Abstract
Weinberg and coworkers have used serial transduction of a human, primary fibroblast cell line with the catalytic domain of human telomerase, large T antigen, small T antigen, and an oncogenic allele of H-ras to study stages leading toward a fully transformed cancerous state. We performed a three-dimensional screening experiment using 4 cell lines, 5 small-molecule perturbagens (2-deoxyglucose, oxamate, oligomycin, rapamycin, and wortmannin), and a large number of metabolic measurements. Hierarchical clustering was performed to obtain signatures of the 4 cell lines, 24 cell states, 5 perturbagens, and a number of metabolic parameters. Analysis of these signatures and sensitivities of the cell lines to the perturbagens provided insights into the bioenergetic states of progressively transformed cell lines, the effect of oncogenes on small-molecule sensitivity, and global physiological responses to modulators of aerobic and anaerobic metabolism. We have gained insight into the relationship between two models of carcinogenesis, one (the Warburg hypothesis) based on increased energy production by glycolysis in cancer cells in response to aberrant respiration, and one based on cancer-causing genes. Rather than being opposing models, the approach described here suggests that these two models are interlinked. The cancer-causing genes used in this study appear to increase progressively the cell's dependence on glycolytic energy production and to decrease its dependence on mitochondrial energy production. However, mitochondrial biogenesis appears to have a more complex dependence, increasing to its greatest extent at an intermediate degree of transduction rather than at the fully transformed state.
Keywords: cancer, metabolic profiling, metabolism, small molecules, warburg effect
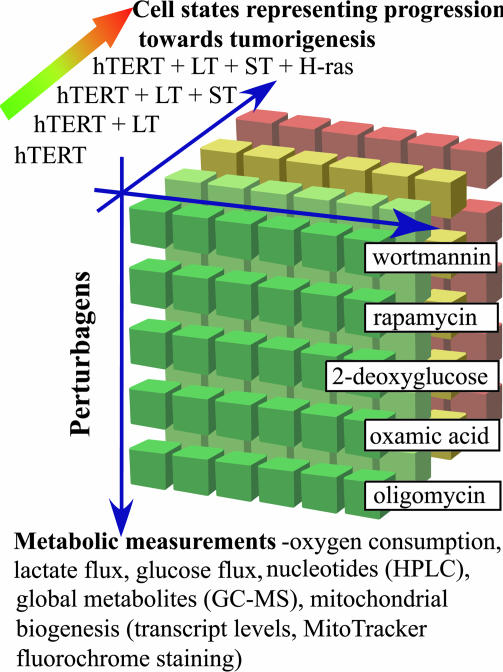
Cancer cells override mechanisms for controlling cellular proliferation, differentiation, and death during malignant transformation. Weinberg and coworkers (1) have shown that ectopic expression of the telomerase catalytic subunit (hTERT) in combination with simian virus 40 large T antigen (LT), small T antigen (ST), and an oncogenic allele of H-ras results in the tumorigenic conversion of normal human epithelial and fibroblast cells (1, 2). In this study, we used the four BJ fibroblast cell lines named 1-[hTERT], 2-[hTERT + LT], 3-[hTERT + LT + ST], and 4-[hTERT + LT + ST + H-ras], which we abbreviate here as CL1, CL2, CL3, and CL4, respectively. To study physiological changes on the path toward tumorigenic conversion, we performed a three-dimensional screening experiment that yielded a matrix of data derived from variations in cell states, cell measurements, and small molecules (Fig. 1). To assess the bioenergetic status of each cell line CL1-CL4, we selected small-molecule inhibitors of metabolic and nutrient-sensing pathways. Each cell line was incubated individually with (i) DMSO; (ii) oxamic acid, an inhibitor of lactate dehydrogenase, which is an enzyme involved in anaerobic glycolysis; (iii) 2-deoxyglucose, an inhibitor of the glycolysis enzyme hexokinase; (iv) oligomycin, an inhibitor of mitochondrial ATP synthase; (v) wortmannin, an inhibitor of phosphatidylinositol 3-kinase; or (vi) rapamycin, an inhibitor of the nutrient-response signaling protein mTOR. Many metabolic cell measurements were made by using each of the resulting 24 cell states. Here, we adopted an approach based on GC-MS and HPLC. We also measured glucose and oxygen consumption, lactate production, and mRNA levels of five genes known to play a central role in mitochondrial function. The transcripts, measured by quantitative RT-PCR, were mitochondrial transcription factor A (TFAM), a key factor in mitochondrial DNA transcription and replication (3); cytochrome c (CYCS, a component of the electron transport chain); nuclear respiratory factor 1 (NRF-1, a nuclear transcription factor for respiratory genes) (4); peroxisome proliferator-activated receptor gamma coactivator (PGC-1α) (5); and ε subunit of F1 ATP synthase (ATP5E) (6). The resulting matrix of data yielded signatures for the 24 cell states and, together with the measured sensitivity of the cell lines to the perturbagens, provided insights into the relationship between cell physiology, especially concerning metabolism and bioenergetics, and cancer-causing genes.
Fig. 1.
Three-dimensional screening. A matrix of data was obtained by altering cell states, small-molecule perturbagens, and cell measurements. Four cell lines, which model tumorigenic conversion and were generated by Weinberg and coworkers (BJ fibroblast cells serially transduced with the four indicated oncogenes), were treated with five small-molecule perturbagens (wortmannin, rapamycin, 2-deoxyglucose, oxamic acid, and oligomycin). Biochemical signatures of the members of this matrix were calculated by using metabolic measurements such as oxygen consumption, glucose consumption, lactate production, and metabolite measurements, using GC-MS or HPLC.
Materials and Methods
Cell Lines. Cell lines serially transduced with the indicated oncogenes [hTERT, simian virus 40 LT, ST, and H-ras] were obtained from William C. Hahn (Dana-Farber Cancer Institute, Boston). Cells were cultured in DMEM containing 1× medium 199 (Invitrogen) and 15% inactivated FBS at 37°C and 5% CO2.
Small Molecules. Oligomycin, rapamycin, wortmannin, 2-deoxyglucose, and oxamic acid were purchased from Sigma-Aldrich and used at 10 μM, 20 nM, 100 nM, 50 mM, and 50 mM, respectively.
Metabolite Extraction. Cells were grown to a density of 6 × 105 cells per 15-mm2 dish and treated with small molecules for 3 h. The extraction protocol was adapted from www.mpimp-golm.mpg.de/fiehn/blatt-protokoll-e.html (7), and ribitol was added as the internal standard.
GC-MS. The dry samples were derivatized, and the GC method was implemented as described in ref. 7.
HPLC. For analysis of nucleotides, the dried samples were reconstituted in ddH2O, and HPLC was performed on an Agilent Technologies 1100 by using a Supelcosil LC-18-DB column from Sigma-Aldrich. Two eluents were used: 100 mM potassium dihydrogen phosphate containing 4 mM tetraammonium bisulfate at pH 6 (solvent A) and 100 mM potassium dihydrogen phosphate containing 4 mM tetraammonium bisulfate at pH 7.2 with 30% methanol (solvent B). An 18-min run, using a gradient from 0% to 100% solvent B, was used, and detection was performed at 254 nm.
Small-Molecule Sensitivity. Cells were seeded in 96-well plates at 2,000 cells per well and treated with varying concentrations of oligomycin (0, 10, 50, 100, and 200 μM), 2-deoxyglucose (0, 10, 100, 500, and 1,000 mM), and oxamic acid (0, 10, 100, 500, and 100 mM). Cells were grown for 48 h, and the IC50 was assayed by using the CyQUANT cell proliferation assay kit (Molecular Probes). A mean graph was constructed according to the definition offered by the Developmental Therapeutics Program of the National Cancer Institute (http://dtp.nci.nih.gov).
Oxygen-Consumption Assay. Cells were transferred to 96-well O2 Biosensor plates (BD Biosciences) with cyclodextran beads, at a density of 500,000 cells per well, and treated with small molecules. After 30 min, fluorescence was measured by using a Spectramax Gemini XS plate reader (Molecular Devices) at an excitation of 485 nm and an emission of 630 nm, followed by subsequent readings every 1 h for 4-5 h.
Glucose-Uptake Assay. Cells were grown and treated as described for the oxygen assay. A 4-μl sample of medium was taken after 30 min and again after 4 h, and was diluted 100-fold. The assay was performed by using the Amplex Red/Glucose Oxidase kit (Molecular Probes). Fluorescence was measured at an excitation of 563 nm and an emission of 587 nm, and absorbance was measured by using a Spectramax Plus 384 plate reader (Molecular Devices) at 563 nm.
Lactate-Production Assay. Lactate oxidase was substituted for glucose oxidase, and the assay was performed as described above.
Data Analysis. Triplicate samples were used to calculate the standard deviation for each metabolite measurement. For GC-MS spectra, metabolites were identified by using electron impact spectra library generated by us and from the metabolomic analysis group at the Max Plank Institute (www.mpimp-golm.mpg.de/fiehn). Relative metabolite levels were normalized to the concentration of internal standard and total ion current. Peaks identified by HPLC were quantified by using total UV absorbance of each peak and normalized based on the total protein concentration of cells. Oxygen, glucose, and lactate measurements were normalized based on the total protein concentration of the cells. For hierarchical clustering and heat map generation, all measurements were expressed as fold change compared to a reference state. Changes with a z value ≥2.2 were considered significant. Average linkage hierarchical clustering was performed by using the gene cluster program, and heat maps and dendrograms were constructed by using the treeview program (both available at http://rana.lbl.gov).
RNA Isolation and Quantitative RT-PCR. Cells were grown and treated with small molecules for 3 h. RNA was isolated by using the RNeasy kit (Qiagen). Quantitative RT-PCR was performed by using the Quantitect SYBR Green kit (Qiagen) on an Opticon instrument (MJ Research). Primers were designed by using the primer3 program (available at http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). Relative quantitation was done by using the ΔCT method by taking the difference (ΔCT) between the CT of β-actin and CT of each transcript and computing 2-ΔCt. CL1 was used as the calibrator cell line, and the fold change was defined as the ratio of the level of transcript in a sample over that in CL1.
Results
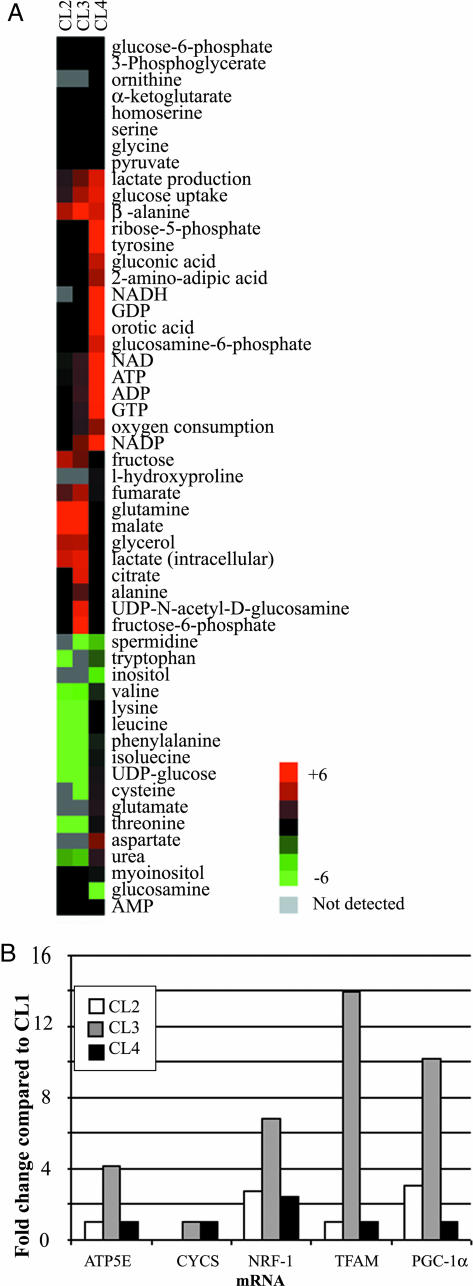
Profiling of the Basal States of the BJ Cell Lines. To quantify the effects of oncogenes on cellular bioenergetics, we compared metabolic measurements of four cell lines that constitute a defined path toward tumorigenic conversion (Fig. 1). In this analysis, the immortalized cell line CL1 was used as the reference state. The metabolic measurements from the cell lines were expressed as the fold change over their respective measurements in the reference CL1 cell line. The heat-map signatures of these changes are shown in Fig. 2A. A progressive increase in oxygen consumption occurred from CL2 to CL4 such that the fully transformed CL4 cells consumed three times the amount of oxygen as the reference state. This was accompanied by a steeper progressive increase in the rates of glucose uptake and lactate production, up to 6-fold in CL4. Levels of ATP, GTP, NAD+, and NADP+ also increased for both CL3 and CL4 cells. CL4 had high levels of ribose-5-phosphate and orotic acid, both of which are involved in nucleotide biosynthesis. CL3 had high levels of citric acid cycle metabolites, such as citrate, malate, and fumarate, but CL4 did not show these changes from the reference state. Similarly, CL3 had high levels of several transcripts, such as PGC-1α, NRF-1, TFAM, and ATP5E, which are important for mitochondrial biogenesis (Fig. 2B), as well as increased mitochondrial mass as determined by mitotracker dye staining of mitochondria (data not shown), but CL4 had low levels of all these transcripts and did not have increased mitochondrial mass.
Fig. 2.
Biochemical signatures of a cell-line model of tumorigenesis. (A) Heat map showing the relative levels of the metabolites in the four cell lines. The metabolic measurements were expressed as the fold change relative to the same measurement in the CL1 cell line. (B) The relative levels of the TFAM, CYCS, ATP5E, PGC-1α, and NRF-1 transcripts, expressed as the fold change over that in the CL1 cell line, are plotted in the bar graph.
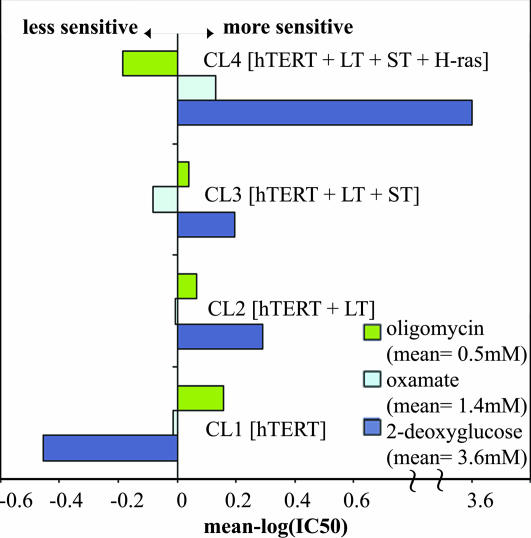
Sensitivity of the BJ Cell Lines to Small-Molecule Modulators of Aerobic and Anaerobic Metabolism. We measured the growth inhibition of cells after treatment with three small molecules that perturb enzymes involved in cellular metabolism (oligomycin, 2-deoxyglucose, and oxamic acid). Oligomycin is an inhibitor of mitochondrial ATP synthase, 2-deoxyglucose is a substrate analog inhibitor of hexokinase, and oxamic acid is a substrate analog inhibitor of lactate dehydrogenase. The concentrations that inhibit the growth of each cell line by 50% (IC50) were measured as described in Materials and Methods, and a mean graph of small-molecule sensitivities was constructed (Fig. 3). We observed a continuous decline in oligomycin sensitivity as cells increased their tumorigenic potential from CL1 to CL4. This trend was reversed with 2-deoxyglucose, with CL4 being the most sensitive and CL1 being the least sensitive. CL4 was also the most sensitive to oxamic acid, but in this case CL3 was the least sensitive.
Fig. 3.
Mean graph of the small-molecule sensitivities of the four cell lines to oligomycin, oxamate, and 2-deoxyglucose. The mean graph was constructed as defined by the Developmental Therapeutics Program of the National Cancer Institute (http://dtp.nci.nih.gov). The mean graph consists of positive (more sensitive) and negative (less sensitive) “delta” values, generated from a set of IC50 values by using a three-step calculation. The IC50 values for each of the cell lines against the small molecules were converted to log(IC50) values. For each small molecule, the log(IC50) values of the cell lines are averaged. Finally, the individual IC50 values are subtracted from this average to create a delta. The mean small-molecule sensitivities of oligomycin, oxamate, and 2-deoxyglucose are 0.5, 1.4, and 3.6 mM, respectively. Therefore, for cell line CL4, a delta value of 3.6 for 2-deoxyglucose indicates that it is ≈1,000-fold more sensitive than the average sensitivities of the four cell lines.
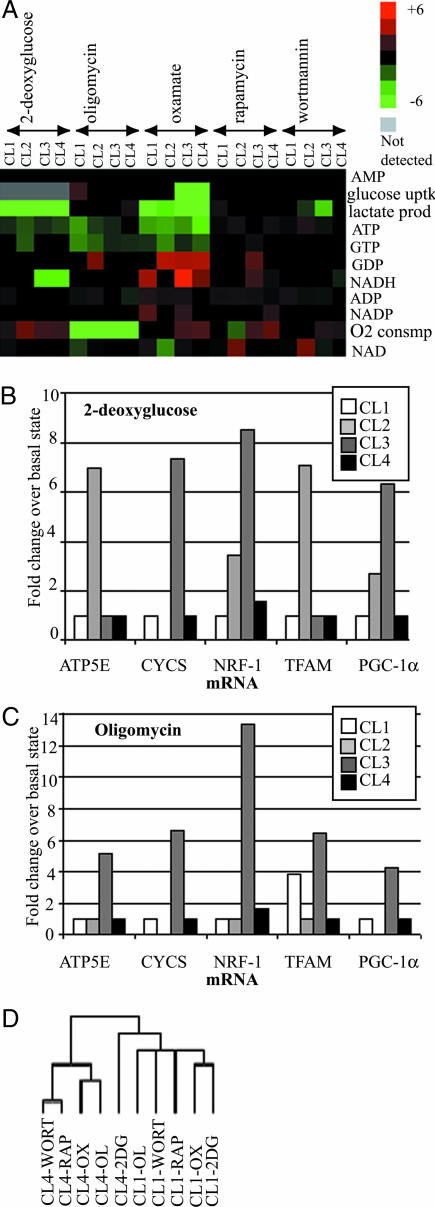
Perturbational Profiling of BJ Cell Lines by Using Small Molecules. To examine the effects of small-molecule perturbations in more detail, we obtained bioenergetic signatures of each cell line after treatment with the five small molecules. In this analysis, we used measurements for each cell line in the basal state as the reference for comparison with the small-molecule treated state (Fig. 4A). Oligomycin treatment caused decreases in ATP levels that were progressively less pronounced in the sequence from CL1 to CL4, such that CL1 had 60% less ATP, CL2 and CL3 had 30% less ATP, and CL4 showed no significant change. Oligomycin also inhibited oxygen consumption in all cell lines, but CL1 was the only cell line to respond by increasing glucose uptake. 2-Deoxyglucose treatments caused decreases in ATP and increases in oxygen flux for all cell lines. This increase was high for CL2, CL3, and CL4 but minor for CL1. Oxamic acid inhibited lactate production in all cell lines. Glucose uptake was also inhibited in CL3 and CL4, but oxygen flux was increased in these cell lines. Oxamic acid caused the greatest decreases in ATP of any small molecule, and CL4 was most severely affected, with a 70% decrease compared with 50% for the other cell lines. Rapamycin and wortmannin caused only small changes in the levels of most metabolites. However, both small molecules led to an increase in oxygen flux in CL3 and CL4.
Fig. 4.
Perturbational profiling of cell-line models of tumorigenesis. The four cell lines were perturbed by using 2-deoxyglucose, oligomycin, oxamate, rapamycin, and wortmannin as described in Materials and Methods. (A) The fold change of the normalized levels of metabolic measurement (with respect to that of the unperturbed basal states of the respective cell lines) was used to construct heat maps. The columns in the heat map are grouped by the small-molecule perturbagen. (B) The relative levels of TFAM, CYCS, ATP5E, PGC-1α, and NRF-1 transcripts, prepared from the four cell lines after treatment with 2-deoxyglucose, were expressed as fold change over their respective levels in the unperturbed basal state and plotted on the bar graph. (C) Same analysis as in B, after the four cell lines were treated with oligomycin. (D) Hierarchical clustering of cell states and metabolites, with additional metabolite measurements from GC-MS. The columns representing the cell states are labeled by the name of the cell line followed by the perturbation. 2DG, 2-deoxyglucose; OL, oligomycin; OX, oxamate; RAP, rapamycin; WORT, wortmannin.
We also measured transcript levels in each cell line after treatment with 2-deoxyglucose and oligomycin. 2-Deoxyglucose caused 5- to 13-fold increases in all five transcripts for CL3 relative to the basal state (Fig. 4B). Oligomycin caused 6- to 8-fold increases in NRF-1, PGC-1α, and CYCS. CL4 only showed a small increase in NRF-1 in both cases (Fig. 4C).
Average linkage hierarchical clustering was performed on the metabolic data (Fig. 4D). The global physiological responses to 2-deoxyglucose of CL1 and CL4 were shown by cluster analysis to be highly dissimilar, whereas their responses to oligomycin are more similar.
Discussion
Over 70 years ago, Warburg performed experiments on tumor and normal tissues that led to a proposal for the pathogenesis of cancer (8). He reported that tumor tissues convert glucose to lactate, via the reduction of pyruvate, even in the presence of oxygen, whereas normal tissues use pyruvate, derived from glycolysis, plus oxygen to produce ATP via mitochondrial respiration. Furthermore, cancer cells were proposed to produce ATP from glycolysis, deriving the requisite NAD+ from the conversion of pyruvate into lactate, in amounts comparable to that derived from oxygen-dependent respiration in normal cells, which were thought to produce 100-fold less ATP from glycolysis relative to respiration. Thus, cancer was proposed to derive from an impairment of cellular respiration (9). Although cancer is now viewed as a disease resulting from cancer-causing genes that deregulate cellular proliferation, differentiation, and death, the relationship between these genes and the deregulation of energy production is only partially understood. The results described below, made possible by the combined use of stepwise transformed cell lines and metabolic profiling, suggest that the two views are intimately linked and fully consonant.
The two views of cancer have been studied in the case of the oncogene Akt by Thompson and colleagues (10). Akt has been shown to promote the malignant transformation of hematopoietic cells (11), and it has been shown that the phosphatidylinositol 3-kinase/Akt pathway can induce glucose uptake and glycolysis in cells. Elstrom and colleagues (10) demonstrated that Akt can promote aerobic glycolysis without up-regulating oxidative phosphorylation in a dose-dependent fashion, suggesting its role in the Warburg effect. The above studies support the possibility that metabolic transformations play an important role in oncogenesis. The high glycolytic activity in cancer cells has been shown to be associated with an increased expression of glycolytic enzymes and glucose transporters (12), and has been linked to the role of the oncogene c-myc and hypoxia-inducible factor 1α (HIF-1α) (13). Higher levels of hexokinase, in hepatomas, mediate this high rate of glycolysis (14). Of the oncogenes used in the matrix experiment (Fig. 1), Ras is the only one that has been studied in some detail for its impact on cellular metabolism. For example, in yeast the Ras-cAMP pathway can activate the glycolytic enzyme 6-phosphofructo-1-kinase via the cAMP-dependent protein kinase (PKA) (15), and the Ras cascade has been shown to increase the levels of mitochondrial oxidative phosphorylation complexes (16). In glioblastoma cells, inhibition of H-ras resulted in diminished glycolysis and cell death (17). Biagalow et al. (18) showed that in rat embryo cells, H-ras stimulates glycolysis and inhibits oxygen consumption. Young et al. (19) used two ovarian cell lines, one transduced with hTERT, LT, and ST, and the other with hTERT, LT ST, and H-ras, to compare immortalized and transformed states. Using two-dimensional electrophoretic separation of cellular proteins, they showed that five proteins involved in redox balance were up-regulated, and that Ras activity was responsible for resistance to oxidative stress-mediated apoptosis.
The mitochondria are known to play a central role in the regulation of cellular energy metabolism; however, the role of mitochondria in oncogenesis is not well understood. Cuveza et al. (20) have measured the expression levels of β-F1-ATPase relative to that of mitochondrial Hsp60 in liver, colon, and renal carcinomas. They showed a general down-regulation of mitochondrial components in liver tumors and a specific down-regulation of β-F1-ATPase in colon and renal tumors (21). Rossignol and colleagues (22) have shown that the mitochondria are structurally and functionally dynamic and can adapt their oxidative capacity to meet the energy requirements of cancer cells. Such alterations in energy metabolism and mitochondrial function may play a role in oncogenesis.
Recently, Nolan and colleagues (23) classified cancers based on their response to perturbation, using five cytokines to perturb the phosphorylation state of six signaling proteins, and used these states to distinguish between leukemic cell lines. In the present study, we have explored the differences in mitochondrial biogenesis and metabolism in a well characterized set of cell lines with serially transduced oncogenes combined with a set of five small-molecule perturbagens. A set of metabolic measurements provided a metabolic signature for the different stages of tumorigenesis (Fig. 1); in addition, metabolic profiles obtained after small-molecule perturbation generated signatures of the response to these perturbagens (heat maps in Figs. 2 and 4A). Hierarchical clustering was performed on the metabolic data (obtained from HPLC and GC-MS analysis) to help visualize the differential effects of cancer genes and the perturbagens on cellular physiology (Fig. 4D).
The glycolysis inhibitor 2-deoxyglucose elicited the maximal difference in sensitivities between CL1 and CL4 (Fig. 3). The dendrogram in Fig. 4D indicates that signatures of these two cell lines upon treatment with 2-deoxyglucose are maximally dissimilar, suggesting that biochemical signatures obtained by using metabolic measurements correlate well with sensitivity to the corresponding small molecules (Fig. 3). Fig. 4D also illustrates that within each cell line, the perturbational profiles of oxamate, another inhibitor of glycolysis, and 2-deoxyglucose are most similar to each other, as are the profiles of rapamycin and wortmannin, small molecules that target elements of the nutrient-response signaling network.
During serial transduction of these cell lines, there is an increased rate of oxygen consumption and an increase in glucose uptake and lactate production, as seen in the heat map in Fig. 2. CL4 is distinguished by its high rates of aerobic and anaerobic metabolism relative to CL1, as well as by high levels of key nucleotides, such as ATP, GTP, NAD+, NADH, and NADP+. There is no concomitant increase in the levels of citric-acid cycle metabolites, suggesting the possibility that increased levels of NADH, necessary for increased oxidative phosphorylation, may come from sources extrinsic to the citric acid cycle. CL2 and CL3, which are intermediate in the progression to the most transformed state, had bioenergetic signatures intermediate between CL1 and CL4.
As the cell lines progress toward a more tumorigenic state, they become more sensitive to 2-deoxyglucose and less sensitive to the mitochondrial ATP synthase inhibitor oligomycin; furthermore, the fully transformed cell line is most dependent on glycolysis and least dependent on the mitochondrial machinery for ATP synthesis. Such responses to small-molecule perturbagens suggest differences in the cellular physiology of cells during tumorigenesis, as judged by the mean graph in Fig. 3 and the heat map in Fig. 4A. Oligomycin sensitivity declined progressively from CL1 to CL4 (Fig. 3), as did the changes in cellular ATP levels caused by oligomycin treatment (Fig. 4A). CL4 is least sensitive to oligomycin and showed no decrease in ATP levels upon treatment with oligomycin, consistent with predominant ATP production via glycolysis in CL4, whereas CL1 is most sensitive and showed a large decrease in ATP levels. CL2 and CL3 are intermediate in both parameters. These results suggest that CL4 is least dependent on the mitochondrial machinery for ATP production. In contrast, CL4 is the most sensitive to both 2-deoxyglucose and oxamic acid. Consistent with the Warburg effect, we found that the inhibition of anaerobic ATP production with 2-deoxyglucose, as opposed to inhibition of aerobic ATP production with oligomycin, induced greater reductions in cellular ATP and proved more lethal in the maximally transformed state represented by CL4.
We imagine two plausible explanations for an increase in ATP production and an increase in the relative contribution to ATP synthesis from glycolysis in cells with high tumorigenic potential. Such cells are either making little ATP by respiration or they are more able to switch readily to glycolytic (anaerobic) ATP synthesis after exposure to an ATP synthase inhibitor. Consistent with the latter explanation, Salomon et al. (24) have shown that Jurkat cells, which are ordinarily insensitive to another inhibitor of mitochondrial ATP synthase, apoptolidin, can be strongly sensitized toward apoptolidin by treatment with 2-deoxyglucose.
It is intriguing that cells with greater tumorigenic potential consume more oxygen and yet exhibit diminished oxygen-dependent (aerobic) ATP synthesis. It is possible that such cells use the mitochondrial electron transport chain and oxidative phosphorylation for reasons other than the production of ATP; for example, for producing heat or reactive oxygen species by allowing leakage of the membrane potential. A caveat to this explanation is that we observe a diminution in mitochondrial biogenesis after the addition of H-ras to CL4. It is also possible that cells with greater tumorigenic potential use mitochondria and oxygen for pyrimidine synthesis (facilitating DNA synthesis) rather than for ATP synthesis, as judged by CL4 having higher levels of ribose-5-phosphate and orotic acid in the basal state (Fig. 2 A). Ribose-5-phosphate is the starting reactant for the formation of 5-phosphoribosyl-1-pyrophosphate (PRPP), which is the ribose phosphate donor in nucleotide biosynthesis. Orotic acid reacts with PRPP in a biosynthetic route that produces pyrimidine nucleotides. Orotic acid itself is produced in a mitochondrial electron transport chain (EC)-dependent manner by dihydroorotate dehydrogenase, which has been shown to be associated with EC enzymes in the mitochondria (25). These results indicate a possible coupling of nucleotide biosynthesis with the mitochondrial machinery to achieve the high rates of cell proliferation in the fully transformed state. This coupling suggests that modulators of mitochondrial metabolism and biogenesis may have a therapeutic window as anticancer agents. Noteworthy in this regard is the discovery by small-molecule screening of a small molecule that selectively induces cell death in mammary epithelial cells overexpressing the neu oncogene. This small molecule targets mitochondria having high membrane potential (26). In the future it will be important to determine whether oxygen might be consumed in cells with greater tumorigenic potential by nonmitochondrial mechanisms, for example, by peroxisomes.
Treatment with small-molecule perturbagens revealed that the inhibition of certain elements of signaling networks produced differential responses in the activities of metabolic pathways and in a manner dependent on the oncogenic background. Rapamycin and wortmannin do not effect the rate of oxygen consumption, and thus, presumably, oxidative phosphorylation, after the introduction of hTERT (CL1) and hTERT plus LT (CL2). But after the additional introduction of ST (CL3), both rapamycin and wortmannin cause an increase in oxygen consumption. This effect is maintained after the additional introduction of H-ras (CL4).
As cell lines progress toward greater tumorigenic potential via the introduction of hTERT, LT, and ST (CL1-CL3), they have increasing levels of mitochondrial biogenesis. However, after the introduction of H-ras (CL4) there is a decline in mitochondrial biogenesis, as seen in the bar graphs in Figs. 2B and 4 B and C. In the basal state, CL3 had the highest levels of PGC-1α, NRF-1, and TFAM transcripts, all of which play key roles in mitochondrial biogenesis. CL3 also has the highest levels of citric-acid cycle metabolites, such as citrate, malate, and fumarate (Fig. 2 A), suggesting an increased supply of reducing equivalents for aerobic metabolism. Together with an increased mitochondrial mass, these results indicate an induction of a mitochondrial biogenesis program by the specific combination of oncogenic elements present in CL3. This effect is even more pronounced when CL3 is treated with 2-deoxyglucose, which caused a severalfold increase over the basal state in all transcripts tested. In contrast, CL4 had low levels of all transcripts in the basal state and only a small increase in NRF-1 in response to 2-deoxyglucose. We therefore infer that the introduction of H-ras in CL4 attenuates the mitochondrial biogenesis response conferred by the combination of hTERT, LT, and ST.
Global biochemical profiles in combination with modulators of bioenergetic and signaling pathways can illuminate the influence of oncogenes on cellular physiology. Using a three-dimensional screen of cell states, cell measurements, and smallmolecule perturbagens, we have discovered differential metabolic responses to small molecules and differential dependencies of tumorigenic states on specific metabolic pathways. The data and our analyses show that the cancer-gene and Warburg concepts are fully consonant but, presumably, with cancer genes as the cause and metabolic alterations as an effect.
Acknowledgments
We thank William C. Hahn for providing the cell lines; Bimal Desai for guidance in initiating this work; Aravind Subramanium, Vamsi Mootha, and Bridget Wagner for advice and discussions; and Matt Wong, Joanne Kelleher, and Gregory Stephanapoulos for use of and training on GC-MS instrumentation. This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases Diabetes Genome Anatomy Project of the Joslin Diabetes Center in Boston. S.L.S. is an Investigator of the Howard Hughes Medical Institute.
Author contributions: A.R., C.W., and S.L.S. designed research; A.R. and C.W. performed research; A.R. and C.W. contributed new reagents/analytic tools; A.R. and C.W. analyzed data; A.R., C.W., and S.L.S. wrote the paper; and S.L.S. is the principal investigator in the laboratory in which this research was performed.
Abbreviations: LT, large T antigen; ST, small T antigen.
References
- 1.Hahn, W. C., Counter, C. M., Lundberg, A. S., Beijersbergen, R. L., Brooks, M. W. & Weinberg, R. A. (1999) Nature 400, 464-468. [DOI] [PubMed] [Google Scholar]
- 2.Dolma, S., Lessnick, S. L., Hahn, W. C. & Stockwell, B. R. (2003) Cancer Cell 3, 285-296. [DOI] [PubMed] [Google Scholar]
- 3.Norrbom, J., Sundberg, C. J., Ameln, H., Kraus, W. E., Jansson, E. & Gustafsson, T. (2004) J. Appl. Physiol. 96, 189-194. [DOI] [PubMed] [Google Scholar]
- 4.Leung, L., Kwong, M., Hou, S., Lee, C. & Chan, J. Y. (2003) J. Biol. Chem. 278, 48021-48029. [DOI] [PubMed] [Google Scholar]
- 5.Mootha, V. K., Lindgren, C. M., Eriksson, K. F., Subramanian, A., Sihag, S., Lehar, J., Puigserver, P., Carlsson, E., Ridderstrale, M., Laurila, E., et al. (2003) Nat. Genet. 34, 267-273. [DOI] [PubMed] [Google Scholar]
- 6.Tu, Q., Yu, L., Zhang, P., Zhang, M., Zhang, H., Jiang, J., Chen, C. & Zhao, S. (2000) Biochem. J. 347, 17-21. [PMC free article] [PubMed] [Google Scholar]
- 7.Fiehn, O., Kopka, J., Dormann, P., Altmann, T., Trethewey, R. N. & Willmitzer, L. (2000) Nat. Biotechnol. 18, 1157-1161. [DOI] [PubMed] [Google Scholar]
- 8.Warburg, O. (1924) Biochem. Z. 152, 319-344. [Google Scholar]
- 9.Warburg, O. (1956) Science 124, 269-270. [PubMed] [Google Scholar]
- 10.Elstrom, R. L., Bauer, D. E., Buzzai, M., Karnauskas, R., Harris, M. H., Plas, D. R., Zhuang, H., Cinalli, R. M., Alavi, A., Rudin, C. M., et al. (2004) Cancer Res. 64, 3892-3899. [DOI] [PubMed] [Google Scholar]
- 11.Karnauskas, R., Niu, Q., Talapatra, S., Plas, D. R., Greene, M. E., Crispino, J. D. & Rudin, C. M. (2003) Oncogene 22, 688-698. [DOI] [PubMed] [Google Scholar]
- 12.Flier, J. S., Mueckler, M. M., Usher, P. & Lodish, H. F. (1987) Science 235, 1492-1495. [DOI] [PubMed] [Google Scholar]
- 13.Lu, H., Forbes, R. A. & Verma, A. (2002) J. Biol. Chem. 277, 23111-23115. [DOI] [PubMed] [Google Scholar]
- 14.Rencurel, F., Munoz-Alonso, M. J., Girard, J. & Leturque, A. (1998) J. Biol. Chem. 273, 26187-26193. [DOI] [PubMed] [Google Scholar]
- 15.Dihazi, H., Kessler, R. & Eschrich, K. (2003) Biochemistry 42, 6275-6282. [DOI] [PubMed] [Google Scholar]
- 16.Dejean, L., Beauvoit, B., Bunoust, O., Guerin, B. & Rigoulet, M. (2002) Biochem. Biophys. Res. Commun. 293, 1383-1388. [DOI] [PubMed] [Google Scholar]
- 17.Blum, R., Jacob-Hirsch, J., Amariglio, N., Rechavi, G. & Kloog, Y. (2005) Cancer Res. 65, 999-1006. [PubMed] [Google Scholar]
- 18.Biaglow, J. E., Cerniglia, G., Tuttle, S., Bakanauskas, V., Stevens, C. & McKenna, G. (1997) Biochem. Biophys. Res. Commun. 235, 739-742. [DOI] [PubMed] [Google Scholar]
- 19.Young, T. W., Mei, F. C., Yang, G., Thompson-Lanza, J. A., Liu, J. & Cheng, X. (2004) Cancer Res. 64, 4577-4584. [DOI] [PubMed] [Google Scholar]
- 20.Cuezva, J. M., Krajewska, M., de Heredia, M. L., Krajewski, S., Santamaria, G., Kim, H., Zapata, J. M., Marusawa, H., Chamorro, M. & Reed, J. C. (2002) Cancer Res. 62, 6674-6681. [PubMed] [Google Scholar]
- 21.Cuezva, J. M., Chen, G., Alonso, A. M., Isidoro, A., Misek, D. E., Hanash, S. M. & Beer, D. G. (2004) Carcinogenesis 25, 1157-1163. [DOI] [PubMed] [Google Scholar]
- 22.Rossignol, R., Gilkerson, R., Aggeler, R., Yamagata, K., Remington, S. J. & Capaldi, R. A. (2004) Cancer Res. 64, 985-993. [DOI] [PubMed] [Google Scholar]
- 23.Irish, J. M., Hovland, R., Krutzik, P. O., Perez, O. D., Bruserud, O., Gjertsen, B. T. & Nolan, G. P. (2004) Cell 118, 217-228. [DOI] [PubMed] [Google Scholar]
- 24.Salomon, A. R., Voehringer, D. W., Herzenberg, L. A. & Khosla, C. (2000) Proc. Natl. Acad. Sci. USA 97, 14766-14771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loffler, M., Jockel, J., Schuster, G. & Becker, C. (1997) Mol. Cell. Biochem. 174, 125-129. [PubMed] [Google Scholar]
- 26.Fantin, V. R., Berardi, M. J., Scorrano, L., Korsmeyer, S. J. & Leder, P. (2002) Cancer Cell 2, 29-42. [DOI] [PubMed] [Google Scholar]