Abstract
Mercury (Hg) is a ubiquitous heavy metal that originates from both natural and anthropogenic sources and is transformed in the environment to its most toxicant form, methylmercury (MeHg). Recent studies suggest that MeHg exposure can alter epigenetic modifications during embryogenesis. In this study, we examined associations between prenatal MeHg exposure and levels of cord blood DNA methylation (DNAm) by meta-analysis in up to seven independent studies (n = 1462) as well as persistence of those relationships in blood from 7 to 8 year-old children (n = 794). In cord blood, we found limited evidence of differential DNAm at cg24184221 in MED31 (β = 2.28 × 10−4, p-value = 5.87 × 10−5) in relation to prenatal MeHg exposure. In child blood, we identified differential DNAm at cg15288800 (β = 0.004, p-value = 4.97 × 10−5), also located in MED31. This repeated link to MED31, a gene involved in lipid metabolism and RNA Polymerase II transcription function, may suggest a DNAm perturbation related to MeHg exposure that persists into early childhood. Further, we found evidence for association between prenatal MeHg exposure and child blood DNAm levels at two additional CpGs: cg12204245 (β = 0.002, p-value = 4.81 × 10−7) in GRK1 and cg02212000 (β = −0.001, p-value = 8.13 × 10−7) in GGH. Prenatal MeHg exposure was associated with DNAm modifications that may influence health outcomes, such as cognitive or anthropometric development, in different populations.
Keywords: DNA methylation, Mercury, Methylmercury, Prenatal exposure, PACE, HELIX study, ALSPAC
1. Introduction
Mercury (Hg) is a ubiquitous environmental toxicant that originates from both natural and anthropogenic sources (United Nations, 2018). Most of the inorganic Hg present in the atmosphere comes from point sources such as mining operations or industrial activities (Hintelmann, 2010). Atmospheric inorganic Hg is deposited in the earth surface and transformed to the organic form, methylmercury (MeHg), by the action of some bacteria present in the aquatic sediments (Parks et al., 2013). MeHg is accumulated and biomagnified through the food chain, reaching the highest concentrations in big, oily, predatory fishes and marine mammals (Hintelmann, 2010). Seafood consumers around the world have recorded high mercury concentrations linked to this consumption (Sheehan et al., 2014). Total Hg (THg) in hair and THg in blood are both validated biomarkers of MeHg intake (80% and 90% of THg is considered MeHg, respectively (Llop et al., 2017)) and correlated with seafood consumption in general human populations (Clarkson and Magos, 2006; Berglund et al., 2005).
Prenatal exposure to Hg has been associated with impaired foetal growth (Murcia et al., 2016a; Ballester et al., 2018a; Drouillet-Pinard et al., 2010) and adverse birth outcomes, such as low placental weight (Murcia et al., 2016b; Al-Saleh et al., 2014), newborn’s anthropometry (Murcia et al., 2016b; Gustin et al., 2020), and length of gestation (Ballester et al., 2018b; Dallaire et al., 2013). Additionally, perinatal and early childhood exposure to MeHg have been associated with adverse outcomes later in life, such as postnatal growth (Murcia et al., 2016b; Kim et al., 2011). Regarding effects in children’s neuropsychological development, consistent results have been observed at high levels of exposure (Karagas et al., 2012; Grandjean et al., 2014; Sharma et al., 2019); however, at low-moderate levels the results were heterogeneous (Ha et al., 2017). No clear pattern has been observed among the limited number of studies that assessed the association between early exposure to mercury and cardiovascular effects (Gallego-Viñas et al., 2019), although recent studies showed some evidence (Farzan et al., 2021; Ying Chan et al., 2021; Zhang et al., 2021).
There are several mechanisms of MeHg toxicity suggested, involving several biological processes. These processes include increased lipid peroxidation, reactive oxygen species (ROS) generation and glutathione (GSH) depletion in vivo and in vitro (Crespo-López et al., 2009), reduced cell membrane integrity (Polunas et al., 2011), altered cell signalling (Worth et al., 2001), mitochondrial impacts (Dreiem et al., 2005), altered DNA repair (Pieper et al., 2014), immunomodulatory impacts (Li et al., 2014), affected regulation of Ca2+ (Aschner et al., 2007), and changed DNA methylation (DNAm), as well as other epigenetics marks (Farina et al., 2011; Cardenas et al., 2017a; Khan et al., 2019). As a consequence of the epigenetics changes, several genes involved in response to oxidative stress, stress response, metabolism, transport, gene regulation, inflammatory response, apoptosis, and hormone regulation, have been associated with differential expression in human cells exposed to MeHg (Yang et al., 2020). Also, some research has suggested that these perturbations during embryogenesis can establish new DNAm patterns that may persist during foetal development and childhood (Khan et al., 2019; Perera and Herbstman, 2011a).
To date, studies assessing the association between prenatal exposure to MeHg and DNAm are scarce. As far as we aware, only four studies have been conducted relating prenatal Hg and epigenome-wide DNAm changes in cord blood with sample sizes between 138 and 321 (Cardenas et al., 2015, 2017a, 2017b; Bakulski et al., 2015). These studies have identified methylation changes in specific CpG sites in relation to Hg exposure. Among them, only one study evaluated the persistence of these epigenetic changes, observing that DNAm at the Paraoxonase 1 gene (PON1) in cord blood found at birth persisted until early childhood and was attenuated in mid-childhood blood (Cardenas et al., 2017b).
The overall aim of this study was to investigate the association between prenatal MeHg exposure and DNAm in cord blood, as well the persistence of these associations in blood from 7 to 8 years-old children. To do this, we performed a fixed effects meta-analysis of up to seven independent studies that are members of the Pregnancy And Childhood Epigenetics (PACE) consortium (Felix et al., 2018) and/or the Human Early Life Exposome (HELIX) study (Maitre et al., 2018). We also aimed to gain insights into the potential biological and health-related impacts of these associations by performing functional enrichment. This study represents the largest examination to date of prenatal MeHg associations with cord and child blood DNAm in human populations and provides novel insights of MeHg toxicity through the fetal period and childhood.
2. Materials and methods
2.1. Participating cohorts
Cohorts that are members of the PACE consortium were identified for participation in the meta-analysis if they had existing prenatal total mercury (THg) measures (in cord blood, hair, or maternal blood), DNAm data quantified in cord or child blood via the Illumina Infinium HumanMethylation450 BeadChip or Infinium MethylationEPIC BeadChip arrays, and if they had information on covariates (see below). The cohorts that participated in the epigenome-wide association study (EWAS) of cord blood were: Avon Longitudinal Study of Parents and Children (ALSPAC) (Golding et al., 2001), Hokkaido Study on Environment and Children’s Health, Sapporo cohort (SAPPORO-HOKKAIDO) (Kishi et al., 2011), Environment and Childhood Project (INMA) (Guxens et al., 2012), KOREAN Exposome (Park et al., 2020), and Project VIVA (VIVA) (Oken et al., 2015). Human Early Life Exposome study (HELIX) (Maitre et al., 2018) included three other on-going European cohorts with child blood DNAm data, and for this particular study we analysed data from: INMA (Guxens et al., 2012), Mother, Father and Child Cohort Study (MoBa) (Magnus et al., 2016), and the Mother-Child Cohort in Crete (RHEA) (Chatzi et al., 2017). ALSPAC (Golding et al., 2001), as well as the cohorts taking part the HELIX study, participated in the EWAS of child blood methylation data at ages 7–8 years. Only the largest ethnicity group in each cohort was considered to maintain the homogenous effect assumption (Hong et al., 2016). Characteristics for individuals of the included cohorts and covariates definitions are shown in Table 1. See Supplementary Methods file for more details on design and study population, collection of samples, DNAm data acquisition, and mercury and covariates measurements in each cohort.
Table 1.
Characteristics of the study participants in the included cohorts.
| Cohort | Units | INMA | KOREA | VIVA | SAPPORO-HOKKAIDO | ALSPAC | MoBa | RHEA |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Country | Spain | Korea | United States | Japan | United Kingdom | Norway | Greece | |
| Main ethnicity | ||||||||
| European | % | 90 | 76 | 100 | 95.8 | 100 | ||
| Asian | % | 100 | 3.3 | 100 | 2.8 | |||
| African | % | 8.6 | ||||||
| Other | % | 10 | 11.8 | 1.4 | ||||
| Prenatal THg matrix | Cord blood | Maternal blood | Maternal blood | Maternal hair | Maternal blood | Maternal blood | Maternal blood | |
| Cord THg* | GM (CI95%), μg/L | 8.70 (8.24, 9.19) | 5.33 (5.19, 5.47) | 5.74 (5.16, 6.39) | 9.30 (6.18–12.42) | 2.01 (0.79, 4.89) | 6.02 (5.72, 6.35) | 7.13 (6.71, 7.59) |
| N children with cord THg + cord blood DNA methylation | N | 304 | 329 | 338 | 268 | 223 | No | No |
| N children with blood THg + blood DNA methylation | N | 182 | No | No | No | 312 | 192 | 197 |
| Maternal age | Mean (SD), years | 30.4 (4.1) | 30.4 (3.6) | 32.7 (4.9) | 30.1 (5.0) | 29.4 (4.3) | 32.8 (3.7) | 30.8 (4.8) |
| Parity | % nulliparous | 37.1 | 56.5 | 49.7 | 46.3 | 49.3 | 52.8 | 60.0 |
| Child sex | % male | 51.2 | 48.0 | 52.1 | 45.5 | 47.1 | 53.7 | 55.5 |
| Maternal education | ||||||||
| Up to primary | % | 26.8 | 22.2 | 5.6 | 3.3 | 8.1 | 0 | 4.6 |
| Secondary | % | 42.4 | 16.4 | 22.3 | 41.0 | 68.6 | 20.1 | 55.8 |
| University | % | 307 | 61.4 | 71.9 | 55.5 | 23.3 | 79.9 | 39.6 |
| Smoking during pregnancy | %, yes | 29.2 | 10.9 | 11.5 | 16.4 | 17.9 | 4.4 | 21.2 |
| Fish consumption | Mean (SD), servings/week | 5.0 (2.3) | 4.2 (4.1) | 0.2 (0.2) | 3.5 (5.0) | 0.2 (0.2) | 5.9 (7.9) | 1.2 (0.9) |
| Correlation with Cord log2THg | Spearman coefficient | 0.30 | 0.16 | 0.46 | 0.22 | 0.31 | 0.31 | 0.18 |
GM: geometric mean; THg: total mercury;
Hg cord blood (μg/L) = 1.7*Hg maternal blood (μg/L); Hg in whole blood (μg/mL) = 250* maternal hair (μg/g).
2.2. Mercury measurements
Cord blood THg concentrations were defined as the exposure variable for each cohort (Table 1). In order to make THg concentrations from cohorts with measurements taken in maternal hair or maternal blood comparable to those performed in cord blood, maternal hair and blood THg levels were transformed according to conversion factors established by the U.S. Environmental Protection Agency in a comprehensive review of 21 studies cross-tissue studies (Stern and Smith, 2003). The cross-study central estimate for the ratio of cord blood to maternal blood identified was 1.7. Further, human hair-mercury concentrations (in μg/g) were observed to average about 250 × whole-blood mercury concentrations (in μg/mL) (ater Quality Cri, 2001). These ratios were used to convert to concentrations measured in maternal whole-blood or hair in the participant cohorts to cord blood levels, accordingly. Cord blood THg concentrations were log2-transformed to avoid skewed distributions. The analytical methods used for measurements of THg in each cohort are described in the Supplementary Methods file.
2.3. Profiling of DNA methylation
Cord blood and child blood DNAm in each respective cohort were assessed with the Infinium Human-Methylation450 array (Illumina, San Diego, CA USA) except in KOREAN exposome cohort which used the MethylationEPIC BeadChip array (Illumina, San Diego, CA USA). Raw methylation data was processed within each cohort following their preferred pipeline and normalization method. However, all pipelines considered sample and probe quality control checks. Technical batch effect correction was addressed as decided by each cohort. Methylation data is expressed as the beta value, that goes from 0 (unmethylated) to 1 (completely methylated). See Supplementary Methods file for extra details. DNAm extreme outliers (<25th percentile – 3*IQR or >75th percentile + 3*IQR across all the samples) were trimmed out before running the EWAS. Cell type composition in cord blood was estimated using the Bakulski reference panel (Bakulski et al., 2016), while child cell type proportions were calculated with the Salas reference panel (Gervin et al., 2019).
2.4. Epigenome-wide association study (EWAS)
Within each cohort, robust linear regression from the mass package (Venables and Ripley, 2002) was used to account for potential heteroscedasticity. The outcome variables were normalized DNAm beta values at each CpG (at birth or childhood) and the exposure variable was prenatal log2-transformed cord blood equivalent THg measures (log2Hg). Models were adjusted for maternal age (years), parity (number of deliveries), education (up to primary, secondary, university), smoking during pregnancy (yes/no), fish consumption (mean daily servings), child sex, as well as cellular heterogeneity, which reflects the differentiation of epigenetic status among cells. A second set of models was run in each cohort also adjusting for maternal fish consumption (servings/week) since it is the main Hg source from diet and its content in nutrients interacting with Hg. Effect size was reported as the difference in percentage units in DNAm for each log2Hg increased unit. In linear regression models with a log2 transformed exposure, a doubling of the exposure is associated with a difference in the DNAm determined by the regression coefficient. Positive regression coefficients indicate hypermethylation and negative regression coefficients indicate hypomethylation.
2.5. Fixed effects meta-analysis
Summarized results from each model in each cohort (regression coefficient for log2Hg), standard error (SE) and p-values for each CpG), were sent to the leading team to carry out parallel quality control and meta-analyses in two independent centres. Quality control in received results from cohorts involved several steps. First, the absence of single nucleotide polymorphisms (SNP) and cross hybridizing probes was confirmed by using the same proposed protocol (Chen et al., 2013). Then, probes were excluded if they lacked ‘cg’ or ‘ch’ naming, were not available on both the Infinium Human-Methylation450 and MethylationEPIC BeadChip arrays, were technical probes, were duplicated, were CpH probes, or were annotated to sex chromosomes. Also, outcomes with NAs or SE > 0.1 were excluded for the following analyses. CpGs associated with Hg (unadjusted p-value<10−5) were annotated by using the IlluminaHumanMethylation450kanno.ilmn12. hg19 package (Hansen, 2016) and only those associated with a gene were retained. CpGs not retained in at least 3 cohorts were also filtered out. Inflation in the distribution of observed p-values was assessed by quantile–quantile plot and lambda value (Supplemental Figure S1 for DNAm cord blood cohorts and Supplemental Figure S2 for DNAm child blood cohorts). Quality control parameters of the models run in each cohort prior to meta-analysis are shown in Supplemental Table S1.
Following quality control, all cohort-specific results of the cord blood (with and without adjustment for fish consumption) and child blood (with and without adjustment for fish consumption) analyses were included in four inverse variance-weighted fixed-effects meta-analyses using the multivariate genome-wide-association meta-analysis (GWAMA) tool (Mägi and Morris, 2010). Full meta-analyses were followed by a leave-one-out analyses to assess heterogeneity modifications (by means of I2 random-effects tests, considering low heterogeneity <25% (Higgins et al., 2003)), as well as differences in Asian cohorts, due to the ethnicity differences (Table 1). Significant changes in meta-analyses results were not observed. Given DNAm patterns across the genome are known to be correlated (Strimmer, 2008), we used the false discovery rate (FDR) procedure to account for multiple testing rather than the more stringent Bonferroni adjustment which assumes independent effects across all CpG sites. CpG sites with FDR <0.1 were considered differentially methylated. CpGs with unadjusted p-values<10−5 were also retained for enrichment analyses. Forest plots to assess the effect of each differentially methylated CpG were obtained by using the meta library. Shadow meta-analyses were conducted independently at the University of Bristol, to verify results.
2.6. Enrichment analyses
To identify plausible pathways associated with THg exposure, functional enrichment analysis was performed for genes where CpGs were observed to be associated with THg in the meta-analyses using the arbitrary cut-off of p < 10−5. Gene ontology tests were carried out using the gometh function of missMethyl package (Phipson et al., 2016), which maps CpGs to genes, take into account the differing number of probes per gene, performs a hypergeometric test and corrects for multiple-testing with FDR for each gene ontology category.
3. Results
3.1. Study population
Table 1 shows the characteristics of participants in the included cohorts. Five cohorts contributed to the cord blood EWAS representing a total of 1462 samples. This included 865 from children of European ancestry (ALSPAC, INMA, VIVA cohorts) and 597 from children of Asian ancestry (HOKKAIDO, KOREA cohorts). Four cohorts contributed to the child blood EWAS (794 samples, 739 of them with fish consumption), all of them of primarily European ancestry (ALSPAC, INMA, MoBa, RHEA cohorts). Only two cohorts (INMA and ALSPAC) contributed data to both the cord blood and child blood studies. Geometric mean cord blood Hg concentrations for the participating cohorts ranged from 2.01 to 9.30 μg/L, where the Japanese cohort showed the highest THg concentrations, followed by the Mediterranean cohorts (INMA and RHEA). Maternal fish consumption was highest in the MoBa cohort, followed by INMA, showing low correlation with cord blood Hg concentrations across cohorts (Dormann et al., 2013). Maternal age at delivery was on average between 29.4 and 32.8 years. The percentage of firstborn children also varied across cohorts, ranging from 37.1 to 60%. Maternal education was coded similarly across the cohorts, with a low percentage of highly educated mothers in ALSPAC and INMA (23.6–30.7%) and highest in VIVA and MoBa (71.9–79.9%). Maternal smoking during pregnancy varied across cohorts, with the most prevalent self-report in INMA (29.2%) and RHEA (21.2%), and the least prevalent in MoBa (4.4%).
3.2. EWAS meta-analysis
Up to 373,251 CpGs were evaluated in each meta-analysis modeling the associations between prenatal MeHg exposure and DNAm. Table 2 summarizes the statistics of the four performed meta-analyses (two for cord blood, with and without adjustment for maternal fish consumption, and two more for child blood, with and without adjustment for maternal fish consumption). Five American, Asian, and European studies contributed to the meta-analyses linking prenatal MeHg exposure to cord blood DNAm, including ALSPAC, SAPPORO-HOKKAIDO, INMA, KOREA, and VIVA cohorts, whereas four European cohorts (ALSPAC, INMA, MoBa, and RHEA) participated in the meta-analyses relating prenatal MeHg exposure to child blood DNAm at age 7–8. Generally, models showed low heterogeneity. The quantile–quantile plots did not reveal inflation in the distribution of observed p-values (lambdas ranged from 0.869 to 0.974) (Supplemental Figure S3).
Table 2.
Summarized statistics of multivariate genome-wide-association meta-analysis.
| Model | Lambda | Sample (N) | Cohorts (N) | Cohorts (names) | QCed CpGs | I2 index (mean) | CpGs BN ≤ 0.1 (N) | CpGs FDR≤0.1 (N) | CpGs Pmeta< 10−5 (N) | Annotated Genes for CpGs Pmeta< 10−5 (N) |
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Cord blood model adjusted for covariates, without maternal fish consumption | 0.894 | 1462 | 5 | ALSPAC, SAPPORO-HOKKAIDO, INMA, KOREA, VIVA | 373,251 | 0.1563 | 0 | 0 | 41 | 31 |
| Cord blood model adjusted for covariates, with maternal fish consumption | 0.869 | 1462 | 5 | ALSPAC, SAPPORO-HOKKAIDO, INMA, KOREA, VIVA | 373,250 | 0.1438 | 0 | 0 | 28 | 25 |
| Child blood model adjusted for covariates, without maternal fish consumption | 0.945 | 794 | 4 | ALSPAC, INMA, MOBA, RHEA | 373,183 | 0.1443 | 0 | 2 | 56 | 44 |
| Child blood model adjusted for covariates, with maternal fish consumption | 0.974 | 739 | 4 | ALSPAC, INMA, MOBA, RHEA | 373,183 | 0.1408 | 1 | 1 | 60 | 46 |
BN: Bonferroni test; FDR: false discovery rate test; I2 heterogeneity index; Pmeta: unadjusted p-value.
Results for differentially methylated sites (Pmeta<10−5) in each model are shown in Supplemental Tables S2 to S5. Among them, two differentially methylated CpGs (Table 3) found in child blood DNAm had p-values more extreme than the FDR adjusted p-value threshold (≤0.1): The first was cg12204245, located in G protein-coupled receptor kinase 1 (GRK1), where a doubling of THg was observed to be associated with 0.2% higher DNAm (β = 0.002, p-value = 4.81 × 10−7). A similar effect estimate was observed when adjusting for maternal fish consumption (β = 0.002, p-value = 1.31 × 10−7). The second was cg02212000, in the gamma-glutamyl hydrolase (GGH) gene, which was only observed to be differentially methylated in the model without fish consumption (β = −0.001, p-value = 8.13 × 10−7). Two more CpGs, cg24184221 and cg15288800 located in Mediator Complex Subunit 31 (MED31), showed nominal association between prenatal MeHg concentrations and DNA methylation in both cord blood (β = 2.28 × 10–4, p-value = 5.87 × 10−5 adjusted for fish) and child blood (β = 0.004, p-value = 4.97 × 10−5 adjusted for fish). Figs. 1–3 show the direction and weights of the effect by cohort of each CpG on DNAm: 1) hypermethylation of cg12204245 (GRK1), as well as cg24184221 and cg15288800 (MED31); and 2) hypomethylation of cg02212000 (GGH). The observed direction of effect for each cohort was consistent for all differentially methylated CpGs.
Table 3.
Prenatal MeHg exposure and DNA methylation. Meta-analysis of epigenome-wide association study including highlighted CpG sites with a Pmeta<10−5.
| Model | CpG site | Beta | SE | p-value | I2 | Studies | Sample | Effects | FDRa | Chr. | Position | UCSC RefGene Name | Location |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||
| Child blood DNAm - Without fish | cg02212000 | −0.0008 | 0.0002 | 8.13E-07 | 0 | 4 | 794 | - | 0.1011 | chr8 | 63951669 | GGH | TSS200 |
| Child blood DNAm - With fish | g02212000 | −0.0008 | 0.0002 | 3.65E-06 | 0 | 4 | 739 | - | 0.4540 | chr8 | 63951669 | GGH | TSS200 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Cord blood DNAm - With fish | cg24184221 | 0.0002 | 0.0001 | 5.87E-05 | 0 | 5 | 1462 | +-+++ | 1.0000 | chr17 | 6,555,443 | MED31; C17orf100 | Exon |
| Child blood DNAm - With fish | cg15288800 | 0.0036 | 0.0009 | 4.97E-05 | 0.3014 | 4 | 739 | ++++ | 0.5133 | chr17 | 6,555,742 | MED31; C17orf100 | 3’ UTR |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Child blood DNAm - Without fish | cg12204245 | 0.0020 | 0.0004 | 4.81E-07 | 0.6585 | 4 | 794 | ++++ | 0.0898 | chr13 | 114,321,214 | GRK1 | TSS1500 |
| Child blood DNAm - With fish | cg12204245 | 0.0022 | 0.0004 | 1.31E-07 | 0.4678 | 4 | 739 | ++++ | 0.0489 | chr13 | 114,321,214 | GRK1 | TSS1500 |
Shown are the 4 highlighted CpGs ordered by p-value. Results presented per 1 μg/L increase in prenatal MeHg exposure (cord blood total Hg concentration or child total Hg concentration). Column heads: Beta: regression coefficient; SE: standard error for regression coefficient; p-value: unadjusted p-value for the regression coefficient; I2: heterogeneity statistics; Studies: number of studies involved; Sample: sample size in the model; Effects: direction of effect across cohorts included in the statistical model (ALSPAC, INMA, MoBa, and RHEA): Hg exposure during pregnancy was associated with increased (+) or decreased (−) methylation, or missing (?) result; FDR: false discovery rate; Chr: chromosome; Position: chromosomal position based on NCBI human reference genome; UCSC RefGene Name: UCSC annotated gene.
All models were adjusted for maternal age, parity, education, smoking during pregnancy, and child sex, and cellular heterogeneity.
Mapped from the array to the hg19 genome provided by Illumina. Location of differentially methylated sites has been added according to the IlluminaHumanMethylation450kanno.ilmn12.hg19 R package.
TSS: Transcription Start Site; UTR: untranslated region.
Epigenome-wide significance threshold (FDR p ≤ 0.1).
Fig. 1.
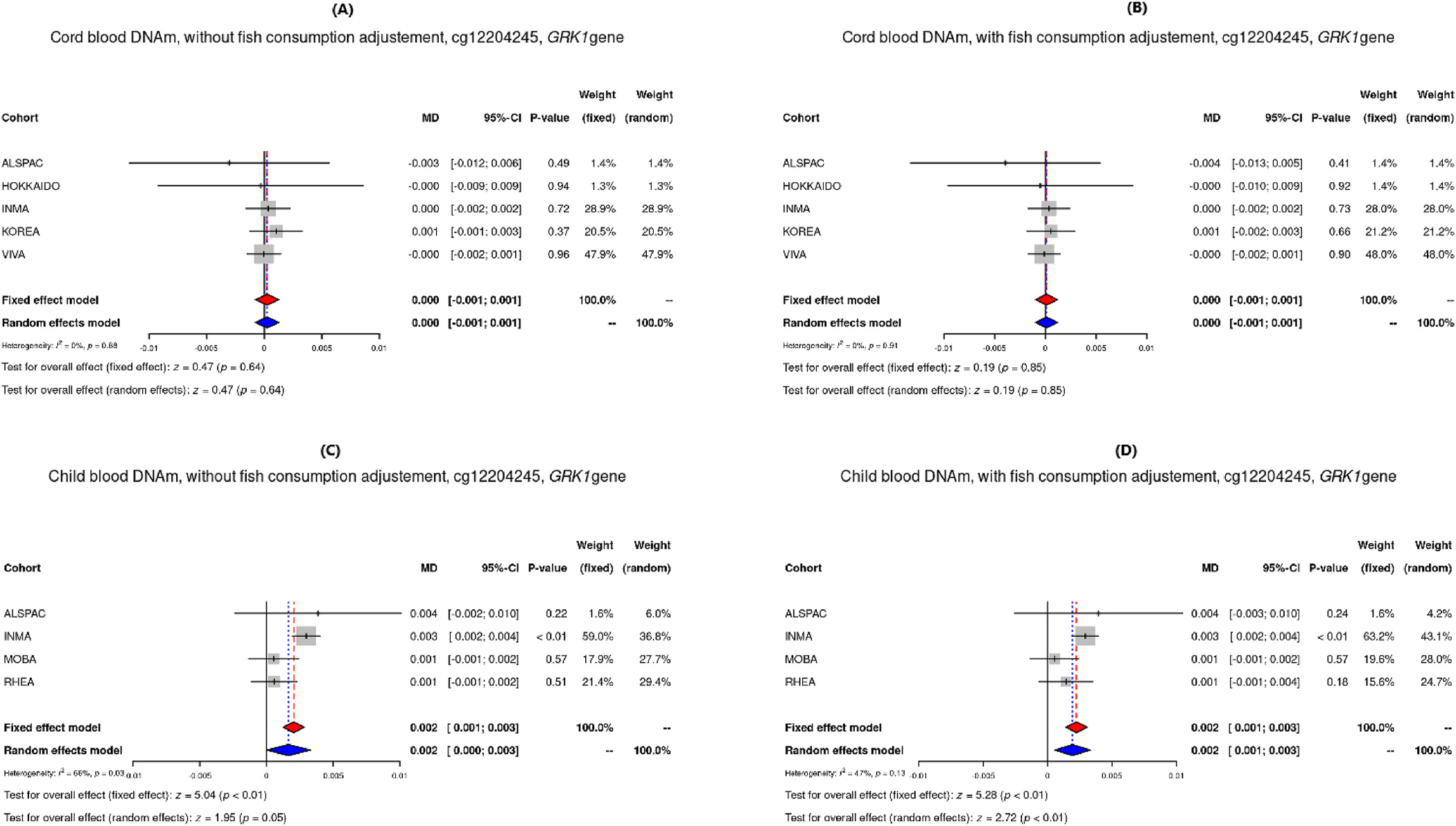
Forest plots of DNAm of the CpG site cg12204245 in GRK1 from models adjusted for maternal age, parity, education, smoking, child sex and cellular heterogeneity, for cord blood without and with adjustment for fish consumption (A and B), and child blood without and with adjustment for fish consumption (C and D).
Fig. 3.
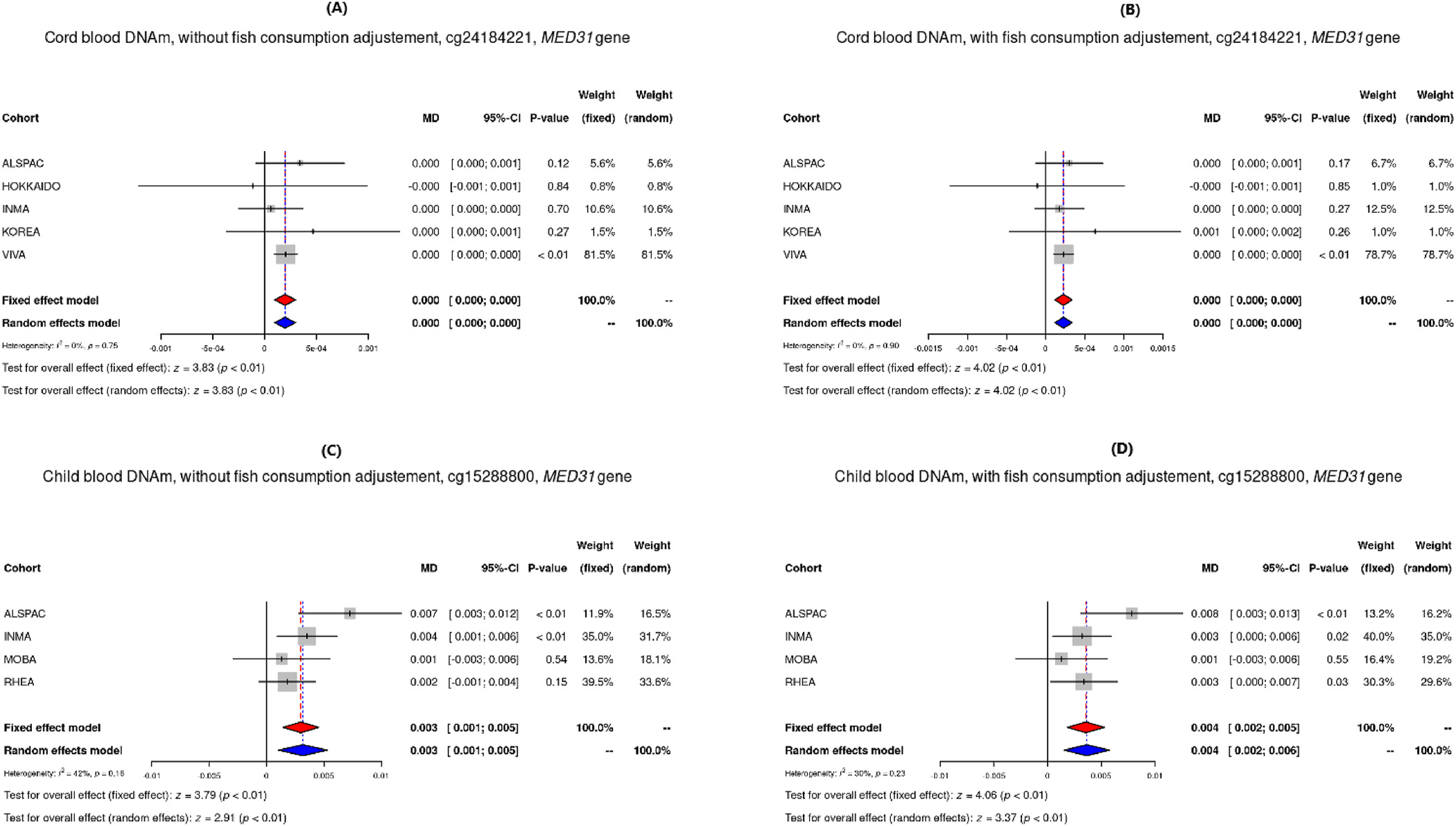
Forest plots of DNAm of the CpG sites in MED31 from models adjusted for maternal age, parity, education, smoking, child sex and cellular heterogeneity, for cord blood without and with adjustment for fish consumption for cg24184221 site (A and B), and child blood without and with adjustment for fish consumption for cg15288800 site (C and D). these genes is linked to MeHg toxicity warrants further functional studies.
The results identified at cg02212000, cg24184221, and cg15288800 were located in genes previously linked to inorganic and organic Hg compounds (GGH, MED31, and MED31 respectively) (Supplemental Table S6b). In addition, other CpGs differentially methylated at the Pmeta<10−5 level (see Supplemental Tables S2 to S5) were identified both in models with and without fish consumption adjustment and their associated genes were also related to Hg compounds in previous studies (Supplemental Tables S6a and b).
The site cg10555307, annotated within catenin delta 2 (CTNND2), showed hypomethylation in ALSPAC and INMA, and hypermethylation in SAPPORO-HOKKAIDO, KOREA and VIVA cohorts in the cord blood methylation model without fish consumption adjustment, but hypermethylation in all cohorts in the adjusted model for fish consumption. Also, cg17452301 in PWWP domain containing 2B (PWWP2B), and cg19374305 in dynein axonemal heavy chain 7 (DNAH7), showed hypermethylation in child blood methylation models in all cohorts (ALSPAC, INMA, MoBa, and RHEA). In these cases, associations did not persist between cord and child blood models.
Functional enrichment analyses among the top CpG sites (Pmeta<10−5) did not show robust results because of the lower insufficient probes and genes by identified pathway after correction for multiple testing (FDR ≈ 1) in any model (Supplemental Table S7).
4. Discussion
This study represents so far the largest-scale epigenome-wide meta-analysis evaluating the association between prenatal MeHg exposure and DNAm in new-borns and children aged 7–8 years old. The combined results showed evidence for associations of prenatal MeHg exposure during pregnancy with differential methylation in MED31 at both time points during child development, suggesting that methylation levels in MED31 may be perturbed in response to in utero MeHg exposure in a manner that persists into early childhood. MED31 is widely expressed in the human body (www.proteinatlas.org (Uhlén et al., 2015)), and is involved in lipid metabolism and RNA Polymerase II transcription function (Beadle et al., 2018). We also identified a link between prenatal MeHg exposure and child blood DNA methylation of GRK1, encoding the protein rhodopsin kinase GRK1, mainly expressed in the retina and involved in retina function (Fan et al., 2010); and GGH, widely expressed in the body and related to glutathione metabolism and innate immune system pathways (Gibson et al., 2011). How methylation of
Differential expression of MED31 and GGH have been previously related to exposure to inorganic and organic Hg compounds (Comparative Toxicogenomics Database: www.ctdbase.org (Grondin et al., 2021)). In zebrafish, increasing mercury chloride concentrations were associated with a decrease in MED31 expression in the liver (Ung et al., 2010). In rats, 4-hydroxymercuribenzoate induced decreased GGH activity, resulting in an increased hydrolysis of folic acid (Shafizadeh and Halsted, 2007). Additionally, methylmercuric chloride was related to decreased expression of GGH in human embryonic stem cells (Waldmann et al., 2017). These studies showed increased gene expression in response to exposure which could be consistent with the hypomethylation we observed in CpG within these genes (see Figs. 2 and 3). The differentially methylated CpG site cg12204245 in GRK1 perhaps represents a novel target gene in the context of MeHg exposure. However, GRK6, a paralog of the GRK1, has been related in vitro to methylmercuric chloride human exposure resulting in dose-dependent increased expression of GRK6 (Waldmann et al., 2017). We identified other previously Hg-associated genes with persistent differential methylation, such as CTNND2 and PWWP2B, where expression has been shown to be decreased in association with mercuric bromide, p-chloromercuribenzoic acid, phenylmercuric acetate and methylmercuric chloride in human cells studies (Waldmann et al., 2017; Rempel et al., 2015; Shinde et al., 2017). Both genes showed general consistent hypermethylation in the affected CpGs sites in our models, which seems to be consistent with these previous studies. Conversely, DNAH7, which also showed hypermethylation in all our cohorts, has been previously related to an increased expression due to MeHg exposure in fish (Klaper et al., 2006).
Fig. 2.
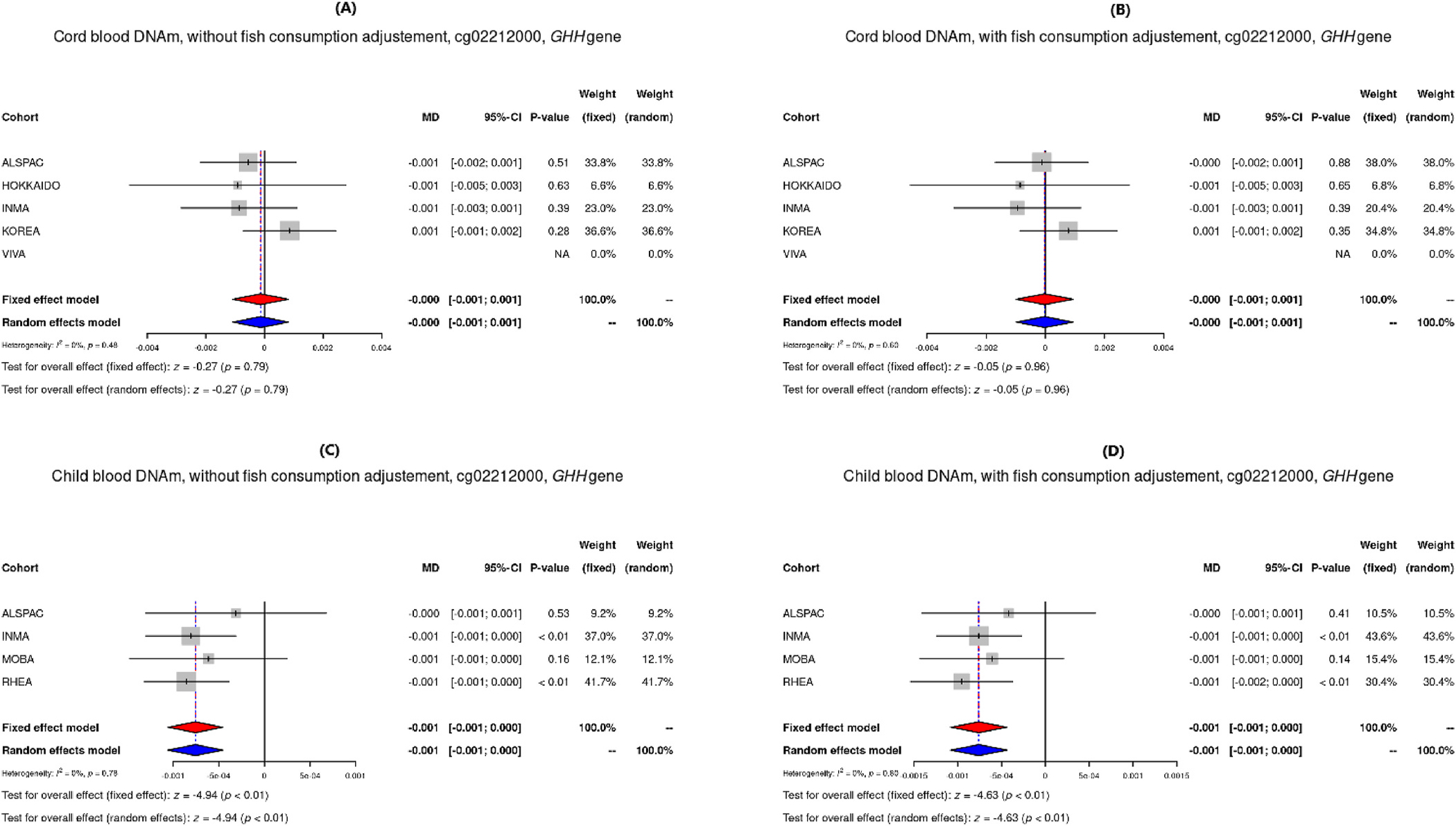
Forest plots of DNAm of the CpG site cg02212000 in GGH from models adjusted for maternal age, parity, education, smoking, child sex and cellular heterogeneity, for cord blood without and with adjustment for fish consumption (A and B), and child blood without and with adjustment for fish consumption (C and D).
Some studies have characterized DNAm changes in cord and child blood relative to prenatal MeHg exposure. These studies observed a differentiated DNAm in cord (Cardenas et al., 2017a, 2017b; Bakulski et al., 2015) as well as child blood (Cardenas et al., 2017a, 2017b). The period of foetal development has been shown to be specially sensitive to toxicants exposure, likely because of the dramatic DNAm changes and epigenomic remodelling that takes place early during embryogenesis, giving rise to differentiated cells and tissues with specific DNAm patterns (Perera and Herbstman, 2011a). The ability of MeHg to cross the placenta during development makes it a candidate toxicant for the disruption of foetal programming events that could propagate through different germ layers during embryogenesis and contribute to postnatal health consequences. The specific mechanisms of these effects remain unclear, but it has been proposed that DNAm may act as a response mechanism to MeHg exposure or a long-term mediator of MeHg-associated effects (Cediel Ulloa et al., 2021). However, studies evaluating the persistence of these changes into childhood are scarce (Vaiserman, 2015). Assessing the persistence of these epigenetic modifications is critical as DNAm is a dynamic process that can drift with age (Acevedo et al., 2015). Work by Cardenas et al. in the US-based Project Viva study previously evaluated the persistence of these changes in 321 cord blood samples followed-up in blood samples taken at 10 years of age, finding that higher DNAm levels of the PON1 region were associated with lower cognitive test scores in early childhood for both sexes (Cardenas et al., 2017b).
A challenge for our observational analysis was to evaluate the potentially confounding influence of fish consumption on our effect of interest, the association between MeHg exposure and altered patterns of DNAm (Perera and Herbstman, 2011b). Fish consumption showed low correlation (Dormann et al., 2013) with cord blood THg concentrations across cohorts in the present study, but it can be a source of other pollutants apart from Hg, such as polychlorinated biphenyls (PCBs), for which synergic effects of both compounds have been suggested (Ballester et al., 2018b; Boucher et al., 2010). Additionally, fish is a source of nutrients, including selenium or poly-unsaturated fatty acids (PUFA), which have been purported to be protective against the toxic effects of MeHg (Ralston and Raymond, 2018; Ginsberg et al., 2015). Comparing multivariate models adjusted and not adjusted for fish consumption allows identification of genes independently affected by MeHg exposure, such as GRK1 and GGH observed in this study. Other studies have included the variable fish consumption in the multivariate models of DNAm MeHg-association analysis. Cardenas et al. (2017b) included adjustments for mean weekly fish intake during pregnancy in its single-cohort study assessing the persistent DNAm associated with prenatal MeHg exposure, but did not consider the contrast between models with and without adjustment for fish consumption.
The main strength of this study is the large sample size achieved by analysing data from all cohorts and combining the results in a meta-analysis allowing to identify only robust results. By using identical analysis protocols and harmonized script across all cohorts we have reduced bias due to heterogeneity. Shadow replication of meta-analysis results has minimized the possibility of coding errors. Also, the inclusion of different THg concentrations and fish consumptions make results more generalizable to other populations. To avoid the violation of the homogeneous effect assumption across studies only the largest ethnic group in each cohort was included. However, including different cohorts with different main ethnicities allowed to check the homogeneity of the epigenetic MeHg effects.
Our study also has some limitations. The most important is that only INMA and ALSPAC cohorts participated with prospective cord-child blood DNAm data. Also, although the sample size was large, some cohorts could not participate in the meta-analyses, such as KANC cohort from HELIX study because it had no fish consumption data. Moreover, most cord blood THg concentrations from different cohorts were obtained by means of transformation factors from maternal hair or blood according to literature. On the other hand, loci and genes identified in cord and child DNAm were, although with limited power, associated with pathways that are critical to foetal growth and development: vascular permeability to maintain physiological tissue homeostasis, astrocyte activation and microtubule nucleation that play a crucial role in orchestrating neural development by coordinating synapse formation and function (Sloan and Barres, 2014; Sánchez-Huertas et al., 2016), as well as titin binding, involved in muscular development (Letourneau and Wright, 2018). Therefore, further experimental research is needed for understanding the functional effects of MeHg-related epigenetic effects of the identified genes in the present study.
5. Conclusions
We have conducted the largest epigenome-wide meta-analysis to date evaluating the association between prenatal Hg exposure and DNAm in new-borns and 7-year-old children. This study adds some evidence that MED31 could serve as a potential target of MeHg exposure, providing a potential epigenetic signature of in utero exposure which persists into early childhood, independently of fish consumption and other possible confounders. DNAm modifications leading to functional genomic changes could help explain heterogeneous findings observed for prenatal mercury exposure and health outcomes such as cognitive or anthropometric development in different populations. Our study also contributes to further understanding of potential underlying mechanisms of the negative health effects of MeHg exposure by highlighting the implications of DNAm in several genes involved in growth and cell cycle processes during foetal development.
Supplementary Material
Acknowledgments
ALSPAC.
ALSPAC study is extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. We would like to acknowledge Oliver Lyttleton, Hashem Shihab, Nabila Kazmi and Geoff Woodward for their earlier contribution to the generation of ARIES data (ALSPAC methylation data).
INMA
The authors would like to thank all the participants for their generous collaboration. full roster of the INMA project investigators can be found at http://www.proyectoinma.org/presentacion-inma/listado-investigadores/en_listado-investigadores.html.
MoBa
The authors would also like to thank Ingvild Essen for thorough field work, Heidi Marie Nordheim for biological sample management and the MoBa administrative unit (MoBa). The MoBa Cohort Study is supported by the Norwegian Ministry of Health and Care Services and the Ministry of Education and Research. We are grateful to all the participating families in Norway who take part in this on-going cohort study.
Project Viva
Project Viva researchers would like to thank all mothers, children and families for their ongoing participation.
RHEA
The authors would like to thank Georgia Chalkiadaki and Danai Feida for biological sample management, to Eirini Michalaki, Mariza Kampouri, Anny Kyriklaki and Minas Iakovidis for field study performance and to Maria Fasoulaki for administrative assistance.
SAPPORO-HOKKAIDO
We would like to express our appreciation to all of the study participants of the Hokkaido Study on Environment and Children’ Health. We also express our profound gratitude to all personnel in the hospitals and clinics that collaborated with the study, including Sapporo Toho Hospital, Keiai Hospital, Endo Kikyo Maternity Clinic, Shiroishi Hospital, Memuro Municipal Hospital, Aoba Ladies Clinic, Obihiro-Kyokai Hospital, Akiyama Memorial Hospital, Sapporo Medical University Hospital, Hokkaido University Hospital, Kitami Red Cross Hospital, Hoyukai Sapporo Hospital, Gorinbashi Hospital, Hashimoto Clinic, Asahikawa Medical College Hospital, Hakodate Central General Hospital, Ohji General Hospital, Nakashibetsu Municipal Hospital, Sapporo Tokushukai Hospital, Asahikawa Red Cross Hospital, Wakkanai City Hospital, Kushiro Rosai Hospital, Sapporo-Kosei General Hospital, Shibetsu City General Hospital, Nikko Memorial Hospital, Sapporo City General Hospital, Kohnan Hospital, Hakodate City Hospital, Hokkaido Monbetsu Hospital, Tenshi Hospital, Hakodate Goryoukaku Hospital, Nakamura Hospital, Kin-ikyo Sapporo Hospital, Kitami Lady’s Clinic, Engaru-Kosei General Hospital, Kushiro Red Cross Hospital, Nayoro City General Hospital, and Obihiro-Kosei General Hospital. We also deeply express our gratitude to the staff of The Hokkaido Study on Environment and Children’s Health for their considerable efforts to support our study, including N. Goto, O. Hayashi, T. Hikichi, N. Kanda, K. Kunita, K. Miura, Y. Shibasaki, K. Sugawara, K. Tanaka, E. Toyama, and T. Yoshikawa. Special thanks to Ms. Mimi Takahashi on her effort to support writing this manuscript.
Funding
ALSPAC
ALSPAC study was funded by the UK Medical Research Council and the Wellcome Trust (grant ref: 102215/2/13/2) and the University of Bristol provide core support for ALSPAC. This publication is the work of the authors, PY and CR, who will serve as guarantors for the contents of this paper. A comprehensive list of grants funding is available on the ALSPAC website (http://www.bristol.ac.uk/alspac/external/documents/grant-acknowledgements.pdf). The Accessible Resource for Integrated Epigenomics Studies (ARIES), was funded by the UK Biotechnology and Biological Sciences Research Council (BB/I025751/1 and BB/I025263/1). Additional epigenetic profiling of ALSPAC was supported by the UK Medical Research Council (MRC) Integrative Epidemiology Unit and the University of Bristol (MC_UU_12013_1, MC_UU_12013_2, MC_UU_12013_5 and MC_UU_12013_8), the Wellcome Trust (WT088806) and the United States National Institute of Diabetes and Digestive and Kidney Diseases (R01 DK10324). PY and CR are supported by the UK MRC Integrative Epidemiology Unit (MC_UU_00011/5).
INMA
Main funding of the epigenetic studies in INMA were grants from Instituto de Salud Carlos III (Red INMA G03/176, CB06/02/0041), Spanish Ministry of Health (FIS-PI04/1436, FIS-PI08/1151 including FEDER funds, FIS-PI11/00610, FIS-FEDER-PI06/0867, FIS-FEDER-PI03–1615, FIS-FEDER PI16/1288, FIS-FEDER PI19/1338; Miguel Servet FEDER 15/0025 and 20/0006; FIS-FSE: 17/00260), Generalitat de Catalunya-CIRIT 1999SGR 00241, Generalitat Valenciana BEST/2020/059, Fundacio ´La Marato ´de TV3 (090,430), EU Commission (261357-MeDALL: Mechanisms of the Development of ALLergy), and European Research Council (268479-BREATHE: Brain dEvelopment and Air polluTion ultrafine particles in school childrEn).
Korean exposome study
This study was supported by the Korean Environment Industry & Technology Institute (KEITI) through “the Environmental Health Action Program”, funded by Korea Ministry of Environment (2017001360005).
MoBa
The Norwegian Mother, Father and Child Cohort Study is supported by the Norwegian Ministry of Health and Care Services and the Ministry of Education and Research, NIH/NIEHS (contract no N01-ES-75558), NIH/NINDS (grant no.1 UO1 NS 047537–01 and grant no.2 UO1 NS 047537–06A1). MoBa1 and MoBa 2 are supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (Z01-ES-49019) and Norwegian Research Council/BIOBANK (grant no 221097). The work in MoBa3 was supported in part by a Postdoctoral Fellowship grant from the Ulleval Hospitals Research Council (now under Oslo University Hospital) and travel grants from the Unger-Vetlesens foundation and the Norwegian American Womens Club, all to M.C.M.K. MoBa3 epigenomics data analyses were funded by INCA/Plan Cancer-EVA-INSERM, France, and the International Childhood Cancer Cohort Consortium (I4C), and performed by the Epigenetics Group at the International Agency for Research on Cancer (IARC, Lyon, France), A.G. was supported by the grant from INCA/Plan Cancer-EVA-INSERM (France, 2015) to Z.H. and also by the IARC Postdoctoral Fellowship, partially supported by the EC FP7 Marie Curie Actions-People-Co-funding of regional, national and international programmes (COFUND).
Project Viva
The Project Viva cohort is funded by NIH grants R01HL111108, R01NR013945, and R37 HD034568.
RHEA
The Rhea project was financially supported by European projects, and the Greek Ministry of Health (Program of Prevention of Obesity and Neurodevelopmental Disorders in Preschool Children, in Heraklion district, Crete, Greece: 2011–2014; ‘Rhea Plus’: Primary Prevention Program of Environmental Risk Factors for Reproductive Health, and Child Health: 2012–2015). The work was also supported by MICINN (MTM2015–68140-R), Centro Nacional de Genotipado-CEGEN-PRB2-ISCIII, the H2020-EU.3.1.2. - Preventing Disease Programme (grant agreement no 874583) (ATHLETE project), and from the European Union’s Horizon 2020 research and innovation programme (grant Agreement number: 733206) (Early Life stressors and Lifecycle Health (LIFECYCLE)).
SAPPORO-HOKKAIDO
The Hokkaido-Sapporo Study is supported by Grant-in-Aid for Scientific Research from the Japanese Ministry of Health, Labour and Welfare (JPMH14427175, JPMH19189425, JPMH17932352); the Japan Society for the Promotion of Science; the Ministry of Education, Culture, Sports, Science and Technology (JP16H02645, JP19H01071, JP18H03035, JP15H04780, JP19K10576, JP19K10636, JP18K10042, JP18K10022, JP19K22730, JP19K20457); and an Environment Research and Technology Development fund (JPMEERF20145054, JPMEERF20175053, JPMEERF20205002); AMED (JPMH19188595, JPMH18950314); and Ministry of Internal Affairs and Communications (JPMI10001).
Footnotes
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envres.2021.112093.
References
- Acevedo N, Reinius LE, Vitezic M, Fortino V, Söderhäll C, Honkanen H, et al. , 2015. Age-associated DNA methylation changes in immune genes, histone modifiers and chromatin remodeling factors within 5 years after birth in human blood leukocytes. Clin. Epigenet. 7, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Saleh I, Shinwari N, Mashhour A, Rabah A, 2014. Mar 1. Birth outcome measures and maternal exposure to heavy metals (lead, cadmium and mercury) in Saudi Arabian population. Int. J. Hyg Environ. Health 217 (2), 205–218. [DOI] [PubMed] [Google Scholar]
- Aschner M, Syversen T, Souza DO, Rocha JBT, Farina M, 2007. Involvement of glutamate and reactive oxygen species in methylmercury neurotoxicity. Braz. J. Med. Biol. Res. 40 (3), 285–291. [DOI] [PubMed] [Google Scholar]
- US EPA, 2001. Water Quality Criteria: Notice of Availability of Water Quality Criterion for the Protection of Human Health: Methylmercury [Internet]. Federal Register [cited 2020 Nov 28]. Available from: https://www.federalregister.gov/documents/2001/01/08/01-217/water-quality-criteria-notice-of-availability-of-water-quality-criterion-for-the-protection-of-human.
- Bakulski KM, Lee HJ, Feinberg JI, Wells EM, Brown S, Herbstman JB, et al. , 2015. Prenatal mercury concentration is associated with changes in DNA methylation at TCEANC2 in newborns. Int. J. Epidemiol. 44 (4), 1249–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakulski KM, Feinberg JI, Andrews SV, Yang J, Brown S, McKenney S L, et al. , 2016. DNA methylation of cord blood cell types: applications for mixed cell birth studies. Epigenetics 11 (5), 354–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballester F, Iñiguez C, Murcia M, Guxens M, Basterretxea M, Rebagliato M, et al. , 2018a. Prenatal exposure to mercury and longitudinally assessed fetal growth: relation and effect modifiers. Environ. Res. 160, 97–106. [DOI] [PubMed] [Google Scholar]
- Ballester F, Iñiguez C, Murcia M, Guxens M, Basterretxea M, Rebagliato M, et al. , 2018b. Prenatal exposure to mercury and longitudinally assessed fetal growth: relation and effect modifiers. Environ. Res. 160, 97–106. [DOI] [PubMed] [Google Scholar]
- Beadle EP, Straub JA, Bunnell BA, Newman JJ, 2018. Oct 1. MED31 involved in regulating self-renewal and adipogenesis of human mesenchymal stem cells. Mol. Biol. Rep. 45 (5), 1545–1550. [DOI] [PubMed] [Google Scholar]
- Berglund M, Lind B, Björnberg KA, Palm B, Einarsson O, Vahter M, 2005. Oct 3. Inter-individual variations of human mercury exposure biomarkers: a cross-sectional assessment. Environ Health Glob Access Sci Source 4, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher O, Bastien CH, Saint-Amour D, Dewailly E, Ayotte P, Jacobson JL, et al. , 2010. Aug. Prenatal exposure to methylmercury and PCBs affects distinct stages of information processing: an event-related potential study with Inuit children. Neurotoxicology 31 (4), 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas A, Koestler DC, Houseman EA, Jackson BP, Kile ML, Karagas MR, et al. , 2015. Differential DNA methylation in umbilical cord blood of infants exposed to mercury and arsenic in utero. Epigenetics 10 (6), 508–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas A, Rifas-Shiman SL, Godderis L, Duca R-C, Navas-Acien A, Litonjua AA, et al. , 2017a. Prenatal exposure to mercury: associations with global DNA methylation and hydroxymethylation in cord blood and in childhood. Environ. Health Perspect. 125 (8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas A, Rifas-Shiman SL, Agha G, Hivert M-F, Litonjua AA, DeMeo DL, et al. , 2017b. Persistent DNA methylation changes associated with prenatal mercury exposure and cognitive performance during childhood. Sci. Rep. 7 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cediel Ulloa A, Gliga A, Love TM, Pineda D, Mruzek DW, Watson GE, et al. , 2021. Feb 1. Prenatal methylmercury exposure and DNA methylation in seven-year-old children in the Seychelles Child Development Study. Environ. Int. 147, 106321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzi L, Leventakou V, Vafeiadi M, Koutra K, Roumeliotaki T, Chalkiadaki G, et al. , 2017. Oct 1. Cohort profile: the mother-child cohort in Crete, Greece (Rhea study). Int. J. Epidemiol. 46 (5), 1392–1393k. [DOI] [PubMed] [Google Scholar]
- Chen Y-A, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, et al. , 2013. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 8 (2), 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson TW, Magos L, 2006. Sep. The toxicology of mercury and its chemical compounds. Crit. Rev. Toxicol. 36 (8), 609–662. [DOI] [PubMed] [Google Scholar]
- Crespo-López ME, Macêdo GL, Pereira SID, Arrifano GPF, Picanço-Diniz DLW, Nascimento JLM.d., et al. , 2009. Mercury and human genotoxicity: critical considerations and possible molecular mechanisms. Pharmacol. Res. 60 (4), 212–220. [DOI] [PubMed] [Google Scholar]
- Dallaire R, Dewailly É, Ayotte P, Forget-Dubois N, Jacobson SW, Jacobson JL, et al. , 2013. Exposure to organochlorines and mercury through fish and marine mammal consumption: associations with growth and duration of gestation among Inuit newborns. Environ. Int. 54, 85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann CF, Elith J, Bacher S, Buchmann C, Carl G, Carré G, et al. , 2013. Collinearity: a review of methods to deal with it and a simulation study evaluating their performance. Ecography 36 (1), 27–46. [Google Scholar]
- Dreiem A, Gertz CC, Seegal RF, 2005. The effects of methylmercury on mitochondrial function and reactive oxygen species formation in rat striatal synaptosomes are age-dependent. Toxicol. Sci. 87 (1), 156–162. [DOI] [PubMed] [Google Scholar]
- Drouillet-Pinard P, Huel G, Slama R, Forhan A, Sahuquillo J, Goua V, et al. , 2010. Oct. Prenatal mercury contamination: relationship with maternal seafood consumption during pregnancy and fetal growth in the ‘EDEN mother-child’ cohort. Br. J. Nutr. 104 (8), 1096–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Sakurai K, Chen C-K, Rohrer B, Wu BX, Yau K-W, et al. , 2010. Feb 17. Deletion of GRK1 causes retina degeneration through a transducin-independent mechanism. J. Neurosci. 30 (7), 2496–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina M, Aschner M, Rocha JBT, 2011. Oxidative stress in MeHg-induced neurotoxicity. Toxicol. Appl. Pharmacol. 256 (3), 405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzan SF, Howe CG, Chen Y, Gilbert-Diamond D, Korrick S, Jackson BP, et al. , 2021. Prenatal and postnatal mercury exposure and blood pressure in childhood. Environ. Int. 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix JF, Joubert BR, Baccarelli AA, Sharp GC, Almqvist C, Annesi-Maesano I, et al. , 2018. Cohort profile: pregnancy and childhood epigenetics (PACE) consortium. Int. J. Epidemiol. 47 (1), 22–23u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Viñas G, Ballester F, Llop S, 2019. Chronic mercury exposure and blood pressure in children and adolescents: a systematic review. Environ. Sci. Pollut. Res. 26 (3), 2238–2252. [DOI] [PubMed] [Google Scholar]
- Gervin K, Salas LA, Bakulski KM, Van Zelm MC, Koestler DC, Wiencke JK, et al. , 2019. Systematic evaluation and validation of reference and library selection methods for deconvolution of cord blood DNA methylation data. Clin. Epigenet. 11 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson TM, Brennan P, Han S, Karami S, Zaridze D, Janout V, et al. , 2011. Comprehensive evaluation of one-carbon metabolism pathway gene variants and renal cell cancer risk. PloS One 6 (10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg GL, Toal BF, McCann PJ, 2015. Oct 3. Updated risk/benefit analysis of fish consumption effects on neurodevelopment: implications for setting advisories. Hum. Ecol. Risk Assess. 21 (7), 1810–1839. [Google Scholar]
- Golding G, Pembrey P, Jones J, The AST, 2001. ALSPAC - the Avon longitudinal study of Parents and children I. Study methodology. Paediatr. Perinat. Epidemiol. 15 (1), 74–87. [DOI] [PubMed] [Google Scholar]
- Grandjean P, Weihe P, Debes F, Choi AL, Budtz-Jørgensen E, 2014. Neurotoxicity from prenatal and postnatal exposure to methylmercury. Neurotoxicol. Teratol. 43, 39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondin CJ, Davis AP, Wiegers JA, Wiegers TC, Sciaky D, Johnson RJ, et al. , 2021. Predicting molecular mechanisms, pathways, and health outcomes induced by Juul e-cigarette aerosol chemicals using the Comparative Toxicogenomics Database. Curr. Res. Toxicol. 2, 272–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin K, Barman M, Stråvik M, Levi M, Englund-Ögge L, Murray F, et al. , 2020. Low-level maternal exposure to cadmium, lead, and mercury and birth outcomes in a Swedish prospective birth-cohort. Environ. Pollut. 265. [DOI] [PubMed] [Google Scholar]
- Guxens M, Ballester F, Espada M, Fernández MF, Grimalt JO, Ibarluzea J, et al. , 2012. Aug. Cohort profile: the INMA—INfancia y medio ambiente—(environment and childhood) project. Int. J. Epidemiol. 41 (4), 930–940. [DOI] [PubMed] [Google Scholar]
- Ha E, Basu N, Bose-O’Reilly S, Dórea JG, McSorley E, Sakamoto M, et al. , 2017. Jan. Current progress on understanding the impact of mercury on human health. Environ. Res. 152, 419–433. [DOI] [PubMed] [Google Scholar]
- Hansen K, 2016. IlluminaHumanMethylation450kanno.ilmn12.hg19: annotation for Illumina’s 450k methylation arrays. R package version 0.6.0. [Google Scholar]
- Higgins JPT, Thompson SG, Deeks JJ, Altman DG, 2003. Sep 4. Measuring inconsistency in meta-analyses. BMJ 327 (7414), 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintelmann H, 2010. Organomercurials. Their formation and pathways in the environment. Met. Ions Life Sci. 7, 365–401. [DOI] [PubMed] [Google Scholar]
- Hong J, Lunetta KL, Cupples LA, Dupuis J, Liu C-T, 2016. Evaluation of a two-stage approach in trans-ethnic meta-analysis in genome-wide association studies. Genet. Epidemiol. 40 (4), 284–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagas MR, Choi AL, Oken E, Horvat M, Schoeny R, Kamai E, et al. , 2012. Evidence on the human health effects of low-level methylmercury exposure. Environ. Health Perspect. 120 (6), 799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan F, Momtaz S, Abdollahi M, 2019. Mar 1. The relationship between mercury exposure and epigenetic alterations regarding human health, risk assessment and diagnostic strategies. J. Trace Elem. Med. Biol. 52, 37–47. [DOI] [PubMed] [Google Scholar]
- Kim B-M, Lee B-E, Hong Y-C, Park H, Ha M, Kim Y-J, et al. , 2011. Dec 1. Mercury levels in maternal and cord blood and attained weight through the 24months of life. Sci. Total Environ. 410–411, 26–33. [DOI] [PubMed] [Google Scholar]
- Kishi R, Sasaki S, Yoshioka E, Yuasa M, Sata F, Saijo Y, et al. , 2011. Cohort profile: the hokkaido study on environment and Children’s Health in Japan. Int. J. Epidemiol. 40 (3), 611–618. [DOI] [PubMed] [Google Scholar]
- Klaper R, Rees CB, Drevnick P, Weber D, Sandheinrich M, Carvan MJ, 2006. Sep. Gene expression changes related to endocrine function and decline in reproduction in fathead minnow (Pimephales promelas) after dietary methylmercury exposure. Environ. Health Perspect. 114 (9), 1337–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letourneau AG, Wright NT, 2018. Structural insights on the obscurin-binding domains in Titin. Protein Pept. Lett. 25 (11), 973–979. [DOI] [PubMed] [Google Scholar]
- Li X, Xu Y, Kou Q, 2014. Molecular phylogeny of Parapenaeopsis Alcock, 1901 (Decapoda: penaeidae) based on Chinese materials and 16S rDNA and COI sequence. J. Ocean Univ. China 13 (1), 104–114. [Google Scholar]
- Llop S, Ballester F, Murcia M, Forns J, Tardon A, Andiarena A, et al. , 2017. Prenatal exposure to mercury and neuropsychological development in young children: the role of fish consumption. Int. J. Epidemiol. 46 (3), 827–838. [DOI] [PubMed] [Google Scholar]
- Mägi R, Morris AP, 2010. May 28. GWAMA: software for genome-wide association meta-analysis. BMC Bioinf. 11 (1), 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnus P, Birke C, Vejrup K, Haugan A, Alsaker E, Daltveit AK, et al. , 2016. Apr 1. Cohort profile update: the Norwegian mother and child cohort study (MoBa). Int. J. Epidemiol. 45 (2), 382–388. [DOI] [PubMed] [Google Scholar]
- Maitre L, Bont J de, Casas M, Robinson O, Aasvang GM, Agier L, et al. , 2018. Sep 1. Human Early Life Exposome (HELIX) study: a European population-based exposome cohort. BMJ Open 8 (9), e021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murcia M, Ballester F, Enning AM, Iñiguez C, Valvi D, Basterrechea M, et al. , 2016. Nov 1. Prenatal mercury exposure and birth outcomes. Environ. Res. 151, 11–20. [DOI] [PubMed] [Google Scholar]
- Murcia M, Ballester F, Enning AM, Iñiguez C, Valvi D, Basterrechea M, et al. , 2016. Nov 1. Prenatal mercury exposure and birth outcomes. Environ. Res. 151, 11–20. [DOI] [PubMed] [Google Scholar]
- Oken E, Baccarelli AA, Gold DR, Kleinman KP, Litonjua AA, Meo DD, et al. , 2015. Cohort profile: project viva. Int. J. Epidemiol. 44 (1), 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Kwon SO, Kim S-H, Kim SJ, Koh EJ, Won S, et al. , 2020. Methylation quantitative trait loci analysis in Korean exposome study. Mol. Cell. Toxicol. 16 (2), 175–183. [Google Scholar]
- Parks JM, Johs A, Podar M, Bridou R, Hurt RA Jr., Smith SD, et al. , 2013. Mar 15. The genetic basis for bacterial mercury methylation. Science 339 (1095–9203) (Electronic)):1332–5. [DOI] [PubMed] [Google Scholar]
- Perera F, Herbstman J, 2011. Apr. Prenatal environmental exposures, epigenetics, and disease. Reprod. Toxicol. 31 (3), 363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera F, Herbstman J, 2011. Apr. Prenatal environmental exposures, epigenetics, and disease. Reprod. Toxicol. 31 (3), 363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipson B, Maksimovic J, Oshlack A, 2016. Jan 15. missMethyl: an R package for analyzing data from Illumina’s HumanMethylation450 platform. Bioinform. Oxf. Engl. 32 (2), 286–288. [DOI] [PubMed] [Google Scholar]
- Pieper I, Wehe CA, Bornhorst J, Ebert F, Leffers L, Holtkamp M, et al. , 2014. Mechanisms of Hg species induced toxicity in cultured human astrocytes: genotoxicity and DNA-damage response. Metallomics 6 (3), 662–671. [DOI] [PubMed] [Google Scholar]
- Polunas M, Halladay A, Tjalkens RB, Philbert MA, Lowndes H, Reuhl K, 2011. Role of oxidative stress and the mitochondrial permeability transition in methylmercury cytotoxicity. Neurotoxicology 32 (5), 526–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston NVC, Raymond LJ, 2018. Nov 1. Mercury’s neurotoxicity is characterized by its disruption of selenium biochemistry. Biochim. Biophys. Acta BBA Gen. Sub. 1862 (11), 2405–2416. [DOI] [PubMed] [Google Scholar]
- Rempel E, Hoelting L, Waldmann T, Balmer NV, Schildknecht S, Grinberg M, et al. , 2015. Sep 1. A transcriptome-based classifier to identify developmental toxicants by stem cell testing: design, validation and optimization for histone deacetylase inhibitors. Arch. Toxicol. 89 (9), 1599–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Huertas C, Freixo F, Lüders J, 2016. Principles of microtubule organization: insight from the study of neurons. The Microtubule Cytoskeleton: Organisation, Function and Role in Disease, pp. 79–116. [Google Scholar]
- Shafizadeh TB, Halsted CH, 2007. May. gamma-Glutamyl hydrolase, not glutamate carboxypeptidase II, hydrolyzes dietary folate in rat small intestine. J. Nutr. 137 (5), 1149–1153. [DOI] [PubMed] [Google Scholar]
- Sharma BM, Sáňka O, Kalina J, Scheringer M, 2019. Apr. An overview of worldwide and regional time trends in total mercury levels in human blood and breast milk from 1966 to 2015 and their associations with health effects. Environ. Int. 125, 300–319. [DOI] [PubMed] [Google Scholar]
- Sheehan MC, Burke TA, Navas-Acien A, Breysse PN, McGready J, Fox MA, 2014. Apr 1. Global methylmercury exposure from seafood consumption and risk of developmental neurotoxicity: a systematic review. Bull. World Health Organ. 92 (4), 254–269F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde V, Hoelting L, Srinivasan SP, Meisig J, Meganathan K, Jagtap S, et al. , 2017. Feb 1. Definition of transcriptome-based indices for quantitative characterization of chemically disturbed stem cell development: introduction of the STOP-Toxukn and STOP-Toxukk tests. Arch. Toxicol. 91 (2), 839–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan SA, Barres BA, 2014. Aug. Mechanisms of astrocyte development and their contributions to neurodevelopmental disorders. Curr. Opin. Neurobiol. 27, 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern AH, Smith AE, 2003. Sep. An assessment of the cord blood:maternal blood methylmercury ratio: implications for risk assessment. Environ. Health Perspect. 111 (12), 1465–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strimmer K, 2008. Jun 15. fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinform. Oxf. Engl. 24 (12), 1461–1462. [DOI] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. , 2015. Jan 23. Proteomics. Tissue-based map of the human proteome. Science 347 (6220), 1260419. [DOI] [PubMed] [Google Scholar]
- Ung CY, Lam SH, Hlaing MM, Winata CL, Korzh S, Mathavan S, et al. , 2010. Mar 30. Mercury-induced hepatotoxicity in zebrafish: in vivo mechanistic insights from transcriptome analysis, phenotype anchoring and targeted gene expression validation. BMC Genom. 11 (1), 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- United Nations, 2018. Global Mercury Assessment [Internet] [cited 2021 Mar 4]. Available from: https://sdg.iisd.org:443/news/unep-publishes-2018-global-mercury-assessment/.
- Vaiserman A, 2015. Sep 11. Epidemiologic evidence for association between adverse environmental exposures in early life and epigenetic variation: a potential link to disease susceptibility? Clin. Epigenet. 7 (1), 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables WN, Ripley BD, 2002. Modern Applied Statistics with S. Springer. [Google Scholar]
- Waldmann T, Grinberg M, König A, Rempel E, Schildknecht S, Henry M, et al. , 2017. Apr 17. Stem cell transcriptome responses and corresponding biomarkers that indicate the transition from adaptive responses to cytotoxicity. Chem. Res. Toxicol. 30 (4), 905–922. [DOI] [PubMed] [Google Scholar]
- Worth RG, Esper RM, Warra NS, Kindzelskii AL, Rosenspire AJ, Todd III RF, et al. , 2001. Mercury inhibition of neutrophil activity: evidence of aberrant cellular signalling and incoherent cellular metabolism. Scand. J. Immunol. 53 (1), 49–55. [DOI] [PubMed] [Google Scholar]
- Yang L, Zhang Y, Wang F, Luo Z, Guo S, Strähle U, 2020. Apr 1. Toxicity of mercury: molecular evidence. Chemosphere 245, 125586. [DOI] [PubMed] [Google Scholar]
- Ying Chan PH, Kwok KM, Ming Chan MH, Li AM, Shuen Chan IH, Fok TF, et al. , 2021. Jul 23. Prenatal methylmercury exposure is associated with decrease heart rate variability in children. Environ. Res. 111744. [DOI] [PubMed] [Google Scholar]
- Zhang M, Liu T, Wang G, Buckley JP, Guallar E, Hong X, et al. , 2021. Jun. In utero exposure to heavy metals and trace elements and childhood blood pressure in a U.S. Urban, low-income, minority birth cohort. Environ. Health Perspect. 129 (6), 67005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


