Figure 1.
PCID2 is enriched at the latent HIV-1 LTR and is a repressor of HIV-1 gene expression during latency
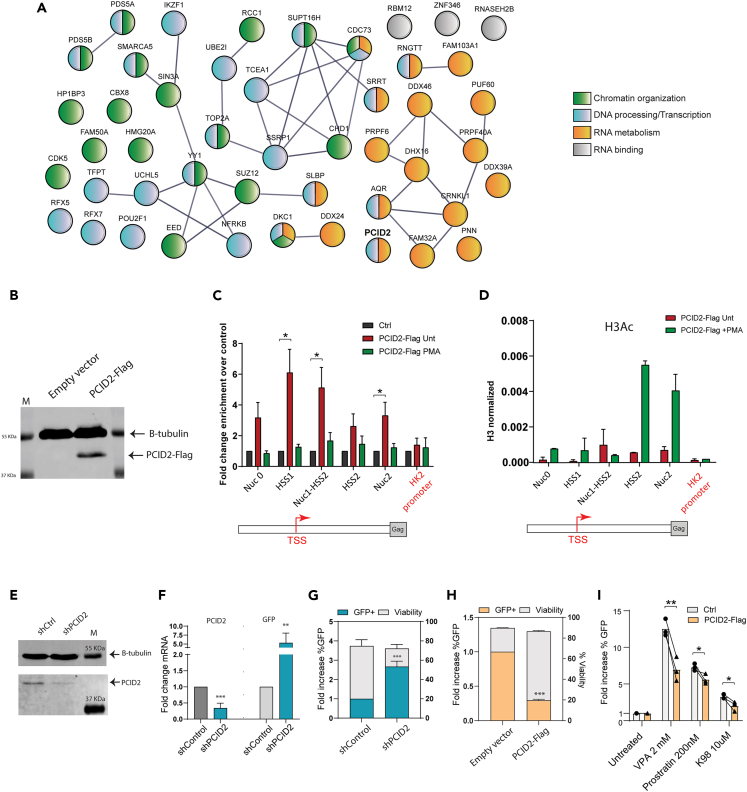
(A) STRING network of proteins identified to be enriched in the latent HIV-1 LTR by the Catchet-MS pipeline as published in Ne, Crespo et al. 2022 Nuc Acid Res. Proteins are colored based on GO analysis enrichment as indicated in the figure. Gray lines represent known interaction (experiments) as reported in the STRING database.
(B) Western blot of PCID2-Flag in control or stably PCID2-Flag expressing J-Lat 11.1. Beta tubulin was used as loading control.
(C) Enrichment of PCID2-Flag at the HIV-1 LTR expressed as fold enrichment in control and untreated and PMA treated PCID2-Flag expressing J-Lat 11.1 lines as assessed by chromatin immunoprecipitation (ChIP) coupled with quantitative PCR. Primers spanning across the HIV-1 promoter were used to assess relative enrichment of PCID2-Flag at sequential regions of the LTR. HK2 promoter was used as a control genomic region. Relative enrichment was normalized to Input. Mean and SD correspond to 4 ChIP replicates with independent chromatin preparations for control, untreated, and PMA treated cells.
(D) ChIP-qPCR analysis of acetylated histone 3 (F) enrichment at the HIV-1 promoter in untreated and PMA treated stably PCID2-Flag expressing J-Lat 11.1 lines. H3Ac enrichment at the HIV-1 LTR is represented as relative enrichment normalized to Total H3 as shown in Figure S1C. ChIP-qPCR corresponds to one chromatin preparation, bars and error lines represent, respectively, mean and SD of technical duplos.
(E) Western blot analysis of PCID2 in control and PCID2 knockdown J-Lat 11.1. Beta-tubulin was used as a loading control. Cells were infected with a VSV-pseudotyped lentivirus containing a scramble shRNA (shControl) or a PCID2 mRNA-targeting shRNA (shPCID2).
(F) Gene expression analysis of shRNA-mediated knockdown of PCID2 and GFP mRNA fold induction in shPCID2 cells relative to shControl and normalized to cyclophilin A. Bars represent mean of 5 independent shRNA-mediated knockdown experiments and error lines represent SEM (n = 4). Statistical significance was determined by t test; ∗∗p < 0.01, ∗∗∗p < 0.001.
(G) Fold increase in the % of GFP (left y axes) and viability (right y axes) in shControl or shPCID2 J-Lat 11.1 cells as measured by flow cytometry. Bars represent mean of 4 independent shRNA-mediated knockdown experiments and error lines represent SEM. Statistical significance was determined by t-test; ∗∗∗p < 0.001. Raw values for percentage of GFP are available in Table S2.
(H) Fold decrease in the % of GFP (left y axes) and viability (right y axes) in control or transiently PCID2-Flag overexpressing J-Lat 11.1 cells as measured by flow cytometry. Bars represent mean of 3 collections and error lines represent SEM. Statistical significance was determined by t test; ∗∗∗p < 0.001. Raw values for percentage of GFP are available in Table S2.
(I) Fold change in % of GFP in control or PCID2-Flag overexpressing J-Lat 11.1 after a 48 h treatment of with latency reversing agents valproic acid (VPA), prostratrin, and K98 as measured by flow cytometry. Fold increase in % GFP was assessed by normalization to untreated control. Bars represent mean and error lines represent SEM. Statistical significance was determined by t test; ∗∗p < 0.01. Raw values for percentage of GFP are available in Table S2.