
| Al851658 | KLF3 | −6.5 |
| U25096 | LKLF | −10† |
| M12848 | Myb | −12.9† |
Total RNA was isolated from triplicate samples of sorted CD8+DbNP366-specific CTL and used to probe Affymetrix U74A gene arrays. A total of 90 genes were identified as being differentially expressed. A partial list is presented. Complete data sets are available upon request from S.J.T.
Genes called as present in BAL but absent in spleen samples.
Genes called as present in spleen but absent in BAL.
The finding that only 25 of 90 genes (Fig. 2 and Table 1) were more highly expressed in BAL than in spleen CD8+DbNP366+ T cells was surprising in view of the hierarchy in functional activation associated with lymphocytes recovered from these two anatomical sites (Fig. 1). Of those 25 genes, integrin αE and Chitinase-3-like protein were the most highly expressed. The Chitinase-like protein may play a role in tissue remodeling (19), perhaps enabling more efficient trafficking of antigen-specific CTLs to the site of inflammation. The expression of integrin αE, which is normally found on gut-associated intraepithelial lymphocytes (20), by BAL but not splenic CD8+DbNP366+ T cells may indicate a role in trafficking to mucosal sites.
The most abundant of the genes that were more highly expressed in the spleen (65 of 90) was the transcription factor Myb, which is also known as c-myb. Other transcription factors that were both prominent in splenic CD8+DbNP366+ T cells and are known to play a role in the lymphocyte function include LKLF, T cell factor-7 (TCF-7, also known as TCF-1), GA-binding protein, and Tob, which is generally thought of as a negative regulator of T cell activation (21). Additional genes with established roles in T cell immunity included CD7, a cell-surface marker associated with the differentiated state of various memory T cell populations (22), and Stat5a, which is involved in cytokine signaling and CD8+ T cell homeostasis (23).
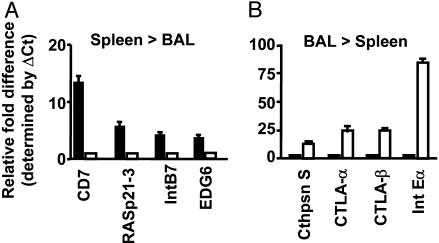
Quantitation by Real-Time PCR. Several genes were selected for SYBR green real-time PCR analysis (Fig. 3), both to validate the Affymetrix array profiles and to provide further quantitative data. It was confirmed (see Fig. 2 and Table 1) that CD7 (13-fold), RAS-p21-3 (5.6-fold), integrin β7 (3.1-fold), and the EDG receptor-6 (2.9-fold) were more highly expressed in CD8+DbNP366+ T cells from spleen versus BAL (Fig. 3A). In contrast, the mRNA for Cathepsin S (4.7-fold), CTLA-2α (7.7-fold), CTLA-2β (7.9-fold), and integrin αE (84-fold) was found, in agreement with the Affymetrix profiles (Fig. 2 and Table 1), to be more prominent in the BAL set (Fig. 3B).
Fig. 3.
Validation by quantitative PCR for a subset of the genes identified by the Affymetrix analysis (Fig. 2 and Table 1) as differentially expressed in CD8+DbNP366+ T cells from the BAL or spleen. Total RNA was extracted from ex vivo-sorted T cells from either the BAL (□) or spleen (▪), and real-time PCR was performed by using SYBR chemistries. The fold differences in mRNA expression for subsets of mRNAs conforming to the spleen > BAL (A) or BAL > spleen (B) profiles were determined by using the ΔCt method, as described in Materials and Methods.
Integrin αE Analysis by Single-Cell PCR. The preceding functional (Fig. 1) and mRNA analyses by microarray and real-time PCR used bulk T cell populations either characterized (Fig. 1) or selected (Figs. 2 and 3 and Table 1) by tetramer staining. As a consequence, the measures of CTL activity (Fig. 1A), cytokine production (Fig. 1B), and mRNA levels (Figs. 2 and 3 and Table 1) reflect average values for what are likely to be heterogeneous lymphocyte populations. The analysis was thus refined for integrin αE to determine whether the mRNA in question was expressed in every CD8+DbNP366+ T cell (Table 2). Single, sorted CD8+DbNP366+ T cells from secondarily challenged mice were characterized by RT-PCR as expressing mRNA for CD8α and integrin αE (Table 2). The single-cell PCR analysis for integrin αE also confirmed the Affymetrix results (Table 1), with the frequency of mRNA expression in CD8+DbNP366+ T cells being higher in the BAL than in the spleen (Table 2).
Table 2. Single-cell frequency of CD8+DbNP366-specific T cells expressing integrin αE mRNA.
| Mouse no. | Spleen | BAL |
|---|---|---|
| 1 | 13 of 58 | 62 of 67 |
| 2 | 10 of 52 | 33 of 60 |
| 3 | 11 of 53 | 52 of 67 |
cDNA from single-cell-sorted CD8+DbNP366-specific CTLs was used to detect both CD8 (positive control) and integrin αE-specific mRNA. Shown is the frequency of CD8+ cells that were positive for integrin αE.
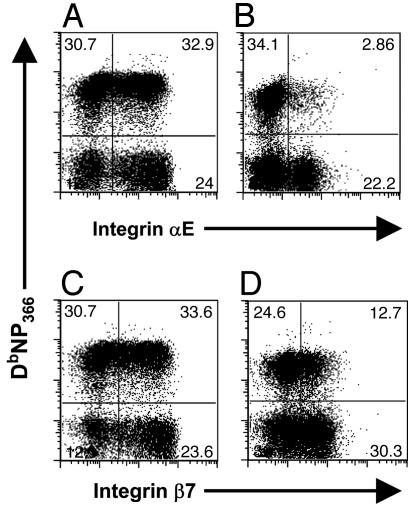
Patterns of Protein Expression for the αE and β7 Integrins. Integrin αE, which pairs with integrin β7 to form a heterodimer (20), is normally expressed on intraepithelial lymphocytes from the gut and lungs (20, 24). Patterns of integrin αE (Fig. 4 A and B) and integrin β7 (Fig. 4 C and D) protein expression were compared for BAL (Fig. 4 A and C) and splenic (Fig. 4 B and D) CD8+DbNP366+ T cells on day 9 after secondary challenge. Although comparable profiles of αE and β7 staining were found for the CD8+DbNP366+ T cells recovered from the lung lumen by BAL, the values for the spleen were clearly discordant (Fig. 4). The fact that a higher percentage of T cells express integrin β7 than αE in the spleen may, of course, reflect that β7 is capable of pairing with other integrin chains (20). The staining comparisons for integrin β7 (Fig. 4) are broadly in accord with the Affymetrix data (Table 1) but differ from the quantitative PCR results (Fig. 3), although the fold comparisons are not large in either case.
Fig. 4.
Integrins αE and β7 are more highly expressed on CD8+DbNP366+ T cells recovered by BAL of the infected lung. Lymphocyte populations were isolated from the BAL (A and C) and spleen (B and D) of HKx31 → PR8-infected mice and costained with the DbNP366 tetramer and anti-integrin αE(A and B) or anti-integrin β7 (C and D).
Discussion
The present analysis probes global patterns of gene expression in functionally different T cells of the same lineage by using Affymetrix array technology. The experiments compared the recall response in spleen and lung for the predominant CD8+DbNP366+ population in mice with acute influenza pneumonia. Detailed T cell receptor analysis has shown that the CD8+DbNP366+ sets recovered from the spleen and the virus-infected respiratory tract by BAL are comprised of exactly the same clonotypes (17). Any difference in molecular profiles is thus likely to relate to the site of anatomical localization and/or the relative antigen load, which is known to be much higher in the respiratory tract.
As might be expected, a number of genes were more highly expressed in T cells that had localized to the site of virus-induced pathology, the lymphocyte set recovered by BAL of the infected respiratory tract. The integrins αE and β7 pair to constitute a prominent receptor on intraepithelial T cells (20, 24, 25). It was initially proposed that expression of αEβ7 integrin functions to retain lymphocytes in close proximity to epithelial cells in the gut and lung by binding E-cadherin (20). Interestingly, it has been demonstrated that expression of αEβ7 integrin is not required for migration of and retention of T cells in the lamina propria (26). The present findings for the CD8+DbNP366+ BAL set are thus in accord with what might be expected, although the precise role of αEβ7 integrin expression virus-specific CTL localized to the BAL still remains to be determined.
Another gene that was more highly expressed in the CD8+DbNP366+ BAL set was Zfp265, which is known to be associated with posttranscriptional processing and mRNA stabilization (27). Although the precise role of Zfp265 expression still remains to be determined, it is possible that Zfp265 facilitates stabilization and, therefore, more efficient protein expression, particularly of cytokines or granzymes. This hypothesis may explain the observed functional superiority of the BAL effectors (Fig. 1 and refs. 9, 11, and 12). A further BAL-associated gene was DNA-damage-induced-4 (also known as dexamethosone-induced gene-2; dig-2). Expression of dig-2 is induced in response to hypoxic conditions (28, 29) or other cellular stresses such as the induction of apoptosis (30). The biological function of dig-2 expression can vary, depending on the cell type. For example, dig-2 can protect rapidly dividing cells from the detrimental effects of hypoxia (29) and can function to slow down cell death (30). Conversely, in terminally differentiated cells, dig-2 expression can actually increase the sensitivity of cells to ischemic injury and oxidative stress (28, 29). The role of dig-2 expression by BAL CTL thus remains to be determined.
Of the 90 genes identified as showing differential expression profiles in the Affymetrix analysis, approximately two-thirds were more highly expressed in the “less-activated” CD8+DbNP366+ T cells recovered from the spleen, a finding that is somewhat counterintuitive. It thus seems that the overall spectrum of gene expression is diminished in the highly activated CTL population recovered from the virus-infected lung. This impression of a narrowing, or “focusing,” of gene transcription to emphasize the expression of essential effector molecules makes sense when the BAL T cells are regarded as a more differentiated, even a terminally differentiated, set. The molecular emphasis is on achieving efficient CTL function at the site of pathology, perhaps at the expense of long-term viability. The down-regulation (in the BAL set) of genes identified as highly expressed in splenic CD8+DbNP366+ T cells, such as Txk kinase, TG-interacting factor, KLRG1, and Tob, that are all known to be involved in dampening cellular responses, may be thought to reflect this tradeoff.
Although expression of the inhibitory receptor KLRG1 on antigen-specific T cells may not directly limit cytokine production or cytotoxicity (31-33), there is evidence that KLRG1 may operate to dampen early T cell receptor signaling events (34). The presence of KLRG1 may thus contribute to increasing the threshold for CTL activation, which could function to protect tissues that do not have a substantial pathogen load from bystander damage. Similarly, LKLF expression is exclusive to quiescent cells (35), and it is possible that LKLF drives the up-regulation of Tob (36). Tob acts as negative inhibitor of T cell proliferation by interacting with members of the TGF-β signaling pathway, Smad 2, and Smad 4 to bind the negative regulatory element of the IL-2 promoter (21). Again, this limits T cell activation and proliferation.
It is also the case that some of the CD8+DbNP366+ T cells in the spleen will be on a path that leads to clonal preservation and long-term maintenance in the memory compartment. Levels of mRNA for the GA-binding protein and Stat5a, which are known to be involved in IL-7R expression and signaling (37, 38), were higher in the splenic CD8+DbNP366+ set. A determining factor of CTL memory differentiation is the expression of IL-7R during the acute phase of infection (39). Genes like the Txk kinase and c-myb that are most commonly associated with thymocyte differentiation (40-42) may perhaps also act to facilitate T cell survival in the responding lymph nodes and spleen (42-44). Additional evidence for active transcriptional pathways in the splenic CD8+DbNP366+ set that could contribute to T cell survival comes from the identification of genes like pim1, which interacts with c-myb (45) and can enhance c-myb transcriptional activity (46).
Analysis using this “broad-brush” array approach has thus provided the important insight that, although effector T cells operating in a site of pathology have a more activated phenotype, there is a concomitant narrowing in the spectrum of gene expression that may perhaps be associated with diminished survival in the longer term. Such experiments with tetramer+ T cells recovered ex vivo from, for instance, a site of viral pneumonia, have limited application because of the mRNA amounts (and lymphocyte numbers) that are required. It would, of course, be highly desirable to do “global” kinetic experiments of this type, especially in the transition phases through the acute immune response to the subsequent development of immune memory. As it is, the analysis to date has identified some intriguing molecular targets that can be followed by using, for example, quantitative and single-cell PCR approaches.
Acknowledgments
This work was supported by a Burnet Award (to P.C.D.) and an R. D. Wright Fellowship (to S.J.T.) provided by the National Health and Medical Research Council of Australia; Science and Technology funds from the Government of Victoria, Australia; U.S. Public Health Service National Institutes of Health Grant AI29579; and ALSAC at St. Jude Children's Research Hospital.
Author contributions: D.R.M. and S.J.T. designed research; D.R.M., E.O., N.L.L.G., A.G., D.G.W., S.W., C.C., and S.J.T. performed research; E.O. and G.A.N. contributed new reagents/analytic tools; D.R.M., S.A., N.L.L.G., G.A.N., A.G., D.G.W., S.W., C.C., P.C.D., and S.J.T. analyzed data; and D.R.M., G.A.N., P.C.D., and S.J.T. wrote the paper.
Abbreviations: B6, C57BL/6J; BAL, bronchoalveolar lavage; CTL, cytotoxic T lymphocyte; H, hemagglutinin; N, neuraminidase; i.n., intranasal; PR8, influenza A virus A/Puerto Rico/34/8; LKLF, lung Krüppel-like factor.
References
- 1.Allan, W., Tabi, Z., Cleary, A. & Doherty, P. C. (1990) J. Immunol. 144, 3980-3986. [PubMed] [Google Scholar]
- 2.Baumgarth, N., Brown, L., Jackson, D. & Kelso, A. (1994) J. Virol. 68, 7575-7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumgarth, N., Egerton, M. & Kelso, A. (1997) J. Immunol. 159, 1182-1191. [PubMed] [Google Scholar]
- 4.Flynn, K. J., Belz, G. T., Altman, J. D., Ahmed, R., Woodland, D. L. & Doherty, P. C. (1998) Immunity 8, 683-691. [DOI] [PubMed] [Google Scholar]
- 5.Marshall, D. R., Turner, S. J., Belz, G. T., Wingo, S., Andreansky, S., Sangster, M. Y., Riberdy, J. M., Liu, T., Tan, M. & Doherty, P. C. (2001) Proc. Natl. Acad. Sci. USA 98, 6313-6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flynn, K. J., Riberdy, J. M., Christensen, J. P., Altman, J. D. & Doherty, P. C. (1999) Proc. Natl. Acad. Sci. USA 96, 8597-8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Townsend, A. R., Rothbard, J., Gotch, F. M., Bahadur, G., Wraith, D. & McMichael, A. J. (1986) Cell 44, 959-968. [DOI] [PubMed] [Google Scholar]
- 8.Belz, G. T., Xie, W., Altman, J. D. & Doherty, P. C. (2000) J. Virol. 74, 3486-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belz, G. T., Xie, W. & Doherty, P. C. (2001) J. Immunol. 166, 4627-4633. [DOI] [PubMed] [Google Scholar]
- 10.Chen, W., Bennink, J. R., Morton, P. A. & Yewdell, J. W. (2002) J. Virol. 76, 10332-10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.La Gruta, N. L., Turner, S. J. & Doherty, P. C. (2004) J. Immunol. 172, 5553-5560. [DOI] [PubMed] [Google Scholar]
- 12.Turner, S. J., La Gruta, N. L., Stambas, J., Diaz, G. & Doherty, P. C. (2004) Proc. Natl. Acad. Sci. USA 101, 3545-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baumgarth, N. & Kelso, A. (1996) Eur. J. Immunol. 26, 2189-2197. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, B. J., Costelloe, E. O., Fitzpatrick, D. R., Haanen, J. B., Schumacher, T. N., Brown, L. E. & Kelso, A. (2003) Proc. Natl. Acad. Sci. USA 100, 2657-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaech, S. M., Hemby, S., Kersh, E. & Ahmed, R. (2002) Cell 111, 837-851. [DOI] [PubMed] [Google Scholar]
- 16.Liu, G., Loraine, A. E., Shigeta, R., Cline, M., Cheng, J., Valmeekam, V., Sun, S., Kulp, D. & Siani-Rose, M. A. (2003) Nucleic Acids Res. 31, 82-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kedzierska, K., Turner, S. J. & Doherty, P. C. (2004) Proc. Natl. Acad. Sci. USA 101, 4942-4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turner, S. J., Diaz, G., Cross, R. & Doherty, P. C. (2003) Immunity 18, 549-559. [DOI] [PubMed] [Google Scholar]
- 19.Hakala, B. E., White, C. & Recklies, A. D. (1993) J. Biol. Chem. 268, 25803-25810. [PubMed] [Google Scholar]
- 20.Shimizu, Y., Rose, D. M. & Ginsberg, M. H. (1999) Adv. Immunol. 72, 325-380. [DOI] [PubMed] [Google Scholar]
- 21.Tzachanis, D., Freeman, G. J., Hirano, N., van Puijenbroek, A. A., Delfs, M. W., Berezovskaya, A., Nadler, L. M. & Boussiotis, V. A. (2001) Nat. Immunol. 2, 1174-1182. [DOI] [PubMed] [Google Scholar]
- 22.Aandahl, E. M., Sandberg, J. K., Beckerman, K. P., Tasken, K., Moretto, W. J. & Nixon, D. F. (2003) J. Immunol. 170, 2349-2355. [DOI] [PubMed] [Google Scholar]
- 23.Kelly, J., Spolski, R., Imada, K., Bollenbacher, J., Lee, S. & Leonard, W. J. (2003) J. Immunol. 170, 210-217. [DOI] [PubMed] [Google Scholar]
- 24.Masopust, D., Vezys, V., Marzo, A. L. & Lefrancois, L. (2001) Science 291, 2413-2417. [DOI] [PubMed] [Google Scholar]
- 25.Masopust, D., Vezys, V., Usherwood, E. J., Cauley, L. S., Olson, S., Marzo, A. L., Ward, R. L., Woodland, D. L. & Lefrancois, L. (2004) J. Immunol. 172, 4875-4882. [DOI] [PubMed] [Google Scholar]
- 26.Lefrancois, L., Parker, C. M., Olson, S., Muller, W., Wagner, N., Schon, M. P. & Puddington, L. (1999) J. Exp. Med. 189, 1631-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morris, B. J., Adams, D. J. & van der Weyden, L. (2001) Clin. Exp. Pharmacol. Physiol. 28, 1044-1047. [DOI] [PubMed] [Google Scholar]
- 28.Brafman, A., Mett, I., Shafir, M., Gottlieb, H., Damari, G., Gozlan-Kelner, S., Vishnevskia-Dai, V., Skaliter, R., Einat, P., Faerman, A., et al. (2002) Mol. Cell. Biol. 22, 2283-2293.11884613 [Google Scholar]
- 29.Shoshani, T., Faerman, A., Mett, I., Zelin, E., Tenne, T., Gorodin, S., Moshel, Y., Elbaz, S., Budanov, A., Chajut, A., et al. (2002) Mol. Cell. Biol. 22, 2283-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, Z., Malone, M. H., Thomenius, M. J., Zhong, F., Xu, F. & Distelhorst, C. W. (2003) J. Biol. Chem. 278, 27053-27058. [DOI] [PubMed] [Google Scholar]
- 31.Wojtasiak, M., Jones, C. M., Sullivan, L. C., Winterhalter, A. C., Carbone, F. R. & Brooks, A. G. (2004) Int. Immunol. 16, 1333-1341. [DOI] [PubMed] [Google Scholar]
- 32.McMahon, C. W., Zajac, A. J., Jamieson, A. M., Corral, L., Hammer, G. E., Ahmed, R. & Raulet, D. H. (2002) J. Immunol. 169, 1444-1452. [DOI] [PubMed] [Google Scholar]
- 33.Robbins, S. H., Terrizzi, S. C., Sydora, B. C., Mikayama, T. & Brossay, L. (2003) J. Immunol. 170, 5876-5885. [DOI] [PubMed] [Google Scholar]
- 34.Beyersdorf, N. B., Ding, X., Karp, K. & Hanke, T. (2001) Eur. J. Immunol. 31, 3443-3452. [DOI] [PubMed] [Google Scholar]
- 35.Buckley, A. F., Kuo, C. T. & Leiden, J. M. (2001) Nat. Immunol. 2, 698-704. [DOI] [PubMed] [Google Scholar]
- 36.Hua, X. & Thompson, C. B. (2001) Nat. Immunol. 2, 1097-1098. [DOI] [PubMed] [Google Scholar]
- 37.Xue, H. H., Bollenbacher, J., Rovella, V., Tripuraneni, R., Du, Y. B., Liu, C. Y., Williams, A., McCoy, J. P. & Leonard, W. J. (2004) Nat. Immunol. 5, 1036-1044. [DOI] [PubMed] [Google Scholar]
- 38.Rosenthal, L. A., Winestock, K. D. & Finbloom, D. S. (1997) Cell. Immunol. 181, 172-181. [DOI] [PubMed] [Google Scholar]
- 39.Kaech, S. M., Tan, J. T., Wherry, E. J., Konieczny, B. T., Surh, C. D. & Ahmed, R. (2003) Nat. Immunol. 4, 1191-1198. [DOI] [PubMed] [Google Scholar]
- 40.Sommers, C. L., Rabin, R. L., Grinberg, A., Tsay, H. C., Farber, J. & Love, P. E. (1999) J. Exp. Med. 190, 1427-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaeffer, E. M., Broussard, C., Debnath, J., Anderson, S., McVicar, D. W. & Schwartzberg, P. L. (2000) J. Exp. Med. 192, 987-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lieu, Y. K., Kumar, A., Pajerowski, A. G., Rogers, T. J. & Reddy, E. P. (2004) Proc. Natl. Acad. Sci. USA 101, 14853-14858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomlinson, M. G., Kane, L. P., Su, J., Kadlecek, T. A., Mollenauer, M. N. & Weiss, A. (2004) Mol. Cell. Biol. 24, 2455-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takeba, Y., Nagafuchi, H., Takeno, M., Kashiwakura, J. & Suzuki, N. (2002) J. Immunol. 168, 2365-2370. [DOI] [PubMed] [Google Scholar]
- 45.Winn, L. M., Lei, W. & Ness, S. A. (2003) Cell Cycle 2, 258-262. [PubMed] [Google Scholar]
- 46.Katakami, N., Kaneto, H., Hao, H., Umayahara, Y., Fujitani, Y., Sakamoto, K., Gorogawa, S., Yasuda, T., Kawamori, D., Kajimoto, Y., et al. (2004) J. Biol. Chem. 279, 54742-54749. [DOI] [PubMed] [Google Scholar]