Summary
Gastrointestinal immune cells, particularly muscularis macrophages (MM) interact with the enteric nervous system and influence gastrointestinal motility. Here we determine the human gastric muscle immunome and its changes in patients with idiopathic gastroparesis (IG). Single cell sequencing was performed on 26,000 CD45+ cells obtained from the gastric tissue of 20 subjects. We demonstrate 11 immune cell clusters with T cells being most abundant followed by myeloid cells. The proportions of cells belonging to the 11 clusters were similar between IG and controls. However, 9/11 clusters showed 578-11,429 differentially expressed genes. In IG, MM had decreased expression of tissue-protective and microglial genes and increased the expression of monocyte trafficking and stromal activating genes. Furthermore, in IG, IL12 mediated JAK-STAT signaling involved in the activation of tissue-resident macrophages and Eph-ephrin signaling involved in monocyte chemotaxis were upregulated. Patients with IG had a greater abundance of monocyte-like cells. These data further link immune dysregulation to the pathophysiology of gastroparesis.
Subject areas: Gastroenterology, Immunology, Transcriptomics
Graphical abstract
Highlights
-
•
Eleven immune clusters in human gastric muscle. T-cells and myeloid largest clusters
-
•
Decreased tissue-protective and increased monocyte trafficking genes in gastroparesis
-
•
Greater abundance of monocyte-like macrophage clusters in gastroparesis
-
•
IL12 mediated JAK-STAT and Eph-ephrin signaling increased in gastroparesis
Gastroenterology; Immunology; Transcriptomics
Introduction
The gastrointestinal (GI) tract is the largest immune organ in the human body and several studies, including those using single cell transcriptional profiling, have highlighted the distribution and function of immune cells in the intestinal mucosa.1,2,3 The muscularis propria of the GI tract houses the bulk of the enteric nervous system (ENS), which along with interstitial cells of Cajal (ICC) and smooth muscle cells, mediates propulsive oral-caudal motility.4 Animal models have highlighted immune interactions to play a critical role in maintaining5 and protecting enteric neuronal6,7 and ICC8,9 function with resulting effects on motility in the intestinal tract. Macrophages residing in the muscularis propria are called muscularis macrophages (MM). In the mouse colon, MM regulates peristaltic motility by secreting bone morphogenetic protein 2 (BMP2) which likely interacts with enteric neurons.5 Recently, MM were found to elicit colonic contractility through prostaglandin E210 release via the TRPV4 signaling pathway. In contrast to the lamina propria (LP) macrophages, which have a pro-inflammatory phenotype, MM in the small intestine exhibit tissue-protective genes such as Retnla, Mrc1, Cd163, Il10, and CD86 and a distinct morphology.11 Furthermore, β-2 adrenergic signaling in intestinal MM was found to augment the anti-inflammatory phenotype12 as well as protect against enteric infection mediated neuronal loss.6 Traditionally, in mice, Ly6Chi monocytes attaining maturation were known to be the primary source of tissue-resident macrophages.13 However, using elegant fate mapping studies, a distinct population of self-maintaining macrophages (Pcdhbhi, Igf2bp3hi, Ly6c2low, Nos1low, and TNFRSF19low) were found to occupy embryonic niches in both LP and myenteric plexus with functional effects on neuronally mediated intestinal secretion and motility.14 A recent study characterized macrophages (CD45+HLA-DR+CD14+) from LP and MM of the human colon using single cell sequencing and demonstrated the presence of unique MM subsets with proinflammatory/angiogenic and neuronal homeostatic properties.15 Together, these studies provide a strong rationale for the role of MM in the maintenance of peristaltic motility as well as in the prevention of neuronal loss in response to pathogen-mediated injury. However, an atlas of the types of macrophages and other immune cells in the muscularis propria of the human stomach has not been carried out.
A prototypical disorder of dysmotility in the GI tract is gastroparesis, defined by a delay in gastric emptying (GE), that results in clinical morbidity.16 A robust body of literature identified dysfunction in enteric pacemaking (ICC) and neuromuscular apparatus as the primary triggers of the pathogenesis of gastroparesis.17,18 Although frequently associated with diabetes, the majority of gastroparesis cases remain idiopathic with some studies suggesting the possibility of an enteric infection leading to the onset of symptoms.19 Our prior work using bulk RNA-seq of gastric muscle tissue has shown macrophage-based immune dysregulation in patients with idiopathic gastroparesis (IG).20 Additionally, using aptamer-based proteomics21 as well as immuno-histochemical analysis, we have shown loss of anti-inflammatory (MRC1hi) MM in human IG.22,23 There are no animal models for IG, making it important to delve into further analysis of human tissues. Our aim in this study was to characterize the immune cell populations in human gastric muscle and determine changes in patients with idiopathic gastroparesis.
Results
Gastric muscle immune cell atlas from controls and patients with idiopathic gastroparesis
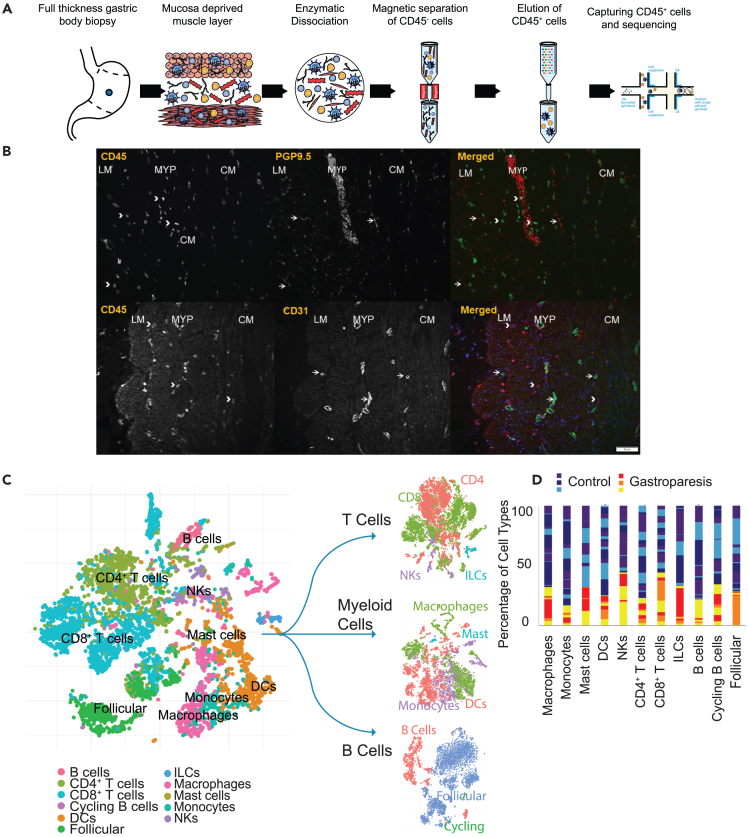
Using dissociated gastric muscle tissue with the mucosa and submucosa removed, from 13 control participants (Mean ± SD age 46 ± 15 years, all female) and 7 patients with IG (Mean ± SD age 49 ± 12 years, all female), a total of 26,596 CD45+ cells were analyzed (Mean 1330 cells/participant) (Figure 1A). Of the 26,596 total cells, 20,522 were from controls (mean 1579) and 6,074 (mean 868) from patients with IG (p = 0.4, Mann Whitney test). A mean of 292,912 reads was obtained per cell (Table S1). No significant associations between processing day (same or next day following procurement) and any of the 10x QC metrics were identified on Hierarchical-all-against-all (HAllA) clustering (FDR>0.05).24 Considering immune cell profiling of human gastric muscle tissue has not been performed to date, we sought to examine if CD45+ cells, were present within the muscle layers and myenteric plexus or only inside the blood vessels. Co-labelling was performed using CD31, as a blood vessel marker (PECAM-1, Platelet endothelial cell adhesion molecule) and CD45. Some CD45+ cells were found adjacent to CD31+ cells, however, a significant proportion of CD45+ cells were not close to CD31+ cells suggesting their presence in the muscle layers as well as myenteric plexus. Additionally, a proportion of CD45+ cells were in proximity to the PGP9.5+ nerve fibers and neurons (Figure 1B).
Figure 1.
Gastric Muscle Immune Cell Atlas from Healthy Volunteers and Patients with Idiopathic Gastroparesis
(A) Schematic representing the dissociation and isolation of CD45+ cells from mucosa deprived muscle layer for scRNA sequencing.
(B) Immunohistochemistry showing the distribution of CD45+ cells (arrowheads) and CD31 (PECAM1) immunoreactivity (arrows). CD45+ cells were noted both in proximity to and away from CD31 immunoreactivity. Also shown is colabeling with PGP9.5, a marker for enteric neurons and nerve fibers. Scale bar = 50 μm.
(C) T-sne graph of all called cell types in the cohort, 13 controls and 7 patients with gastroparesis. Cell types called represented as T cells, myeloid, and B cell compartments. T-sne graph of T cells colored by CD4+ T cells (red), CD8+ T cells (green), natural killers (NKs, purple), and ILCs (blue). T-sne graph of myeloid cells colored by macrophages (green), mast cells (blue), monocytes (purple), and dendritic cells (DC, red). T-sne graph of B cells color by B cells (red), follicular (blue), and cycling B cells (green).
(D) Percentage of patient contribution to each cell type colored by disease status (control patients are blue, dark blue, and purple; patients with gastroparesis are red, orange, and yellow). Color shades in the respective bars are to represent contribution of unique patient samples.
See also Figures S1–S3 and Tables S1, S2, and S3.
Eleven clusters of immune cell subsets identified in human gastric muscle
Using a library of established2 and new gene expression markers (544 genes) predominantly derived from genes expressed by GI LP immune cells, we identified 11 cell clusters (Table S2; Figure 1C). These were macrophages, dendritic cells (DCs), monocytes, mast cells, natural killer (NKs) cells, innate lymphoid cells (ILCs), CD4+ T cells, CD8+ T cells, follicular cells, cycling B cells, and B cells. T cells constituted the largest cluster with 11,949 cells (44.7% of overall cells), followed by the myeloid cell population (26.6% divided between DC, 11.1%; macrophages, 9.8%; monocytes, 5.7%). Clustering was also done by combining the markers and displaying broader categories of myeloid cells, T cells, and B cells. The t-distributed stochastic neighbor embedding (tSNE) distribution of the clusters in IG and controls separately is shown in Figure S1. Top conserved genes for the clusters representing macrophages (CTSD, Z score 21; CREG1, GPNMB, FPR3, GFRA2, and CSF1R), mast cells (TM6SF1, Z score 130; TIMP3, SIGLEC10, CD9 and TREM1), monocytes (CSTA, Z score 1921; MNDA, EREG, CCL20, MS4A4A, MS4A7, and CSF3R), NK cells (FGFBP2, Z score 20; KLRB1, KLRF1, CHST12, FCGR3A, and CD160), DC (ZNF385A, Z score 61557; WNT10A, ZBTB32, LGALS2, SLC4A10, and TMPRSS3), CD4+ T cells (CD247, Z score 33067; SLC2A3, BIRC3, C15orf53, PTPN7, RARRES3, and CD40LG), CD8+ T cells (CD8B, Z score 1653; DTHD1, EOMES, KLRC4, CTSW, and GZMA), B cells (IGHA2, Z score 14; GNG7, ISG20, IGHA1, C16orf74 and IFGN-AS1), cycling B cells (AURKB, Z score 15859; GINS2. CDC45, CENPW, NUSAP1, BIRC5, and BUB1), ILC (AREG, Z score 13; ICAM1, IL18R1, IL1RN, CXCL2, and IL26), and follicular cells (ARHGAP24, Z score 51; AFF3, CLECL1, FAM177B, HLA-DRB5, and CD40) (Table S2). Cell type classifications were further validated by performing unsupervised clustering of all cells and identifying clusters expressing genes specific to immune subsets as reported by the tissue-specific, single-cell database derived from the Tabula Sapiens cohort (Figure S2).25 The Tabula Sapiens expression profile for immune subsets from the small intestine was used as that tissue had most number of immune cells and was the most relevant comparison for our study. Of the 27 unsupervised clusters, 23 had directly matching Tabula Sapiens classifications to our most abundant cell type for the cluster, suggesting high concordance between two separate methods. Additionally, the proportion of cells assigned to the myeloid, T and B cell compartments was similar between the two approaches (Table S3).
We then searched the gene expression data against a different set of established canonical genes in the literature for monocytes (GLUL, Z score 7.8; FCGR2B, 5.6; FCGR1A, 5.5; F13A1, 5.2; IL10, 1.8), DC (CLEC4E, Z score 550; CTSC, 129; MERTK, 8), macrophages (FCGR2B; Z score 1.5), NK cells (FCGR3A, Z score 3.3), CD4+ T cells (IL10, Z score 4.8; FCGR3A, 2.1),5,11,26,27 cycling B cells (CD19, Z score 35),26 and follicular cells (HLA-DRB5, Z score 13).28 Markers for cDC1 (BATF3, IRF8, FLT3) and cDC2 (CD1D, CLEC10A, FCGR2B) shown in other studies were also found to be among the top expressed markers for those cell types in this dataset.1,29 The expression of additional genes known to be important for enteric function was assessed. Although mast cells were richest in KIT (CD117) expression (Z score 3.2), a similar expression was also seen in DC1 (Z score 2.9) and slightly weaker expression in ILC (Z score 2.4).30 CD11b (ITGAM) expression was present in myeloid cells including DC (Z score 2.2), macrophages (Z score 1.7) and monocytes (Z score 1.3). CX3CR1 had the strongest expression in the DC2 cell population in this dataset (Z score 3.1, followed by myeloid and NK cells). The DC compartment is known to be heterogeneous from an ontogenetic and functional standpoint. A recent study of human jejunal LP immune cells showed that monocyte derived cells (CSF1R+FLT3-) contribute ∼50% of cells in the subset that is conventionally categorized as DC.31 However, these are functionally distinct from DC due to their lower migratory potential and ineffective activation of T cells. A representative set of canonical genes for the 11 subsets is highlighted in the heatmap (Figure S3). All 11 cell subsets were present in each of the 20 samples in variable proportions (Figure 1D).
Distribution and molecular signatures for the immune cell clusters
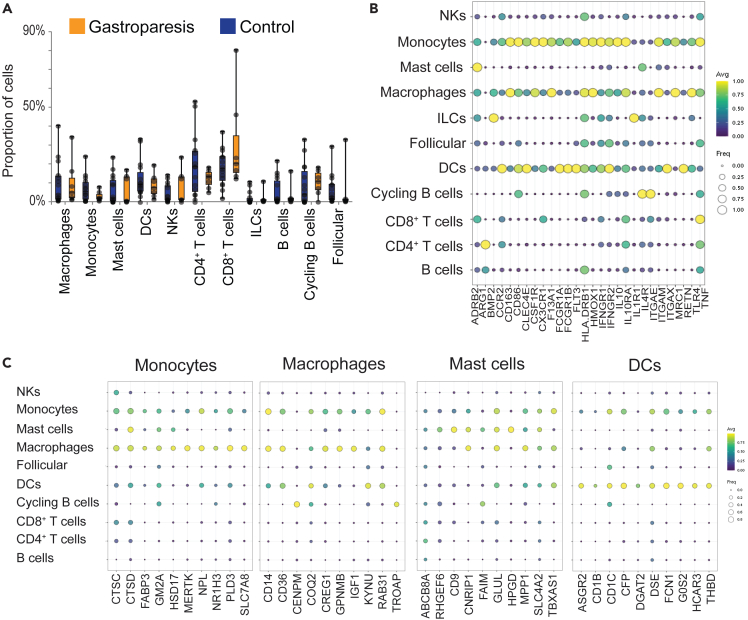
Control and IG groups did not demonstrate a statistically significant difference in the proportion of cells in any of the 11 clusters; perhaps due to the inter-individual variability and small number in the IG group. Among the largest clusters, CD8+ T cells represented 30.5 (24.7)% of cells in the IG group vs. 18.6 (9.9)% in controls, p = 0.14. The NK cells were 7.7 (9.1)% in the IG vs. 3.1 (3.6)% in controls, p = 0.12 and CD4+ T cells were 21.9 (16.1)% in the controls vs. 12.2 (4.8)% in gastroparesis, p = 0.14 (Figure 2A). Considering the significance and interest in myeloid cells in the maintenance and functional regulation of enteric neurons,5,6,12 ICC,8,32 and smooth muscle cells,10,33 we used a broad set of canonical gene markers for myeloid cells (Table S4) and determined their expression in the 11 clusters identified. The most robust expression of these markers was seen in myeloid cells (monocytes, macrophages, DC) (Figure 2B). Inducible NOS (iNOS, NOS2) expression was not found in any of the cell types. Although a marker for anti-inflammatory macrophages in mice, this gene is epigenetically suppressed in humans serving as a negative control in this human dataset.34 Beta-2 adrenergic receptor gene, recently shown to be important for signaling in anti-inflammatory macrophages,6 was expressed on monocytes (TPM10Kp1 0.36) and macrophages (TPM10Kp1 0.36). However, the most robust expression for this gene was in mast cells (TPM10Kp1 0.74). Additionally, interleukin (IL) receptor 1 expression was strongest on innate lymphoid cells and ITGAE or CD103 expression on cycling B cells. Lastly, as expected, TNF expression was most robust on CD8+ T cells (TPM10Kp1 1.12) closely followed by monocytes (1.05), macrophages (0.85) and CD4+ T cells (0.83). A low level of Arginase 1 expression was noted on macrophages and monocytes (TPM10Kp1 0.0001) with the highest expression on CD4+ T cells (0.01). CX3CR1 expression was present on 38% of macrophages but CX3CR1 was expressed on 100% monocytes, 67% DC, 20% NK and on 0% mast cells (Table S4 has the frequency and expression of these markers on all identified cell types).
Figure 2.
Immune Cell Composition Changes in Idiopathic Gastroparesis and Markers for Myeloid Cells
(A) The proportion of cells making up an individual’s total cell count for either control (blue) or patients with gastroparesis (orange). Dots represent individual patients. p > 0.05 for all cell-types, heteroscedastic unpaired Student’s t test.
(B) Bubble plot showing the expression (color, yellow high, purple low) of genes previously associated with myeloid cells and fraction of cells expressing those genes (log TP10K+1 > 0.5, size of bubble). Monocytes, macrophages and DCs show the highest average expression level and the expression in largest proportion of cells. The data represents expression for cells from both IG and controls.
(C) Bubble plots of genes associated with specific cell types, in order: monocytes, macrophages, mast cells, dendritic cells. Expression (log TP10K+1) is represented by color (yellow high, purple low) and fraction of cells expressing (log TP10K+1 > 0.5) is represented by size of the bubble. Genes were selected by calculating a Z score for the associated cell type against all other cell types, and genes with the highest Z score were plotted.
Canonical genes for the 4 myeloid clusters (Figure 2C; Table S5) and remaining cell types (Figure S4) are presented.
Gastric immunome highlighting gene expression changes in immune cell subsets between controls and idiopathic gastroparesis
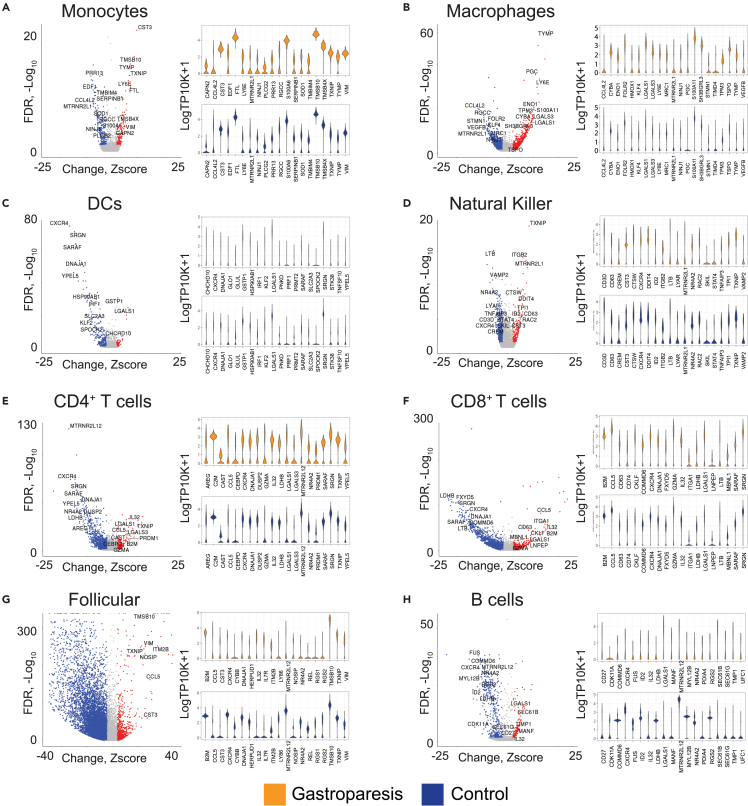
Differential expression analysis showed 578-11,429 differentially abundant genes (FDR < 0.05) in 9 of 11 clusters (Table S6); while no genes were differentially expressed in two clusters (mast cell and cycling B cell) between controls and IG. The follicular cell subset showed the greatest number of differentially expressed genes (n = 11,429) and the monocyte subset showed the least (n = 578). The top over-expressed genes in IG were for macrophages (TYMP, MT2A, LY6E), DC (RPLP1, LGALS1, GIMAP7), monocytes (CST3, TMSB10, CD74), NK cells (TXNIP, LCP1, MT2A), follicular (RPL10, MALAT1, RPL28), CD4+ T cells (MALAT1, TXNIP, IL32), CD8+ T cells (MT.CYB, MT.CO1, MT.CO3), and B cells (RPL41, RPL11, SEC61B) (Figure 3). Top under-expressed genes in macrophages (CCL4L2, RGCC, MYADM, LYVE1), DC (CXCR4, YPEL5, SRGN), monocytes (PRR13, PET100, SNX6), NK cells (TRBC2, TRAC, CD3E), follicular (HLA.DRB5, CD79A, HLA.DQB1), CD4+ T cells (FAM118A, MTRNR2L12, CXCR4), CD8+ T cells (IGKC, SLC25A6, CD74), and B cells (HLA.DRB5, SRSF2, CDC42) (Figure 3).
Figure 3.
Gene Expression Differences between Healthy Volunteers and Patients with Idiopathic Gastroparesis
Volcano and violin plot of differentially expressed genes between patients with IG and controls.
(A–H) Selected genes of interest (either based on differential expression or functional significance) are highlighted with text. The y axis represents transformed false discovery rates (FDR) with colored dots requiring an FDR <0.05. The x axis represents a Z score, representing a pseudo fold change with colored dots requiring a Z score +/−1. Respective violin plots for each cell type represent log TP10K+1 expression for highlighted genes in corresponding volcano plots. Violin plots are separated by disease status (control blue, gastroparesis orange).
See also Table S6.
Tissue protective genes
Macrophage genes associated with tissue-protective effects were under-expressed in IG (MRC1, Z score −6.9; HMOX1, -6.7; FCGR2B, −6.2; CSF1R, −3.2; IL10RA, −1.3, CD163L1, -1.3). Genes associated with apoptotic clearance were also under-expressed in gastroparesis (C1QC, Z score −4.5; C1QA, −3.9; C1QB, −1.8).
Inflammatory response genes
Genes associated with inflammatory responses (IL1R2, Z score 2; IFNγR1, 0.2) and lymphocyte and stromal activating cytokine genes were overexpressed (IL6ST, Z score 2.4; TNFAIP3, 2.6, TNFSF13B, 1.9) in IG. Additionally, CCR2, a marker for monocyte migration (Z score 2.5), Ly6E, a core monocyte marker (Z score 12.5), and MHCII genes, required for antigen processing and presentation (CD74, HLA (DRB1, DRA, DPA1)), were overexpressed in macrophages from IG.
Neuro-immune crosstalk genes
The C1Q genes expressed in cells with microglia properties, and expression of other microglia-like genes was decreased in IG (GPR34, Z score −5.2; GAS6, -3.9; HEXB, −2.9).14 A recent study showed that MHCIIhiLyve1lo resident tissue macrophages occupy a niche close to nerves and MHCIIloLyve1hi reside alongside blood vessels.35 Interestingly, in addition to the high MHCII expression, the expression of Lyve1 was significantly reduced in macrophages from patients with IG (Z score −9.4), suggesting a specific loss of blood vessel-associated macrophages. Timd4, a marker for long-term residence in LP macrophages was underexpressed in macrophages from IG.14 This suggests that the key homeostatic role of macrophages involving removal of cellular and neuronal debris might be impaired and monocyte trafficking to the gastric tissue enhanced in IG.
Patients with idiopathic gastroparesis have enriched signaling pathways in macrophages
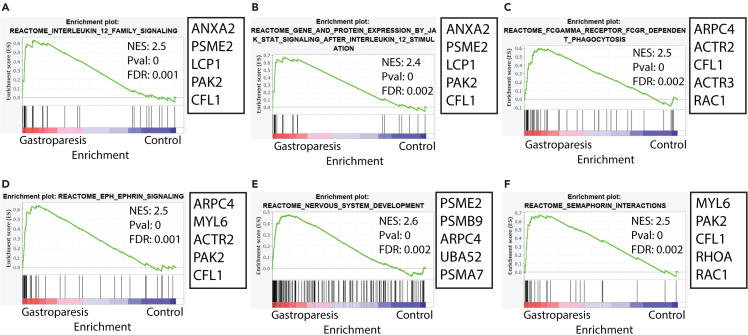
We identified that among the Reactome pathways (https://reactome.org/) linking genes associated with macrophages, only the upregulated pathways were statistically significant (FDR < 0.05). The top 6 most significant pathways are displayed (Figure 4; Table S7). We found the upregulation of the IL12 signaling pathway and the downstream JAK-STAT signaling pathway. IL12 is a potent proinflammatory cytokine that promotes Th1 response and induces IFNγ production by T cells and NK cells.36 IL-12/23 signaling has been targeted for the treatment of inflammatory conditions such as psoriasis37 and inflammatory bowel disease.38 Upregulation of the Fcγ receptor-dependent phagocytosis pathway was also noted in macrophages from patients with IG. Eph-ephrin signaling is involved in monocyte chemotaxis to inflammatory sites, adhesion, and transmigration across the vascular endothelium.39 Members of EphA and EphB receptors are also expressed with variable receptor patterns on subsets of DC.40,41 Similarly, the EphA family of proteins has been shown to play a role in T cell trafficking to inflammatory sites.42 Signaling associated with semaphorin interactions was also upregulated in IG. A study showed the semaphorin 4D expressed by tumor stroma associated macrophages facilitated tumor angiogenesis and metastatic potential.43 Additionally, semaphorins and Eph-ephrin signaling genes are also involved in axon guidance which likely resulted in the “nervous system development” pathway being unregulated in IG. However, it is unclear if these genes may mediate interactions between macrophages and nerves in the muscularis propria.
Figure 4.
Differentially Abundant Macrophage Specific Signaling Pathways
Gene Set Enrichment Analysis (GSEA) showed multiple upregulated signaling pathways in macrophages (FDR<0.05). The top 6 Reactome pathways (https://reactome.org/) are displayed. No statistically significant downregulated pathways were observed.
(A) IL12 Family Signaling.
(B) JAK-STAT Signaling After IL12 Stimulation.
(C) FC Gamma Receptor FCGR Dependent Phagocytosis.
(D) EPH-ephrin Signaling.
(E) Nervous System Development.
(F) Semaphorin Interactions.
See also Table S7.
Distinct macrophage clusters were observed in idiopathic gastroparesis and controls
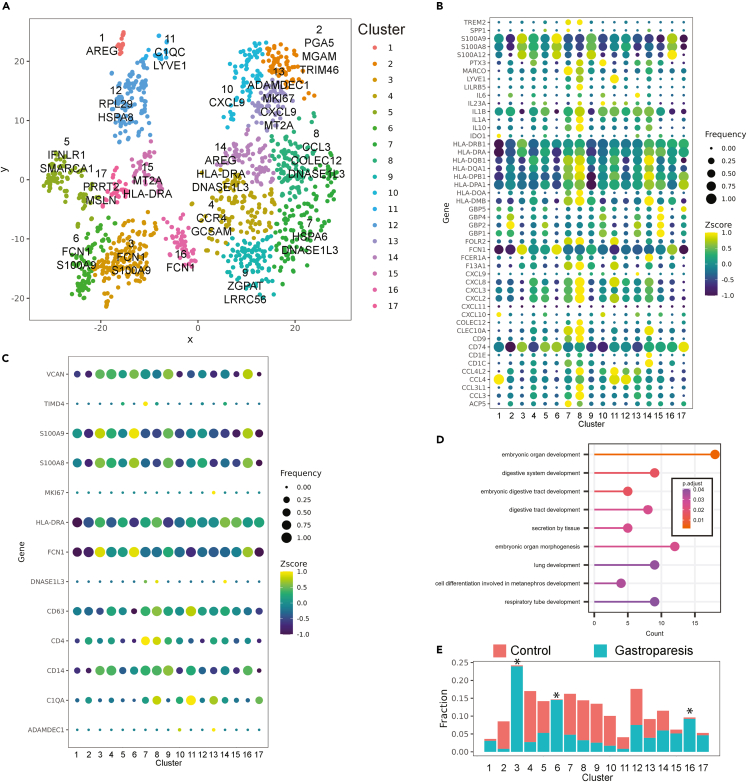
Unsupervised clustering of HLA-DR+CD14+ cells using the PhenoGraph algorithm demonstrated 17 distinct clusters44 (Figure 5A). Clusters 9, 4, and 14 demonstrate the highest expression of canonical markers associated with DCs (ASRG2, GOS2, HCAR3, and FCN1). Cluster 6 had the highest expression of S100A9 and S100A8 followed by cluster 16 which also had the highest expression of S100A12 (Figure 5B). Both clusters had low expression of CXCL chemokines and HLA. The broader markers expressed by clusters 6 and 16 along with cluster 3 (S100A4, S100A11, CSF3, and FCN1) demonstrate monocyte-like identity to these clusters. Cluster 8 had the highest expression of HLA-genes (DQB1, DQA1, DPB1, DPA1), chemokines (CXCL8, CXCL2, CXCL3, CCL3, and CCL3L1), interleukins (IL1A and IL1B), CCL3, CLEC10A, COLEC12, and FOLR2. Interestingly, this cluster also had the highest expression of PMP22 and EMP2, genes mainly expressed in Schwann cells and involved in neuronal protection.45 An adjacent cluster 14 was also enriched in HLA genes, chemokines, interleukins, and CLEC10A. In addition, this cluster was also high for FCER1A. Cluster 7 also had high expression of PMP22 and EMP2. In addition, it had among the highest expressions for MARCO, CD1C, Lyve1, COLEC12, and CCL3. These clusters also had high expression of selected genes associated with embryonically derived macrophages such as CD63 (clusters 8 and 11), TIMD4 (cluster 7), and FOL2R (clusters 7, 8, and 11)46 (Figure 5C). Gene Ontology enrichment terms for canonical genes in clusters 7, 8, and 14 revealed pathways associated with embryonic digestive tract development and embryonic organ morphogenesis (Figure 5D). The macrophage clusters 8 and 7 in this study show similarities to clusters MM5 and MM11 identified in the study by Domanska et al. with high and low proinflammatory properties respectively. Both clusters expressed neuroprotective genes such as PMP2, EMP2 with cluster 7 also expressing high levels of homeostatic genes such as MARCO, Lyve1, and COLEC12. Clusters 3, 6 and 16 had significantly higher number of cells from patients with IG compared to the controls (Figure 5E; Table S8). Clusters 6 and 16 with high expression of S100A family of proteins resembled cluster MM0 with properties associated with cellular response to bacterium, LPS, and antigen presentation. Cluster 11 had the highest expression of Lyve1 and C1QC. This cluster also had high expression of CCL4, CCL4L2, and FOLR2 but lower expression of expression of HLA and CXCL chemokine genes. Cluster 8 had the next highest expression of Lyve1 and C1QC. Clusters 1 and 11 had the highest expression of HSP proteins. Of these, cluster 1 had low expression of interleukins and cytokines. When CD68+ Lyve1+ cells were compared, patients with gastroparesis demonstrated lower abundance of these cells compared to controls (2.7% vs. 11.6%, Chi square = 33, 2-tailed p value<0.0001).
Figure 5.
Unsupervised clustering of Gastric Muscularis Propria macrophages
(A) Unsupervised clustering of HLA-DR+CD14+ cells reveal 17 distinct clusters.
(B) Bubble plot demonstrating Z score expression of selected genes in 17 clusters (color) as well as percentage of cells expressing the genes (size).
(C) Bubble plot showing Z score expression of selected genes associated with embryonic-derived macrophages in the 17 clusters (color) as well as percentage of cells expressing the genes (size).
(D) Statistically significant Gene Ontology enrichment terms for canonical genes in clusters 7, 8 and 14.
(E) Clusters 3, 6 and 16 are overrepresented by cells from gastroparesis (blue) samples than controls (red, FDR<0.05 for all three).
See also Table S8.
Discussion
The presented atlas showed a diverse repertoire of immune cells in human gastric muscle layers with T cells making the largest fraction followed by myeloid cells. While several canonical markers based on rodent and other human studies were confirmed, many additional markers were identified. Examples are high KIT expression in DC2 and CX3CR1 in the DC1 compartment. Only 38% macrophages but 67% DC and 100% monocytes expressed CX3CR1. Mast cells had the highest expression of the β-2 adrenergic receptor, in addition to macrophages where this receptor plays a role in neuroprotection.6 Macrophages expressed a low level of ITGAX (CD11c), which is consistent with observations in another study where ∼75% of MM lacked CD11c expression.3 Similar to that study, we also show a high level of CD11c expression in DCs. Arginase 1, a marker for an anti-inflammatory macrophage spectrum in rodents had low expression in this dataset, and iNOS was not seen, confirming observations made in human endothelial cells.34 Single cell characterization of human muscularis propria immune cells provides a crucially needed resource for studying a wide variety of diseases involving muscle layers in the GI tract such as malignancy, inflammatory bowel disease as well as motility disorders.
In a non-obese diabetic mice model, diabetes is associated with the appearance of the HO1+ anti-inflammatory spectrum of macrophages in gastric muscularis propria.47 Development of delayed GE following diabetes did not alter the total macrophage number but was associated with loss of macrophages expressing canonical anti-inflammatory markers (CD206, HO1, MRC1) and mice refractory to developing delayed GE retained these macrophages.47 Furthermore, macrophage-deficient Csf1op/op mice were protected from injury to ICC and development of delayed GE even in the presence of diabetes,9 a phenotype that reversed after the administration of CSF1 that caused macrophage infiltration into the tissues.8 Conditioned medium from macrophages exposed to oxidative injury caused damage to cultured ICC, which was preventable by the inhibition of TNF and IL6R.8 This suggests a possible role of paracrine mediators underlying immune-ICC interactions. A recent study also showed MM alter the proportion of nitrergic myenteric neurons.48 Our data using human cells indicated lower expression of genes associated with tissue-protection (MRC1, HMOX1, FCGR2B, CSF1R, IL10RA, CD163L1) and neuronal function (BMP2, CCDC141, NPTX2) in MM from patients with gastroparesis.5 In contrast, monocyte-trafficking (CCR2, Ly6E) and stromal activating genes (IL6ST, TNFAIP3, TNFSF13B) were increased in gastroparesis, consistent with data in mice. Additionally, there was an increase in the subset of HLA-DR+CD14+ cells with monocyte like expression in the tissue suggesting possibly increased monocyte trafficking in gastroparesis. Furthermore, we found the upregulation of IL12 and the downstream JAK-STAT signaling pathway in gastroparesis. Additionally, there was upregulation of the Eph-ephrin mediated signaling pathway, which is involved in monocyte trafficking to the tissues.
The leading-edge genes regulating the IL12 signaling pathway, which was upregulated in patients with IG were Annexin A2 (ANXA2), Proteasome Activator Subunit 2 (PSME2), Lymphocyte Cytosolic Protein 1 (LCP1), PAK2 (P21 (RAC1) Activated Kinase 2) and Cofilin 1 (CFL1). ANXA2, a Ca2+ binding protein associated with macrophage phagocytosis. ANXA2 tetramer was found to activate macrophages with enhanced bacterial phagocytic ability, and it also caused the phosphorylation of several MAP kinases with increased production of TNFα, IL-1β, and IL-6 (which can enhance the recruitment of leukocytes).49 PSME2 modulates proteasomal activity and was associated with M1 gene signature.50 Proteasome 20S Subunit Beta (PSMB) 8 and 9 are immunoproteosomes that replace constitutively expressing PSMB5 and 6 under inflammatory conditions (such as upon induction by IFNγ and TNFα) which mediate the production of HLA peptides.51 PAK2 disruption increases hematopoietic progenitor cell sensitivity to GM-CSF, driving commitment to granulocyte-monocytes and promoting the development of myeloid derived suppressor cells which are hyperproliferative and apoptosis resistant.52 PAK signaling is involved in chemokine-induced macrophage migration.53 Cofilin-1 was found to promote monocyte differentiation into fibrocytes in pulmonary fibrosis and plays a role in transendothelial motility.54 Actin related protein 2/3 complex subunit 4 (ARPC4), another leading-edge gene in IL-12 pathway, mediates actin filament binding leading to the formation of phagocytic cup in the macrophages.55 Activation of IL12 signaling pathway in gastroparesis suggests an influence on macrophage maturation, migration, and activation.
The JAK-STAT signaling pathway in macrophages can induce a pro- (IFNγ) or anti-inflammatory (IL4, IL10, IL13) phenotype based on the activating mediator.56 LPS was shown to induce IL12 and iNOS synthesis in macrophages via the TLR4/NF-kB dependent pathway.57 Interferon regulatory factor 8 (IRF8) deficient macrophages had a significantly impaired ability to produce IL12.58 IRF5 was found to enhance the IFNγ/JAK/STAT-1-dependent production of IL12.59 The JAK-STAT signaling observed in IG was downstream of IL12. EphA-ephrin A, another signaling pathway activated in IG, promotes monocyte adhesion via integrin activation and formation of protrusions.60 A study showed ephrin-A1 induced EphA4 forward signaling promotes monocyte adhesion to endothelial cells.61 Monocytes demonstrated preferential adhesion to endothelial cells overexpressing full-length ephrin-B2 compared to those with cytoplasmically truncated ephrin-B2. Additionally, adhesion of EphB4-receptor-overexpressing monocytes was further augmented. Monocyte-to-macrophage differentiation was associated with an increase in EphB2 expression, and these cells interacted with ephrin-B2 receptor on endothelial cells resulting in its phosphorylation and expression of proinflammatory cytokines such as IL8 and CCL2.62 This suggests a role for Eph-ephrin interactions in monocyte adhesion, transmigration through vascular endothelium, and activation of pro-inflammatory responses.63
The contribution of embryonically (yolk-sac and fetal liver)-seeded and circulating monocyte-derived cells toward tissue resident macrophages is an area of significant interest and investigation across various organs.64 Numerous intrinsic factors (age, sex, species, genetic background) and local niche-associated factors (Kupffer cell macrophages in the liver, microglia in the brain, red pulp macrophages in spleen, alveolar macrophages) play a role in governing the identity of tissue resident macrophages. Furthermore, their identity and transcriptional profile dramatically changes during homeostasis and in response to an acute and chronic injury. For example, one study showed that there is a 90% variance of transcriptional profile with pro-inflammatory polarization; however, only 9% variance was observed during anti-inflammatory polarization.65 Using lineage traced CX3CR1+ macrophages in mouse ileum, distinct transcriptional differences were noted in LP between self-maintained and monocyte-derived macrophages.14 In contrast, these differences were much less pronounced in the muscularis externa with no specific discriminatory markers identified. Detailed studies of adult human organs are lacking particularly in areas that are harder to safely access such as the muscle layers of the intestinal tract. The available studies have shown that relying on isolated markers across different sites and species may not allow true appreciation of cellular identity for a specific organ of interest. The current study provides the panel of markers that will provide both sensitivity as well as specificity for characterizing unique immune cell types. For example, we identify macrophage clusters that are suggestive of embryonically derived macrophages.
In conclusion, this study provides immune cell characterization of human gastric muscle validating several existing as well as unraveling gene markers for eleven different immune cell clusters. These can be used in future studies, including using flow cytometry which requires a library of a priori protein targets. The results provide changes in gastroparesis further substantiating a myeloid cell-based immune dysregulation as one of the central drivers of its pathogenesis. Furthermore, our results elucidate alterations in IL12 and monocyte trafficking signaling which can serve as prime targets for disease modification in gastroparesis. Further studies will need to examine these pathways and their influence on ICC, other cells of the ENS, and the smooth muscle fibers that regulate propulsive gastrointestinal motility.
Limitations of the study
A limitation of the study is smaller sample size, particularly of the IG group. Inclusion of only females is another limitation. This study does not determine protein expression to validate identified markers. Furthermore, the distribution of various macrophage subsets and proximity to nerve fibers vs. blood vessels will need to be determined using dedicated spatial expression techniques. Lastly, even though the focus of current study was on macrophages, gastric immunome is comprised of dense plethora of other immune cells which may have plausible role in immune dysregulation associated with IG. Further studies including those using data provided in this atlas will need to examine that.
Consortia
The members of the NIDDK Gastroparesis Clinical Research Consortium are T. Thomas Abell, Amirah Abdullah, Margaret Adamo, Emerald Adler, Guillermo Barahona Hernandez, Lynn Baxter, Patricia Belt, Cheryl Bernard, Cynthia Bouette, Margaret Breen-Lyles, Anya Brown, Robert Bulat, Robert Burns, Jorge Calles-Escandon, Bridget Cannon, Heather Charron, Bruno Chumpitazi, Sean Connery, Kelly Cooper, Nata DeVole, John Dodge, Michelle Donithan, Karen Earle, Karina Espino, Gianrico Farrugia, Liz Febo-Rodriguez, Marvin Friedman, Madhusudan Grover, Sherry Hall, Frank Hamilton, William Hasler, William Herman, John Hollier, Milana Isaacson, Stephen James, Madeline Kane, Kjersti Kirkeby, Kenneth Koch, Andrew Kraftson, Braden Kuo, Candice Lee, Linda Lee, Mimi Lin, Zubair Malik, Alan Maurer, Catherine McBride, Richard W. McCallum, Lindsay McElmurray, Megan McKnight, Jill Meinert, April Mendez, Laura Miriel, Linda Nguyen, Samuel Nurko, Chiara Orlando, Amiya Palit, Henry P. Parkman, Pankaj Jay Pasricha, Amy E. Rothberg, Irene Sarosiek, Jose Serrano, Emily Sharkey, Robert Shulman, Casey Silvernale, Jacqueline Smith, Michael Smith, William Snape, Kyle Staller, Alice Sternberg, Abigail Stocker, Paula Stuart, Andrea Thurler, James Tonascia, Rebecca Torrance, Doug Troyer, Mark Van Natta, Denise Vasquez, Natalia Vega, Christopher Velez, Anna von Bakonyi, Annette Wagoner, Stephanie Wall, Kent Williams, Laura A. Wilson, Frederick Woodley, Sophanara Wootten, Goro Yamada, Katherine P. Yates, and Lina Yossef-Salameh.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD45 | AbD Serotec | Cat# MCA87; RRID: AB_871979 |
| PGP9.5 | GeneTex | Cat# GTX82567; RRID: AB_11179128 |
| CD31 | Abcam | Ab28364; RRID: AB_726362 |
| Biological samples | ||
| Gastric muscle biopsies | Human | NA |
| Chemicals, peptides, and recombinant proteins | ||
| MACS® Tissue Storage Solution | Miltenyi Biotec | 130-100-008 |
| Dead Cell Removal Kit | Miltenyi Biotec | 130-090-101 |
| autoMACS® Rinsing Solution | Miltenyi Biotec | 130-091-222 |
| MACS® BSA Stock Solution | Miltenyi Biotec | 130-091-376 |
| Deposited data | ||
| Gene expression | GEO | GSE252126 |
| Software and algorithms | ||
| Custom codes | Zenodo | 10366426 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Madhusudan Grover (grover.madhusudan@mayo.edu).
Materials availability
No unique reagents or relevant chemical materials were developed as a part of this study.
Data and code availability
-
•
The gene expression data has been uploaded to Gene expression Omnibus (GEO; accession number: GSE252126).
-
•
Custom codes are uploaded to Zenodo (https://zenodo.org/records/10366426).
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Tissue biopsy specimens
Full thickness gastric body biopsies were obtained from 13 control subjects undergoing bariatric surgery at the Mayo Clinic and, from 7 IG patients undergoing surgery for the implantation of a gastric electrical stimulation device at a clinical site of the National Institute of Diabetes and Digestive and Kidney Diseases Gastroparesis Clinical Research Consortium (GpCRC). All participants were females due to higher prevalence of gastroparesis in females and all were Caucasian. Mean ± SD age for controls was 46 ± 15 years for IG patients was 49 ± 12 years. The gastric body specimens from control subjects were collected in MACS® Tissue Storage Solution (Miltenyi Biotec, 130-100-008) and processed immediately after arrival in the lab or stored overnight at −80 C. The tissue samples from IG patients obtained at the University of Louisville, KY or the Temple University, PA, were collected in MACS® Tissue Storage Solution and shipped overnight. Upon arrival at Mayo Clinic, the tissues were processed immediately using standardized protocols. All control subjects and IG patients provided oral and written informed consent respectively for the procurement and use of gastric tissue. The study was approved by Mayo Clinic IRB 07-003371.
Gastric emptying test
A GE test was performed by using a standardized protocol.66 Briefly, the patients were administered radiolabeled low-fat egg white meal and scintigraphy was done at 0, 1, 2, and 4 hours after ingestion. All IG patients exhibited gastric retention of > 60% at 2 hours or > 10% at 4 hours as determined by scintigraphy test and showed no evidence of obstruction of the gastric outlet.
Method details
Tissue dissociation and isolation of CD45+ immune cells
The mucosal layer of the gastric body biopsies was removed and the muscularis layer was used for dissociation. The tissue dissociation was performed using Multi Tissue Dissociation kit 1 (Miltenyi Biotec, 130-110-201) as per manufacturer’s instructions with slight modifications. Briefly, the muscle layer was precut into ∼4mm pieces and transferred into gentleMACS C Tubes (Miltenyi Biotec, 130-093-237) containing 2.5ml DMEM (Gibco, 11965-092) supplemented with the enzymes of Multi Tissue Dissociation kit 1. These gentleMACS C Tubes were transferred onto gentleMACS™ Octo Dissociator (Miltenyi Biotec, 130-096-427) and the 37°C_h_Stomach_01 program was run after attaching the heating elements, for 40 minutes. The dissociated cell suspension was then filtered using MACS SmartStrainers, 70 μm (Miltenyi Biotec, 130-098-462) and the cells were washed with DMEM by centrifugation at 300xg for 7 minutes. The viable cells were isolated from the cell suspension using the Dead Cell Removal Kit (Miltenyi Biotec, 130-090-101) as per kit protocol. The cells were then washed and re-suspended in PEB buffer containing phosphate buffered saline (PBS), pH 7.2, 2 mM EDTA (autoMACS® Rinsing Solution, Miltenyi Biotec, 130-091-222), and 0.5% bovine serum albumin (MACS® BSA Stock Solution, Miltenyi Biotec, 130-091-376).
The CD45+ immune cells were isolated from the above viable cell suspension by staining the cells with CD45 (TIL) MicroBeads, human (Miltenyi Biotec, 130-118-780) for 15 minutes at 4°C. The stained cells were passed through LS Columns (Miltenyi Biotec, 130-042-401) in the presence of magnetic field (QuadroMACS Separator, Miltenyi Biotec, 130-090-976) to remove CD45- cells.
The CD45+ cells were then eluted from column using PEB buffer, after taking out the LS column from magnetic field. The cells were then pelleted by centrifugation, re-suspended in phosphate buffered saline (PBS), pH 7.2, 2 mM EDTA, and 0.04% bovine serum albumin, and immediately submitted to the single cell core facility at Mayo Clinic.
Single-cell RNA
Cell capture and library preparation
The CD45+ cells were counted and measured for viability using Vi-Cell XR Cell Viability Analyzer (Beckman-Coulter). The barcoded Gel Beads were thawed, and the cDNA master mix was prepared using Chromium Single Cell 3’ v3 library kit (10x Genomics) as per manufacturer’s instructions. Based on the desired number of cells to be captured, a volume of live cells was mixed with the cDNA master mix. The cell suspension and master mix, thawed Gel Beads and partitioning oil were added to a Chromium Single Cell B chip. The filled chip was loaded into the Chromium Controller, where each sample was processed and the individual cells within the sample were captured into uniquely labeled Gel Beads-In-Emulsion (GEMs). The GEMs were collected from the chip and taken to the bench for reverse transcription, GEM dissolution, and cDNA clean-up. The resulting cDNA contained a pool of uniquely barcoded molecules. A portion of the cleaned and measured pooled cDNA continued on to library construction, where standard Illumina sequencing primers and a unique i7 Sample index were added to each cDNA pool. All cDNA pools and resulting libraries were measured using Qubit High Sensitivity assays (Thermo Fisher Scientific), Agilent Bioanalyzer High Sensitivity chips (Agilent) and Kapa DNA Quantification reagents (Kapa Biosystems).
Sequencing
Libraries were sequenced at 60,000 fragment reads per cell following Illumina’s standard protocol using the Illumina cBot and HiSeq 3000/4000 PE Cluster Kit. The flow cells were sequenced as 100 X 2 paired end reads on an Illumina HiSeq 4000 using HiSeq 3000/4000 sequencing kit and HCS v3.3.52 collection software. Base-calling was performed using Illumina’s RTA version 2.7.3.
Alignment and feature quantification
Raw BCL files were demultiplexed into fastq sequencing files using the Cell Ranger version 3.1 suite from 10xGenomics. Cell Ranger was then used to produce aligned bam files and a raw feature count matrix for each sample. The raw feature count matrix was normalized across patients using the R package Seurat. Events were required to contain ≥200 features and less than 40% mtRNA counts of all RNA counts. Features were filtered by requiring at least 3 cells to have identified a feature. Normalized read counts were converted to logTP10K+1 expression values.
Quality control
For the 20 samples, 9 were processed on the same day as collection and 11 were processed on the day following collection. Hierarchical-all-against-all (HAllA) clustering was performed using the 10X Genomics QC metrics as the x-dataset and the processing date group (same day or next day) as the y-dataset. An FDR cut-off of 0.05 was used to determine significance of processing date and QC metric comparison.
Quantification and statistical analysis
Classification by cell type
Unique barcodes corresponding to individual cells were classified using sets of genes characteristic to cell types.2 Two levels of cell definitions from Broad database were used, each with cell-type-gene associations, one containing 31 more detailed cell types (M.Mast, T.CD4, CD69- Mast, CD4+ PD1+, Cycling Monocytes, CD4+ Memory, Cycling T, B.Bcells, CD8+ IL17+, M.Myeloid, M.DCs, Tregs, CD8+ IELs, Follicular, T.Tcells, ILCs, DC2, Inflammatory Monocytes, GC, DC1, Macrophages, CD4+ Activated Fos-hi, NKs, CD8+ LP, M.Monocytes, Cycling B Plasma, CD4+ Activated Fos-lo, T.CD8, CD69+ Mast, MT-hi) and a second containing 11 reduced cell types (B cells, CD4+ T cells, CD8+ T cells, Cycling B cells, DCs, Follicular, ILCs, Macrophages, Mast cells, Monocytes, Natural Killers). The strategy for reduction of 31 to 11 clusters is outlined in Table S2. We removed genes that were assigned to multiple cell types to get a list of unique genes for each cell type. The Z-score of expression for each gene for each cell type was calculated as the number of standard deviations from the mean of all cells. Cells were then clustered using the R package Rphenograph (from cytofkit v1.48) based only on the genes in the unique lists. Finally, classification occurred by assigning the cell type to the cluster with the largest proportion of significant genes associated with a specific cell type. The expression of known canonical genes (Table S4) for each cell type was used to verify that classification was successful.
Comparison to immune cell clusters from Tabula Sapiens cohort
All cells were submitted for unsupervised clustering using the Seurat package. The expression Z-score for each gene was calculated across the 27 clusters to identify genes enriched or repressed in each cluster. The tissue-specific, single-cell expression database Tabula Sapiens (https://tabula-sapiens-portal.ds.czbiohub.org/) was used to identify the top differentially expressed genes for each cell type in small intestine (B cells, CD4+ T cells, CD8+ T cells, Cycling B cells, DCs, Follicular, ILCs, Macrophages, Mast cells, Monocytes, Natural Killers). The large intestine and stomach datasets were limited in the number of defined cell types and were not utilized. The differentially expressed genes from Tabula sapiens were compared to the expression Z-scores of the clusters to define the cell type associated with each cluster. Independently, the most common cell type, as defined by the supervised gene list classification, in each cluster was compared to Tabula sapiens classification (Table S3).
Gene expression for cell types
The Z-score calculated for each gene from the log TP10K+1 expression data for each cell type was plotted on a heatmap. The order of genes was fixed to show the progression of genes across each cell type.
Expression of canonical genes
Canonical genes for myeloid or specific cell types were plotted using bubble color to represent expression and size to represent the fraction of cells with detectable expression (logTP10K+1 > 0). Expression and fraction was scaled from 0 to 1 for each gene to allow for each gene to have an appropriate visual range, with 1 representing the cell type with the highest expression and 0 with the lowest. All cell types were plotted for each gene list to show unique enrichment.
t-SNE plots of different cell groups
The clustering and distribution of cells based on gene expression for all cells, myeloid cells, T cells, B cells and marker intensity plots were performed using the Rtsne package and plotted using the ggplot2 package from R.
Proportion of cell types in gastroparesis and control cohorts
The proportion of each cell type making up the population of cells in individual patients was compared between IG and control cohorts. P-values were calculated by performing an unpaired t-test for each cell type. Individual patient data is represented along with average.
Differential gene expression analyses
Differential gene expression analyses were performed using the R package MAST (1.16.0). Each cluster was compared to all other cells, acting as the control population. Z-scores, which approximated a fold change magnitude, direction and false discovery rates were reported.
Volcano plots of differential expression
Differential gene expression data from the MAST output was plotted into volcano plots to show the relative change (Z-score from MAST) compared to the significance of expression change (-log10 P-value). The x-axis represent a Z-score . The y-axis represent a -log10 FDR-value. Key, biologically relevant genes were displayed via text.
Violin plots of differential expression
The cell-level distribution of expression (logTPK10+1) between IG and control cells was plotted for the biologically relevant genes highlighted in the volcano plots.
Gene set enrichment analyses
GSEA software (version 4.0.0) was used. The output from MAST was used to calculate the GSEA pre-rank as the -log10(p-value)∗sign(Zscore). Pre-ranked lists were generated for each cell type, and each myeloid unsupervised cluster. Ribosomal genes were omitted from the pre-ranked list. Biologically relevant pathways were highlighted.
Unsupervised clustering of macrophages
Unsupervised clustering was performed on macrophage called cells (CD45+HLA-DR+CD14+) using Rphenograph (R package v0.99.1) with default options and using all genes as an input. The fraction of control or gastroparesis cells contributing to each cluster was calculated by summation of all cells associated with patients from each group, normalized for each group’s initial contribution to the total number of macrophage cells. The Z-score expression for each gene across the 17 identified clusters was calculated based on expression distribution of all macrophage cells, such that a positive Z-score represented increased expression of a gene in a cluster and a negative Z-score represented reduced expression of a gene in a cluster. Comparison of clusters to previously identified macrophage subsets from Domanska et. al. was performed by identifying clusters that expressed reported subset-specific genes. Clusters without a corresponding subset were analyzed as separate subsets.
Gene ontology term enrichment
R package ClusterProfiler v4.4.1 (https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html) was utilized to perform gene ontology enrichment for the category of biological process.67 For the respective scRNA-seq clusters, enrichment data were obtained using the built-in enrichGO function referencing the R package “org.Hs.eg.db” v3.17 (https://bioconductor.org/packages/release/data/annotation/html/org.Hs.eg.db.html) and Benjamini-Hochberg FDR set to < 0.05. R package GGplot2 v3.4.3 (https://cran.r-project.org/web/packages/ggplot2/index.html) was used to visualize the resulting GO terms.68
Study approval
The study was approved by Institutional Review Boards at the University of Louisville, Temple University, and Mayo Clinic. A written informed consent was obtained from all patients prior to participation.
Acknowledgments
FundingSources: U01DK112194 (R.S., B.C.), U01DK073983 (P.P.), U01DK112193 (B.K.), U01DK073975 (H.P.), U01DK074035 (R.M., I.S.), U01DK074007 (T.A.), U01DK073974 (K.K.), U01DK074008 (M.G., G.F., J.T.). M.G. and G.F. are supported by DK127992. M.G. is also supported by DK127998.
Authors thank Ms. Kristy Zodrow for administrative assistance.
Author contributions
LLC: Collected and analyzed the data, interpreted the results of experiments, and edited and revised the article.
EJ: Collected and analyzed the data, interpreted the results of experiments, and edited and revised the article.
∗The co-authorship order was determined considering the involvement in tissue collection, processing, cell isolation, sequencing (LLC) and bioinformatics, generation of figures (EJ).
CEB: Collected the data and edited and revised the article.
WKEP: Interpreted the results of experiments and edited and revised the article.
MBL: Collected the data and edited and revised the article.
GC: Interpreted the results of experiments and edited and revised the article.
SRP: Interpreted the results of experiments and edited and revised the article.
YI: Collected and analyzed the data and edited and revised the article.
SA: Collected and analyzed the data and edited and revised the article.
LW: Collected the data and edited and revised the article.
KLK: Interpreted the results of experiments and edited and revised the article.
BK: Interpreted the results of experiments and edited and revised the article.
RJS: Interpreted the results of experiments and edited and revised the article.
BPC: Interpreted the results of experiments and edited and revised the article.
TJM: Collected the data, interpreted the results of experiments, and edited and revised the article.
TAK: Collected data, interpreted the results of experiments, and edited and revised the article.
JT: Interpreted the results of experiments and edited and revised the article.
FAH: Interpreted the results of experiments and edited and revised the article.
IS: Interpreted the results of experiments and edited and revised the article.
RM: Interpreted the results of experiments and edited and revised the article.
HPP: Collected the data and edited and revised the article.
PJP: Interpreted the results of experiments and edited and revised the article.
TLA: Collected the data, interpreted the results of experiments, and edited and revised the article.
GF: Interpreted the results of experiments and edited and revised the article.
SD: Analyzed the data, interpreted the results of experiments, prepared figures, and edited and revised the article.
MG: Conceived and designed the research, analyzed the data, interpreted the results of experiments, prepared figures, drafted the article, and edited and revised the article.
All authors approved final version of the article.
Declaration of interests
Madhusudan Grover has received investigator-initiated grant funding from Takeda pharmaceuticals and clinical trial funding from Alexza and Donga pharmaceuticals. He serves as a consultant for Alfasigma, Ardelyx and Evoke pharmaceuticals. Bruno P. Chumpitazi receives research funding from Cairn Diagnostics, is a consultant for Ironwood Pharmaceuticals, and receives royalties from the Rome Foundation. These entities did not support the presented work. No disclosures for other co-authors.
Published: January 23, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.108991.
Supplemental information
Z scores represent the enrichment of gene expression relative to other cell types.
Proportion of cells belong to the supervised 11 clusters between the supervised and unsupervised calling.
References
- 1.Martin J.C., Chang C., Boschetti G., Ungaro R., Giri M., Grout J.A., Gettler K., Chuang L.S., Nayar S., Greenstein A.J., et al. Single-Cell Analysis of Crohn's Disease Lesions Identifies a Pathogenic Cellular Module Associated with Resistance to Anti-TNF Therapy. Cell. 2019;178:1493–1508.e20. doi: 10.1016/j.cell.2019.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smillie C.S., Biton M., Ordovas-Montanes J., Sullivan K.M., Burgin G., Graham D.B., Herbst R.H., Rogel N., Slyper M., Waldman J., et al. Intra- and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell. 2019;178:714–730.e22. doi: 10.1016/j.cell.2019.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bujko A., Atlasy N., Landsverk O.J.B., Richter L., Yaqub S., Horneland R., Øyen O., Aandahl E.M., Aabakken L., Stunnenberg H.G., et al. Transcriptional and functional profiling defines human small intestinal macrophage subsets. J. Exp. Med. 2018;215:441–458. doi: 10.1084/jem.20170057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vanner S.J., Greenwood-Van Meerveld B., Mawe G.M., Shea-Donohue T., Verdu E.F., Wood J., Grundy D. Fundamentals of Neurogastroenterology: Basic Science. Gastroenterology. 2016;150:1280–1291. doi: 10.1053/j.gastro.2016.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller P.A., Koscsó B., Rajani G.M., Stevanovic K., Berres M.L., Hashimoto D., Mortha A., Leboeuf M., Li X.M., Mucida D., et al. Crosstalk between muscularis macrophages and enteric neurons regulates gastrointestinal motility. Cell. 2014;158:300–313. doi: 10.1016/j.cell.2014.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matheis F., Muller P.A., Graves C.L., Gabanyi I., Kerner Z.J., Costa-Borges D., Ahrends T., Rosenstiel P., Mucida D. Adrenergic Signaling in Muscularis Macrophages Limits Infection-Induced Neuronal Loss. Cell. 2020;180:64–78.e16. doi: 10.1016/j.cell.2019.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farro G., Stakenborg M., Gomez-Pinilla P.J., Labeeuw E., Goverse G., Di Giovangiulio M., Stakenborg N., Meroni E., D'Errico F., Elkrim Y., et al. CCR2-dependent monocyte-derived macrophages resolve inflammation and restore gut motility in postoperative ileus. Gut. 2017;66:2098–2109. doi: 10.1136/gutjnl-2016-313144. [DOI] [PubMed] [Google Scholar]
- 8.Cipriani G., Gibbons S.J., Miller K.E., Yang D.S., Terhaar M.L., Eisenman S.T., Ördög T., Linden D.R., Gajdos G.B., Szurszewski J.H., Farrugia G. Change in Populations of Macrophages Promotes Development of Delayed Gastric Emptying in Mice. Gastroenterology. 2018;154:2122–2136.e12. doi: 10.1053/j.gastro.2018.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cipriani G., Gibbons S.J., Verhulst P.J., Choi K.M., Eisenman S.T., Hein S.S., Ordog T., Linden D.R., Szurszewski J.H., Farrugia G. Diabetic Csf1(op/op) mice lacking macrophages are protected against the development of delayed gastric emptying. Cell. Mol. Gastroenterol. Hepatol. 2016;2:40–47. doi: 10.1016/j.jcmgh.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo J., Qian A., Oetjen L.K., Yu W., Yang P., Feng J., Xie Z., Liu S., Yin S., Dryn D., et al. TRPV4 Channel Signaling in Macrophages Promotes Gastrointestinal Motility via Direct Effects on Smooth Muscle Cells. Immunity. 2018;49:107–119.e4. doi: 10.1016/j.immuni.2018.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Viola M.F., Boeckxstaens G. Intestinal resident macrophages: Multitaskers of the gut. Neuro Gastroenterol. Motil. 2020;32 doi: 10.1111/nmo.13843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gabanyi I., Muller P.A., Feighery L., Oliveira T.Y., Costa-Pinto F.A., Mucida D. Neuro-immune Interactions Drive Tissue Programming in Intestinal Macrophages. Cell. 2016;164:378–391. doi: 10.1016/j.cell.2015.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bain C.C., Bravo-Blas A., Scott C.L., Perdiguero E.G., Geissmann F., Henri S., Malissen B., Osborne L.C., Artis D., Mowat A.M. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat. Immunol. 2014;15:929–937. doi: 10.1038/ni.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Schepper S., Verheijden S., Aguilera-Lizarraga J., Viola M.F., Boesmans W., Stakenborg N., Voytyuk I., Schmidt I., Boeckx B., Dierckx de Casterlé I., et al. Self-Maintaining Gut Macrophages Are Essential for Intestinal Homeostasis. Cell. 2018;175:400–415.e13. doi: 10.1016/j.cell.2018.07.048. [DOI] [PubMed] [Google Scholar]
- 15.Domanska D., Majid U., Karlsen V.T., Merok M.A., Beitnes A.C.R., Yaqub S., Bækkevold E.S., Jahnsen F.L. Single-cell transcriptomic analysis of human colonic macrophages reveals niche-specific subsets. J. Exp. Med. 2022;219 doi: 10.1084/jem.20211846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grover M., Farrugia G., Stanghellini V. Gastroparesis: a turning point in understanding and treatment. Gut. 2019;68:2238–2250. doi: 10.1136/gutjnl-2019-318712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grover M., Farrugia G., Lurken M.S., Bernard C.E., Faussone-Pellegrini M.S., Smyrk T.C., Parkman H.P., Abell T.L., Snape W.J., Hasler W.L., et al. Cellular changes in diabetic and idiopathic gastroparesis. Gastroenterology. 2011;140:1575–1585.e8. doi: 10.1053/j.gastro.2011.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grover M., Bernard C.E., Pasricha P.J., Lurken M.S., Faussone-Pellegrini M.S., Smyrk T.C., Parkman H.P., Abell T.L., Snape W.J., Hasler W.L., et al. Clinical-histological associations in gastroparesis: results from the Gastroparesis Clinical Research Consortium. Neuro Gastroenterol. Motil. 2012;24:531–e249. doi: 10.1111/j.1365-2982.2012.01894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bityutskiy L.P., Soykan I., McCallum R.W. Viral gastroparesis: a subgroup of idiopathic gastroparesis--clinical characteristics and long-term outcomes. Am. J. Gastroenterol. 1997;92:1501–1504. [PubMed] [Google Scholar]
- 20.Grover M., Gibbons S.J., Nair A.A., Bernard C.E., Zubair A.S., Eisenman S.T., Wilson L.A., Miriel L., Pasricha P.J., Parkman H.P., et al. Transcriptomic signatures reveal immune dysregulation in human diabetic and idiopathic gastroparesis. BMC Med. Genom. 2018;11:62. doi: 10.1186/s12920-018-0379-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grover M., Dasari S., Bernard C.E., Chikkamenahalli L.L., Yates K.P., Pasricha P.J., Sarosiek I., McCallum R., Koch K.L., Abell T.L., et al. Proteomics in gastroparesis: unique and overlapping protein signatures in diabetic and idiopathic gastroparesis. Am. J. Physiol. Gastrointest. Liver Physiol. 2019;317:G716–G726. doi: 10.1152/ajpgi.00115.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grover M., Bernard C.E., Pasricha P.J., Parkman H.P., Gibbons S.J., Tonascia J., Koch K.L., McCallum R.W., Sarosiek I., Hasler W.L., et al. Diabetic and idiopathic gastroparesis is associated with loss of CD206-positive macrophages in the gastric antrum. Neuro Gastroenterol. Motil. 2017;29 doi: 10.1111/nmo.13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernard C.E., Gibbons S.J., Mann I.S., Froschauer L., Parkman H.P., Harbison S., Abell T.L., Snape W.J., Hasler W.L., McCallum R.W., et al. Association of low numbers of CD206-positive cells with loss of ICC in the gastric body of patients with diabetic gastroparesis. Neuro Gastroenterol. Motil. 2014;26:1275–1284. doi: 10.1111/nmo.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghazi A.R., Sucipto K., Rahnavard A., Franzosa E.A., McIver L.J., Lloyd-Price J., Schwager E., Weingart G., Moon Y.S., Morgan X.C., et al. High-sensitivity pattern discovery in large, paired multiomic datasets. Bioinformatics. 2022;38:i378–i385. doi: 10.1093/bioinformatics/btac232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tabula Sapiens Consortium∗. Jones R.C., Karkanias J., Krasnow M.A., Pisco A.O., Quake S.R., Salzman J., Yosef N., Bulthaup B., Brown P., et al. The Tabula Sapiens: A multiple-organ, single-cell transcriptomic atlas of humans. Science. 2022;376 doi: 10.1126/science.abl4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen M., Giladi A., Gorki A.D., Solodkin D.G., Zada M., Hladik A., Miklosi A., Salame T.M., Halpern K.B., David E., et al. Lung Single-Cell Signaling Interaction Map Reveals Basophil Role in Macrophage Imprinting. Cell. 2018;175:1031–1044.e18. doi: 10.1016/j.cell.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 27.De Schepper S., Stakenborg N., Matteoli G., Verheijden S., Boeckxstaens G.E. Muscularis macrophages: Key players in intestinal homeostasis and disease. Cell. Immunol. 2018;330:142–150. doi: 10.1016/j.cellimm.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friebel E., Kapolou K., Unger S., Núñez N.G., Utz S., Rushing E.J., Regli L., Weller M., Greter M., Tugues S., et al. Single-Cell Mapping of Human Brain Cancer Reveals Tumor-Specific Instruction of Tissue-Invading Leukocytes. Cell. 2020;181:1626–1642.e20. doi: 10.1016/j.cell.2020.04.055. [DOI] [PubMed] [Google Scholar]
- 29.Brown C.C., Gudjonson H., Pritykin Y., Deep D., Lavallée V.P., Mendoza A., Fromme R., Mazutis L., Ariyan C., Leslie C., et al. Transcriptional Basis of Mouse and Human Dendritic Cell Heterogeneity. Cell. 2019;179:846–863.e24. doi: 10.1016/j.cell.2019.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moon T.C., St Laurent C.D., Morris K.E., Marcet C., Yoshimura T., Sekar Y., Befus A.D. Advances in mast cell biology: new understanding of heterogeneity and function. Mucosal Immunol. 2010;3:111–128. doi: 10.1038/mi.2009.136. [DOI] [PubMed] [Google Scholar]
- 31.Richter L., Landsverk O.J.B., Atlasy N., Bujko A., Yaqub S., Horneland R., Øyen O., Aandahl E.M., Lundin K.E.A., Stunnenberg H.G., et al. Transcriptional profiling reveals monocyte-related macrophages phenotypically resembling DC in human intestine. Mucosal Immunol. 2018;11:1512–1523. doi: 10.1038/s41385-018-0060-1. [DOI] [PubMed] [Google Scholar]
- 32.Eisenman S.T., Gibbons S.J., Verhulst P.J., Cipriani G., Saur D., Farrugia G. Tumor necrosis factor alpha derived from classically activated "M1" macrophages reduces interstitial cell of Cajal numbers. Neuro Gastroenterol. Motil. 2017;29 doi: 10.1111/nmo.12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji S., Traini C., Mischopoulou M., Gibbons S.J., Ligresti G., Faussone-Pellegrini M.S., Sha L., Farrugia G., Vannucchi M.G., Cipriani G. Muscularis macrophages establish cell-to-cell contacts with telocytes/PDGFRalpha-positive cells and smooth muscle cells in the human and mouse gastrointestinal tract. Neuro Gastroenterol. Motil. 2021;33 doi: 10.1111/nmo.13993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dreger H., Ludwig A., Weller A., Baumann G., Stangl V., Stangl K. Epigenetic suppression of iNOS expression in human endothelial cells: A potential role of Ezh2-mediated H3K27me3. Genomics. 2016;107:145–149. doi: 10.1016/j.ygeno.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 35.Chakarov S., Lim H.Y., Tan L., Lim S.Y., See P., Lum J., Zhang X.M., Foo S., Nakamizo S., Duan K., et al. Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science. 2019;363 doi: 10.1126/science.aau0964. [DOI] [PubMed] [Google Scholar]
- 36.Tait Wojno E.D., Hunter C.A., Stumhofer J.S. The Immunobiology of the Interleukin-12 Family: Room for Discovery. Immunity. 2019;50:851–870. doi: 10.1016/j.immuni.2019.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thibodaux R.J., Triche M.W., Espinoza L.R. Ustekinumab for the treatment of psoriasis and psoriatic arthritis: a drug evaluation and literature review. Expet Opin. Biol. Ther. 2018;18:821–827. doi: 10.1080/14712598.2018.1492545. [DOI] [PubMed] [Google Scholar]
- 38.Ramos G.P., Faubion W.A., Papadakis K.A. Targeting Specific Immunologic Pathways in Crohn's Disease. Gastroenterol. Clin. N. Am. 2017;46:577–588. doi: 10.1016/j.gtc.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 39.Darling T.K., Lamb T.J. Emerging Roles for Eph Receptors and Ephrin Ligands in Immunity. Front. Immunol. 2019;10:1473. doi: 10.3389/fimmu.2019.01473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Saint-Vis B., Bouchet C., Gautier G., Valladeau J., Caux C., Garrone P. Human dendritic cells express neuronal Eph receptor tyrosine kinases: role of EphA2 in regulating adhesion to fibronectin. Blood. 2003;102:4431–4440. doi: 10.1182/blood-2003-02-0500. [DOI] [PubMed] [Google Scholar]
- 41.Rissoan M.C., Duhen T., Bridon J.M., Bendriss-Vermare N., Péronne C., de Saint Vis B., Brière F., Bates E.E.M. Subtractive hybridization reveals the expression of immunoglobulin-like transcript 7, Eph-B1, granzyme B, and 3 novel transcripts in human plasmacytoid dendritic cells. Blood. 2002;100:3295–3303. doi: 10.1182/blood-2002-02-0638. [DOI] [PubMed] [Google Scholar]
- 42.Sharfe N., Freywald A., Toro A., Dadi H., Roifman C. Ephrin stimulation modulates T cell chemotaxis. Eur. J. Immunol. 2002;32:3745–3755. doi: 10.1002/1521-4141(200212)32:12<3745::AID-IMMU3745>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 43.Sierra J.R., Corso S., Caione L., Cepero V., Conrotto P., Cignetti A., Piacibello W., Kumanogoh A., Kikutani H., Comoglio P.M., et al. Tumor angiogenesis and progression are enhanced by Sema4D produced by tumor-associated macrophages. J. Exp. Med. 2008;205:1673–1685. doi: 10.1084/jem.20072602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levine J.H., Simonds E.F., Bendall S.C., Davis K.L., Amir E.a.D., Tadmor M.D., Litvin O., Fienberg H.G., Jager A., Zunder E.R., et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor V., Welcher A.A., Program A.E., Suter U. Epithelial membrane protein-1, peripheral myelin protein 22, and lens membrane protein 20 define a novel gene family. J. Biol. Chem. 1995;270:28824–28833. doi: 10.1074/jbc.270.48.28824. [DOI] [PubMed] [Google Scholar]
- 46.Cahill T.J., Sun X., Ravaud C., Villa Del Campo C., Klaourakis K., Lupu I.E., Lord A.M., Browne C., Jacobsen S.E.W., Greaves D.R., et al. Tissue-resident macrophages regulate lymphatic vessel growth and patterning in the developing heart. Development. 2021;148 doi: 10.1242/dev.194563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi K.M., Kashyap P.C., Dutta N., Stoltz G.J., Ordog T., Shea Donohue T., Bauer A.J., Linden D.R., Szurszewski J.H., Gibbons S.J., Farrugia G. CD206-positive M2 macrophages that express heme oxygenase-1 protect against diabetic gastroparesis in mice. Gastroenterology. 2010;138:2399–2409.e1. doi: 10.1053/j.gastro.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cipriani G., Terhaar M.L., Eisenman S.T., Ji S., Linden D.R., Wright A.M., Sha L., Ordog T., Szurszewski J.H., Gibbons S.J., Farrugia G. Muscularis Propria Macrophages Alter the Proportion of Nitrergic but Not Cholinergic Gastric Myenteric Neurons. Cell. Mol. Gastroenterol. Hepatol. 2019;7:689–691.e4. doi: 10.1016/j.jcmgh.2019.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swisher J.F.A., Khatri U., Feldman G.M. Annexin A2 is a soluble mediator of macrophage activation. J. Leukoc. Biol. 2007;82:1174–1184. doi: 10.1189/jlb.0307154. [DOI] [PubMed] [Google Scholar]
- 50.Qureshi N., Morrison D.C., Reis J. Proteasome protease mediated regulation of cytokine induction and inflammation. Biochim. Biophys. Acta. 2012;1823:2087–2093. doi: 10.1016/j.bbamcr.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kalaora S., Lee J.S., Barnea E., Levy R., Greenberg P., Alon M., Yagel G., Bar Eli G., Oren R., Peri A., et al. Immunoproteasome expression is associated with better prognosis and response to checkpoint therapies in melanoma. Nat. Commun. 2020;11:896. doi: 10.1038/s41467-020-14639-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zeng Y., Hahn S., Stokes J., Hoffman E.A., Schmelz M., Proytcheva M., Chernoff J., Katsanis E. Pak2 regulates myeloid-derived suppressor cell development in mice. Blood Adv. 2017;1:1923–1933. doi: 10.1182/bloodadvances.2017007435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weiss-Haljiti C., Pasquali C., Ji H., Gillieron C., Chabert C., Curchod M.L., Hirsch E., Ridley A.J., Hooft van Huijsduijnen R., Camps M., Rommel C. Involvement of phosphoinositide 3-kinase gamma, Rac, and PAK signaling in chemokine-induced macrophage migration. J. Biol. Chem. 2004;279:43273–43284. doi: 10.1074/jbc.M402924200. [DOI] [PubMed] [Google Scholar]
- 54.Guo W., Guo T., Zhou Q., Long Y., Luo M., Shen Q., Duan W., Ouyang X., Peng H. Cofilin-1 promotes fibrocyte differentiation and contributes to pulmonary fibrosis. Biochem. Biophys. Res. Commun. 2021;565:43–49. doi: 10.1016/j.bbrc.2021.05.085. [DOI] [PubMed] [Google Scholar]
- 55.Tu Y., Zhang L., Tong L., Wang Y., Zhang S., Wang R., Li L., Wang Z. EFhd2/swiprosin-1 regulates LPS-induced macrophage recruitment via enhancing actin polymerization and cell migration. Int. Immunopharm. 2018;55:263–271. doi: 10.1016/j.intimp.2017.12.030. [DOI] [PubMed] [Google Scholar]
- 56.Malyshev I., Malyshev Y. Current Concept and Update of the Macrophage Plasticity Concept: Intracellular Mechanisms of Reprogramming and M3 Macrophage "Switch" Phenotype. BioMed Res. Int. 2015;2015 doi: 10.1155/2015/341308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fung E., Tang S.M.T., Canner J.P., Morishige K., Arboleda-Velasquez J.F., Cardoso A.A., Carlesso N., Aster J.C., Aikawa M. Delta-like 4 induces notch signaling in macrophages: implications for inflammation. Circulation. 2007;115:2948–2956. doi: 10.1161/CIRCULATIONAHA.106.675462. [DOI] [PubMed] [Google Scholar]
- 58.Liu J., Guan X., Tamura T., Ozato K., Ma X. Synergistic activation of interleukin-12 p35 gene transcription by interferon regulatory factor-1 and interferon consensus sequence-binding protein. J. Biol. Chem. 2004;279:55609–55617. doi: 10.1074/jbc.M406565200. [DOI] [PubMed] [Google Scholar]
- 59.Krausgruber T., Blazek K., Smallie T., Alzabin S., Lockstone H., Sahgal N., Hussell T., Feldmann M., Udalova I.A. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat. Immunol. 2011;12:231–238. doi: 10.1038/ni.1990. [DOI] [PubMed] [Google Scholar]
- 60.Mukai M., Suruga N., Saeki N., Ogawa K. EphA receptors and ephrin-A ligands are upregulated by monocytic differentiation/maturation and promote cell adhesion and protrusion formation in HL60 monocytes. BMC Cell Biol. 2017;18:28. doi: 10.1186/s12860-017-0144-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jellinghaus S., Poitz D.M., Ende G., Augstein A., Weinert S., Stütz B., Braun-Dullaeus R.C., Pasquale E.B., Strasser R.H. Ephrin-A1/EphA4-mediated adhesion of monocytes to endothelial cells. Biochim. Biophys. Acta. 2013;1833:2201–2211. doi: 10.1016/j.bbamcr.2013.05.017. [DOI] [PubMed] [Google Scholar]
- 62.Braun J., Hoffmann S.C., Feldner A., Ludwig T., Henning R., Hecker M., Korff T. Endothelial cell ephrinB2-dependent activation of monocytes in arteriosclerosis. Arterioscler. Thromb. Vasc. Biol. 2011;31:297–305. doi: 10.1161/ATVBAHA.110.217646. [DOI] [PubMed] [Google Scholar]
- 63.Pfaff D., Héroult M., Riedel M., Reiss Y., Kirmse R., Ludwig T., Korff T., Hecker M., Augustin H.G. Involvement of endothelial ephrin-B2 in adhesion and transmigration of EphB-receptor-expressing monocytes. J. Cell Sci. 2008;121:3842–3850. doi: 10.1242/jcs.030627. [DOI] [PubMed] [Google Scholar]
- 64.Blériot C., Chakarov S., Ginhoux F. Determinants of Resident Tissue Macrophage Identity and Function. Immunity. 2020;52:957–970. doi: 10.1016/j.immuni.2020.05.014. [DOI] [PubMed] [Google Scholar]
- 65.Martinez F.O., Gordon S., Locati M., Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J. Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 66.Rao S.S.C., Camilleri M., Hasler W.L., Maurer A.H., Parkman H.P., Saad R., Scott M.S., Simren M., Soffer E., Szarka L. Evaluation of gastrointestinal transit in clinical practice: position paper of the American and European Neurogastroenterology and Motility Societies. Neuro Gastroenterol. Motil. 2011;23:8–23. doi: 10.1111/j.1365-2982.2010.01612.x. [DOI] [PubMed] [Google Scholar]
- 67.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wickham H. Springer-Verlag; 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Z scores represent the enrichment of gene expression relative to other cell types.
Proportion of cells belong to the supervised 11 clusters between the supervised and unsupervised calling.
Data Availability Statement
-
•
The gene expression data has been uploaded to Gene expression Omnibus (GEO; accession number: GSE252126).
-
•
Custom codes are uploaded to Zenodo (https://zenodo.org/records/10366426).
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.