Figure 2.
PCID2 is a repressor of HIV-1 transcription initiation during viral latency
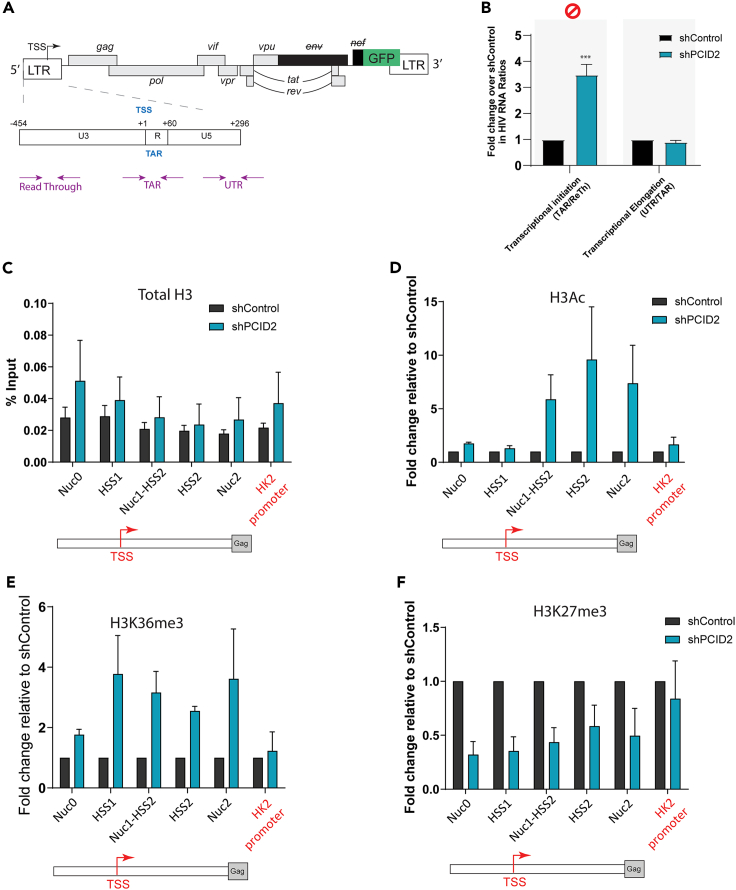
(A) Schematic figure of the HIV-1 genome in J-Lat 11.1. Arrows represent primer location and amplicons across the HIV-1 genome used in the transcriptional profiling assay.
(B) Transcriptional profiling assay of shControl and shPCID2 J-Lat 11.1. Gene expression blocks at transcriptional initiation, elongation, and post-transcriptional steps are assessed by calculating the ratio of the relative abundance of HIV-1 RNA species as shown in the figure. Data are presented as fold change in HIV-1 RNA ratios in shPCID2 relative to shControl. Bars represent mean of 5 independent shRNA-mediated knockdown experiments and error lines represent SEM. Statistical significance was determined by t test; ∗p < 0.05, ∗∗∗p < 0.001.
(C–F) ChIP-qPCR analysis of total histone 3 (C), acetylated histone 3 (D), Histone 3 K36-trimethyl (E), and Histone 3 K27-trimethyl (F) enrichment at the HIV-1 promoter in control (shControl) and PCID2-knockdown (shPCID2) J-Lat 11.1. Total H3 at the HIV-1 LTR is represented as % input. H3Ac, H3K36me3, and H3K27me3 enrichment at the HIV-1 LTR is represented as relative enrichment normalized to Total H3 as shown in A and relative to control. Bars and error lines represent, respectively, mean and SEM of four ChIP-qPCR experiments in four independent chromatin preparations and shRNA-mediated knockdown experiments except for H3K36me3 mark where ChIP-qPCR was performed three times.