Abstract
Mutations at five loci named PEANUT1-5 (PNT) were identified in a genetic screen for radially swollen embryo mutants. pnt1 cell walls showed decreased crystalline cellulose, increased pectins, and irregular and ectopic deposition of pectins, xyloglucans, and callose. Furthermore, pnt1 pollen is less viable than the wild type, and pnt1 embryos were delayed in morphogenesis and showed defects in shoot and root meristems. The PNT1 gene encodes the Arabidopsis thaliana homolog of mammalian PIG-M, an endoplasmic reticulum–localized mannosyltransferase that is required for synthesis of the glycosylphosphatidylinositol (GPI) anchor. All five pnt mutants showed strongly reduced accumulation of GPI-anchored proteins, suggesting that they all have defects in GPI anchor synthesis. Although the mutants are seedling lethal, pnt1 cells are able to proliferate for a limited time as undifferentiated callus and do not show the massive deposition of ectopic cell wall material seen in pnt1 embryos. The different phenotype of pnt1 cells in embryos and callus suggest a differential requirement for GPI-anchored proteins in cell wall synthesis in these two tissues and points to the importance of GPI anchoring in coordinated multicellular growth.
INTRODUCTION
The plant extracellular matrix is composed of polysaccharides, proteins, and glycoproteins and plays a crucial role in morphogenesis and development of plants. The synthesis and organization of cellulose and xyloglucan polymers largely determines the mechanical characteristics of the wall, and the pectin network is crucial for cell adhesion and wall porosity. Cellulose microfibrils are synthesized by plasma membrane enzyme complexes and are extruded directly into the cell wall (reviewed in Williamson et al., 2002). By contrast, the other polysaccharides and proteins of the cell wall are synthesized in the endoplasmic reticulum (ER) or Golgi and reach the cell wall through the secretory pathway (Gibeaut and Carpita, 1994). The mechanisms by which these polymers are assembled into functional networks are unknown.
Many proteins found in the cell wall are posttranslationally modified. The importance of N-glycosylation for cellulose biosynthesis has been demonstrated (Lukowitz et al., 2001; Burn et al., 2002; Gillmor et al., 2002). Another posttranslational modification of cell wall–localized proteins is the addition of a glycosylphosphatidylinositol (GPI) membrane anchor, which is synthesized by enzyme complexes associated with the ER membrane. Proteins to be modified with a GPI anchor are cotranslationally inserted into the ER and typically contain a single C-terminal transmembrane domain that is proteolytically cleaved during transfer of the protein to the GPI anchor (Kinoshita and Inoue, 2000). The final destination of GPI-anchored proteins is the plasma membrane, where the anchor allows hydrophilic polypeptides to stably associate with the extracellular face of the membrane. The GPI moiety can be cleaved by specific phospholipases, releasing the polypeptide into the extracellular matrix in a regulated manner. In addition, evidence exists for the association of GPI-anchored proteins into lipid rafts, higher order structures that can coordinate enzymatic functions or signaling events (Ferguson, 1999; Peskan et al., 2000; Borner et al., 2005).
GPI-anchored proteins (GAPs) have been implicated in many processes. The coat of trypanosomes consists of GPI-anchored surface glycoproteins, which serve to make these parasites antigenically variable (Ferguson, 1999). In animals, mutations in a class of GPI-anchored glycoproteins called glypicans cause defects in cell division and tissue morphogenesis (Selleck, 2000). The genome of the yeast Saccharomyces cerevisiae encodes for ∼50 GAPs, many of which are essential for cell wall synthesis and organization or for cell–cell signaling in the mating response (Caro et al., 1997; Kapteyn et al., 1999). Recent studies have focused on a genome-wide identification of the total set of GAPs in Arabidopsis thaliana (Borner et al., 2002, 2003; Eisenhaber et al., 2003). Based on their sequences, the 250 or so predicted GAPs of Arabidopsis most likely participate in cell wall deposition and remodeling, defense responses, and cell signaling. Arabinogalactan proteins (AGPs), a class of heavily glycosylated cell wall proteins, have been the subject of study for several years and are known to be modified by the addition of a GPI anchor (Youl et al., 1998). AGPs are thought to play roles in growth and differentiation, though a mechanism for their action has not been defined (Gaspar et al., 2001).
Only a few predicted or demonstrated GAPs of plants have been functionally characterized. Conditional mutations in the COBRA (COB) gene were isolated based on their root swelling phenotype, and it was proposed that COB is required for oriented cellulose deposition (Schindelman et al., 2001). BC1, a homolog of COB, is required for normal cellulose levels in secondary cell walls of rice (Oryza sativa) plants (Li et al., 2003). Loss of SOS5, a protein with fasciclin and arabinogalactan domains, causes reduced root growth and root swelling (Shi et al., 2003). Mutations in SKU5 alter the growth properties of roots and affect root waving and cell expansion (Sedbrook et al., 2002). The PMR6 gene, which encodes a putative pectate lyase, was identified in a genetic screen for mutants with noncompatible interactions with the powdery mildew fungus (Erysiphe cichoracearum) (Vogel et al., 2002). The Arabidopsis classical AGP, AGP18, was shown to be required for female gametophyte development (Acosta-García and Vielle-Calzada, 2004). Overexpression of the tomato (Lycopersicon esculentum) AGP, LeAGP-1, caused reduced plant growth and seed size (Sun et al., 2004a). Of the above proteins, only SKU5 and LeAGP-1 were experimentally shown to be GPI anchored (Sedbrook et al., 2002; Sun et al., 2004b). A recent article also describes the effect on pollen of loss-of-function mutations in genes of the GPI anchor biosynthesis pathway. SETH1 and SETH2 genes were shown to encode homologs of mammalian PIG-C and PIG-A proteins, which are components of the GPI-N-acetylglucosaminyltransferase complex. Mutations in SETH1 and SETH2 affect pollen germination and growth in vitro and result in abnormal callose deposition in pollen tubes (Lalanne et al., 2004). The above studies all underscore the importance of GAPs for cell wall organization and function.
This work describes mutations in five genes, designated PEANUT1-5 (PNT), required for accumulation of GAPs. The mutants were recovered in a genetic screen for radially swollen embryos (Gillmor et al., 2002). PNT1 encodes a predicted mannosyltransferase with sequence similarity to human PIG-M, an ER-localized mannosyltransferase that is required for synthesis of the GPI anchor (Maeda et al., 2001). pnt1 embryos show a large reduction in crystalline cellulose, an increase in pectin, as well as ectopic deposition of xyloglucan, pectin, and callose. Although pnt1 mutants are embryo lethal, callus differentiated from pnt1 embryos can grow for a limited time. Ectopic callose deposition is observed in pnt1 callus, but, in contrast with pnt1 embryos, cell wall structure appears otherwise normal. Our results highlight the importance of GAPs for the synthesis and secretion of cell wall polymers and point to a reduced requirement for GAPs in the absence of coordinated multicellular development.
RESULTS
pnt Mutants Are Radially Swollen during Embryogenesis
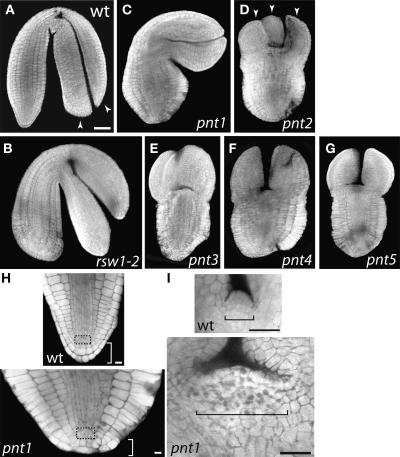
Five pnt mutants were recovered by microscopic analysis of embryo and seed morphology (Gillmor et al., 2002). The gene name refers to the characteristic shape and wrinkled seed coat of mutant seeds. Mutant embryos exhibit a radially swollen phenotype that is similar to strong alleles of the cellulose-deficient rsw1 mutants (Beeckman et al., 2002; Gillmor et al., 2002), in that they show altered polarity of cell expansion in outer cell layers during embryogenesis (Figure 1).
Figure 1.
pnt Mutants Are Radially Swollen.
Confocal images of the wild type (A), rsw1-2 (B), pnt1 (C), pnt2 (D), pnt3 (E), pnt4 (F), and pnt5 (G) at the late bent cotyledon stage of embryogenesis. Confocal sections of the wild type and pnt1 root (H) and shoot (I) meristems of dissected late bent cotyledon stage embryos. (A) to (G) are shown at the same magnification. Arrowheads in (A) and (D) point to cotyledons. Root meristem region is boxed in (H); shoot meristem region is indicated with a bracket in (I). Bars = 50 μm in (A) and 25 μm in (H) and (I).
Complementation analysis and mapping demonstrated that the five pnt mutants were in five distinct genes, named PNT1 to PNT5. PNT1 was mapped between the markers T10F18(A) and MWD9(A) and was subsequently identified as gene At5g22130 (see below). PNT2 was mapped to ∼30 centimorgan (cM) on chromosome 5 between the markers nga151 and P01 (in 350 meiotic events, five recombinations between nga151 and pnt2 and three between pnt2 and P01 were found). PNT3 was mapped to ∼39 cM on chromosome 1 between the markers F21M12 and ciw12 (in 210 meiotic events, 10 recombinations between F21M12 and pnt3 and 10 between pnt3 and ciw12 were found). PNT4 was mapped to ∼45 cM on chromosome 3 between the markers nga162 and ciw4 (in 210 meiotic events, 29 recombinations between nga162 and pnt4 and 36 between pnt4 and ciw4 were found). PNT5 was mapped to ∼60 cM on chromosome 4 between the markers g4539 and CH42 (in 240 meiotic events, two recombinations between g4539 and pnt5 and one between pnt5 and CH42 were found). All pnt mutations were recessive but segregated significantly <25% homozygous mutants upon selfing. Reciprocal crossing experiments with pnt1 demonstrated that the altered pnt1 segregation ratio was due to poor transmission through pollen, with no significant effect on egg transmission (Table 1).
Table 1.
Reduced Transmission of the pnt1-1 Allele through Pollen
|
pnt1/pnt1 or pnt1/PNT1 Segregation in F1a
|
||
|---|---|---|
| Cross (Male × Female) | Observed | Expectedb |
| pnt1/PNT1 × pnt1/PNT1 (selfed) | 2.7% Top half silique | 12.5% Top half silique |
| 2.6% Bottom half silique | 12.5% Bottom half silique | |
| 5.3% Total pnt1/pnt1 (n = 1132) | 25% Total pnt1/pnt1 | |
| pnt1/PNT1 × PNT1/PNT1 | 7% pnt1/PNT1 (n = 100) | 50% pnt1/PNT1 |
| PNT1/PNT1 × pnt1/PNT1 | 46.3% pnt1/PNT1 (n = 108) | 50% pnt1/PNT1 |
For the pnt1/PNT1 × pnt1/PNT1 cross, F1 progeny were scored directly by counting mutant embryos in the upper and lower halves of the silique. For pnt1/PNT1 × PNT1/PNT1 and PNT1/PNT1 × pnt1/PNT1 crosses, the genotype of F1 plants was ascertained by scoring the segregation of F2 mutant embryos in the siliques of selfed F1 plants.
Expected segregation for a recessive nuclear gene with normal transmission.
Similar to rsw1-2 (Figure 1B), the root, hypocotyls, and cotyledons of all five classes of pnt mutants are swollen. Cells of the outer layers of the embryonic root and hypocotyl are brick shaped (Figures 1C to 1G), as opposed to the typical cube-shaped epidermal and endodermal cells of wild-type embryos (Figure 1A, magnification of the root in Figure 1H). Unlike mutations in rsw1, which primarily cause defects in cell elongation, pnt mutations also cause developmental defects. The shoot apical meristem of pnt embryos is greatly enlarged compared with that of the wild type (Figure 1I). As seen in Figure 1D (arrowheads), pnt mutants commonly have three or four cotyledons, compared with two cotyledons in wild-type embryos (Figure 1A). Furthermore, pnt embryos lack most cells of the root cap, and the cells of the quiescent center and initials of the endodermis are disorganized (Figure 1H). Because of the similarity of all five pnt mutants, we chose to characterize the pnt phenotype by studying pnt1 in detail.
Delayed Morphogenesis and Abnormal Cell Divisions in pnt1 Embryos
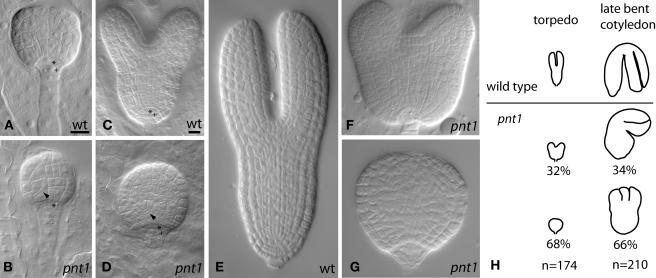
The appearance of cotyledons during the progression from the globular to heart stage of embryo development constitutes a transition from radial to bilateral symmetry (cf. Figures 2A and 2C). pnt1 embryos are consistently delayed in this transition. Compared with the wild-type heart stage embryo in Figure 2C, the pnt1 embryo shown in Figure 2D remains radially symmetrical, although it has increased considerably in size compared with the globular stage pnt1 embryo shown in Figure 2B. Later in development at the torpedo stage, two classes of pnt1 embryos are apparent: those that have made the transition to bilateral symmetry by developing two cotyledons (Figure 2F) and those that have increased in size, but which remain radially symmetrical (Figure 2G). A comparison of the percentage of pnt1 embryos with delayed morphogenesis at the torpedo stage and the number of late bent cotyledon stage pnt1 embryos with three or more cotyledons is shown in Figure 2H. At the torpedo stage of development, 32% of pnt1 embryos have developed cotyledon primordia, whereas 68% remain round (n = 174). At the late bent cotyledon stage of development, 34% of pnt1 embryos have two cotyledons, whereas 66% have three or more cotyledons (n = 210). Thus, there is a good correlation between a delay in the initiation of cotyledon primordia and the development of extra cotyledons (see also Torres-Ruíz and Jürgens, 1994).
Figure 2.
Delayed Morphogenesis and Abnormal Cell Divisions in pnt1 Embryos.
(A) to (G) Wild type ([A], [C], and [E]) and pnt1 ([B], [D], [F], and [G]) embryos shown at the late globular ([A] and [B]), late heart ([C] and [D]), and torpedo ([E] to [G]) stages of embryogenesis. Embryos within ovules ([A] to [D]) and dissected embryos ([E] to [G]) were cleared in Hoyer's solution and viewed with Nomarski optics. Arrowheads, oblique cell division; asterisk, apical daughter of hypophyseal cell; cross, basal daughter of hypophyseal cell. Bars = 10 μm. (A) and (B) are the same magnification. (C) to (G) are the same magnification.
(H) Tracings of typical wild-type and pnt1 embryos at the wild-type torpedo (left) and late bent cotyledon (right) stages. The percentage of pnt1 embryos with bilateral (top) or radial (bottom) symmetry at the wild-type torpedo stage is shown at the left. The percentage of pnt1 embryos with two cotyledons (top) or three or more cotyledons (bottom) at the late bent cotyledon stage is shown at the right.
In addition to delaying morphogenesis, pnt1 mutations also interfere with the normal pattern of cell division in the early embryo. For example, the globular stage pnt1 embryo shown in Figure 2B has several irregular oblique divisions in the proembryo (arrowhead), and the hypophysis, the cell supporting the proembryo, remains undivided. This is in contrast with corresponding wild-type embryos (Figure 2A), where the hypophysis divides to produce a lens-shaped derivative that will form the quiescent center of the root meristem and a basal derivative that forms the central root cap (Jürgens and Mayer, 1994). By the heart stage, the pnt1 embryo shown in Figure 2D has produced a cell similar in size and shape to a wild-type lens cell, but the basal derivative of the hypophysis is unusually large and has an irregular shape. Furthermore, continued oblique divisions in the proembryo make it difficult to distinguish elongated vascular precursor cells (visible in the wild type, Figure 2C). Thus, the anatomical defects observed in the root meristem and root cap of late-stage pnt1 embryos (Figure 1H) are correlated with abnormal cell divisions of the hypophyseal cell and its derivatives during early embryogenesis.
Seedling-Lethal pnt1 Mutants Are Able to Proliferate as Callus
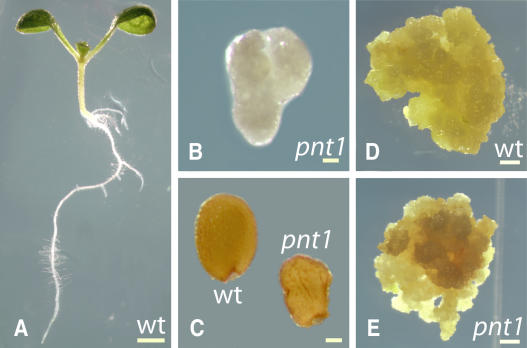
A wild-type Arabidopsis seedling grown for 5 d on tissue culture medium is shown in Figure 3A. By this stage, the cotyledons have expanded, the first true leaves are apparent, and the root system has begun to develop. By contrast, pnt1 seedlings of similar age show little sign of postembryonic development (Figure 3B). After seed imbibition, pnt1 embryos expand slightly and emerge from the seed coat (cf. with pnt1 seed shown in Figure 3C) but soon become necrotic. In contrast with the lethality of pnt1 seedlings, when green late-stage pnt1 embryos were dissected out of the ovule and placed on callus-inducing media, pnt1 calli (Figure 3E) were able to grow for 4 to 6 weeks, with a growth rate similar to calli from wild-type embryos (Figure 3D), before undergoing necrosis (dark brown tissue in Figure 3E). This result suggests that the undifferentiated growth typical of callus tissue is more permissive for loss of the PNT1 gene product than the coordinated multicellular growth required for seedling development.
Figure 3.
pnt1 Mutants Are Seedling Lethal, but pnt1 Callus Is Able to Proliferate.
Bright-field images of wild-type (A) and pnt1 (B) seedlings after 5 d of growth on solidified agar medium plus sucrose. Mature wild-type and pnt1 seeds are shown in (C). Six-week-old callus induced from wild-type (D) and pnt1 (E) late stage immature embryos. Bars = 1 mm in (A), 100 μm in (B) and (C), and 1 mm (D) and (E).
Decreased Cellulose, Increased Pectin, and Neutral Sugars in pnt1 Embryos
The phenotypic similarity of pnt1 and rsw1-2 phenotypes during embryogenesis suggested that pnt1 mutants might also have cell wall alterations. To test this hypothesis, we quantified the sugar composition of cell wall polysaccharides in late bent cotyledon stage wild-type and pnt1 embryos. The results of these analyses are shown in Table 2. pnt1 embryos had a 40% reduction in crystalline cellulose (117 nmol glucose/mg dry weight) compared with 196 nmol/mg in wild-type embryos. By contrast, pnt1 embryos contained 93 nmol/mg dry weight of uronic acid (the principal component of pectin), more than twice as much as wild-type embryos (41 nmol/mg). Other noncellulosic polymers were quantified by measuring their neutral sugar constituents. Wild-type embryos had 483 nmol total neutral sugars/mg dry weight, whereas pnt1 embryos were found to have 782 nmol/mg dry weight, a 60% increase. Although the total amount of neutral sugars differed between wild-type and pnt1 embryos, the ratio of the amounts of the different neutral sugars did not vary significantly (Table 2).
Table 2.
Cell Wall Composition of Wild-Type and pnt1 Embryosa
| Cell Wall Component | Wild Type | pnt1 | ||
|---|---|---|---|---|
| Celluloseb | 196 ± 17 | 117 ± 11 | ||
| Pectinc | 41 ± 3 | 93 ± 4 | ||
| Neutral sugarsd | ||||
| Rhamnose | 35 ± 3 | (7%) | 62 ± 2 | (8%) |
| Fucose | 18 ± 2 | (4%) | 20 ± 1 | (3%) |
| Arabinose | 248 ± 29 | (51%) | 416 ± 17 | (53%) |
| Xylose | 104 ± 13 | (22%) | 139 ± 4 | (18%) |
| Mannose | 18 ± 5 | (4%) | 16 ± 2 | (2%) |
| Galactose | 60 ± 6 | (12%) | 129 ± 10 | (16%) |
| Total nmol neutral sugars | 483 | 782 | ||
| Average dry weight per embryo | 4.8 μg | 3 μg |
Expressed as nmol sugar/mg dry weight of embryo. Value is the average of at least three measurements, with standard deviation.
Cellulose measured as acetic/nitric acid–insoluble glucose.
Pectin measured as total uronic acid after trifluoroacetic acid hydrolysis.
Amount of each neutral sugar as a percentage of the total neutral sugars is listed in parentheses.
Chemical analysis of pnt1 cell walls illustrated that, although pnt1 and rsw1-2 have similar radially swollen phenotypes, the two mutants have significantly different cell wall compositions. The primary effect of the rsw1-2 mutation was a 75% reduction in crystalline cellulose, concomitant with a slight, possibly compensatory, increase in pectin (Gillmor et al., 2002). Whereas the cellulose decrease in pnt1 embryos (40%) is less dramatic than that of rsw1-2, the pnt1 mutation has a much greater effect on the other polysaccharide constituents of the wall.
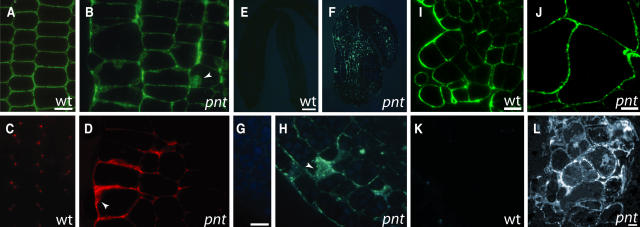
Ectopic Accumulation of Xyloglucan, Pectin, and Callose in pnt1 Embryos and Ectopic Callose in pnt1 Callus
The increased amount of uronic acid and neutral sugars measured in pnt1 embryos suggested that the distribution or deposition of noncellulosic structural polysaccharides might also be altered. Reagents that specifically identify various polysaccharides were used to determine the spatial distribution of xyloglucan, pectin, and callose in wild-type and pnt1 embryos. The CCRC M1 antibody recognizes a terminal α-(1,2)-linked fucosyl residue present on xyloglucans (Puhlmann et al., 1994). As shown in Figure 4A, CCRC M1 recognizes epitopes throughout the wild-type cell wall, with the exception of triangular areas at cell junctions. CCRC M1 epitopes are also present throughout pnt1 embryos, though staining is more diffuse than in the wild type (Figure 4B). In addition, ectopic patches of signal are seen in many cells. The monoclonal antibody JIM5 recognizes esterified pectins (Knox et al., 1990). In sections of wild-type embryos, the strongest JIM5 signal is seen in a triangular area at three cell junctions. JIM5 signal is also seen at lower intensity in a thin line corresponding to the middle lamella between adjacent cell walls (Figure 4C). In pnt1 embryos, JIM5 epitopes are more abundant than in the wild type and are not confined to the middle lamella and three cell junctions (Figure 4D). Ectopic patches of JIM5 staining, similar to the patches seen with the CCRC M1 antibody, are frequent.
Figure 4.
Ectopic Deposition of Xyloglucan, Pectin, and Callose in pnt1 Embryos and Callose Deposition in pnt1 Callus.
Sections of wild-type ([A], [C], [E], and [G]) and pnt1 ([B], [D], [F], and [H]) embryos at the late bent cotyledon stage of development. Sections of 2-week-old callus differentiated from wild-type ([I] and [K]) and pnt1 ([J] and [L]) seeds. Sections in (A), (B), (I), and (J) were incubated with the CCRC-M1 antibody, which recognizes terminal α-(1,3)-linked fucose residues on xyloglucans (Puhlmann et al., 1994). Sections in (C) and (D) were incubated with JIM5 antibody, which recognizes esterified pectins (Knox et al., 1990). Sections in (E) to (H) were stained with the callose-specific analine blue fluorochrome Sirofluor, which fluoresces yellow-white when bound to callose. Sections in (K) and (L) were incubated with anti-β-1,3 glucan antibody. Arrowheads in (B), (D), and (H) indicate ectopic patches of signal. Bars = 5 μm in (A) and (G), 50 μm in (E), and 10 μm (I) and (L). (A) to (D), (G) and (H), (E) and (F), (I) and (J), and (K) and (L) are shown at equal magnification.
The abnormal chemical composition of pnt1 cell walls, and the aberrant localization of pectin and xyloglucan epitopes, suggested that the mechanical integrity of pnt1 cell walls might be compromised. Production of the 1,3-β-glucan, callose, is a common stress response and has been observed in the cellulose-deficient mutant cyt1 (Nickle and Meinke, 1998; Lukowitz et al., 2001). Callose is synthesized in pollen tubes and transiently during cell wall formation but is not normally found in most plant cell walls (Delmer, 1987). Thus, ectopic callose deposition can be interpreted as a marker of compromised cell wall integrity. Sirofluor, a fluorophore that specifically binds callose (Stone et al., 1984), was used to determine if pnt1 embryos accumulate callose during embryogenesis. As shown in Figure 4E, late bent cotyledon stage wild-type embryos have no appreciable sirofluor staining. This is in marked contrast with the ectopic accumulation of callose throughout the pnt1 embryo shown in Figure 4F. At higher magnification (Figure 4H), callose staining is seen in a punctate pattern throughout pnt1 cell walls, as well as in large patches.
We also examined the cell wall phenotype of wild-type and pnt1 callus cells. Staining of wild-type and pnt1 callus with CCRC M1 showed that both exhibit a similar pattern of fluorescence associated with xyloglucan in the cell walls (Figures 4I and 4J). Interestingly, pnt1 callus cells lack the patches of ectopic xyloglucan epitope seen in pnt1 embryo cells. We used a 1,3-β-glucan antibody to visualize callose deposition in wild-type and pnt1 calli. No staining was detected in wild-type callus cells (Figure 4K), whereas abundant signal was detected in the pnt1 callus (Figure 4L). The abundance of callose in pnt1 callus is consistent with the frequent occurrence of cell death in pnt1 callus (Figure 3E; data not shown).
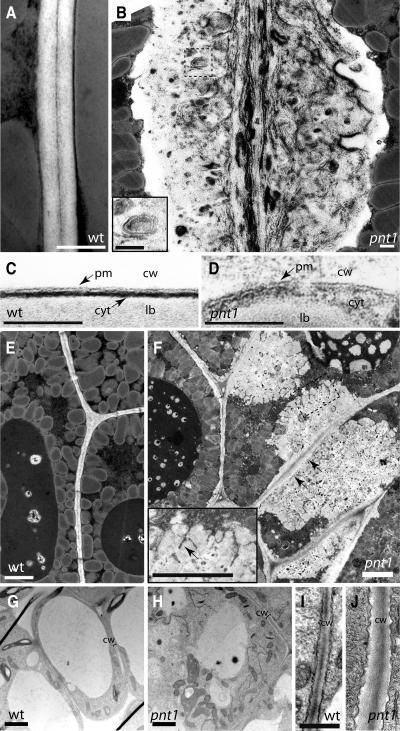
Our light microscopic analysis revealed that the deposition pattern of cell wall polymers was aberrant in pnt1 embryos. To determine the consequences of these abnormalities for the structural organization of pnt1 cell walls, ultrathin sections of wild-type and pnt1 embryos and callus were examined by transmission electron microscopy. Wild-type embryo cell walls have a uniform thickness and appearance. The thin band of electron-dense staining of the middle lamella seen in between adjacent wild-type walls is characteristic of pectic polysaccharides, whereas the electron-translucent areas of the wall are composed mainly of cellulose and xyloglucans (Figures 5A, 5C, and 5E). As expected from our previous findings, pnt1 embryo cells produce and secrete larger amounts of cell wall material. Two types of wall ultrastructure predominate. The first is an altered primary wall (Figure 5B). This primary wall conserves some elements of wild-type wall structure, with a concentration of electron-dense, presumably pectic material in the center of the wall. However, fibrillar electron-dense material as well as thick electron-dense inclusions and swirls are distributed throughout the wall in an apparently random fashion. In addition, islands of electron-dense material surrounded by a membrane are also observed (hatched box in Figure 5B, enlarged in inset) and point to a secretory defect in pnt1 mutants. As in the wild type, the contents of these altered primary walls are outside of the cell, on the opposite side of the plasma membrane from the cytoplasm (cf. wild-type cell wall, plasma membrane, and cytoplasm in Figure 5C with pnt1 in Figure 5D). Accumulation of lipid bodies and protein bodies appeared normal in pnt1 embryos, and defects in the morphology of cellular organelles were not apparent (data not shown).
Figure 5.
Disorganized and Ectopic Deposition of Cell Wall Material in pnt1 Embryos and Normal Wall Structure in pnt1 Callus.
(A) to (F) Transmission electron microscopy images of wild-type ([A], [C], and [E]) and pnt1 ([B], [D], and [F]) embryo cell walls. pnt1 (B) primary cell walls are disorganized compared with the wild type (A). Inset in (B), electron dense material surrounded by double membrane. Primary cell wall in pnt1 (D) is outside the plasma membrane, as in the wild type (C). pnt1 cells (F) secrete large amounts of ectopic cell wall material that is appressed to more normal primary wall. Inset in (F), cell wall material with blobby morphology. In (B) and (F), contents of hatched box are magnified in inset. pm, plasma membrane; cw, cell wall; cyt, cytoplasm; lb, lipid body. Arrowheads in (F), junction of primary and ectopic wall; arrowhead in (F) inset, invagination of cytoplasm. Bars = 250 nm in (A) and (B), 100 nm in (C) and (D), and 2 μm in (E) and (F).
(G) to (J) Transmission electron microscopy images of cell walls of 2-week-old callus differentiated from wild-type ([G] and [I]) and pnt1 ([H] and [J]) seed. (I) and (J) are shown at equal magnification. Bars = 2 μm in (G) and (H) and 500 nm in (I).
The second type of wall structure seen in pnt1 embryos can best be described as massive, ectopic accumulations of wall material. This material has a heterogeneous appearance and often appears appressed to, but distinct from, the primary wall (arrows in Figure 5F point to junction of primary wall and ectopic wall). Invaginations of cytoplasm are visible within these blobs of wall material (inset, Figure 5F), making it often difficult to determine which parts were topologically outside of the cell and which were in the process of being secreted. Similar to the pnt1 wall structure seen in Figure 5B, the ectopic wall material contains inclusions of varying electron densities, but fibrillar electron dense material is less commonly observed.
The ultrastructural abnormalities observed in pnt1 embryo cell walls were, for the most part, not seen in cell walls of pnt1 callus. As observed by comparing Figures 4I and 4J, pnt1 callus cells are many times larger than wild-type callus cells. Electron micrographs of wild-type (Figure 5G) and pnt1 (Figure 5H) callus cells at low magnification show that pnt1 cells are more highly cytoplasmic than their wild-type counterparts. At higher magnification, pnt1 callus cell walls are structurally similar to wild-type cell walls, except that they have increased electron-dense staining in the lamella, suggesting increased pectin content, and are slightly thicker than wild-type walls. Notably, the ectopic depositions of wall material and drastic alteration in wall structure in pnt1 embryos were not observed in pnt1 callus cells (cf. Figure 5B with 5J and Figure 5F with 5H).
In conclusion, pnt1 embryo cell walls contain increased amounts of cell wall material that are, at least in part, found in large patches appressed to the primary wall. This strongly suggests that pnt1 mutants are impaired in integrating secreted polysaccharides into a functional wall. It appears as if wall components are initially incorporated into the primary wall in a somewhat disorganized fashion, whereas later the secreted wall material is not integrated into the wall at all, and instead accumulates ectopically. In contrast with pnt1 embryo cell walls, pnt1 callus cell walls have an almost normal structure, with only an apparent slight increase in the electron-dense staining characteristic of pectin.
PNT1 Encodes the Arabidopsis Homolog of ER Mannosyltransferase PIG-M
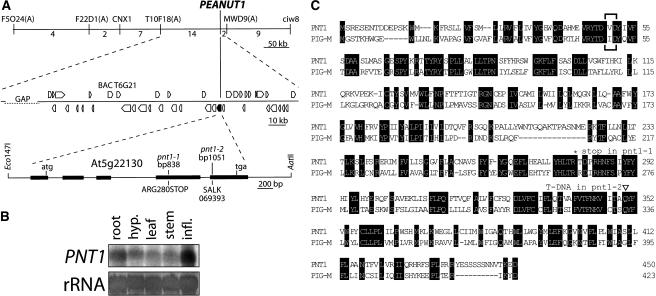
The PNT1 gene was identified based on its map position and found to correspond to At5g22130 (see Figure 6A for details). The pnt1-1 allele was found to have a cytosine-to-thymine change at bp 838 of the At5g22130 coding sequence, resulting in a stop at codon 280. The identity of PNT1 was confirmed by complementation of the pnt1-1 mutation with a wild-type At5g22130 genomic clone (Figure 6A). A second mutant allele of the PNT1 gene, pnt1-2, was found in the SALK T-DNA insertional line SALK_069393. This second allele, generated in the Columbia ecotype, segregated an embryo mutant with a similar phenotype to pnt1-1. The left border of the T-DNA in SALK_069393 was found to be inserted at bp 1051 of the PNT1 coding sequence. Allelism was confirmed by complementation test. Pollen transmission of the pnt1-2 allele was slightly increased compared with the pnt1-1 allele, suggesting that the pnt1-2 allele may have more enzymatic activity than the ethyl methanesulfonate–induced pnt1-1 allele. This conclusion is consistent with the fact that the T-DNA insertion in pnt1-2 occurs downstream of the stop codon in pnt1-1 (Figure 6A).
Figure 6.
PNT1 Encodes the Arabidopsis Homolog of PIG-M.
(A) Identification of the PNT1 gene based on its map position at ∼40 cM on chromosome V. Top line, genetic interval used to identify PNT1. PCR-based simple sequence length polymorphism markers are shown above the line; the numbers of recombinants recovered between each marker (out of 2010 meiotic products) are shown below the line. Middle line, the ∼140-kb physical interval between markers T10F18(A) and MWD9(A), containing 40 predicted genes. Genes are shown as pentagons; PNT1 is in black. Bottom line, the 3641-bp Eco147I/AatII genomic fragment (bp 108026 to 104386 of BAC T6G21) used to complement the pnt1-1 mutation. PNT1 exons are represented by thick lines. The position of the cytosine-to-thymine change at bp 838 of the PNT1 coding sequence in pnt1-1 is noted. This nucleotide change results in a translational stop at codon 280. The position of the T-DNA insertion in SALK 069393 at bp 1051 of the PNT1 coding sequence is also noted. The full-length PNT1 cDNA sequence is available from GenBank/EMBL/DDBJ under accession number BX831719.
(B) Expression analysis of PNT1 in different tissue types. PNT1 is expressed at low levels in 7-d-old seedling roots, 7-d-old seedling hypocotyls and cotyledons, 4-week-old leaves, and 4-week-old inflorescence stems and at higher levels in flowers and inflorescences from 4-week-old plants. rRNA stained with methylene blue is shown as a loading control.
(C) The predicted PNT1 protein is 38% identical to human PIG-M. Alignment of PNT1 and PIG-M predicted protein sequences. Identical residues are boxed in black. The signature DxD glycosyltransferase motif is shown in brackets (Maeda et al., 2001). The position of the stop codon in pnt1-1 is marked with an asterisk, and the position of the T-DNA insertion in pnt1-2 is marked with a triangle.
Using a PNT1 cDNA obtained by RT-PCR as a probe, PNT1 mRNA was found to be present at low levels in roots, hypocotyls, leaves, and stems and at higher levels in the inflorescence meristem (Figure 6B). The predicted PNT1 protein is 38% identical to the human enzyme PIG-M (Figure 6C), an ER membrane–localized 1-4-α-mannosyl transferase that is required for synthesis of the GPI anchor within the ER lumen (Maeda et al., 2001). The PNT1 gene encodes the only PIG-M homolog in the Arabidopsis genome (data not shown).
GAPs Are Absent in pnt Mutants
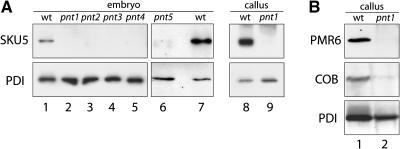
The similarity of the PNT1 protein to PIG-M suggested that the primary defect of pnt mutants was in GPI anchor biosynthesis. To test this hypothesis, we used antibodies against demonstrated and predicted GAPs to test for the presence of these proteins in pnt embryos. The SKU5 protein has been shown to be GPI anchored based on the presence of C-terminal consensus sequences and its release from membrane preparations after treatment with phospholipase C (Sedbrook et al., 2002). Figure 7A shows the results of protein gel blot detection of SKU5 protein from wild-type, pnt1, pnt2, pnt3, pnt4, and pnt5 embryo extracts. SKU5 protein was present in wild-type embryos but undetectable in pnt1-pnt4. Two faint bands of slightly lower molecular weight are visible on the pnt5 blot, suggesting that pnt5-1 is a leaky allele or that loss of the PNT5 gene product does not lead to the degradation of all detectable SKU5 protein. Similar to embryos, SKU5 is abundant in wild-type callus but undetectable in pnt1 callus (Figure 7A). Figure 7B shows the detection of PMR6 and COBRA protein from extracts of wild-type and pnt1 callus. Based on the presence of GPI-anchoring sequence motifs, both PMR6 and COB have been proposed to be GPI anchored (Schindelman et al., 2001; Vogel et al., 2002). PMR6 protein is abundant in wild-type extracts and undetectable in pnt1 extracts, whereas COB is present in the wild type and strongly reduced in pnt1 extracts.
Figure 7.
Absence of GAPs in pnt Mutants.
(A) Protein gel blot detection of SKU5 and protein disulfide isomerase from extracts of wild-type and pnt1, pnt2, pnt3, pnt4, and pnt5 embryos and from wild-type and pnt1 callus tissue. Protein disulfide isomerase (PDI) is shown as a loading control.
(B) Protein gel blot detection of PMR6, COB, and protein disulfide isomerase from wild-type and pnt1 callus extracts. Protein disulfide isomerase (PDI) is shown as a loading control.
The high sequence identity of PNT1 and PIG-M and the demonstration that the GAP SKU5 is absent in pnt1 embryos provide strong evidence that PNT1 functions as the first mannosyltransferase in GPI anchor biosynthesis in Arabidopsis. The absence or strong decrease of SKU5 in the other four pnt mutants suggests that they have biochemical defects related to that in pnt1, and it seems likely that they encode other functions in the pathway for GPI anchor biosynthesis.
DISCUSSION
Functional evidence for the role of GAPs in plant physiology and development is largely lacking (Borner et al., 2003). In this work, analysis of mutations in the Arabidopsis PNT genes demonstrates the importance of GAPs in cell wall synthesis and development. Surprisingly, the extreme defects in cell wall ultrastructure seen in pnt1 embryos were not seen in callus tissue generated from pnt1 embryos. This result suggests that GAPs play a more important role in cell wall synthesis during coordinated multicellular growth than in proliferation of undifferentiated tissue, such as callus.
pnt Mutants Lack GAPs
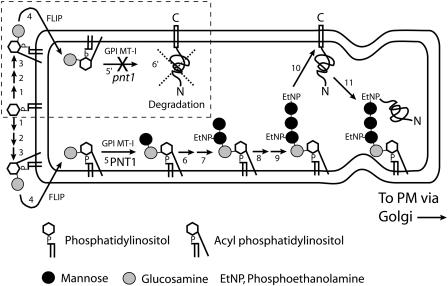
The enzymatic steps involved in production of the GPI anchor are represented in schematic form in Figure 8. Although some details vary among animals, yeast, trypanosomes, and plants, synthesis of the core structure is essentially conserved. In mammalian cells, loss of any of the functions required for making the GPI anchor results in termination of anchor synthesis (reviewed in Kinoshita and Inoue, 2000). Furthermore, lack of transfer of the membrane-associated protein precursor to a functional GPI anchor results in ER retention and degradation of the untransferred protein (Field et al., 1994). This degradation can be accomplished via a pathway specific for ER-localized proteins (reviewed in Bonifacio and Lippincott-Schwartz, 1991). Using antibodies to the Arabidopsis GAP SKU5 (Sedbrook et al., 2002) and to the predicted GAPs COBRA and PMR6 (Schindelman et al., 2001; Vogel et al., 2002), we have shown that these proteins are diminished or undetectable in pnt1 embryos and callus. We propose that the loss of the GAPs SKU5, COB, and PMR6 in pnt1 is due to degradation of these proteins in the absence of their transfer to a functional GPI anchor (hatched box, Figure 8). In view of the effect of the pnt1 mutation on these three proteins, it is likely that the lack of GPI mannosyltransferase I activity in pnt1 mutants affects the targeting and stability of most or all GPI-anchored proteins.
Figure 8.
Synthesis of the GPI Anchor and Transfer of the Anchor to Proteins in Wild-Type and pnt1 Cells.
For simplicity, some intermediates are not shown. Enzymatic steps are as follows: 1, generation of GlcNAc-PI from UDP-GlcNAc and phosphatidylinositol; 2, deacetylation of GlcNAc-PI to GlcN-PI; 3, acylation of GlcN-PI; 4, GlcN-acyl-PI is flipped from cytosolic to lumenal side of ER; 5, first mannose transferred to GlcN-acyl-PI by PIG-M enzyme; 6, phosphoethanolamine side chain added to Man-GlcN-acyl-PI; 7 and 8, second and third mannoses added to EtNP-Man-GlcN-acyl-PI; 9, EtNP added to third mannose of Man-Man-(EtNP)Man-GlcN-acyl-PI; 10, transamidation transfer of C-terminal membrane bound protein precursor to GPI anchor; 11, secretion of GAP (by way of Golgi) to plasma membrane. Hatched box: proposed GPI anchor synthesis pathway in pnt1 cells (steps 1 to 6′). In the pnt1 mutant, steps 1 to 4 occur as in wild-type cells, but in the absence of PNT1 function, the first mannose is not added to GlcN-acyl-PI, and GPI-anchor biosynthesis is arrested (step 5′). In the absence of a complete GPI anchor, the C-terminal membrane-bound protein precursor cannot be transferred to a GPI anchor and is eventually degraded (step 6′). Figure modified from Kinoshita and Inoue (2000).
The phenotype of pnt1 embryos demonstrates that GAPs are required for cell wall biosynthesis and proper development of the shoot and root meristem during embryogenesis. Lalanne et al. (2004) have also recently described mutants of Arabidopsis with defects in GPI anchoring. The seth1 and seth2 mutants were recovered as male sterile plants in which pollen tube growth is defective. SETH1 corresponds to PIG-C, phosphatidylinositol glucan synthase subunit C, and SETH2 corresponds to PIG-A, GPI-N-acetylglucosaminyltransferase. Both proteins are part of the GPI-N-acetylglucosaminyltransferase enzyme complex that is required for addition of GlcNAc to the phosphatidylinositol membrane anchor, the first step in GPI anchor biosynthesis (step 1 in Figure 8; Kinoshita and Inoue, 2000). Thus, both SETH genes act at an earlier step in GPI anchor biosynthesis than PNT1. Because the seth mutants are essentially completely male sterile, the phenotype of homozygous embryos was not investigated by Lalanne et al. (2004). We expect that homozygous seth embryos would have the same phenotype as the pnt mutants.
That the pnt1-1 allele is transmitted through the pollen (albeit at a low frequency) may indicate that this allele encodes some residual GPI-mannosyltransferase I activity. In support of this hypothesis, the stop codon in the pnt1-1 allele occurs after the signature DxD glycosyltransferase motif shown by Maeda et al. (2001) to be essential for PIG-M enzyme activity, and we detected a low amount of COB protein in pnt1 embryos. Analysis of pnt1-2, a T-DNA–induced allele of PNT1, did not help to resolve this issue because the T-DNA insertion in pnt1-2 occurs near the C terminus of the protein, downstream of the stop codon in pnt1-1 and further downstream of the DxD glycosyltransferase motif. Indeed, the pnt1-2 allele was seen to have a weaker pollen transmission defect than the pnt1-1 allele.
Importance of GAPs for Cell Wall Biosynthesis
pnt1 embryos have a significant decrease in crystalline cellulose, coupled with increases in cell wall neutral sugars and pectin. Furthermore, pnt1 embryos also show considerable amounts of ectopic deposition of cell wall components, including callose. How can these observations be interpreted? It is likely that the effect on the cell wall of loss of PNT1 function is indirect: the cell wall phenotypes of pnt1 embryos are the result of the absence of GAPs that are required for cell wall synthesis and assembly. Recent mutational analysis of a small number of GAPs, the prediction of the total set of GAPs in Arabidopsis, and some inhibitor studies allow the identification of candidate proteins and processes responsible for the cell wall defects observed in pnt1 embryos.
Conditional mutations in the COB gene result in a decrease in crystalline cellulose of ∼30% in seedling roots (Schindelman et al., 2001). The BC1 gene of rice is a homolog of COB, and bc1 plants have a 30% reduction in crystalline cellulose in vascular bundles and ground tissue (Li et al., 2003). Although the biochemical role of COB in cellulose synthesis is unknown, this genetic evidence demonstrates that COB-like genes are required for normal levels of cellulose. We found that COB was undetectable in embryo extracts but present in callus tissue, where levels of COB protein are strongly reduced in pnt1 compared with the wild type. COB is one of an 11-member gene family, several of which are expressed during embryogenesis, and all of which have been either predicted or shown to be GPI anchored (Roudier et al., 2002; Borner et al., 2003). The magnitude of cellulose decrease in cob roots (∼30%) is similar to that seen in pnt1 embryos (40%). Thus, a likely explanation for the cellulose decrease seen in pnt1 embryos is that lack of a functional GPI anchor in pnt1 mutants leads to degradation or mislocalization of the COB family members that are normally required for cellulose synthesis or deposition during embryogenesis.
Genetic and inhibitor studies also point to an important role for AGPs in cell expansion and cell wall assembly. AGPs are extracellular proteoglycans that are often expressed in a cell- or tissue-specific manner (Maewska-Sawka and Nothnagel, 2000). All 16 classical AGPs of Arabidopsis are predicted to be GPI anchored (Schultz et al., 2000). Growth of Arabidopsis seedlings on Yariv reagent, a synthetic phenyl glycoside that binds AGPs, resulted in seedlings with short, swollen roots (Willats and Knox, 1996). In addition, the root epidermal bulger1 mutant of Arabidopsis has decreased amounts of AGPs in the root epidermal cells (Ding and Zhu, 1997; Seifert et al., 2002). Loss of SOS5, a plasma membrane–localized protein with AGP and fasciclin domains that is predicted to be GPI anchored, results in seedlings with swollen root tips when grown under salt stress conditions. Cell walls of the sos5 mutant have decreased electron-dense staining in transmission electron micrographs, characteristic of decreased amounts of pectin (Shi et al., 2003).
Perhaps the strongest evidence for the role of AGPs in contributing to cell wall structure comes from a study by Roy et al. (1998), where lily (Lilium longiflorum) pollen tubes were grown in the presence of Yariv reagent. Ultrastructural examination of treated pollen tubes revealed that Yariv reagent interfered with cell wall assembly; secretory vesicles released their contents into the periplasmic space, but these vesicle contents, primarily pectin and callose, the main carbohydrates of pollen walls, were not integrated into the cell wall in a normal manner. Instead, large amounts of these materials accumulated randomly in the extracellular space, in a manner strikingly similar to what has been observed in pnt1 embryos. Roles for AGPs in chaperoning cell wall polymers through the secretory pathway and in integrating these polymers into the cell wall have previously been proposed (Gibeaut and Carpita, 1994; Kieliszewski and Lamport, 1994). Thus, the abnormal deposition of pectin and xyloglucans in pnt1 mutants might be due to the absence of AGPs. This could also explain the increased amounts of cell wall material deposited in pnt1 embryos: a lack of properly integrated pectins and xyloglucans might prompt the cells to attempt to compensate for this deficiency by increasing the synthesis and export of these polymers.
Ectopic callose deposition is characteristic of the pnt1 phenotype. Large amounts of ectopic callose production during embryogenesis were previously observed in the cyt1 mutant, which shares several phenotypic aspects with pnt1, including reduced crystalline cellulose and abnormal accumulation of cell wall material (Nickle and Meinke, 1998; Lukowitz et al., 2001). This phenotypic similarity is not a coincidence because the CYT1 gene encodes an enzyme that is required for the production of GDP-mannose, an essential component of the core GPI anchor as well as the core N-glycan (Lukowitz et al., 2001). pnt1 and cyt1 embryos are unique among cell wall mutants of plants in the extent of their uncontrolled deposition of xyloglucans and pectins into the extracellular space. This uncontrolled wall deposition, coupled with a decrease in cellulose, likely compromises the mechanical integrity of the wall and could therefore trigger callose synthesis as part of a stress or wound response. A second, nonexclusive explanation for the accumulation of callose in the mutants is suggested by the experiments showing that treatment of Arabidopsis cell cultures with Yariv reagent induces callose production and programmed cell death (Gao and Showalter, 1999; Guan and Nothnagel, 2004). These experiments with Yariv reagent also suggest that a lack of AGPs in pnt1 embryos and cells might be responsible for the seedling lethality of pnt1 and necrosis seen in pnt1 callus. It is intriguing that callus grown from pnt1 embryos lacks the massive ectopic deposition of cell wall material seen in pnt1 embryos. This suggests that there is a fundamental difference in the importance of GAPs to cell wall synthesis in embryos and callus. The difference might arise because of a different composition of the structural components of the cell wall in callus cells, as suggested by analysis of sugar composition of different tissues of Arabidopsis (Richmond and Somerville, 2001).
As in Arabidopsis, GPI anchor synthesis mutants in yeast are lethal (Leidich et al., 1994). Yeast GAPs are required for maintaining the architecture of the yeast cell wall, and the GPI anchor itself is often incorporated along with the GAP as a structural component of the wall (Kapteyn et al., 1999). Our analysis of pnt1 embryos shows that in plants, like in yeast, GAPs play an important role in organizing cell wall structure. However, only a subset of the GAPs of Arabidopsis are predicted to be involved in cell wall synthesis or assembly (Borner et al., 2003). Many of the remaining GAPs of Arabidopsis likely play essential roles in cell–cell signaling and multicellular development.
METHODS
Plant Material, Growth Conditions, and Genetic Mapping
All mutant lines, isolated in the genetic screens described by Gillmor et al. (2002), were generated in the Landsberg erecta ecotype. The pnt1-1 line P13 (ABRC stock number CS6120) and pnt2-1 line 2-1 (ABRC stock number CS6121) were generated by ethyl methanesulfonate mutagenesis. The pnt2-2 line FN70, the pnt3-1 line FN45 (ABRC stock number CS6122), the pnt4-1 line FN72 (ABRC stock number CS6123), and the pnt5-1 line FN105 were generated by fast neutron mutagenesis. SALK_069393 containing the T-DNA allele pnt1-2 in the Columbia ecotype was obtained from the ABRC. Mutant lines are available from the ABRC through The Arabidopsis Information Resource at www.arabidopsis.org. Plants in soil and seedlings on tissue culture plates were grown as described by Gillmor et al. (2002). Callus was produced from wild-type and pnt1 embryos dissected from seed coats at the late bent cotyledon stage, while the embryos were still green, and grown on media described by Encina et al. (2001). Mapping of PNT genes was performed as described by Lukowitz et al. (2000). Primer sequences for all PCR-based genetic markers are available from The Arabidopsis Information Resource at www.arabidopsis.org.
Confocal, Light, and Electron Microscopy
Preparation of embryos for confocal and electron microscopy, collection of data, and processing of data were performed as described by Gillmor et al. (2002). For fluorescent immunostaining of embryo sections, osmium tetroxide–stained semithin sections were first etched in 0.5% H2O2 for 10 min, followed by two rinses with distilled water for 5 min, then 100 mM HCl for 10 min, followed by two rinses with distilled water for 5 min. Staining with CCRC M1, JIM5, and anti-β-1,3 glucan (Biosupplies, Parkville, Australia) monoclonal antibodies was then performed as described by Rhee and Somerville (1998). Callose staining with sirofluor was performed as described by Lukowitz et al. (2001). Fluorescence and immunofluorescence embryo images were acquired with a Leica DMRB compound microscope (Wetzlar, Germany) with 20× air and 100× oil objectives using a SPOT camera (Diagnostic Instruments, Sterling Heights, MI). Immunofluorescence images of callose were aquired with a Nikon Eclipse E600 microscope (Tokyo, Japan) at 40× magnification using a Spot RT Slider camera (Diagnostic Instruments). Confocal images of embryos were obtained with a Bio-Rad MRC 1024 confocal system (Hercules, CA) with a Nikon microscope using 20× and 40× air objectives. Light and confocal microscopy were performed at room temperature (∼22°C). Transmission electron microscopy images were obtained with Philips 410 (Eindhoven, The Netherlands) and Jeol 1230 transmission electron microscopes (Peabody, MA).
Molecular Biology
The PNT1 gene (At5g22130) was sequenced directly from gel-purified PCR products amplified from genomic DNA prepared from wild-type and pnt1-1 embryos, using an ABI 310 sequencer (Perkin-Elmer Applied Biosystems, Foster City, CA). For complementation of the pnt1 phenotype, a 3641-bp Eco147I/AatII genomic fragment was excised from BAC T6G21 (obtained from the ABRC) and subcloned into the pCAMBIA 3300 binary vector, modified to contain Eco147I and AatII sites, to produce plasmid pCS702. Plasmid pCS702 was then transformed into Agrobacterium tumefaciens and used to transform a mixed population of pnt1/PNT1 and PNT1/PNT1 plants. pnt1/pnt1 homozygous plants were identified in the T1 generation, indicating complementation of the pnt1 mutation. RNA gel blot analysis of PNT1 gene expression was performed according to standard protocols using the full-length PNT1 cDNA labeled with 32P and total RNA extracted with Trizol reagent (Sigma-Aldrich, St. Louis, MO).
Protein Gel Blot Analysis
Wild-type and pnt embryo and callus tissue were ground in a micro mortar and pestle in 50 μL of grinding buffer (50 mM sodium phosphate buffer, pH 8.0, and 300 mM NaCl) plus 1× Calbiochem protease inhibitor cocktail set I (San Diego, CA). Proteins were quantified using a Bradford protein assay kit (Bio-Rad). Ten micrograms of total protein extract was loaded per lane, separated by 10% SDS-PAGE, and electroblotted using semidry transfer to polyvinylidene difluoride membrane. SKU5 primary antibody was used at a dilution of 1:1000, protein disulfide isomerase antibody (http://rosebiotech.com/) was used at a dilution of 1:2500, COB antibody used at a dilution of 1:3000, and PMR6 antibody used at a dilution of 1:3000. Primary antibodies were visualized by incubation with goat anti-rabbit IgG secondary antibodies conjugated to horseradish peroxidase (Bio-Rad) followed by a chemiluminescence reaction (SuperSignal; Pierce Chemical, Rockford, IL).
Cell Wall Analysis
Cell wall analysis of wild-type and pnt1 embryos was performed as described by Gillmor et al. (2002).
Acknowledgments
The expert technical assistance of R. Gillmor (Carnegie Institution, Stanford, CA) is gratefully acknowledged. Thanks to F. Roudier and P. Benfey (Duke University, Durham, NC) for the kind gift of COBRA antibody, to J. Vogel (USDA, Albany, CA) and S. Somerville (Carnegie Institution) for the PMR6 antibody, to P. Knox (Leeds University, Leeds, UK) for the JIM5 antibody, and to M. Hahn (University of Georgia, Athens, GA) for the CCRC M1 antibody. Thanks to B. Fang and A. Paredez for genetic mapping, to H. Youngs for advice on cell wall chemistry, and to Genoscope (INRA, Evry, France) for the full-length PNT1 cDNA. C.S.G. was supported by a U.S. Department of Energy/National Science Foundation/USDA triagency training grant and by a grant from the U.S. Department of Energy (DE-FG02-03ER20133). W.L. was supported by a fellowship from the Human Frontier Science Program, a Barbara McClintock fellowship from the Carnegie Institution, and by USDA Grant CSREES 00-35304-9394. J.C.S. was supported by a National Institutes of Health postdoctoral fellowship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Chris Somerville (crs@stanford.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.031815.
References
- Acosta-García, G., and Vielle-Calzada, J.-P. (2004). A classical arabinogalactan protein is essential for the initiation of female gametogenesis in Arabidopsis. Plant Cell 16, 2614–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeckman, T., Przemeck, G.H.K., Stamatiou, G., Lau, R., Terryn, N., De Rycke, R., Inzé, D., and Berleth, T. (2002). Genetic complexity of cellulose synthase A gene function in Arabidopsis embryogenesis. Plant Physiol. 130, 1883–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacio, J.S., and Lippincott-Schwartz, J. (1991). Degradation of proteins within the endoplasmic reticulum. Curr. Opin. Cell Biol. 3, 592–600. [DOI] [PubMed] [Google Scholar]
- Borner, G.H., Lilley, K.S., Stephens, T.J., and Dupree, P. (2003). Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A proteomic and genomic analysis. Plant Physiol. 132, 568–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner, G.H., Sherrier, D.J., Stevens, T.J., Arkin, I.T., and Dupree, P. (2002). Prediction of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A genomic analysis. Plant Physiol. 129, 486–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner, G.H., Sherrier, D.J., Weimar, T., Michaelson, L.V., Hawkins, N.D., Macaskill, A., Napier, J.A., Beale, M.H., Lilley, K.S., and Dupree, P. (2005). Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 137, 104–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burn, J.E., Hurley, U.A., Birch, R.J., Arioli, T., Cork, A., and Williamson, R.E. (2002). The cellulose deficient Arabidopsis mutant rsw3 is defective in a gene encoding a putative glucosidase II, an enzyme processing N-glycans during ER quality control. Plant J. 32, 949–960. [DOI] [PubMed] [Google Scholar]
- Caro, L.H.P., Tettelin, H., Vossen, J.H., Ram, A.F.J., Van Den Ende, H., and Klis, F.M. (1997). In silico identification of glycosyl-phosphatidylinositol-anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast 13, 1477–1489. [DOI] [PubMed] [Google Scholar]
- Delmer, D. (1987). Cellulose biosynthesis. Annu. Rev. Plant Physiol. 38, 259–290. [Google Scholar]
- Ding, L., and Zhu, J.-K. (1997). A role for arabinogalactan-proteins in root epidermal cell expansion. Planta 203, 289–294. [DOI] [PubMed] [Google Scholar]
- Eisenhaber, B., Wildpaner, M., Schultz, C.J., Borner, G.H., Dupree, P., and Eisenhaber, F. (2003). Glycosylphosphatidylinositol lipid anchoring of plant proteins. Sensitive prediction from sequence- and genome-wide studies for Arabidopsis and rice. Plant Physiol. 133, 1691–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encina, C.L., Constantin, M., and Botella, J. (2001). An easy and reliable method for establishment and maintenance of leaf and root cell cultures of Arabidopsis thaliana. Plant Mol. Biol. Report. 19, 245–248. [Google Scholar]
- Ferguson, M.A.J. (1999). The structure, biosynthesis and functions of glycosylphosphatidylinositol anchors, and the contributions of trypanosome research. J. Cell Sci. 112, 2799–2809. [DOI] [PubMed] [Google Scholar]
- Field, M.C., Moran, P., Li, W., Keller, G.-A., and Caras, I.W. (1994). Retention and degradation of proteins containing an uncleaved glycosylphosphatidylinositol signal. J. Biol. Chem. 269, 10830–10837. [PubMed] [Google Scholar]
- Gao, M., and Showalter, A.M. (1999). Yariv reagent treatment incudes programmed cell death in Arabidopsis cell cultures and implicates arabinogalactan protein involvement. Plant J. 19, 321–331. [DOI] [PubMed] [Google Scholar]
- Gaspar, Y., Johnson, K.L., McKenna, J.A., Bacic, A., and Schulz, C.J. (2001). The complex structures of arabinogalactan-proteins and the journey towards understanding function. Plant Mol. Biol. 47, 161–176. [PubMed] [Google Scholar]
- Gibeaut, D.M., and Carpita, N.C. (1994). Biosynthesis of plant cell wall polysaccharides. FASEB J. 8, 904–915. [DOI] [PubMed] [Google Scholar]
- Gillmor, C.S., Poindexter, P., Lorieau, J., Palcic, M.M., and Somerville, C. (2002). α-Glucosidase I is required for cellulose biosynthesis and morphogenesis in Arabidopsis. J. Cell Biol. 156, 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, Y., and Nothnagel, E.A. (2004). Binding of arabinogalactan proteins by Yariv phenylglycoside triggers wound-like responses in Arabidopsis cell cultures. Plant Physiol. 135, 1346–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens, G., and Mayer, U. (1994). Arabidopsis. In Embryos: Color Atlas of Development, J.B.L. Bard, ed (London: Wolfe), pp. 7–21.
- Kapteyn, J.C., Van Den Ende, H., and Klis, F.M. (1999). The contribution of cell wall proteins to the organization of the yeast cell wall. Biochim. Biophys. Acta 1426, 373–383. [DOI] [PubMed] [Google Scholar]
- Kieliszewski, M.J., and Lamport, D.T.A. (1994). Extensin: Repetitive motifs, functional sites, post-translational codes, and phylogeny. Plant J. 5, 157–172. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T., and Inoue, N. (2000). Dissecting and manipulating the pathway for glycosylphosphatidylinositol-anchor biosynthesis. Curr. Opin. Chem. Biol. 4, 632–638. [DOI] [PubMed] [Google Scholar]
- Knox, J.P., Linstead, P.J., King, J., Cooper, C., and Roberts, K. (1990). Pectin esterification is spatially regulated both within cell walls and between developing tissues of root apices. Planta 181, 512–521. [DOI] [PubMed] [Google Scholar]
- Lalanne, E., Honys, D., Johnson, A., Borner, G.H.H., Lilley, K.S., Dupree, P., Grossniklaus, U., and Twell, D. (2004). SETH1 and SETH2, two components of the glycosylphosphatidylinositol anchor biosynthetic pathway, are required for pollen germination and tube growth in Arabidopsis. Plant Cell 16, 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidich, S.D., Drapp, D.A., and Orlean, P. (1994). A conditionally lethal yeast mutant blocked at the first step in glycosyl phosphatidylinositol anchor synthesis. J. Biol. Chem. 269, 10193–10196. [PubMed] [Google Scholar]
- Li, Y., Qian, Q., Zhou, Y., Yan, M., Sun, L., Zhang, M., Fu, Z., Wang, Y., Han, B., Pang, X., Chen, M., and Li, J. (2003). BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell 15, 2020–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz, W., Gillmor, C.S., and Scheible, W.-R. (2000). Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol. 123, 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz, W., Nickle, T.C., Meinke, D.W., Last, R.L., Conklin, P.L., and Somerville, C.R. (2001). Arabidopsis cyt1 mutants are deficient in a mannose-1-phosphate guanylyltransferase and point to a requirement of N-linked glycosylation for cellulose biosynthesis. Proc. Natl. Acad. Sci. USA 98, 2262–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, Y., Watanabe, R., Harris, C.L., Hong, Y., Ohishi, K., Kinoshita, K., and Kinoshita, T. (2001). PIG-M transfers the first mannose to glycosylphosphatidylinositol on the lumenal side of the ER. EMBO J. 20, 250–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maewska-Sawka, A., and Nothnagel, E.A. (2000). The multiple roles of arabinogalactan proteins in plant development. Plant Physiol. 122, 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickle, T.C., and Meinke, D.W. (1998). A cytokinesis-defective mutant of Arabidopsis (cyt1) characterized by embryonic lethality, incomplete cell walls, and excessive callose accumulation. Plant J. 15, 321–332. [DOI] [PubMed] [Google Scholar]
- Peskan, T., Westermann, M., and Oelmüller, R. (2000). Identification of low-density Triton X-100-insoluble plasma membrane microdomains in higher plants. Eur. J. Biochem. 267, 6989–6995. [DOI] [PubMed] [Google Scholar]
- Puhlmann, J., Bucheli, E., Swain, M.J., Dunning, N., Albersheim, P., Darvill, A.G., and Hahn, M.G. (1994). Generation of monoclonal antibodies against plant cell-wall polysaccharides. I. Characterization of a monclonal antibody to a terminal α-(1-->2)-linked fucosyl-containing epitope. Plant Physiol. 104, 699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee, S.Y., and Somerville, C.R. (1998). Tetrad pollen formation in quartet mutants of Arabidopsis thaliana is associated with persistence of pectic polysaccharides of the pollen mother cell wall. Plant J. 15, 79–88. [DOI] [PubMed] [Google Scholar]
- Richmond, T., and Somerville, C.R. (2001). Integrative approaches to determining Csl function. Plant Mol. Biol. 47, 131–143. [PubMed] [Google Scholar]
- Roudier, F., Schindelman, G., DeSalle, R., and Benfey, P.N. (2002). The COBRA family of putative GPI-anchored proteins in Arabidopsis: A new fellowship in expansion. Plant Physiol. 130, 538–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, S., Jauh, G.Y., Hepler, P.K., and Lord, E.M. (1998). Effects of Yariv phenylglycoside on cell wall assembly in the lily pollen tube. Planta 204, 450–458. [DOI] [PubMed] [Google Scholar]
- Schindelman, G., Morikami, A., Jung, J., Baskin, T.I., Carpita, N.C., Derbyshire, P., McCann, M.C., and Benfey, P.N. (2001). COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 15, 1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, C.J., Johnson, K.L., Currie, G., and Bacic, A. (2000). The classical arabinogalactan protein gene family of Arabidopsis. Plant Cell 12, 1751–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook, J.C., Carroll, K.L., Hung, K.F., Masson, P.H., and Somerville, C.R. (2002). The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell 14, 1635–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert, G.J., Barber, C., Wells, B., Dolan, L., and Roberts, K. (2002). Galactose biosynthesis in Arabidopsis: Genetic evidence for substrate channeling from UDP-D-Galactose into cell wall polymers. Curr. Biol. 12, 1840–1845. [DOI] [PubMed] [Google Scholar]
- Selleck, S.B. (2000). Proteoglycans and pattern formation. Sugar biochemistry meets developmental genetics. Trends Genet. 16, 206–212. [DOI] [PubMed] [Google Scholar]
- Shi, H., Kim, Y.-S., Guo, Y., Stevenson, B., and Zhu, J.-K. (2003). The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell 15, 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, B.A., Evans, N.A., Bonig, I., and Clarke, A.E. (1984). The application of Sirofluor, a chemically defined fluorochrome from analine blue for the histochemical detection of callose. Protoplasma 122, 191–195. [Google Scholar]
- Sun, W., Kieliszewski, M.J., and Showalter, A.M. (2004. a). Overexpression of tomato LeAGP-1 arabinogalactan-protein promotes lateral branching and hampers reproductive development. Plant J. 40, 870–881. [DOI] [PubMed] [Google Scholar]
- Sun, W., Zhao, Z.D., Hare, M.C., Kieliszewski, M.J., and Showalter, A.M. (2004. b). Tomato LeAGP-1 is a plasma membrane-bound, glycosylphosphatidylinositol-anhored arabinogalactan-protein. Physiol. Plant. 120, 319–327. [DOI] [PubMed] [Google Scholar]
- Torres-Ruíz, R.A., and Jürgens, G. (1994). Mutations in the FASS gene uncouple pattern formation and morphogenesis in Arabidopsis development. Development 120, 2967–2978. [DOI] [PubMed] [Google Scholar]
- Vogel, J.P., Raab, T.K., Schiff, C., and Somerville, S.C. (2002). PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 14, 2095–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willats, W.G.T., and Knox, J.P. (1996). A role for arabinogalactan-proteins in plant cell expansion: Evidence from studies on the interaction of B-glucosyl Yariv reagent with seedlings of Arabidopsis thaliana. Plant J. 9, 919–925. [DOI] [PubMed] [Google Scholar]
- Williamson, R.E., Burn, J.E., and Hocart, C.H. (2002). Towards the mechanism of cellulose synthesis. Trends Plant Sci. 7, 461–467. [DOI] [PubMed] [Google Scholar]
- Youl, J.J., Bacic, A., and Oxley, D. (1998). Arabinogalactan-proteins from Nicotiana alata and Pyrus communis contain glycosylphosphatidylinositol membrane anchors. Proc. Natl. Acad. Sci. USA 95, 7921–7926. [DOI] [PMC free article] [PubMed] [Google Scholar]