Abstract
Voltage-dependent K+ channels rely on precise dynamic protein interactions with surrounding plasma membrane lipids to facilitate complex processes such as voltage sensing and channel gating. Many transmembrane-spanning proteins use palmitoylation to facilitate dynamic membrane interactions. Herein, we demonstrate that the human Kv1.1 ion channel is palmitoylated in the cytosolic portion of the S2-S3 linker domain at residue C243. Through heterologous expression of the human Kv1.1 protein in Sf9 cells, covalent radiolabeling with [3H]palmitate, chemical stability studies of the [3H]-palmitoylated protein, and site-directed mutagenesis, C243 was identified as the predominant site of palmitoylation. The functional sequelae of palmitoylation were examined by analysis of whole cell currents from WT and mutant channels, which identified a 20-mV leftward shift in the current-voltage relationship when palmitoylation at C243 (but not with other cysteine deletions) is prevented by site-directed mutagenesis, implicating a role for palmitoylated C243 in modulating voltage sensing through protein-membrane interactions. Database searches identified an amino acid palmitoylation consensus motif (ACP/RSKT) that is present in multiple other members of the Shaker subfamily of K+ channels and in several other unrelated regulatory proteins (e.g., CD36, nitric oxide synthase type 2, and the mannose-6 phosphate receptor) that are known to be palmitoylated by thioester linkages at the predicted consensus site cysteine residue. Collectively, these results (i) identify palmitoylation as a mechanism for K+ channel interactions with plasma membrane lipids contributing to electric field-induced conformational alterations, and ii) define an amino acid consensus sequence for protein palmitoylation.
Keywords: ion channels, K+ channel, fatty acid, membrane potential
Palmitoylation is increasingly recognized as a frequent and important modification of eukaryotic signaling proteins (for review, see refs. 1 and 2). Palmitoylated proteins include G protein α-subunits and monomeric GTP-binding proteins (e.g., p21ras), G protein-coupled receptors, RGS proteins, and protein kinases (3-7). Unlike prenylation and myristoylation, palmitoylation is a reversible covalent modification, allowing for dynamic regulation of multiple complex cellular systems (1). Protein-bound palmitate turns over continuously in cells and is responsive to alterations in cellular signaling, environment, and nutritional contexts (1, 8). The covalent addition of fatty acid to proteins facilitates targeted membrane association as well as modulates protein conformation, dynamics, and function (9-12). In the case of p21ras and G protein α-subunits, palmitoylation helps anchor the proteins to the plasma membrane and facilitates signal processing through resultant conformational and dynamic alterations (5, 6).
Fatty acid covalent modification of proteins are known to be either cotranslational or posttranslational. Cotranslational modification typically involves myristoylation of an amino terminal glycine (at the second position following the initiator Met) through an amide bond (13). In contrast, posttranslational acylation typically uses palmitic acid and to a lesser extent, other fatty acids including, amongst others, oleic acid, arachidonic acid, and myristic acid (14). The covalent linkage of fatty acid to protein is typically through a thioester or oxyester linkage but also may use amide linkages (15, 16). Thus, posttranslational palmitoylation is dynamic, catalyzed by enzymes (protein fatty acyl transferases and thioesterases) regulating the net occupancy of the palmitoylated sites of specific protein targets (3, 17, 18). Many proteins that are modified by other permanent lipid attachments (e.g., myristoylation and prenylation) are also palmitoylated at a nearby cysteine residue (15). However, a consensus sequence for palmitoylation has not yet been identified.
It has been recognized that fatty acids can covalently directly bind to ion channels such as the nicotine acetylcholine receptor channel, sodium channels, calcium channels, and the viral M protein channel through posttranslational modifications (19-21). The cysteine residues involved in palmitoylation of the glutamate receptor GluR6 and the β2α subunit of Ca2+ channels have been identified (22, 23). In the case of the GluR6 receptor, two acylation sites in the carboxyl terminus have been demonstrated and are implicated in the regulation of the phosphorylation state of the channel and, potentially, modulation of peak current amplitude (24). In contrast, the β2α subunit of the Ca2+ channel contains a palmitoylated doublet of cysteines at the amino terminus whose functional consequences, as revealed by a double mutant, involve altered protein-protein interactions with the α-subunit leading to decreased current amplitude and alterations in channel inactivation (22, 24).
Recent elegant studies (e.g., ref. 25) have raised provocative questions on the mechanism of channel interactions with membrane phospholipids during voltage sensing and gating. In the present study, we sought to determine whether the Kv1.1 ion channel is palmitoylated and the potential consequences of palmitoylation on ion channel function. Herein, we report that the Kv1.1 ion channel is palmitoylated at C243, which modulates voltage sensing and provides a hydrophobic adaptor to facilitate dynamic interactions between the Kv1.1 protein and surrounding lipid constituents during voltage-induced conformational alterations of the channel protein.
Methods
Baculovirus Expression of Human Kv1.1 (Huk1). All studies were performed in a baculovirus expression system by using Spodoptera frugiperda (Sf9) cells. Sf9 cells were maintained in 1× Grace's media supplemented with 10% FBS, 1.7% lactalbumin, 1.7% yeastolate, and 0.9% antibiotic/antimycotic (Invitrogen). Sf9 cells were infected with human brain Kv1.1 channel (Huk1) in the baculovirus transfer plasmid, pVL1393, kindly provided by Alexander Kamb (University of California, San Francisco) (26), at a multiplicity of infection of 1.7 for 72 h by using standard methodology (27).
PCR and Site-Directed Mutagenesis of Kv1.1. Kv1.1 was subcloned into baculovirus vector pBlueBacIII (Invitrogen) for all subsequent studies as follows: PCR was performed in 30 cycles of amplification by using primers engineered to incorporate appropriate restriction sites at the 5′ (m94: 5′-ggaggatccatgacggtgatgtctggg-3′) and 3′ (m95: 5′-gcgaagcttttaaacatcggtcagtagc-3′) ends of Kv1.1-coding sequence. cDNA product (1.5 kb) was subcloned into the BamHI/HindIII sites of the baculovirus vector pBlueBacIII to produce WT Kv1.1-pBlueBacIII. The entire Kv1.1 insert and immediate flanking sequences were sequenced in entirety on both strands by standard dideoxy sequencing techniques. The Kv1.1-pBlueBacIII construct was used as a template for PCR site-directed mutagenesis of Kv1.1 cysteine residues. A C35A, C36A Kv1.1 mutant was generated by using forward primer (m94) paired with reverse primer m146 (5′-cggagatgttgatcaccacgcgctcggctgcctcgtg-3′) in PCR to amplify a 144-nt product, which was cut with BamHI/BclI and ligated to 5-kb BamHI/BclI Kv1.1-pBlueBacIII vector. A C243A Kv1.1 mutant was prepared by PCR with sense primer m154 (5′-ggtgcgcttcttcgccgctcccagcaagacggac-3′) and m95 reverse primer to PCR amplify a 789-nt fragment, which was restriction digested with HhaI/PshAI and ligated to 222-nt ApaI/HhaI Kv1.1 fragment to generate a 1-kb fragment, which was then ligated to ApaI/PshAI digested Kv1.1-pBlueBacIII. A C-terminal deletion mutant D478-495 was prepared as follows: Antisense primer m144 (5′-agctaagctttaattggcagttctg-3′) containing the 3′-stop codon after N477 followed by a HindIII site for cloning was used for PCR with sense primer m143 (5′-gggcctccagatcctgggccagaccctc-3′) to amplify a 516-nt product, which was restriction digested with PflMI/HindIII and ligated to 945-nt BamHI/PflMI Kv1.1 fragment and BamHI/HindIII cut pBlueBacIII vector.
Electrophysiology Measurements in Sf9 Cells. Whole-cell voltage clamp recordings of Sf9 cells were performed by using standard techniques as described (28-31). Sf9 cells lack any depolarization-induced current and, therefore, provide a null background for determination of voltage-activated potassium channels (26, 28). In brief, a series resistance compensation of 80-90% was routinely applied by using fire-polished recording electrodes with a tip resistance of 6-10 MΩ. For whole-cell voltage clamp analysis, infected cells in 35-mm dishes were rinsed with serum-free medium 30 min before recordings then placed in extracellular recording solution [115 mM NaCl/2.5 mM KCl/2 mM MnCl2/10 mM Hepes/1 mM CaCl2/1 mM 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid (BAPTA), pH 7.3] and held at -80 mV followed by depolarizing voltage steps (-40 mV to +40 mV by 10- or 20-mV increments/300-ms duration). The activation/inactivation time constants were determined by bestfit analysis of monexponential equations corresponding to 10-90% rise times.
Incorporation of Palmitic Acid. Sf9 cells (1 × 107 cells) expressing either WT or mutant recombinant Kv1.1 were incubated with 2 mCi of [9,10-3H]palmitic acid resuspended in 10 μl of dimethyl sulfoxide, 100 μl of FBS, and 1 ml of 1× Grace's insect cell culture medium for 2 h at 22°C. After incubation, the cells were recovered by centrifugation, rinsed, and recentrifuged before final resuspension in cell lysis buffer (20 mM Tris·HCl/100 mM NaCl, pH 7.5) including the protease inhibitor mixture [500 μM 4-(2-aminoethyl)benzenesulfonylfluoride hydrochloride (AEBSF)/500 μM EDTA/1 μM E-64/1 μM leupeptin/1 μg/ml aprotinin (Calbiochem)] (14). Cells were ruptured by successive freeze thawing before a 4-h incubation at 4°C in 1:1 dilution of solubilization buffer (lysis buffer containing 2% Triton X-100 and 2× protease inhibitor mixture).
Immunoprecipitation and Western Analysis of Kv1.1. Solubilized labeled membrane fractions were mixed with polyclonal Kv1.1 antiserum (gift of Bruce Tempel, University of Washington School of Medicine, Seattle) overnight at 4°C, incubated with Protein A-coated Sepharose beads (Santa Cruz Biotechnology) for 6 h at 4°C, and pelleted by centrifugation at 3,000 × g for 15 s with two washes using solubilization buffer. A 5× SDS sample buffer was added and the sample boiled for 1 min before SDS/PAGE analysis. Before autoradiographic analysis, gels were fixed for 1 h, incubated in Enlightening solution for 30 min, dried for 2 h at 80°C, and exposed to Kodak X-AR film according to standard methods.
Determination of Hydroxylamine Sensitivity. After immunoprecipitation of WT or mutant Kv1.1, the bead/Ab-antigen complexes were rinsed with PBS (137 mM NaCl/2.6 mM KCl/10 mM Na2HPO4/1.8 mM KH2PO4) and resuspended in 20 μl of PBS and 20 μl of 1 M hydroxylamine (pH 7) or as control 1 M Tris (pH 7) for 2 h at 22°C. Next, SDS sample buffer was added, and the sample was boiled for 1 min before gel electrophoresis.
Results
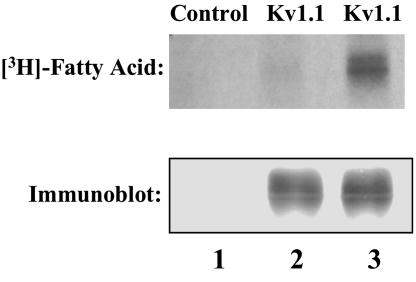
Identification of Covalent Palmitoylation of the Human Kv1.1 Ion Channel Expressed in Sf9 Cells. Cultures of Sf9 cells expressing human Kv1.1 protein were radiolabeled with [3H]palmitic acid, lysed, and immunoprecipitated with an anti-Kv1.1 polyclonal Ab, and proteins were separated by SDS/PAGE as described in Methods. Autoradiography revealed the presence of a radioactive doublet of the anticipated Mr (≈68 kDa) in cells expressing the Kv1.1 protein, whereas control cells (infected with control virus lacking the Kv1.1 insert) did not demonstrate a radiolabeled band (Fig. 1). Identical experiments in the presence of [3H]myristic acid loading an equivalent mass of Kv1.1 protein did not result in the incorporation of [3H]myristate into the Kv1.1 ion channel protein, demonstrating substantial specificity for palmitic acid in the acylation of the Kv1.1 ion channel (Fig. 1).
Fig. 1.
Palmitoylation of the Kv1.1. ion channel. Sf9 cells infected with recombinant Kv1.1-containing virus or control virus were incubated in the presence of 2 mCi of [3H]palmitic or [3H]myristic acid for 2 h at 22°C. Immunoblot analysis of immunoprecipitated solubilized whole cell lysates by using rabbit polyclonal antisera demonstrated equal amounts of immunoreactive Kv1.1 in Kv1.1-infected Sf9 cells (lanes 2 and 3) whereas the control virus-infected cells (lane 1) did not contain any immunoreactive material. Autoradiographs of immunoprecipitated, radiolabeled samples detected a radioactive doublet only in the Kv1.1-infected, palmitate-labeled sample (lane 3) but not in either the control virus-infected, [3H]palmitate-labeled sample (lane 1) or the Kv1.1-infected, [3H]myristate-labeled sample (lane 2). These data are representative of the results of three independent experiments.
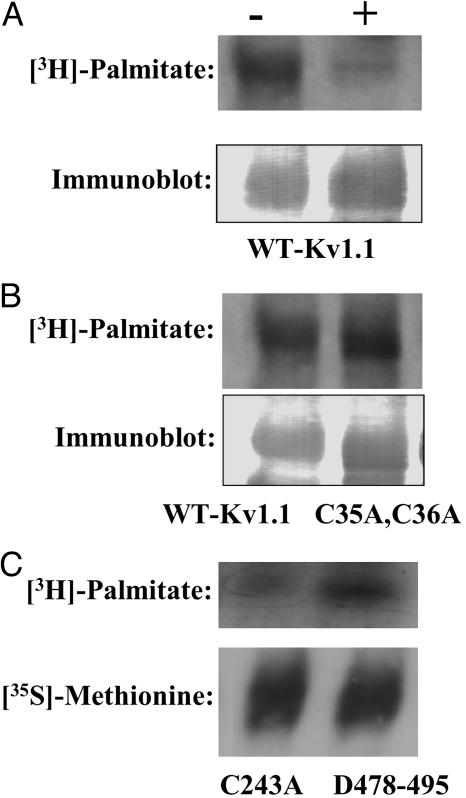
Sensitivity of Palmitoylated Kv1.1 to Treatment with Hydroxylamine. To determine the covalent nature of the chemical linkage (i.e., oxyester or thioester) between the palmitic acid and Kv1.1 protein, samples were subjected to treatment with neutral 1 M hydroxylamine (pH 7), which cleaves thioesterified fatty acids, or 1 M Tris (pH 7) as a control. The majority of the incorporated palmitic acid was cleaved by treatment with 1 M hydroxylamine at pH 7, indicating that the large majority of the bound palmitic acid was thioesterified (Fig. 2A). Thus, Kv1.1 is acylated primarily at cysteine residues, but we cannot rule out that a small amount of acylation may occur at oxyester linkages as well.
Fig. 2.
Hydroxylamine sensitivity and site-directed mutagenesis of the palmitoylated Kv1.1 channel protein. (A) Immunoprecipitated proteins were treated with buffer alone (-) or buffer containing 1 M hydroxylamine at pH 7 (+) before SDS/PAGE and autoradiography (Upper) or Western blotting analysis (Lower). (B) Comparison of palmitoylation of the Kv1.1 WT (WT-Kv1.1) with the C35A, C36A double mutant metabolically labeled with [3H]palmitate. Sf9 cells expressing either WT protein (WT-Kv1.1) or the C35A, C36A double mutant were metabolically labeled with [3H]palmitate, immunoprecipitated, and subjected to SDS/PAGE and autoradiography (Upper) or Western blotting (Lower). (C) Comparison of the palmitoylation of the Kv1.1 C243A mutant to palmitoylation of the C-terminal Kv1.1 deletion mutant D478-495 was performed after metabolic labeling with [3H]palmitate or [35S]methionine, immunoprecipitation, and autoradiography.
Determination of the Sites of Palmitoylation in the Kv1.1 Ion Channel. Because the majority of the palmitic acid was covalently linked to the Kv1.1 channel protein through thioester linkages, we next focused on determining which of the five cytoplasmic cysteines present in the human Kv1.1, ion channel were candidates for acylation. Because Chien et al. (22) demonstrated that an amino terminal cysteine doublet was acylated in the Ca2+ channel β2α subunit, we first constructed a double point mutant C35A, C36A, which lacked thioacylation sites in the N terminus. Sf9 cells were infected with either WT Kv1.1 or the C35A, C36A double mutant and metabolically labeled with [3H]palmitic acid. Immunoprecipitated samples of WT Kv1.1 and the double mutant C35A, C36A displayed no decrease in labeling intensity of the double mutant, demonstrating that neither C35 nor C36 were responsible for the majority of the observed thioacylation (Fig. 2B). Therefore, two additional mutants were constructed including the point mutant C243A, and a mutant in which the residues at carboxyl terminus, D478-495 were truncated thereby removing the remaining two cysteines. Comparison of the intensity of radiolabeled C243A mutant protein to the intensity of the D478-495 deletion mutant demonstrated that the single point mutant C243A largely was responsible for the observed incorporation of [3H]palmitic acid (Fig. 2C). The small, residual incorporation of palmitate present in the C243A mutant suggests that other residues in the Kv1.1 channel protein may be palmitolyated, albeit to a lesser extent than C243.
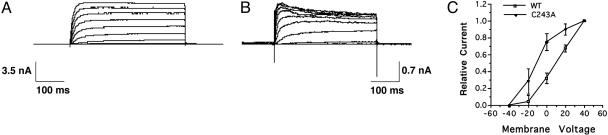
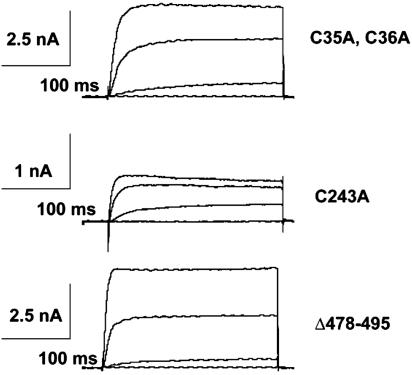
Functional Properties of Cysteine Mutants on K+ Currents and Voltage Sensing. To explore the potential functional role of thioesterified palmitic acid on the electrophysiologic properties of K+ ion channels, we next examined alterations in the voltage-dependence profiles of the expressed currents from Sf9 cells infected with either WT Kv1.1 or the C243A mutant. Notably, the current recorded from cells expressing C243A displayed a 20 mV leftward shift in the current-voltage relationship and reached 80% of peak amplitude near 0 mV compared with only 30% of peak amplitude at 0 mV manifest by WT channels (Fig. 3). In addition, the C243A current demonstrated inactivation at more depolarized potentials, which is not seen in the WT channels. Similarly, previous reports (28, 32, 33) have documented lipid-mediated inactivation of a variety of ion channels. Whereas currents recorded from C243A expressing Sf9 cells showed altered kinetic properties, the currents from the C35A, C36A double mutant and the D478-495 truncation mutant were all similar to WT Kv1.1 (Fig. 4). In addition, although equivalent amounts of mass were present, the magnitudes of C243A currents were four to five times smaller than currents expressed by WT Kv1.1 or the mutants C35A, C36A, and D478-495. This reduction in expressed current likely represents decreased numbers with C243A Kv1.1 ion channels present in the plasma membrane. The most likely explanation is that the disruption of the major site of palmitoylation in Kv1.1 resulted in alterations in palmitoylation-sensitive membrane trafficking as has been documented in other systems (1, 3, 34, 35). Collectively, these results demonstrate that the covalent palmitoylation of the Kv1.1 ion channel at C243 has important functional consequences.
Fig. 3.
Macroscopic current kinetics of the C243A Kv1.1 channel mutant. (A and B) Whole-cell, voltage clamp recordings with depolarizing voltage steps from -40 to +40 mV (10-mV increments) from a holding potential of -80 mV were performed on infected Sf9 cells expressing WT-Kv1.1 (A) or C243A mutant protein (B). (C) Comparisons of WT-Kv1.1 (□) and C243A (▪) current-voltage relationships. Notably, C243A currents demonstrated a leftward shift of 20 mV in the current-voltage relationship in comparison to WT protein. Error bars represent the x̄ ± SE of three independent preparations.
Fig. 4.
Comparison of the macroscopic current kinetics of the Kv1.1 cysteine mutants. Whole-cell voltage-clamp recordings of Sf9 cells expressing each of the cysteine mutants (C35A, C36A; C243A; and D478-495) were compared withWT Kv1.1-expressing Sf9 cells as described in Methods. Peak macroscopic current is reduced 5-fold in the C243A mutant compared withWT-Kv1.1 or the other cysteine mutants. These studies are representative of three separate experiments.
Database Comparisons of Amino Acid Sequence Surrounding the Palmitoylated C243 in the Kv1.1 Channel. Comparisons of the amino acid sequence surrounding the C243 palmitoylation site in the Kv1.1 ion channel with other known palmitoylation sites in other proteins identified the presence of similar sequences in three independent families of membrane-associated regulatory proteins. First, there was a five of six amino acid match with CD36 with the predicted consensus cysteine residue palmitoylated [ACPSKT (Kv1.1) vs. ACRSKT (CD36)] (36). Second, nitric oxide synthase type 2 is palmitoylated at the sequence ACPWK with the consensus cysteine palmitoylated (37). Third, the mannose 6-P receptor contains a CRSK sequence, which is also palmitoylated at the predicted consensus cysteine (38). The close sequence similarity in three unrelated membrane protein families that are each palmitoylated at the predicted cysteine identifies the presence of a palmitoylation motif that is recognized by at least one of the potentially 23 different protein palmitoyl acyl transferases recently described (17, 18). We point out that the majority of proteins that are palmitoylated do not contain this motif and that it is only one of many palmitoylation motifs used by these multiple different protein acyl transferases (similar to the multiple different phosphorylation sequence motifs used by different protein kinases).
Discussion
The results of the present study integrate emerging insights into the chemical mechanisms underlying K+ channel voltage sensing with the importance of protein palmitoylation to collectively identify the role of covalently linked lipids to ion channels in modulating the electrophysiologic characteristics of cellular action potentials. The covalent attachment of a fatty acyl moiety to the Kv1.1 protein results in an expanded repertoire of interactions of the Kv1.1 channel (and its family members) with membrane bilayers including: (i) the ability of the protein to access additional conformational states in the membrane bilayer; (ii) the provision of an interface between the Kv1.1 protein and the membrane to facilitate dynamic bidirectional membrane-protein interactions during the action potential, and (iii) a mechanism to directly exploit hydrophobic forces in the execution of complex cellular voltage sensing functions.
The results demonstrate both the covalent thioesterification of the human Kv1.1 ion channel at C243 as well as the functional sequelae of Kv1.1 palmitoylation in the simplest of biologic electrical contexts (a single nominally nonelectrically active cell unstimulated by chemical moieties subjected to simple whole-cell voltage clamp analysis). Moreover, they identify a consensus sequence for palmitoylation, ACP/RSKT, which is conserved among multiple Kv1 channel family members (Kv1.1, 1.2, 1.3, 1.5, and 1.6) as well as CD36, nitric oxide synthase type 2, and cation-dependent mannose-6-phosphate receptor (CDMPR). Each of these proteins is palmitoylated at the predicted cysteine residues in their consensus sequences, respectively (36-38). The remarkable specificity of distinct palmitoyl transferases for their target proteins has recently been described and it is likely that ACP/RSKT represents one of multiple discrete consensus sequences underlying the selective recognition by distinct palmitoyl transferases (17, 18). It is intriguing to note that ion channels, as well as CD36 and the cation-dependent mannose-6-phosphate receptor (CDMPR), undergo extensive endosomal cycling and that at least some of the enzymes catalyzing the reversible palmitoylation of plasma membrane polypeptides have been indirectly localized to these rapidly recycling endosomal compartments (39).
The presence of a palmitate group at C243 has at least three immediate and obvious consequences. First, structurally, the palmitoylated S2-S3 linker domain is held in close spatial proximity to the membrane interface thereby mandating that the conformation and dynamics of adjacent residues adapt to the constraints of hydrophobic forces placed on this portion of the protein-lipid interface. Recent progress in the structural biology of the voltage gating and sensing mechanisms used by the voltage-dependent K+ channels has generated intense interest because it is the prototypic case representing the foundations of voltage-responsive electrophysiologic functions occurring during action potentials. A model for voltage sensing and gating using a positively charged “moving paddle” structure has been proposed based on the recently obtained crystal structure of an archaebacteria Aeropyrum pernix Kv channel (25). The proposed moving paddle structure differs dramatically from multiple previous studies that suggest that the S4 domain is well insulated from the membrane bilayer by the surrounding protein (ref. 40 and references therein). Recent site-directed cysteine mutagenesis and binding proximity studies have provided additional support for this classical canonical model (40). However, an even more recent electron paramagnetic resonance investigation of local K+ channel structure and dynamics in lipid bilayers has led to a revised structural model that uses the essential characteristics of the moving paddle model, as proposed by MacKinnon et al. (25), but reorients some segments of the protein structure to accommodate the experimental frequency and vector analyses obtained from the spin-labeling experiments using the KvAP protein in membrane bilayers (41). The results of our present study clearly identify the covalent modification of the Kv1.1 ion channel in the S2-S3 spanning loop at residue C243 by thioesterification to palmitate. Thus, the S2-S3 segment must necessarily have cytoplasmic access.
Second, through palmitoylation, a direct hydrophobic interface between the channel and the membrane is created that can, under appropriate conditions, facilitate specific motions promoting precisely predetermined voltage-dependant conformational changes within the K+ channel voltage-sensing domain itself. A large body of work has identified a model of voltage-dependent activation in which the channel protein itself acts to shield the highly charged S4 segment from the hydrophobic membrane environment. The palmitate moiety not only can participate in such shielding by virtue of the fact that it is obligatorily bound to the channel protein itself, but also can participate in orienting lipids around the channel in such a way that motion is facilitated (i.e., by creating defects in the membrane, which domains of the protein can be induced to occupy by field-induced altered conformations of subjacent phospholipid polar head groups). Moreover, it is becoming increasingly clear that voltage-mediated conformational alterations represent an orchestrated interplay between channel proteins and membrane lipids.
Third, the presence of the palmitate group will create and uniquely interact with membrane dipoles. Specifically, the thioester linkage itself creates a new dipole to couple to alterations in the electric field during the action potential. Consideration of the dipole moment present in mammalian phospholipids alone identifies the necessity that the dynamics and conformational space of the membrane are altered during the changing electric fields present during the action potential. It has been demonstrated that the conformation and dipole moment of plasmalogens (a lipid enriched in electrically active membranes) are substantially different from their diacyl phospholipid counterparts. Thus, it is important to consider the specific compositional nature of the lipid environment interacting with the palmitate group and the Kv1.1 channel protein.
The results of the present study demonstrate that palmitoylation is not necessary for voltage sensing because the nonpalmitoylated mutant can be induced to open by depolarizing voltages albeit with different sensitivities and dynamic responses. Thus, the mechanism for voltage sensing is inherent in the primary structure of the ion channel itself, whereas palmitoylation alters the sensitivity, amplitude, and dynamic range of available responses of the channel during changes in membrane potential by the mechanisms discussed above. In this regard, it is intriguing to speculate that other covalent modifications in the Kv1.1 channel could also be present and that such covalent alterations to channel proteins may be a general mechanism for the regulation of voltage sensing (e.g., phosphorylation and sulfhydryl oxidation). The increasing appreciation for the important roles of lipids in modulating ion channel function is an emerging theme that has been emphasized in recent years (42-44). The role of palmitoylation in orchestrating the dynamic interactions of the K+ channel (and other ion channels) with plasma membrane lipids that collectively modulate cellular electrophysiology is still at its earliest stages of understanding.
Acknowledgments
This research was supported by National Institutes of Health Grant 5R01 HL 41250-12.
Author contributions: R.W.G. designed research; R.A.G.-K. performed research; D.J.M. contributed new reagents/analytic tools; R.A.G.-K. and R.W.G. analyzed data; and R.A.G.-K., D.J.M., and R.W.G. wrote the paper.
Abbreviation: CDMPR, cation-dependent mannose-6-phosphate receptor.
References
- 1.Bijlmakers, M. J. & Marsh, M. (2003) Trends Cell Biol. 13, 32-42. [DOI] [PubMed] [Google Scholar]
- 2.Dunphy, J. T. & Linder, M. E. (1998) Biochim. Biophys. Acta 1436, 245-261. [DOI] [PubMed] [Google Scholar]
- 3.Smotrys, J. E. & Linder, M. E. (2004) Annu. Rev. Biochem. 73, 559-587. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein, L. S., Linder, M. E. & Hepler, J. R. (2004) Methods Mol. Biol. 237, 195-204. [DOI] [PubMed] [Google Scholar]
- 5.Qanbar, R. & Bouvier, M. (2003) Pharmacol. Ther. 97, 1-33. [DOI] [PubMed] [Google Scholar]
- 6.Berthiaume, L. G. (2002) Sci. STKE 2002, PE41. [DOI] [PubMed] [Google Scholar]
- 7.Kosugi, A., Hayashi, F., Liddicoat, D. R., Yasuda, K., Saitoh, S. & Hamaoka, T. (2001) Immunol. Lett. 76, 133-138. [DOI] [PubMed] [Google Scholar]
- 8.Yamada, S., Komatsu, M., Sato, Y., Yamauchi, K., Aizawa, T. & Kojima, I. (2003) Endocrinology 144, 5232-5241. [DOI] [PubMed] [Google Scholar]
- 9.Acconcia, F., Ascenzi, P., Bocedi, A., Spisni, E., Tomasi, V., Trentalance, A., Visca, P. & Marino, M. (2005) Mol. Biol. Cell. 16, 231-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gauthier-Campbell, C., Bredt, D. S., Murphy, T. H. & El-Husseini, A.-D. (2004) Mol. Biol. Cell 15, 2205-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiol, A., Davey, P. C., Osterhout, J. L., Waheed, A. A., Fischer, E. R., Chen, C. K., Milligan, G., Druey, K. M. & Jones, T. L. (2003) J. Biol. Chem. 278, 19301-19308. [DOI] [PubMed] [Google Scholar]
- 12.Osterhout, J. L., Waheed, A. A., Hiol, A., Ward, R. J., Davey, P. C., Nini, L., Wang, J., Milligan, G., Jones, T. L. & Druey, K. M. (2003) J. Biol. Chem. 278, 19309-19316. [DOI] [PubMed] [Google Scholar]
- 13.Sefton, B. M. & Buss, J. E. (1987) J. Cell Biol. 104, 1449-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linder, M. E., Kleuss, C. & Mumby, S. M. (1995) Methods Enzymol. 250, 314-330. [DOI] [PubMed] [Google Scholar]
- 15.Manahan, C. L., Patnana, M., Blumer, K. J. & Linder, M. E. (2000) Mol. Biol. Cell 11, 957-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell, D. A., Farh, L., Marshall, T. K. & Deschenes, R. J. (1994) J. Biol. Chem. 269, 21540-21546. [PubMed] [Google Scholar]
- 17.Fukata, M., Fukata, Y., Adesnik, H., Nicoll, R. A. & Bredt, D. S. (2004) Neuron 44, 987-996. [DOI] [PubMed] [Google Scholar]
- 18.Huang, K., Yanai, A., Kang, R., Arstikaitis, P., Singaraja, R. R., Metzler, M., Mullard, A., Haigh, B., Gauthier-Campbell, C., Gutekunst, C., et al. (2004) Neuron 44, 977-986. [DOI] [PubMed] [Google Scholar]
- 19.Holsinger, L. J., Shaughnessy, M. A., Micko, A., Pinto, L. H. & Lamb, R. A. (1995) J. Virol. 69, 1219-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt, J. W. & Catterall, W. A. (1987) J. Biol. Chem. 262, 13713-13723. [PubMed] [Google Scholar]
- 21.Olson, E. N., Glaser, L. & Merlie, J. P. (1984) J. Biol. Chem. 259, 5364-5367. [PubMed] [Google Scholar]
- 22.Chien, A. J., Carr, K. M., Shirokov, R. E., Rios, E. & Hosey, M. M. (1996) J. Biol. Chem. 271, 26465-26468. [DOI] [PubMed] [Google Scholar]
- 23.Pickering, D. S., Taverna, F. A., Salter, M. W. & Hampson, D. R. (1995) Proc. Natl. Acad. Sci. USA 92, 12090-12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hurley, J. H., Cahill, A. L., Currie, K. P. & Fox, A. P. (2000) Proc. Natl. Acad. Sci. USA 97, 9293-9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang, Q. X., Wang, D. N. & MacKinnon, R. (2004) Nature 430, 806-810. [DOI] [PubMed] [Google Scholar]
- 26.Kamb, A., Korenbrot, J. I. & Kitajewski, J. (1992) Methods Enzymol. 207, 423-431. [DOI] [PubMed] [Google Scholar]
- 27.O'Reilly, D., Miller, L. & Luckow, V. (1992) Baculovirus Expression Vectors: A Laboratory Manual (Oxford Univ. Press, New York).
- 28.Gubitosi-Klug, R. A., Yu, S. P., Choi, D. W. & Gross, R. W. (1995) J. Biol. Chem. 270, 2885-2888. [DOI] [PubMed] [Google Scholar]
- 29.Gubitosi-Klug, R. A. & Gross, R. W. (1996) J. Biol. Chem. 271, 32519-32522. [DOI] [PubMed] [Google Scholar]
- 30.Hamill, O. P., Marty, A., Neher, E., Sakmann, B. & Sigworth, F. J. (1981) Pflugers Arch. 391, 85-100. [DOI] [PubMed] [Google Scholar]
- 31.Bizzozero, O. A. (1995) Methods Enzymol. 250, 361-379. [DOI] [PubMed] [Google Scholar]
- 32.Talavera, K, Staes, M., Janssens, A., Droogmans, G. & Nilius, B. (2004) J. Gen. Physiol. 124, 225-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, G. Y., Shin, Y. K., Lee, C. S. & Song, J. H. (2002) Brain Res. 950, 95-102. [DOI] [PubMed] [Google Scholar]
- 34.Resh, M. D. (2004) Subcell. Biochem. 37, 217-232. [DOI] [PubMed] [Google Scholar]
- 35.Sugars, J. M., Cellek, S., Manifava, M., Coadwell, J. & Ktistakis, N. T. (2002) J. Biol. Chem. 277, 29152-29161. [DOI] [PubMed] [Google Scholar]
- 36.Tao, N., Wagner, S. J. & Lublin, D. M. (1996) J. Biol. Chem. 271, 22315-22320. [DOI] [PubMed] [Google Scholar]
- 37.Navarro-Lerida, I., Corvi, M. M., Barrientos, A. A., Gavilanes, F., Berthiaume, L. G. & Rodriguez-Crespo, I. (2004) J. Biol. Chem. 279, 55682-55689. [DOI] [PubMed] [Google Scholar]
- 38.Schweizer, A., Kornfeld, S. & Rohrer, J. (1996) J. Cell Biol. 132, 577-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stockli, J. & Rohrer, J. (2004) Mol. Biol. Cell 15, 2617-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laine, M., Lin, M. C., Bannister, J. P., Silverman, W. R., Mock, A. F., Roux, B. & Papazian, D. M. (2003) Neuron 39, 467-481. [DOI] [PubMed] [Google Scholar]
- 41.Cuello, L. G., Cortes, D. M. & Perozo, E. (2004) Science 306, 491-495. [DOI] [PubMed] [Google Scholar]
- 42.Wong, W. & Schlichter, L. C. (2004) J. Biol. Chem. 279, 444-452. [DOI] [PubMed] [Google Scholar]
- 43.Wong, W., Newell, E. W., Jugloff, D. G., Jones, O. T. & Schlichter, L. C. (2002) J. Biol. Chem. 277, 20423-20430. [DOI] [PubMed] [Google Scholar]
- 44.Takimoto, K., Yang, E. K. & Conforti, L. (2002) J. Biol. Chem. 277, 26904-26911. [DOI] [PubMed] [Google Scholar]