Abstract
A “universal strategy” replacing the full-length CFTR cDNA may treat >99% of people with cystic fibrosis (pwCF), regardless of their specific mutations. Cas9-based gene editing was used to insert the CFTR cDNA and a truncated CD19 (tCD19) enrichment tag at the CFTR locus in airway basal stem cells. This strategy restores CFTR function to non-CF levels. Here, we investigate the safety of this approach by assessing genomic and regulatory changes after CFTR cDNA insertion. Safety was first assessed by quantifying genetic rearrangements using CAST-seq. After validating restored CFTR function in edited and enriched airway cells, the CFTR locus open chromatin profile was characterized using ATAC-seq. The regenerative potential and differential gene expression in edited cells was assessed using scRNA-seq. CAST-seq revealed a translocation in ∼0.01% of alleles primarily occurring at a nononcogenic off-target site and large indels in 1% of alleles. The open chromatin profile of differentiated airway epithelial cells showed no appreciable changes, except in the region corresponding to the CFTR cDNA and tCD19 cassette, indicating no detectable changes in gene regulation. Edited stem cells produced the same types of airway cells as controls with minimal alternations in gene expression. Overall, the universal strategy showed minor undesirable genomic changes.
Keywords: MT: RNA/DNA Editing, cystic fibrosis, CFTR super-exon, CFTR gene regulation, airway basal cell differentiation, genetic rearrangements, airway stem cell therapy, CFTR genome editing, universal CFTR correction
Graphical abstract

Vaidyanathan and colleagues characterize the safety of a gene-corrected airway stem cell therapy to treat cystic fibrosis. They have inserted the CFTR cDNA into the CFTR locus to restore CFTR function. They have characterized undesirable genetic alterations, CFTR gene regulation, and airway stem cell differentiation after gene insertion.
Introduction
Cystic fibrosis (CF) is a life-limiting genetic disorder caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene which encodes a Cl−/HCO3− ion channel. CF is a systemic disorder that affects many organs and causes pancreatic insufficiency, infertility, and chronic lung infections that result in lung failure. Efforts to treat CF have focused mostly on the lungs since lung failure caused by repeated infections is the leading cause of death in people with CF (pwCF).1 Recently, a combination of small-molecule modulators (elexacaftor/tezacaftor/ivacaftor) that facilitate CFTR folding and potentiation have dramatically improved the prognosis of pwCF with at least one copy of the F508del mutation who account for ∼90% of known pwCF.2,3 However, several hundred CF-causing variants have been reported in CFTR, and the prevalence of these mutations varies widely in different races.4 In contrast to White pwCF, only 30%–70% of pwCF from other races carry the F508del mutation in at least one allele.4,5 Even after accounting for other variants responsive to modulators, 30%–50% of pwCF from other races still need a new therapeutic option.5,6 Thus, new therapies that durably restore CFTR function in a mutation-agnostic manner hold the promise for a one-time treatment for all pwCF and thereby promote health equity.
The development of CRISPR-Cas9-based genome editing enables the precise correction of CF-causing variants and thus offers the promise of a durable therapy if the correction is achieved in airway stem cells.7 Cas9 complexed to the appropriate single-guide RNA (sgRNA) can be used to induce a double-stranded break (DSB) at a targeted locus in the genome.7 This DSB is repaired using either nonhomologous end joining or homologous recombination (HR). The HR repair process can be used to correct mutations or insert gene replacement cassettes if a template DNA (HR template) with homology to the DSB site is supplied. Multiple studies have reported the precise correction of CF-causing mutations in airway stem cells, intestinal organoids, and induced pluripotent stem cells using this approach.8,9,10,11 In addition, both the partial and complete replacement of the CFTR cDNA have been reported in airway stem cells.12,13 The insertion of the partial CFTR cDNA in intron 8 partially restored CFTR function and did not alter the open chromatin profile of the CFTR locus in differentiated airway cells.13 In a previous study, the full-length CFTR cDNA along with an enrichment cassette expressing truncated CD19 (tCD19) was inserted into exon 1 of CFTR. This mutation-agnostic universal CFTR correction strategy (universal strategy) restored CFTR function in differentiated cells to levels seen in non-CF controls.12 In addition, the study further reported the lack of significant off-target editing when editing was performed using high-fidelity Cas9 and no enrichment of cells with oncogenic mutations. Although these results are promising, the gene editing process can result in genomic rearrangements such as on-target insertions-deletions (indels) and translocations, which pose safety concerns.14,15,16,17 In addition, both intronic and extragenic enhancers interact with the CFTR promoter to control the regulation of CFTR expression in a cell-type-dependent manner.18,19,20 The impact of inserting the CFTR cDNA in exon 1 on the chromatin architecture of the CFTR locus in airway cells is still unknown. Lastly, there is a concern that the editing process can alter the regenerative potential of the corrected airway basal cells. Our objective in this study was to characterize (1) the incidence of chromosomal rearrangements at the target site, (2) the chromatin architecture of the CFTR locus in edited airway cells after differentiation, and (3) the regenerative potential of the edited airway stem cells.
Chromosomal aberrations analysis by single targeted ligation-mediated PCR sequencing (chromosomal aberrations analysis by single targeted linker-mediated PCR sequencing [CAST-seq]) was recently reported to enable the detection of off-target mediated translocations, large deletions, insertions, inversions, and homology-mediated translocations.17 We used CAST-seq to quantify the incidence of chromosomal aberrations in response to DSBs mediated by Cas9 activity in exon 1 of CFTR. Assay for transposase-accessible chromatin using sequencing (ATAC-seq) is commonly used to investigate chromatin accessibility. The technique has been used to characterize the chromatin architecture of the CFTR locus in unmodified and genome-edited airway cells.13 Omni-ATAC-seq was used to characterize the chromatin architecture of the CFTR locus in differentiated airway cells obtained from airway basal cells edited using the universal strategy. Finally, we characterized the regenerative potential of edited airway stem cells by evaluating the different cell types generated upon differentiation in the air-liquid interface (ALI) using single-cell RNA-seq (scRNA-seq).
Results
Genome editing does not induce significant undesired chromosomal rearrangements
Upper airway basal cells (UABCs) from four different donors were edited using Cas9 and sgRNA alone with no HR template. A 400-bp region spanning exon 1 was amplified, and Cas9 activity was measured by quantifying the percentage of alleles with indels. Indels were observed in 97% ± 2% of alleles by Sanger sequencing and a +1 insertion was observed in ∼90% of modified cells. CAST-seq was performed on these edited cells obtained from four donors in two sequencing orientations to quantify aberrant events such as large indels and translocations (Figure 1A). The most frequently observed outcomes were large indels >50 bp in length which accounted for ∼1% of alleles in UABCs from all four donors (Figure 1B). A translocation with chromosome 5 was observed in 0.001%–0.01% of alleles in all of the donors (Figure 1B). This is an off-target mediated translocation since the region in chromosome 5 corresponds to a known off-target that was previously reported (Figures 1B and 1C).12 Additional translocations within chromosome 7 were reported in ≤0.001% of alleles. However, they were not reproducible between donors. Figure 1D maps the events to the genome visually and indicates translocations with the homologous chromosome, inversions, and deletions that are up to 15 kb away from the target site. Nevertheless, these events cumulatively accounted for only ∼1% of alleles. Thus, the reagents used to target the CFTR locus result in minimal undesired changes in the genome.
Figure 1.
Evaluation of genomic rearrangements using CAST-seq
(A) Scheme depicting CAST-seq strategy. A prey primer binding to the linker and a bait binding the CFTR locus amplify aberrant alleles that do not bind the decoy primers. These amplicons are sequenced to characterize aberrant alleles formed in response to Cas9/sgRNA-induced DSBs. The process is also repeated in the reverse orientation. (B) UABCs from 4 different donors were edited using Cas9/sgRNA alone. Upon quantification, aberrant sequences amounted to only ∼1% of alleles. Apart from on-target indels over 50 bp, a translocation from chromosome 5 was observed in 0.001%–0.01% of alleles. On-target indels under 50 bp were not included. (C) Circos plot highlights the large on-target indels and translocation with chromosome 5. This corresponds to a known off-target site. The sequence alignment of the off-target site is also presented and compared with the on-target site. (D) Integrative Genomics Viewer plots show the aberrant sequences detected using CAST-seq from the ∼25-kb region surrounding the DSB induced by Cas9/sgRNA in the CFTR locus in chromosome 7.
CF human breast epithelial cells (HBECs) corrected and enriched using the universal strategy differentiate to produce differentiated epithelial sheets with restored CFTR function
HBECs from five independent donors with CF were edited using the universal strategy (Figure 2A). A total of 86% ± 11% of tCD19+ cells were observed after editing and enrichment. The edited and enriched HBECs were then differentiated upon ALI cultures. Differentiated cells maintained tCD19 expression (Figure 2B). The integration of the universal CFTR cDNA in the CFTR locus was verified using droplet digital PCR (ddPCR) for donors 4 and 5 (Figure 2C). The percentage of tCD19+ alleles was ∼50% of the tCD19+ cells measured by flow cytometry. This is consistent with monoallelic integration of the universal CFTR cDNA and tCD19 cassette. CFTR function was quantified by measuring short-circuit currents using the Ussing chamber assay (Figures 2D and 2E). A representative example of short-circuit currents measured using the Ussing chamber assay is shown in Figure 2D. Epithelial sheets generated from corrected airway basal cells displayed CFTRinh-172 sensitive short-circuit currents of 15 ± 8 μA/cm2. Unedited control CF samples showed CFTRinh-172 sensitive short-circuit currents of 0.6 ± 0.5 μA/cm2. Non-CF control samples from two different donors showed CFTRinh-172 sensitive short-circuit currents of 10 ± 4 μA/cm2. Table 1 lists CFTRinh-172 responses from individual donors. ALIs from these edited and enriched HBECs that were validated for CFTR function were then used for subsequent ATAC-seq and scRNA-seq experiments.
Figure 2.
Validation of CFTR cDNA insertion and restoration of CFTR function
(A) The universal CFTR correction strategy involves the insertion of the full-length CFTR cDNA in exon 1 of the CFTR gene, along with a cassette expressing truncated CD19 driven by a PGK promoter. (B) HBECs corrected using the universal strategy were differentiated on ALI. In this example from donor 4, differentiated airway cells maintained tCD19 expression. (C) The integration of the CFTR cDNA and tCD19 in the CFTR locus was further verified by ddPCR. The percentage of tCD19+ alleles was ∼50% of the number of tCD19+ cells. This is consistent with each cell containing one corrected allele. (D) CFTR function was measured in the differentiated epithelial sheets using the Ussing chamber assay. Representative traces obtained from epithelial sheets generated from HBECs obtained from a donor with CF before and after correction. The traces from the corrected epithelial sheets show responses to the addition of forskolin and CFTRinh-172. (E) CFTRinh-172 short-circuit currents observed in epithelial ALI cultures generated from corrected CF HBECs (5 individual subjects) are higher than those observed in control ALIs and are comparable to currents observed in non-CF controls (3 subjects).
Table 1.
Summary of percentage of editing in HBEC samples and CFTR responses in differentiated airway epithelial sheets derived from corrected HBECs
| Donor | Genotype | Editing efficiency (% tCD19+ cells) | Raw-inhibitable CFTR current μA/cm2 | Percentage of non-CF inhibitable current |
|---|---|---|---|---|
| Non-CF average: 10 ± 4 μA/cm2 | ||||
| Edited samples | ||||
| 1 | F508del/N1303K | 74.3 | 6.80 | 70 |
| 1 | F508del/N1303K | 74.3 | 25.6 | 266 |
| 1 | F508del/N1303K | 74.3 | 17.5 | 181 |
| 2 | F508del/W1282X | 89.2 | 18.9 | 196 |
| 3 | W1282X/W1282X | 97.2 | 6.30 | 65 |
| 4 | F508del/G542X | 72.7 | 21.7 | 225 |
| 5 | F508del/F508del | 56.1 | 4.60 | 47 |
| Control samples | ||||
| 1 | F508del/N1303K | 0 | 1.02 | 0 |
| 1 | F508del/N1303K | 0 | 0.96 | 0 |
| 2 | F508del/W1282X | 0 | 0.05 | 0 |
| 3 | W1282X/W1282X | 0 | 1 | 0 |
| 4 | F508del/G542X | 0 | 0.23 | 0 |
| 5 | F508del/F508del | 0 | 0.4 | 0 |
Insertion of the CFTR cDNA in exon 1 does not alter the chromatin architecture of the CFTR locus
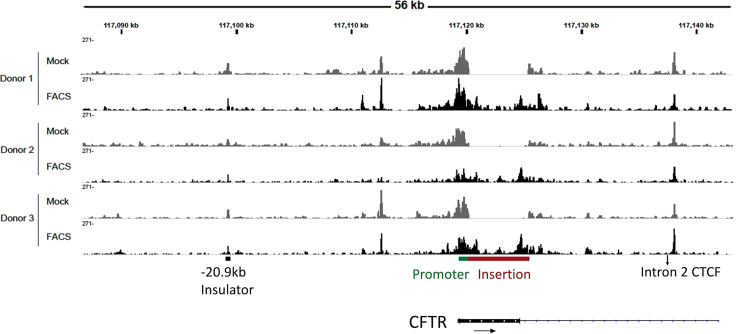
To investigate changes in the regulation of CFTR expression, we characterized the open chromatin profile of the CFTR locus using Omni-ATAC-seq. Omni-ATAC-seq was performed on control mock electroporated HBECs and HBECs corrected using the universal strategy (donors 1–3). Differentiated airway cells obtained from corrected and enriched HBECs were first validated for restored CFTR function (Figure 2; Table 1) before use in Omni-ATAC-seq. Analysis included ∼400 kb encompassing the CFTR topologically associated domain, although only a 56-kb region surrounding exon 1 of CFTR is shown in Figure 3. This includes the −20.9-kb insulator region, which has an important role in chromatin conformation across the locus.21 The chromatin accessibility at the site of the CFTR promoter was similar between control and treated epithelial sheets (Figure 3). There was increased accessibility in the region corresponding to the CFTR cDNA that is evident only in the edited and enriched cells. Of note, the broad peak of open chromatin at the 3′ end of the cDNA corresponds to the location of the 3-phosphoglycerate kinase (PGK) promoter within the construct. Apart from these changes expected from the universal strategy, there were no significant changes in the open-chromatin profile of the CFTR locus in response to gene editing using the universal strategy.
Figure 3.
Open chromatin profile of the CFTR locus was characterized in control and edited and enriched samples (labeled as FACS) from 3 different donors
No noticeable differences were observed between the samples outside the region corresponding to the CFTR cDNA and tCD19 cassette.
Epithelial sheets obtained from corrected and enriched HBECs produce the major cell types reported in airway epithelia
Epithelial sheets were generated by culturing uncorrected CF and corrected and enriched CF HBECs on ALI cultures. ALI cultures from donors 1, 4, and 5 were validated for restored CFTR function (Figure 2; Table 1) before performing scRNA-seq. Data were obtained from a total of 35,176 cells. Uniform manifold approximation and projections (UMAPs) as well as dot plots showing the relative expression of selected genes were generated from control and edited cells separately and after integration to anchor similar cell types (Figures 4A–4G and S1A–S1F). All of the samples had three clusters expressing cytokeratin 5 (KRT5). The cluster with high levels of KRT5 and TP63 were labeled as basal. The cluster with expression of TOP2A and MKI67 in addition to KRT5 and TP63 were labeled cycling basal cells. The third KRT5+ cluster with high expression of KRT13, SERPINB4, and KRT4 were labeled KRT13+ and likely correspond to the previously reported Basal3 cluster (also called suprabasal cells).22 All of the samples had one cluster that was positive for SCGB1A1 and VMO1 and were labeled secretory cells. All of the samples had one cluster that showed high expression of STATH. The cluster was labeled STATH+, and it is unclear how it corresponds to clusters reported in previous publications. All of the samples had one cluster positive for ciliated cell markers such as FOXJ1 and TPPP3. Lastly, one or two CFTR high cells that were also positive for ASCL3 and FOXI1 were observed in all of the samples. However, they were too few to be recognized as a separate cluster. In addition to these common clusters, there were clusters that were present in some but not all of the samples.
Figure 4.
UMAP from control and edited cells (after integration)
(A–C) UMAP from control and edited cells (after integration) obtained from (A) donor 1, (B) donor 4, and (C) donor 5. (D–F) Dot plot indicating genes unique to each cluster for cells from (D) donor 1, (E) donor 4, and (F) donor 5. (G) The number of cells in each cluster plotted for edited and control cells from each donor (1 dot per donor). There was no consistent change in the number of cells in each cluster across donors. The bars represent the mean number of cells in each cluster from the three samples and the error bars represent the standard deviation.
Donors 4 and 5 had a second cluster with high expression of MUC5B, and this second cluster was labeled goblet cells (Figures 4B, 4C, 4E, and 4F). Donors 1 and 4 had one cluster adjacent to secretory cells and STATH+ clusters that had lower levels of SCGB1A1 and STATH. This cluster was labeled immature secretory. Donors 1 and 5 had a cluster that was high in FOXN4 expression, with other markers reported in multiciliated cells such as CCNO, DEUP1, and HES6.23 These were labeled FOXN4+ cells. Donors 4 and 5 showed one ciliated cluster with expression of SAA1 and SAA2, which were labeled SAA+ cilia. In addition, cells from donors 1 and 5 also had one cluster of ciliated cells that had slightly lower levels of ciliated genes and expression of SCGB1A1 or KRT4. These were labeled immature cilia. Donors 1 and 5 showed a cluster positive for ciliated markers and higher levels of SCGB1A1. This cluster was labeled differentiating ciliated. Lastly, control cells from donor 1 had one cluster positive for KRT5, SCGB1A1, FOXJ1, TPPP3, and VMO. This was labeled differentiating basal.
All of the major clusters after integration contained cells from both mock and edited samples, indicating that the editing process did not prevent the differentiation of basal cells into specific cell types. Of note, however, most of the cells in the SAA cilia cluster from donor 4 and differentiating cilia cluster from donor 5 were from mock samples. However, this was unique to these donors since the edited basal cells from the other donor with these cell types (donor 5 and donor 1, respectively) still produced them. We reassembled the sequences from donor 4 to assess whether the edited cells were positive for tCD19 and the universal CFTR cDNA sequences. The universal CFTR cDNA is distinguishable from the native CFTR cDNA because it is associated with a BGH polyadenylation sequence and it is also codon diverged. Most cells were tCD19+ by scRNA-seq (Figure S2) and flow cytometry (Figure 2B). A few cells in the secretory and basal cell clusters were positive for the universal CFTR cDNA (Figure S2). The number of cells from each cluster in the mock and edited samples is presented in Figure 4G and show no consistent difference between mock and edited cells across all of the donors.
The cells from each integrated cluster were further evaluated using the FindMarkers function in Seurat to identify genes with a significant change in expression (adjusted p < 0.05) that was reproducible across two donors (Table 2). There was no gene that was differentially expressed in all three donors. All of the genes that showed significant differential expression (adjusted p < 0.05) are provided in Table S1. In many cases, the cell types in which genes showed reproducible changes in expression accounted for <10% of the transcripts for the gene. Among these genes, BPIFB1 showed reduced expression after editing in three clusters, which accounted for only ∼0.1%–5% of the BPIFB1 transcripts each. A related gene BPIFA1 also showed reduced expression in immature secretory cells from edited cells in both donors who produced this cell type. This cell type accounts for 20%–30% of BPIFA1 transcripts. Basal cells from the edited samples showed increased S100A8 expression and accounted for ∼5% of the S100A8 transcripts. Ciliated cells also showed a reduced expression of ALDH3A1 and GSTA1 after editing and account for 10%–22% of the transcripts of those genes.
Table 2.
Genes with significant change (adjusted p < 0.05) in expression that was reproducible in at least 2 donors
| Cell type/donor | Gene | Avg_log2FCa | Percentage 1 | Percentage 2 | Adjusted p value | % of total transcripts | |
|---|---|---|---|---|---|---|---|
| Basal | |||||||
| 1 | BPIFB1 | 0.28 | 0.795 | 0.333 | 0.00941571 | 0.5 | |
| 5 | BPIFB1 | 1.1 | 0.361 | 0.986 | 5.55E−22 | 0.03 | |
| 4 | S100A8 | −0.32 | 0.191 | 0.404 | 1.41E−17 | 7.0 | |
| 5 | S100A8 | −0.29 | 0.252 | 0.718 | 0.00784068 | 6.2 | |
| Cilia | |||||||
| 1 | BPIFB1 | 0.44 | 0.957 | 0.702 | 2.07E−20 | 4.7 | |
| 4 | BPIFB1 | 0.47 | 0.976 | 0.986 | 0.02828378 | 1.7 | |
| 1 | MSMB | 0.26 | 0.842 | 0.383 | 1.48E−15 | 1.0 | |
| 5 | MSMB | 0.37 | 0.409 | 0.885 | 7.19E−5 | 0.42 | |
| 1 | ALDH3A1 | 0.30 | 0.954 | 0.723 | 2.30E−8 | 20 | |
| 5 | ALDH3A1 | 0.29 | 0.994 | 1 | 0.00058844 | 9.8 | |
| 1 | GSTA1 | 0.36 | 0.788 | 0.557 | 0.00521609 | 22 | |
| 5 | GSTA1 | 0.32 | 0.934 | 0.99 | 0.01368748 | 15 | |
| KRT13 | |||||||
| 1 | SAA1 | 0.27 | 0.475 | 0.126 | 0.00790995 | 1.0 | |
| 5 | SAA1 | 0.26 | 0.251 | 0.797 | 2.94E−5 | 0.26 | |
| Immature secretory | |||||||
| 1 | BPIFA1 | 0.40 | 1 | 0.976 | 2.71E−30 | 20 | |
| 4 | BPIFA1 | 0.39 | 1 | 0.997 | 5.35E−5 | 32 | |
| Immature cilia | |||||||
| 1 | BPIFB1 | 0.28 | 1 | 0.849 | 3.39E−15 | 4.9 | |
| 5 | BPIFB1 | 0.26 | 0.981 | 0.988 | 0.01406199 | 4.1 | |
| Differentiating cilia | |||||||
| 1 | KRT14 | 0.52 | 0.428 | 0.025 | 3.20E−7 | 0.13 | |
| 5 | KRT14 | 0.36 | 0.1 | 1 | 3.19E−14 | 0.45 | |
| 1 | SLPI | 0.31 | 1 | 1 | 2.75E−27 | 14 | |
| 5 | SLPI | 1.50 | 0.991 | 0.938 | 0.02666603 | 3.9 | |
| STATH | No change in gene expression reproducible across donors | ||||||
| FOXN4 | No change in gene expression reproducible across donors | ||||||
Positive avg_log2FC indicates higher expression in control samples and negative indicates higher expression in edited samples.
Discussion
The results of this study confirm and further support our previous findings that the insertion of the CFTR cDNA in exon 1 of the CFTR locus is effective. This strategy can be used to restore CFTR function in pwCF affected by variants throughout the CFTR coding sequence. Alternative approaches may be needed for pwCF affected by biallelic mutations in the promoter region or mutations that overlap with the sgRNA site listed in Table S2. In a previous publication, we demonstrated that the use of high-fidelity Cas9 reduced off-target indels from ∼50% to ∼1% relative to wild-type Cas9.12 In terms of safety, although we detected the presence of translocations and large indels, these events are present in only ∼1% of alleles. Notably, CAST-seq was performed in the absence of the HR templates coding for CFTR and tCD19, since the episomal HR template DNA would compete in the PCR reaction with the genomic sequences, lowering the sensitivity of the assay. The translocation that was reproducible between all four donors corresponds to a previously reported off-target site in chromosome 5 associated with this sgRNA. Thus, the CAST-seq data further highlight the improved safety profile enabled by high-fidelity Cas9. Moreover, the use of Cas9 ribonucleoprotein, which is cleared from cells within a few days, further limits aberrant changes. This window is sufficient for gene correction, while limiting the time in which undesired changes can accumulate. Overall, the methods used to insert the CFTR cDNA exon 1 appears to cause minimal undesired changes.
In addition to not observing aberrant genetic changes, the process did not result in any changes in the open chromatin profile of the CFTR locus. In particular, extragenic enhancers20 at −44 and −35 kb remain unchanged. Previous studies have shown the importance of intronic and extragenic regions in the regulation of CFTR expression.18,19,20 Our approach leaves all of the introns and exons of CFTR (other than exon 1) intact. Although this approach to insert the CFTR cDNA is effective in airway cells, it needs to be validated in other epithelial cells (e.g., intestinal cells) in which the regulation of CFTR is different.18 In this approach, the PGK promoter used in the tCD19 cassette makes the chromatin more accessible. However, the impact is local and does not extend beyond the tCD19 cassette. A previous study investigated the enhancer effects of different promoters and observed PGK to have a low impact.24 Thus, our observations are consistent with previous reports.
While investigating the impact of gene editing on the regenerative potential of HBECs using scRNA-seq, we considered hallmarks of different airway cell types reported in multiple recent studies.22,23,25,26 The cell types identified in our dataset are broadly consistent with these studies. Widely characterized and recognized cell types such as basal cells, ciliated cells, suprabasal cells, secretory cells, and cycling basal cells were present in all of the samples. Consistent with previous studies, not all of the subtypes of each cell reported in tissue were found in the ALI cultures.22 In addition to previously reported cell types, there was one cluster of cells marked by the expression of STATH that was present in all of the samples. It did not overlap with cells expressing high levels of SCGB1A1 (Figure S3). STATH has been reported to be present in salivary glands and nasal and bronchial epithelia.27 In the salivary glands, STATH has been reported to be present in the serous glandular cells.28 STATH has been shown to have antibacterial activity in the oral cavity,29 and the increased expression of STATH has been reported to be associated with milder forms of CF.30 It is unclear whether these cells represent one of the previously reported secretory subtypes. In addition, all of the donors presented some cells that appeared to be in the process of transitioning into secretory or ciliated cells. They were marked by lower levels of secretory or ciliated genes. Donor 1 was unique in having many cell types that appeared to be in the process of differentiation despite being cultured on ALI for the same amount of time as the other donors.
Some cell types (e.g., FOXN4, SAA cilia) were observed in only two out of three samples. Among these was one group of ciliated cells positive for SAA1 and SAA2 present in donors 4 and 5 (SAA cilia). Carraro et al. recently reported a ciliated cluster (ciliated 3) expressing SAA1 and SAA2 when characterizing airway epithelial cells obtained freshly from tissue.22 It is likely that the SAA cilia cells correspond to this cluster. However, the expression of SAA1 and SAA2 was also observed in nonciliated cells, including some secretory cells (Figure S4). Although we detected cells with the signature of ionocytes (FOXI1+, CFTR+), they are too few to be grouped into a separate cluster. We also verified the expression of tCD19 and the codon-diverged CFTR cDNA in one edited sample (Figure S2). Most of the cells were positive for tCD19. The universal CFTR cDNA was also detectable, albeit in only a few cells. Nevertheless, all of the edited samples showed restoration of CFTR function by Ussing chamber analysis. Previous studies have indicated that a low level of CFTR expression may be sufficient for significant CFTR-mediated chloride transport, and these results are consistent with those of these previous studies.31,32 Lastly, we did not observe any pulmonary neuroendocrine cells both in the control and edited samples. Overall, the scRNA-seq results indicate that the editing process does not compromise the regenerative potential of the basal cells.
In addition to the cell types produced, we did not observe drastic changes in gene expression between edited and control samples. One challenge with assessing changes in gene expression using scRNA-seq is the high propensity for false positives.33 Therefore, we only considered genes with differential expression that was consistent across at least two donors and had an adjusted p < 0.05. Among these genes, BPIFA1, BPIFB1, and S100A8 have been previously associated with CF.22,34 All three have innate immune/antimicrobial functions. BPIFA1 and BPIFB1 have been previously reported to be upregulated in secretory cells in pwCF.22,34 The reduced expression of BPIFA1 in immature secretory cells is thus consistent with these studies. Although BPIFB1 showed reduced expression in edited samples, this change in expression was reproducible only in cell types that accounted for <10% of transcripts. Immature secretory cells from donor 1, which account for 16% of BPIFB1 transcripts, did show reduced expression. However, the change was not seen in donor 4, who also produced that cell type. The number of transcripts for both genes was lower in the secretory cell types in edited samples compared to the controls, but they were not identified as differentially expressed. Apart from BPIFA1 and BPIFB1, Carraro et al. also reported the reduced expression of S100 family proteins in pwCF.22 Thus, the increased expression of S100A8 in the edited basal cells is consistent with this report. Approximately 40% of S100A8 transcripts were present in KRT13+ cells in all three donors. KRT13+ cells from the edited samples showed increased S100A8 in two out of three donors. However, the change was not statistically significant. ALDH3A1 and GSTA1 were the other two genes that showed reduced expression in ciliated cells in at least two donors and ciliated cells accounted for 10%–20% of their transcripts. Carraro et al. reported the expression of ALDH3A1 in both secretory cells and ciliated cells and GSTA1 expression in ciliated cells. However, they did not highlight any differential expression in these genes between CF samples and controls.22 Overall, the changes in gene expression highlighted in our results are consistent with restored CFTR function.
In summary, our study investigated the safety of inserting the CFTR cDNA in exon 1 of the CFTR locus by assessing the presence of undesirable genomic changes and the impact of the cDNA insertion on the open chromatin profile of the CFTR locus and on the regenerative potential of the edited basal cells. The edited cells did not exhibit an altered open chromatin profile at the CFTR locus and were able to produce all of the cells produced by the unedited control basal cells. Translocations and large indels were limited to ∼1% of alleles. Thus, the universal strategy to insert the CFTR cDNA in exon 1 of the CFTR locus does not pose significant concerns regarding adverse genomic changes or loss of regenerative potential.
Materials and methods
Airway stem cell culture
Airway stem cells (both UABCs and human bronchial epithelial basal cells) were plated on tissue culture-treated plates in Pneumacult Ex-plus media at a density of 5,000–10,000 cells/cm2. ROCK inhibitor (Y-27632) was added to the media at a concentration of 10 μM. Plates were coated with iMatrix511-silk (recombinant laminin 511, catalog no. 892021, Nacalai USA). Cells were cultured at 37°C, 5% O2, and 5% CO2.
Genome editing of airway stem cells
Airway stem cells were edited after 4–6 days in culture. Cells were resuspended by treatment with Tryple (catalog no. 12605010, Thermo Fisher Scientific) and resuspended in OPTI-MEM (catalog no. 31985070, Thermo Fisher Scientific) at a concentration of 10 million cells/mL. A total of 30 μg of high-fidelity Cas9 (SpyFi Cas9 nuclease, Aldevron) was complexed with 16 μg sgRNA (sequence: TTCCAGAGGCGACCTCTGCA, Synthego) at room temperature for 10 min. The sgRNA used contained 2′O-methyl-3′ phosphorothioate modifications in the last three bases on either end. A total of 100 μL cells in OPTI-MEM was mixed with Cas9 and sgRNA and transferred into a Nucleocuvette (Lonza). Cells were electroporated using the Lonza 4D system with the program CA137 and buffer setting of P3. After electroporation, 400 μL OPTI-MEM was added to the cuvette. Adeno-associated viruses (AAVs) corresponding to the universal strategy were added such that there were 105 vector genomes per cell. The vector genomes were quantified using ddPCR. Cells were plated at a density of 5,000–10,000 cells/cm2 using the same conditions described above. Cells were passaged 4–7 days after editing. Edited cells were enriched 10–14 days after editing using fluorescence-activated cell sorting (FACS).
Enrichment of edited airway stem cells
Edited airway stem cells were resuspended by treatment with Tryple (catalog no. 12605010, Thermo Fisher Scientific). They were stained with anti-human CD19 antibody (HIB19 clone) conjugated with fluorescein isothiocyanate (catalog no. 302206, BioLegend) for 20–30 min at 4°C. Cells were treated with 7-AAD for 10 min. The cells were washed three times with OPTI-MEM and resuspended in FACS buffer. Edited cells were sorted using FACS Aria II SORP (BD Biosciences). Unedited cells and cells treated with AAV but no Cas9 were used to set the gates for identifying tCD19+ cells. Enriched cells were expanded for 4–5 days before differentiation upon ALI cultures.
Differentiation of airway stem cells
Enriched cells and controls were plated in 6.5-mm transwell plated with 0.4-μm pore polyester membrane inserts (catalog no. 3470, Corning) at a density of 30,000–60,000 cells/well. Cells were expanded in Pneumacult Ex-plus until they were confluent (3–7 days). After cells were confluent, they were differentiated using ALI media obtained from the Marsico Lung Institute Tissue Procurement and Cell Culture Core at the University of North Carolina for another 21–28 days. Restoration of CFTR function was validated in the differentiated ALI cultures from 2 to 3 transwells. Cells from replicate transwells were used for ATAC-seq and scRNA-seq.
Measurement of CFTR function
Ussing chamber assay was used to measure CFTR function in differentiated ALI cultures.11 CFTR function was measured in the presence of a chloride gradient using the following solutions—apical (in mM): Na(gluconate) 120, NaHCO3 25, KH2PO4 3.3, K2HPO4 0.8, Ca(gluconate)2 4, Mg(gluconate)2 1.2, and mannitol 10; basolateral (in mM): NaCl 120, NaHCO3, 25, KH2PO4 3.3, K2HPO4 0.8, CaCl2 1.2, MgCl2 1.2, and glucose 10.
The concentrations of ion channel activators and inhibitors were as follows: amiloride (10 μM, apical), forskolin (10 μM, bilateral), VX-770 (10 μM, apical), CFTRinh-172 (20 μM, apical), and uridine triphosphate (100 μM, apical).
scRNA-seq
Cells differentiated on ALI cultures were resuspended by treatment with Tryple on both apical and basolateral sides. scRNA-seq was performed using Chromium Next GEM Single Cell 3′ HT version 3.1 (10X Genomics) and sequencing was performed with MiSeq (Illumina). The data were processed using Cell Ranger 6.0.0 using GRCh38 human genome assembly. Analysis was performed using the Seurat35 pipeline in R Studio. UMAPs were generated using the NormalizeData function and SCTransform was compared. There were no noticeable differences. The data presented here were generated using the NormalizeData function. Differential gene expression was assessed using the FindMarkers function using the Wilcoxon rank-sum test. Genes with a change in expression and adjusted p < 0.05 were considered for further analysis. A custom transcriptome containing the last 93 bases of the CFTR cDNA and tCD19 was used to check transgene expression in donor 4.
The 93 bases preceding BGH polyA to detect Universal CFTR cDNA: TCCTCCAAGTGTAAGAGCAAGCCTCAGATCGCCGCCCTGAAGGAGGAGACCGAGGAGGAGGTGCAGGACACCAGACTGTAGgccccgctgatc.
The 93 bases preceding sv40 polyA to detect tCD19: CCTGCAGAGGGCCCTGGTGCTGAGGAGGAAGAGGAAGAGGATGACCGACCCCACCAGGAGGTTCTGAtaactcgagggcgcgccccgctgatc.
Omni-ATAC-seq
Differentiated cells grown on inserts were collected using Accutase, and biological replicates (n = 2) were collected from individual inserts. Omni-ATAC-seq was performed on 50,000 cells as described previously,36 with minor modifications.20 Library size distributions were visualized by TapeStation (Agilent). Libraries were quantified using the KAPA Library Quantification Kit (Roche) before pooling and sequenced on a NextSeq 550 at high output (Illumina) using 75-bp single reads at the Case Western Reserve University Genomics Core. Raw files were trimmed using Sickle37 and aligned to the hg19 or modified hg19 genomes containing the insertion sequence (except PGK) using the BWA aligner.38 Aligned reads were marked for duplicates using Picard, and coverage tracks were generated using deepTools bamCoverage.39 The analysis of ATAC-seq was done visually, which is the standard practice.
CAST-seq
UABCs from four different donors were edited using high-fidelity Cas9 and sgRNA alone. The cells were cultured for 4 days. The genomic DNA (gDNA) was purified using column purification (GeneJET genomic DNA purification kit, catalog no. K0722, Thermo Fisher Scientific). The region around exon 1 from control and edited samples was amplified using PCR and Sanger sequenced. The presence of on-target indels was verified using the Inference of CRISPR Edits analysis algorithm developed by Synthego. CAST-seq was subsequently performed on the gDNA as described previously.17 The NEB Next Ultra II FS (catalog no. E6177, NEB) was deployed to randomly shear the gDNA in fragment lengths between 200 and 700 bp that were end repaired and ligated to the linker.
The first PCR reaction was performed with Q5 polymerase and the following specific primers: CFTR-Bait: 5′-CAGAGTAGTAGGTCTTTGGC-3' and CFTR-Decoy: 5′-AAAGTTTGGAGACAACGCT-3', whereas the nested PCR used 3 μL from the previous reaction and the following specific primer: CFTR-Nested: 5′-GACTGGAGTTCAGACGTGTGCTCTTCCGATCTATTAGGAGCTTGAGCCCAGAC-3' (CFTR specific sequence in boldface type).
The amplicons were purified with 0.7× AMP pure beads (catalog no. A63882, Beckman Coulter) to remove fragments <200 bp after nested and barcoding PCR performed with NEB Next Multiplex Oligos for Illumina (catalog no. E6440L, NEB). The library was sequenced on Illumina MiSeq platform (MiSeq Reagent Kit version 2, 500 cycles, catalog no. MS-102-2003) and analyzed with the CAST-seq pipeline (https://github.com/AG-Boerries/CAST-Seq).
Data and code availability
The data supporting the findings presented in this study are available on request from the corresponding author, S.V.
Acknowledgments
M.H.P. and S.V. thank the Cystic Fibrosis Research Institute for their support through the New Horizons grant. M.H.P. thanks the Crandall Foundation for a philanthropic gift that supported this work. S.V. was supported by the Cystic Fibrosis Foundation (VAIDYA19F0;VAIDYA22A0-KB) and the NIH (R00HL151900). Research in the A.H lab was supported by the Cystic Fibrosis Foundation (Davis19XX0) and NIH HL094585 and HD068901. A.P. was supported by T32 GM008056. G.T. and A.J.T. were supported by the Wellcome Trust (217112/Z/19/Z), the NIHR Biomedical Research Centre at Great Ormond Street Hospital for Children NHS Foundation Trust, and University College London. G.T. was also supported by the University College London Therapeutic Acceleration Support fund.
Author contributions
Conceptualization, S.V., M.H.P., G.T., A.H., and A.J.T; methodology, S.V., G.T., A.H., M.H.P., J.L.K., A.P., B.L., and T.A.B.; investigation, S.V., V.S., J.L.K., A.P., B.L., and T.A.B.; writing, S.V., J.L.K., A.P., V.S., B.L., T.A.B., A.H., A.J.T., G.T., and M.H.P. All of the authors participated in the design and conception of the experiments and provided editorial feedback on the manuscript.
Declaration of interests
M.H.P. serves on the Board of Directors of Graphite Bio and the Scientific Advisory Board of Allogene and serves as an advisor to Versant Ventures. M.H.P. has equity in CRISPR Tx and has equity in and is a founder of Kamau Therapeutics. The companies had no input on the design or execution of the experiments described here and were also not involved in the data analysis or interpretation.
Declaration of generative AI and AI-assisted technologies in the writing process
During the preparation of this work, S.V. used ChatGPT4 to refine sentence structure and edit the abstract to conform to word limits. After using this tool/service, S.V. reviewed and edited the content as needed. All of the authors have reviewed the manuscript and take full responsibility for the content of the publication.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2024.102134.
Supplemental information
References
- 1.Kapnadak S.G., Dimango E., Hadjiliadis D., Hempstead S.E., Tallarico E., Pilewski J.M., Faro A., Albright J., Benden C., Blair S., et al. Cystic Fibrosis Foundation consensus guidelines for the care of individuals with advanced cystic fibrosis lung disease. J. Cyst. Fibros. 2020;19:344–354. doi: 10.1016/j.jcf.2020.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Keating D., Marigowda G., Burr L., Daines C., Mall M.A., McKone E.F., Ramsey B.W., Rowe S.M., Sass L.A., Tullis E., et al. VX-445–Tezacaftor–Ivacaftor in Patients with Cystic Fibrosis and One or Two Phe508del Alleles. N. Engl. J. Med. 2018;379:1612–1620. doi: 10.1056/NEJMoa1807120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heijerman H.G.M., McKone E.F., Downey D.G., Van Braeckel E., Rowe S.M., Tullis E., Mall M.A., Welter J.J., Ramsey B.W., McKee C.M., et al. Efficacy and safety of the elexacaftor plus tezacaftor plus ivacaftor combination regimen in people with cystic fibrosis homozygous for the F508del mutation: a double-blind, randomised, phase 3 trial. Lancet. 2019;394:1940–1948. doi: 10.1016/S0140-6736(19)32597-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schrijver I., Pique L., Graham S., Pearl M., Cherry A., Kharrazi M. The Spectrum of CFTR Variants in Nonwhite Cystic Fibrosis Patients: Implications for Molecular Diagnostic Testing. J. Mol. Diagn. 2016;18:39–50. doi: 10.1016/j.jmoldx.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Vaidyanathan S., Trumbull A.M., Bar L., Rao M., Yu Y., Sellers Z.M. CFTR genotype analysis of Asians in international registries highlights disparities in the diagnosis and treatment of Asian patients with cystic fibrosis. Genet. Med. 2022;24:2180–2186. doi: 10.1016/j.gim.2022.06.009. [DOI] [PubMed] [Google Scholar]
- 6.McGarry M.E., McColley S.A. Cystic fibrosis patients of minority race and ethnicity less likely eligible for CFTR modulators based on CFTR genotype. Pediatr. Pulmonol. 2021;56:1496–1503. doi: 10.1002/ppul.25285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Porteus M.H. A New Class of Medicines through DNA Editing. N. Engl. J. Med. 2019;380:947–959. doi: 10.1056/NEJMra1800729. [DOI] [PubMed] [Google Scholar]
- 8.Schwank G., Koo B.-K., Sasselli V., Dekkers J.F., Heo I., Demircan T., Sasaki N., Boymans S., Cuppen E., van der Ent C.K., et al. Functional Repair of CFTR by CRISPR/Cas9 in Intestinal Stem Cell Organoids of Cystic Fibrosis Patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Firth A.L., Menon T., Parker G.S., Qualls S.J., Lewis B.M., Ke E., Dargitz C.T., Wright R., Khanna A., Gage F.H., Verma I.M. Functional Gene Correction for Cystic Fibrosis in Lung Epithelial Cells Generated from Patient iPSCs. Cell Rep. 2015;12:1385–1390. doi: 10.1016/j.celrep.2015.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crane A.M., Kramer P., Bui J.H., Chung W.J., Li X.S., Gonzalez-Garay M.L., Hawkins F., Liao W., Mora D., Choi S., et al. Targeted Correction and Restored Function of the CFTR Gene in Cystic Fibrosis Induced Pluripotent Stem Cells. Stem Cell Rep. 2015;4:569–577. doi: 10.1016/j.stemcr.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaidyanathan S., Salahudeen A.A., Sellers Z.M., Bravo D.T., Choi S.S., Batish A., Le W., Baik R., de la O S., Kaushik M.P., et al. High-Efficiency, Selection-free Gene Repair in Airway Stem Cells from Cystic Fibrosis Patients Rescues CFTR Function in Differentiated Epithelia. Cell Stem Cell. 2020;26:161–171.e4. doi: 10.1016/j.stem.2019.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vaidyanathan S., Baik R., Chen L., Bravo D.T., Suarez C.J., Abazari S.M., Salahudeen A.A., Dudek A.M., Teran C.A., Davis T.H., et al. Targeted replacement of full-length CFTR in human airway stem cells by CRISPR-Cas9 for pan-mutation correction in the endogenous locus. Mol. Ther. 2022;30:223–237. doi: 10.1016/j.ymthe.2021.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki S., Crane A.M., Anirudhan V., Barillà C., Matthias N., Randell S.H., Rab A., Sorscher E.J., Kerschner J.L., Yin S., et al. Highly Efficient Gene Editing of Cystic Fibrosis Patient-Derived Airway Basal Cells Results in Functional CFTR Correction. Mol. Ther. 2020;28:1684–1695. doi: 10.1016/j.ymthe.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kosicki M., Tomberg K., Bradley A. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018;36:765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leibowitz M.L., Papathanasiou S., Doerfler P.A., Blaine L.J., Sun L., Yao Y., Zhang C.Z., Weiss M.J., Pellman D. Chromothripsis as an on-target consequence of CRISPR-Cas9 genome editing. Nat. Genet. 2021;53:895–905. doi: 10.1038/s41588-021-00838-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Höijer I., Emmanouilidou A., Östlund R., van Schendel R., Bozorgpana S., Tijsterman M., Feuk L., Gyllensten U., den Hoed M., Ameur A. CRISPR-Cas9 induces large structural variants at on-target and off-target sites in vivo that segregate across generations. Nat. Commun. 2022;13:627. doi: 10.1038/s41467-022-28244-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turchiano G., Andrieux G., Klermund J., Blattner G., Pennucci V., El Gaz M., Monaco G., Poddar S., Mussolino C., Cornu T.I., et al. Quantitative evaluation of chromosomal rearrangements in gene-edited human stem cells by CAST-Seq. Cell Stem Cell. 2021;28:1136–1147.e5. doi: 10.1016/j.stem.2021.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Ott C.J., Blackledge N.P., Kerschner J.L., Leir S.-H., Crawford G.E., Cotton C.U., Harris A. Intronic enhancers coordinate epithelial-specific looping of the active CFTR locus. Proc. Natl. Acad. Sci. USA. 2009;106:19934–19939. doi: 10.1073/pnas.0900946106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith A.N., Barth M.L., McDowell T.L., Moulin D.S., Nuthall H.N., Hollingsworth M.A., Harris A. A Regulatory Element in Intron 1 of the Cystic Fibrosis Transmembrane Conductance Regulator Gene (∗) J. Biol. Chem. 1996;271:9947–9954. doi: 10.1074/jbc.271.17.9947. [DOI] [PubMed] [Google Scholar]
- 20.NandyMazumdar M., Yin S., Paranjapye A., Kerschner J.L., Swahn H., Ge A., Leir S.-H., Harris A. Looping of upstream cis-regulatory elements is required for CFTR expression in human airway epithelial cells. Nucleic Acids Res. 2020;48:3513–3524. doi: 10.1093/nar/gkaa089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang R., Kerschner J.L., Gosalia N., Neems D., Gorsic L.K., Safi A., Crawford G.E., Kosak S.T., Leir S.-H., Harris A. Differential contribution of cis-regulatory elements to higher order chromatin structure and expression of the CFTR locus. Nucleic Acids Res. 2016;44:3082–3094. doi: 10.1093/nar/gkv1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carraro G., Langerman J., Sabri S., Lorenzana Z., Purkayastha A., Zhang G., Konda B., Aros C.J., Calvert B.A., Szymaniak A., et al. Transcriptional analysis of cystic fibrosis airways at single-cell resolution reveals altered epithelial cell states and composition. Nat. Med. 2021;27:806–814. doi: 10.1038/s41591-021-01332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruiz García S., Deprez M., Lebrigand K., Cavard A., Paquet A., Arguel M.J., Magnone V., Truchi M., Caballero I., Leroy S., et al. Novel dynamics of human mucociliary differentiation revealed by single-cell RNA sequencing of nasal epithelial cultures. Development. 2019;146 doi: 10.1242/dev.177428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lombardo A., Cesana D., Genovese P., Di Stefano B., Provasi E., Colombo D.F., Neri M., Magnani Z., Cantore A., Lo Riso P., et al. Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat. Methods. 2011;8:861–869. doi: 10.1038/nmeth.1674. [DOI] [PubMed] [Google Scholar]
- 25.Plasschaert L.W., Žilionis R., Choo-Wing R., Savova V., Knehr J., Roma G., Klein A.M., Jaffe A.B. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature. 2018;560:377–381. doi: 10.1038/s41586-018-0394-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okuda K., Dang H., Kobayashi Y., Carraro G., Nakano S., Chen G., Kato T., Asakura T., Gilmore R.C., Morton L.C., et al. Secretory Cells Dominate Airway CFTR Expression and Function in Human Airway Superficial Epithelia. Am. J. Respir. Crit. Care Med. 2021;203:1275–1289. doi: 10.1164/rccm.202008-3198OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sabatini L.M., Warner T.F., Saitoh E., Azen E.A. Tissue Distribution of RNAs for Cystatins, Histatins, Statherin, and Proline-rich Salivary Proteins in Humans and Macaques. J. Dent. Res. 1989;68:1138–1145. doi: 10.1177/00220345890680070101. [DOI] [PubMed] [Google Scholar]
- 28.Isola M., Cabras T., Inzitari R., Lantini M.S., Proto E., Cossu M., Riva A. Electron microscopic detection of statherin in secretory granules of human major salivary glands. J. Anat. 2008;212:664–668. doi: 10.1111/j.1469-7580.2008.00888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kochańska B., Kedzia A., Kamysz W., Maćkiewicz Z., Kupryszewski G. The effect of statherin and its shortened analogues on anaerobic bacteria isolated from the oral cavity. Acta Microbiol. Pol. 2000;49:243–251. [PubMed] [Google Scholar]
- 30.Wright J.M., Merlo C.A., Reynolds J.B., Zeitlin P.L., Garcia J.G.N., Guggino W.B., Boyle M.P. Respiratory epithelial gene expression in patients with mild and severe cystic fibrosis lung disease. Am. J. Respir. Cell Mol. Biol. 2006;35:327–336. doi: 10.1165/rcmb.2005-0359OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Char J.E., Wolfe M.H., Cho H.J., Park I.H., Jeong J.H., Frisbee E., Dunn C., Davies Z., Milla C., Moss R.B., et al. A little CFTR goes a long way: CFTR-dependent sweat secretion from G551D and R117H-5T cystic fibrosis subjects taking ivacaftor. PLoS One. 2014;9 doi: 10.1371/journal.pone.0088564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Nooijer R.A., Nobel J.M., Arets H.G.M., Bot A.G., van Berkhout F.T., de Rijke Y.B., de Jonge H.R., Bronsveld I. Assessment of CFTR function in homozygous R117H-7T subjects. J. Cyst. Fibros. 2011;10:326–332. doi: 10.1016/j.jcf.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 33.Squair J.W., Gautier M., Kathe C., Anderson M.A., James N.D., Hutson T.H., Hudelle R., Qaiser T., Matson K.J.E., Barraud Q., et al. Confronting false discoveries in single-cell differential expression. Nat. Commun. 2021;12:5692. doi: 10.1038/s41467-021-25960-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bingle L., Wilson K., Musa M., Araujo B., Rassl D., Wallace W.A., LeClair E.E., Mauad T., Zhou Z., Mall M.A., Bingle C.D. BPIFB1 (LPLUNC1) is upregulated in cystic fibrosis lung disease. Histochem. Cell Biol. 2012;138:749–758. doi: 10.1007/s00418-012-0990-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corces M.R., Trevino A.E., Hamilton E.G., Greenside P.G., Sinnott-Armstrong N.A., Vesuna S., Satpathy A.T., Rubin A.J., Montine K.S., Wu B., et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods. 2017;14:959–962. doi: 10.1038/nmeth.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joshi N.A., Fass J.N. 2011. Sickle: A Sliding-Window, Adaptive, Quality-Based Trimming Tool for FastQ Files. [Google Scholar]
- 38.Li H., Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings presented in this study are available on request from the corresponding author, S.V.