Abstract
Cullin-based E3 ubiquitin ligases play important roles in the regulation of diverse developmental processes and environmental responses in eukaryotic organisms. Recently, it was shown in Schizosaccharomyces pombe, Caenorhabditis elegans, and mammals that Cullin3 (CUL3) directly associates with RBX1 and BTB domain proteins in vivo to form a new family of E3 ligases, with the BTB protein subunit functioning in substrate recognition. Here, we demonstrate that Arabidopsis thaliana has two redundant CUL3 (AtCUL3) genes that are essential for embryo development. Besides supporting anticipated specific AtCUL3 interactions with the RING protein AtRBX1 and representative Arabidopsis proteins containing a BTB domain in vitro, we show that AtCUL3 cofractionates and specifically associates with AtRBX1 and a representative BTB protein in vivo. Similar to the AtCUL1 subunit of the SKP1-CUL1-F-box protein–type E3 ligases, the AtCUL3 subunit of the BTB-containing E3 ligase complexes is subjected to modification and possible regulation by the ubiquitin-like protein Related to Ubiquitin in vivo. Together with the presence of large numbers of BTB proteins with diverse structural features and expression patterns, our data suggest that Arabidopsis has conserved AtCUL3-RBX1-BTB protein E3 ubiquitin ligases to target diverse protein substrates for degradation by the ubiquitin/proteasome pathway.
INTRODUCTION
It has become increasingly clear that the regulation of many cellular and developmental processes involves ubiquitin-dependent degradation of critical proteins (Hershko and Ciechanover, 1998; Hellmann and Estelle, 2002; Sullivan et al., 2003). Substrates selected for degradation by this pathway are specifically tagged by a chain of ubiquitin molecules, which directs them to the 26S proteasome. Attachment of ubiquitin molecules to a target protein involves a cascade of three steps, catalyzed by an E1 ubiquitin-activating enzyme, an E2 ubiquitin-conjugating enzyme, and an E3 ubiquitin ligase. Typically, the E2 enzyme associates with an E3 ubiquitin ligase, which is primarily responsible for recognizing the substrate. As the E3 ligase action determines the life or death of a protein, it is critical to understand how these enzymes select their targets.
Ubiquitin E3 ligases come in a large variety of forms, including multisubunit complexes organized around a cullin (CUL) scaffolding protein. The paradigm for this large group of ligases is based on studies of the SCF (for SKP1-CUL-F-box) complex. A typical SCF complex consists of a cullin family member, CUL1, a RING-finger protein RBX1 that interacts with the cullin and the E2 conjugating enzyme, a SKP1-like adaptor protein, and an F-box protein that confers the specificity for the recruitment of the substrate (Schulman et al., 2000; Zheng et al., 2002; Deshaies, 2003). CUL2 forms a similar group of E3 ligases, in which an elongin C-SOCS module replaces the SKP1-F-box protein module (Stebbins et al., 1999). These E3 ligases can target a range of substrates for protein degradation, as determined by the specificity of their associated F-box or SOCS protein.
A series of recent studies in Caenorhabditis elegans (Kurz et al., 2002; Pintard et al., 2003b; Xu et al., 2003), Schizosaccharomyces pombe (Geyer et al., 2003), and mammals (Furukawa et al., 2003; Kobayashi et al., 2004) has shed light on CUL3-based ubiquitin ligases. The findings include identification of an in vivo substrate (Kurz et al., 2002; Pintard et al., 2003a) and a novel type of substrate specificity adaptors for CUL3-based ligases (Geyer et al., 2003; Pintard et al., 2003b; Xu et al., 2003). The substrate specificity factors associate with CUL3 through a Bric a brac, Tramtrack, and Broad complex/Pox virus and Zinc finger (BTB/POZ) domain. Surprisingly, the same BTB protein was found to also bind the substrate, thus combining in one protein the functions of the SKP1 adaptor and the F-box subunits. One of these BTB proteins, MEL-26, binds to the microtubule-severing protein MEI-1/katanin (Pintard et al., 2003b; Xu et al., 2003), which is an essential component of the C. elegans meiotic spindle (Clark-Maguire and Mains, 1994). Although the precise role of the MEI-1 protein is unknown, it has been found that MEI-1 is degraded at the meiosis-to-mitosis transition and that CUL3 is required for its degradation (Pintard et al., 2003a). Genetic studies, together with the finding that MEL-26 physically interacts with MEI-1 (Pintard et al., 2003b; Xu et al., 2003), strongly indicate that a CUL3-based ubiquitin ligase containing MEL-26 might target MEI-1 for degradation. In C. elegans, depletion of CUL3 by RNA interference causes defects in early embryogenesis, resulting in abnormal microfilament and microtubule organization (Kurz et al., 2002). Drosophila melanogaster CUL3 is involved in Hedgehog signaling by regulating the stability of the CUBITUS INTERRUPTUS protein, thus affecting external sensory organ development, pattern formation, and cell growth and survival (Mistry et al., 2004). The mammalian CUL3 ubiquitin ligase has been implicated in the turnover of monomeric forms of cyclin E, and mice lacking cul3 die at embryonic day 6.5 with defects in both embryonic and extraembryonic compartments (Singer et al., 1999; Winston et al., 1999). Plants such as Arabidopsis thaliana have closely related CUL3 proteins (Risseeuw et al., 2003). The presence of CUL3-based multisubunit protein ligases was implied by a recent report of an in vitro interaction between Arabidopsis CUL3 and the BTB protein ETO1, which is involved in ethylene biosynthesis regulation (Wang et al., 2004).
Here, we show the existence in Arabidopsis of CUL3 (AtCUL3)-containing multisubunit E3 ubiquitin ligase complexes and demonstrate that AtCUL3 is essential for embryo development. Furthermore, we show that AtCUL3 interacts in vitro with several proteins containing a BTB domain, but not with the SKP1-like proteins ASK1 or ASK2. The domains responsible for this interaction have been identified both in AtCUL3 and in the BTB domain–containing proteins. In agreement with our in vitro findings, we further demonstrate that the in vivo AtCUL3 complex contains the RING finger protein RBX1 as well as a BTB protein and that the AtCUL3 subunit is subjected to modification by RUB (for Related to Ubiquitin). Together, our studies confirm that plants possess a large number of AtCUL3-RBX-BTB E3 ubiquitin ligase complexes that target proteins for degradation via the ubiquitin/proteasome pathway.
RESULTS
Arabidopsis Has Two Active AtCUL3 Genes
Analysis of the Arabidopsis genome revealed the presence of at least 10 predicted cullin-related genes (Risseeuw et al., 2003). Six of these contain reading frames with apparently intact canonical N- and C-terminal domains. At least five (AtCUL1, AtCUL2, AtCUL3A, AtCUL3B, and AtCUL4) of those six genes are expressed, as indicated by the presence of their corresponding EST clones (Risseeuw et al., 2003). Phylogenetic analysis of cullins from Arabidopsis and from other eukaryotes indicates that AtCUL3A and AtCUL3B are two closely related members of a two-gene family and are clustered in the same clade with CUL3 from Homo sapiens, D. melanogaster, C. elegans, S. pombe, and Saccharomyces cerevisiae (Risseeuw et al., 2003).
Both of the AtCUL3A and AtCUL3B genes have two exons and encode proteins of 732 amino acids residues (GenBank accession numbers CAC87120 and AAG52544, respectively). The predicted protein sequences for AtCUL3A and AtCUL3B share 88% identity (Figure 1A; see Supplemental Figure 1 online), with AtCUL3A being slightly more closely related to its human homolog hCUL3 (52% amino acid identity) (Figure 1A). The greatest similarity between these proteins is in the C-terminal region, which contains both the RUB/NEDD8 conjugation site (del Pozo and Estelle, 1999) and the cullin homology domain (Figure 1A). This domain represents a conserved module and is responsible for connecting an E2 ubiquitin-conjugating enzyme to the ubiquitin ligase (Yu et al., 1998).
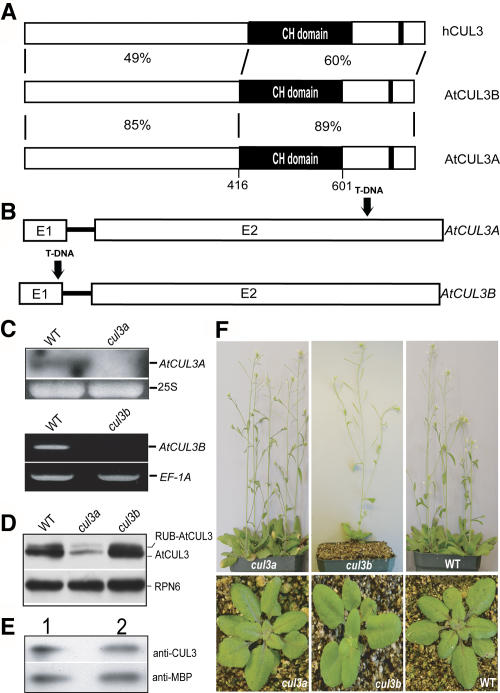
Figure 1.
Characterization of Two Redundant AtCUL3 Genes and Their Mutant Phenotypes.
(A) Schematic comparison of the AtCUL3 proteins with the human ortholog. The black-boxed regions of the AtCUL3 protein denote the cullin homology domain. The small black bars toward the C terminus denote the RUB modification site. The identity percentages of the homologous regions of the Arabidopsis and human proteins are indicated.
(B) Representation of the AtCUL3A and AtCUL3B genes and the T-DNA insertion sites, as indicated by black arrows. Both genes have two exons, denoted E1 and E2.
(C) The AtCUL3A and AtCUL3B mRNAs are not detectable in the cul3a and cul3b mutants, respectively. RNA gel blot analysis was performed using a probe specific for the 3′ untranslated region of the AtCUL3A gene (top), whereas RT-PCR was used to examine AtCUL3B expression (bottom). WT, total RNA from wild-type seedlings; cul3a, total RNA from the cul3a mutant; cul3b, total RNA from the cul3b mutant. The intensities of 25S rRNA and EF-1A were used as equal loading controls for RNA gel blot or RT-PCR, respectively.
(D) Effect of cul3a and cul3b mutations on AtCUL3 protein accumulation. Protein blot analysis was performed using antibodies against AtCUL3A. WT, total proteins from wild-type seedlings; cul3a, total proteins from cul3a mutant seedlings; cul3b, total proteins from cul3b mutant seedlings. An anti-RNP6 antibody was used as an equal loading control. For all samples, 24 μg of total protein from 6-d-old seedlings was used.
(E) Anti-CUL3 antibody has similar affinity for the AtCUL3A and AtCUL3B proteins. MBP-AtCUL3A (lane 1) and MBP-AtCUL3B (lane 2) were expressed in E. coli, and the protein extracts were examined by immunoblot analysis using anti-CUL3 and anti-MBP antibodies.
(F) Both the cul3a and cul3b mutant plants have no obvious change from the wild-type (WT) phenotype. Plants at top are 5 weeks old, and those at bottom are 3 weeks old.
AtCUL3A and AtCUL3B Are Both Expressed and Are Subjected to Posttranslational Modification
As an initial step in studying the biological role of the AtCUL3 proteins, we conducted a database search for Arabidopsis T-DNA insertion mutant lines. We found one T-DNA line that contains an insertion located within the second exon of AtCUL3A, 417 bp upstream of the STOP codon (Figure 1B). A T-DNA insertion line for the AtCUL3B gene was also identified, with the T-DNA inserted 155 bp downstream of the START codon (Figure 1B). Both insertions likely give rise to null mutations, as neither homozygous line expresses the AtCUL3A or AtCUL3B mRNA, respectively (Figure 1C). Protein blot analyses of protein extract from Arabidopsis cul3a mutant seedlings showed a drastic reduction in total AtCUL3 protein level with respect to wild-type plants, whereas no significant differences in overall AtCUL3 level were observed between the cul3b mutants and wild-type siblings (Figure 1D). Because the anti-CUL3 antibody has similar affinity for both AtCUL3 proteins (Figure 1E), the results shown in Figure 1D suggest that AtCUL3A may be predominantly expressed with respect to AtCUL3B in seedlings.
The presence of two specific bands for CUL3 in the protein blot analysis of seedling extracts (Figure 1D) suggests that AtCUL3 might be posttranslationally modified by the attachment of the ubiquitin-like protein RUB (called NEDD8 in mammals and yeast). In fact, it has been shown that CUL3, like other cullins, can be covalently modified by the attachment of RUB/NEDD8 (Pintard et al., 2003a). In the case of the SCF complex, the RUB/NEDD8 modification of CUL1 (rubylation) provides an important positive regulation of the E3 ligase activity, enhancing E2 recruitment and polyubiquitination of the substrate (Wu et al., 2000; Kawakami et al., 2001). In the case of CUL3, rubylation and derubylation have been shown to be required for targeting MEI-1/katanin for degradation in C. elegans (Pintard et al., 2003a).
To test whether Arabidopsis CUL3 is covalently modified by RUB, the same protein extracts probed with anti-CUL3 antibody (Figure 1D) were immunoblotted against anti-RUB antibodies. The results demonstrate that only the upper band is recognized by anti-RUB antibodies (data not shown). Because both AtCUL3 forms were observed in the cul3a and cul3b mutants, as well as in wild-type plants, it is possible to conclude that, like AtCUL1, the AtCUL3A and AtCUL3B proteins are subjected to modification by RUB.
AtCUL3A and AtCUL3B Are Largely Functionally Redundant and Are Essential for Embryo Development
Although each insertion gives rise to a null mutation, when grown under normal growth conditions the single homozygous cul3a and cul3b insertion mutants are virtually indistinguishable from their wild-type siblings throughout their entire life cycles (Figure 1F).
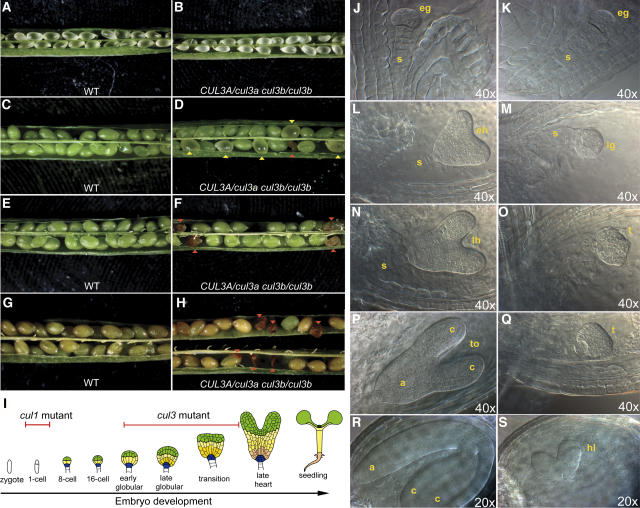
However, no homozygous double mutant plants were recovered, despite multiple attempts using PCR genotyping of 60 segregating progeny obtained from the self-pollination of parental plants homozygous for the mutation in one of the two AtCUL3 loci and heterozygous for the mutation in the other. To further investigate the absence of double homozygous cul3a cul3b mutants, we selected plants that were heterozygous for cul3a and homozygous for cul3b and performed detailed examinations with a dissection stereoscope (Figures 2A to 2H) and a differential interference contrast (DIC) microscope (Figures 2J to 2S). Siliques and developing seeds, at different developmental stages after self-pollination, were analyzed. As shown in Figures 2A and 2B, at early developmental stages the fertilized ovules obtained from the self-pollination of wild-type (Figure 2A) and CUL3A/cul3a cul3b/cul3b (Figure 2B) parental plants are virtually indistinguishable from each other, as they all contain developing embryos at the early globular stage (Figures 2J and 2K). However, after the transition from the globular to the heart stage (Figures 2L to 2O), all ovules from wild-type plants uniformly contain heart-stage embryos (data not shown), whereas the ovules from CUL3A/cul3a cul3b/cul3b plants segregate wild-type–like ovules with heart-stage embryos (Figures 2L and 2N) and ovules with embryos arrested at the globular stage (Figure 2M) or a slightly later stage (Figure 2O). At a later developmental stage, whereas wild-type siliques uniformly contain green seeds (Figures 2C and 2E) that undergo the maturation process (Figure 2G), siliques from CUL3A/cul3a cul3b/cul3b plants segregate both wild-type developing seeds and pale green/yellowish developing seeds, indicated by yellow arrowheads in Figure 2D. DIC microscopy of cleared ovules from these siliques demonstrated that although the green ovules contain mature embryos (Figure 2R), the pale green ovules contain underdeveloped embryos arrested at a late globular-to-heart transition (Figure 2Q) or at a heart-like stage (Figure 2S). During the seed maturation and desiccation processes, the wild-type–like green mature ovules from CUL3A/cul3a cul3b/cul3b plants develop into brown seeds (Figure 2H), indistinguishable from the wild-type ovules (Figure 2G), whereas the pale yellowish ovules of the same plants turned into red shrunken seeds (indicated by red arrowheads in Figure 2H), possibly because the absence of a mature embryo causes the collapse of the seed coat. Similar analyses of developing siliques obtained from the self-pollination of the reciprocal cul3a/cul3a CUL3B/cul3b double mutant gave results identical to those in Figure 2 for the self-pollination of CUL3A/cul3a cul3b/cul3b double mutant plants (data not shown).
Figure 2.
Loss of Function in Both Arabidopsis CUL3 Genes Causes Arrest in Embryogenesis.
(A) and (B) Stereomicroscopic images of siliques obtained from self-pollinated wild-type (WT [A]) or CUL3A/cul3a cul3b/cul3b (B) parental plants at an early developmental stage, when wild-type embryos are at the transition between the globular and heart stages.
(C) and (D) Stereomicroscopic images of siliques obtained from self-pollinated wild-type (C) or CUL3A/cul3a cul3b/cul3b (D) parental plants at a later developmental stage, when wild-type embryos are at the mature stage. Yellow arrowheads indicate pale green/yellow ovules containing arrested embryos. Red arrowheads indicate red shrunken ovules.
(E) to (H) Stereomicroscopic images of siliques obtained from self-pollinated wild-type ([E] and [G]) or CUL3A/cul3a cul3b/cul3b ([F] and [H]) parental plants during the onset of seed maturation. Red arrowheads in (F) and (H) indicate the pale green/yellow ovules (described in [D]) that turned into shrunken seeds.
(I) Scheme of Arabidopsis embryo development, adapted from Laux et al. (2004). Red bars indicate the stages of arrest of Arabidopsis cul1 and cul3 mutant embryos.
(J) to (S) DIC images of cleared ovules obtained from the self-pollination of CUL3A/cul3a cul3b/cul3b parental plants. At an early developmental stage (early globular; [J] and [K]), all of the embryos are uniformly developed. Starting from the transition between the late globular and torpedo stages, the cul3a cul3b embryos arrest at late globular/transition stages ([M], [O], and [Q]), whereas the sibling embryos ([L], [N], and [P]) from the same siliques proceed from globular, to heart, to torpedo, to mature embryos. Occasionally, cul3a cul3b embryos were arrested at a heart-like stage (S).
a, axis; c, cotyledons; eg, early globular-stage embryo; eh, early heart-stage embryo; hl, heart-like embryo; lg, late globular-stage embryo; lh, late heart-stage embryo; s, suspensor; t, transition-stage embryo; to, torpedo-stage embryo.
As shown in Table 1, stereoscopic analyses of a large number of developing siliques from the self-pollination of both CUL3A/cul3a cul3b/cul3b and cul3a/cul3a CUL3B/cul3b parental plants indicate that the frequency of pale green/yellowish ovules that turned into red shrunken seeds corresponds to ∼16.5 and 17% of the total developing seeds, respectively. Because these defective seeds have not been observed in either of the single cul3a or cul3b mutants (Table 1), and because we have been unable to recover homozygous double mutant plants, it is plausible that the defective ovules containing arrested embryos correspond to the double homozygous cul3a cul3b mutants. However, the frequency of these defective double mutant seeds is lower than the expected 25% rate for the double homozygous mutant progeny from a self-pollination of CUL3A/cul3a cul3b/cul3b and cul3a/cul3a CUL3B/cul3b plants. Such a discrepancy might be related to the observation that the siliques obtained from the self-pollination of both reciprocal parental plants (CUL3A/cul3a cul3b/cul3b and cul3a/cul3a CUL3B/cul3b) also contain variable numbers of early-aborted ovules, which might represent unfertilized ovules or ovules arrested at a very early stage after fertilization. The frequency of these early-aborted ovules is higher than that observed in wild-type plants and may represent the missing fraction of double cul3a cul3b mutants. Based on this observation, it is possible that a reduction in the fitness of the cul3a cul3b gametophytes alone, or a very early arrest of the fertilized double mutant ovules, or a combination of both, might be responsible for the early abortion of the ovules. Further studies of the effect of cul3a cul3b double mutations on pollen and ovule fitness will likely clarify this issue.
Table 1.
Segregation of the cul3a cul3b Double Mutant Progeny
| Genotype of Plant Used for Self-Pollinationa | Wild-Type Seeds with Mature Embryosb | Defective Seeds with Arrested Embryosc | Total Number of Seeds Analyzed |
|---|---|---|---|
| Wild type | 99.48% | 0.52% | 580 |
| CUL3A/CUL3A cul3b/cul3b | 99.48% | 0.52% | 764 |
| cul3a/cul3a CUL3B/CUL3B | 99.84% | 0.16% | 637 |
| CUL3A/cul3a cul3b/cul3b | 83.43% | 16.57% | 1316 |
| cul3a/cul3a CUL3B/cul3b | 82.88% | 17.12% | 479 |
Developing seeds from siliques obtained from the self-pollination of plants with the genotypes described were used for the stereoscopic and DIC analyses.
Segregation frequencies of green developing ovules that contain normally developing embryos, which eventually turn into mature brown seeds after the normal maturation/desiccation processes (see Figure 2 and Results for details).
Segregation frequencies of defective pale green/yellowish developing ovules that contain embryos arrested at globular/early heart-like stages. These ovules turn into aborted red shrunken seeds (see Figure 2 and text for details).
The Two AtCUL3 Genes Exhibit Overlapping Expression Patterns
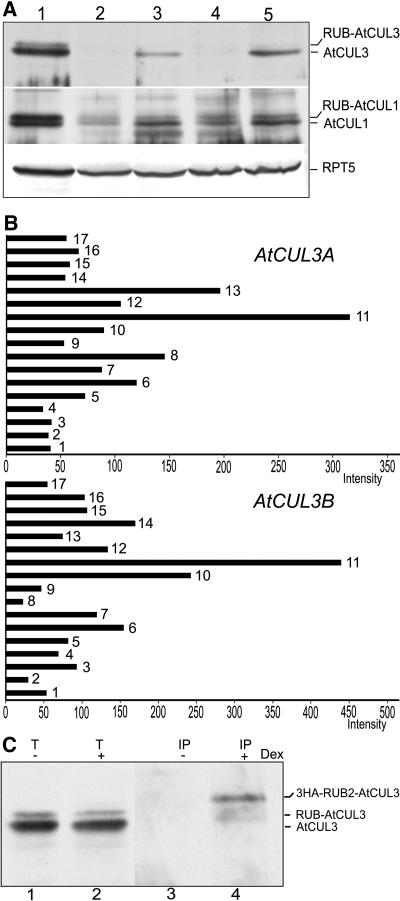
To obtain additional insight into the role of the AtCUL3 proteins beyond embryo development, we surveyed the expression patterns of the two AtCUL3 genes in different tissues by immunoblot analysis (Figure 3A). We found that the AtCUL3 protein is more highly expressed in flowers, stems, and seedlings than in cauline and rosette leaves. Gene-specific 70-mer oligonucleotide probes for the two AtCUL3 genes were used in a microarray analysis of the steady state mRNA levels of a broad set of Arabidopsis tissue and organ types (Figure 3B). Although the difference in the 70-mer oligonucleotide sequences, and thus the hybridization efficiency, prevents us from making a quantitative comparison of the relative mRNA levels of the two genes, the relative levels of each transcript in different organ or tissue types are comparable. We found that the patterns of mRNA expression for the AtCUL3 genes largely overlap but are not exactly the same, with germinated seeds having the highest levels of transcripts of both genes. Notably, we found a good correlation between the relative AtCUL3 protein level and its corresponding mRNA in those organs in which both protein and mRNA were analyzed. For example, both cauline and rosette leaves have low levels of both AtCUL3 mRNAs and proteins.
Figure 3.
Protein and mRNA Expression Patterns of the AtCUL3 Genes and Posttranslational Modification of AtCUL3 Proteins.
(A) The AtCUL3 protein is differentially expressed in Arabidopsis tissues. Total soluble protein extracts from flowers (lane 1), cauline leaves (lane 2), stems (lane 3), rosette leaves (lane 4), and light-grown seedlings (lane 5) were examined by protein gel blot analysis using anti-AtCUL3 and anti-AtCUL1 antibodies. An anti-RPT5 antibody was used as a sample loading control.
(B) Expression profiles of the two AtCUL3 genes in different Arabidopsis tissues/organs derived from a genome-profiling study using a 70-mer oligonucleotide microarray (Ma et al., 2005). The relative expression levels of each tissue/organ considered, shown with normalized intensity, are represented by black bars. Bar 1, cauline leaf; bar 2, rosette leaf; bar 3, pistil, 1 d after pollination; bar 4, pistil, 1 d before pollination; bar 5, silique, 3 d after pollination; bar 6, silique, 8 d after pollination; bar 7, stem; bar 8, sepal; bar 9, stamen; bar 10, petal; bar 11, germinated seed; bar 12, root in dark; bar 13, root in white light; bar 14, hypocotyl in dark; bar 15, hypocotyl in white light; bar 16, cotyledon in dark; bar 17, cotyledon in white light.
(C) The AtCUL3 protein is subjected to modification by RUB2 in vivo. Total protein extracts (T) from 3HA-RUB2 transgenic seedlings with (+) or without (−) dexamethasone (DEX) induction were subjected to immunoprecipitation (IP) with anti-HA antibodies. Protein gel blot analysis was subsequently performed using anti-AtCUL3 antibodies.
Epitope-Tagged RUB2 Can Conjugate with the AtCUL3 Protein in Stable Transgenic Arabidopsis
Arabidopsis has three RUB proteins, with the highly similar RUB1 and RUB2 playing the dominant roles (Bostick et al., 2004). To examine whether RUB is able to conjugate to AtCUL3, a hemagglutinin epitope (HA)-tagged version of RUB2, under a dexamethasone-inducible promoter (Bostick et al., 2004), was used for immunoprecipitation of RUB proteins. Protein blot analysis with anti-AtCUL3 antibodies revealed a slowly migrating band after immunoprecipitation of dexamethasone-treated extracts for 3HA-RUB2 (Figure 3C, lane 4), which should correspond to 3HA-RUB2-AtCUL3. In total extracts, unconjugated AtCUL3 and native RUB-conjugated AtCUL3 were observed (Figure 3C, lanes 1 and 2), whereas 3HA-RUB-AtCUL3 was not observable because of its low abundance. These data confirm that the AtCUL3 proteins are subjected to modification by RUB in planta.
The Arabidopsis Genome Has a Large Number of BTB Proteins
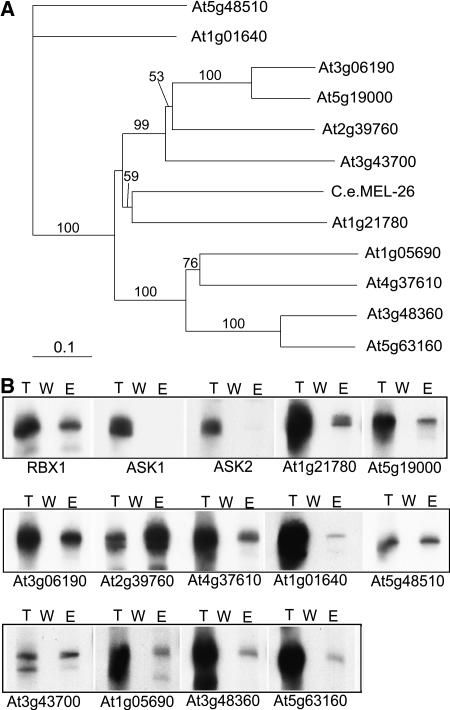
Based on recent studies in C. elegans showing that CUL3 interacts with a protein harboring a BTB domain (Pintard et al., 2003b; Xu et al., 2003), we searched in the PFAM (Protein Families Database of Alignments) domain database (Bateman et al., 2004) and easily identified at least 37 Arabidopsis proteins containing a BTB domain. We also conducted a BLAST search for Arabidopsis proteins with a sequence homologous with the BTB domain of the C. elegans MEL-26 protein (smallest sum probability, P < 2.0e-07). Based on these two searches, we defined and selected 21 high-confidence Arabidopsis BTB proteins for further analysis. Using RT-PCR, we successfully cloned full-coding cDNA for 11 of those Arabidopsis BTB genes. Phylogenetic analysis of the BTB domain of the 11 proteins, together with C. elegans MEL-26, revealed the presence of three main branches, with the BTB domain encoded by At1g21780 being the most similar to the BTB domain of MEL-26, with 35% sequence identity between the two (Figure 4A; see Supplemental Figure 2 online).
Figure 4.
Phylogenetic Comparison of 11 Cloned BTB Protein Genes and Their in Vitro Binding to AtCUL3.
(A) Phylogenetic analysis of the BTB domains in several Arabidopsis proteins. Sequence data for the proteins used in the phylogenetic analysis were taken from the EMBL/GenBank databases using the following accession numbers: At1g21780 (AAM65090), At5g19000 (AAM97116), At3g06190 (AAF30312), At2g39760 (NP_030522), At4g37610 (AAN15363), At1g01640 (AAF78396), At5g48510 (NP_199662), At3g43700 (CAB83071), At1g05690 (AAQ87006), At3g48360 (AAN31824), At5g63160 (AAR24650), and C. elegans MEL-26 (NP_492449). Sequence alignment of the BTB protein families was conducted using ClustalX (version 1.83) software (Thompson et al., 1997). The Gonnet protein weight matrices (Gonnet 40, 80, 120, 160, 250, and 350 matrices [Gonnet et al., 1992]) were selected, and the gap-opening and extension parameters were 10 and 0.2, respectively. Distance (mean standard distance) phylogenetic analysis was performed in PAUP 4.0 (score 3.41; Swofford, 2000). Bootstrap support (Felsenstein, 1985) for nodes was estimated using 1000 replicates under mean standard distance.
(B) AtCUL3A interacts in vitro with several members of the Arabidopsis BTB family but not with SKP1/ASK1 or SKP1/ASK2. An in vitro pull-down assay was performed using purified MBP-AtCUL3A and in vitro–translated Arabidopsis BTB proteins. Lane T, total in vitro–translated BTB protein; lane W, the last wash before elution; lane E, the protein eluted.
AtCUL3A Interacts in Vitro with the BTB Proteins but Not with the SKP1-Like Proteins ASK1 and ASK2
To examine a possible interaction between AtCUL3 and the Arabidopsis BTB proteins, the 11 cloned BTB proteins were subjected to an in vitro pull-down assay with the recombinant Maltose Binding Protein (MBP)-AtCUL3A protein. Briefly, the purified MBP-AtCUL3A was incubated separately with each individual in vitro–translated BTB protein, and its affinity for a given BTB protein was examined after an extensive washing procedure. The results shown in Figure 4B demonstrate that all of the 11 Arabidopsis BTB proteins tested to date were able to interact with MBP-AtCUL3A in the in vitro pull-down assay. As expected, on the basis of previous findings in other eukaryotic organisms, Arabidopsis RBX1 was able to interact with MBP-AtCUL3A in the same pull-down assay, whereas, interestingly, the SKP1-like Arabidopsis proteins ASK1 and ASK2 were not. Based on the highest sequence homology in the BTB domains between At1g21780 and MEL-26, and on the positive interaction observed in our pull-down assay, At1g21780 was selected for further detailed structural and functional analysis.
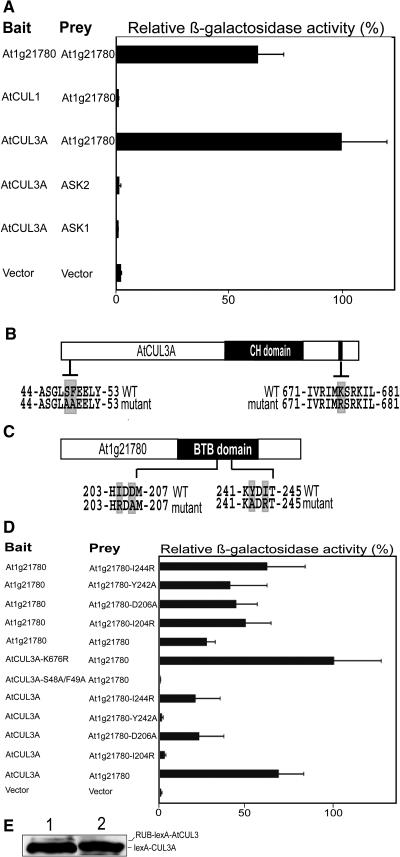
The BTB Protein At1g21780 Is Capable of Dimerization
A yeast two-hybrid assay was used to further verify the interaction between the At1g21780 protein and AtCUL3A (Figure 5A). In agreement with the results obtained from the in vitro pull-down assay, the yeast two-hybrid assay confirmed that AtCUL3A interacts with the At1g21780 BTB protein but not with the SKP1-like ASK1 or ASK2 protein. A control yeast two-hybrid assay also showed that AtCUL1 was unable to interact with At1g21780, further supporting the notion that the interaction between At1g21780 and AtCUL3A is specific and does not apply to the entire cullin family.
Figure 5.
Interaction of the Arabidopsis BTB Protein At1g21780 with Itself and with AtCUL3A and Their Structural Requirements by Yeast Two-Hybrid Assay.
(A) The prey contains the DNA binding domain, and the bait contains the activation domain. The degree of interaction is shown by β-galactosidase activity (black bars). Numbers on the x axis are average values of at least six independent transformants, and error bars represent sd.
(B) Scheme of the AtCUL3 protein, with the regions of site-directed mutagenesis highlighted. The cullin homology (CH) domain at the C terminus of the AtCUL3 protein region are shown in black. WT, wild type.
(C) Scheme of the At1g21780 BTB protein, with the region where site-directed mutagenesis was performed. The BTB domain in the C terminus region is shown in black.
(D) Yeast two-hybrid assay of the effect of point mutations on the BTB protein and AtCUL3A interaction.
(E) AtCUL3A K676R is not modified by RUB/NEDD8 in yeast. AtCUL3A and AtCUL3A K676R were expressed in yeast, and protein extracts were examined by immunoblot analysis using anti-AtCUL3 antibody.
Interestingly, the yeast two-hybrid assay also revealed that At1g21780 is able to interact with itself (Figure 5A). This observation is consistent with the report that BTB proteins are capable of homodimerization and of heterodimerization with other members of the same family (Bardwell and Treisman, 1994; Pintard et al., 2004).
The N-Terminal Region of AtCUL3 and the BTB Domain of At1g21780 Are Critical for the AtCUL3–BTB Protein Interaction
A comparative analysis of CUL3 proteins from several organisms revealed the presence of highly conserved N-terminal elements corresponding to the H2 helix of CUL1 (Zheng et al., 2002; Xu et al., 2003). Indeed, mutagenesis analyses indicated that residues in this predicted helix region are important for the interaction between the C. elegans CUL3 and MEL-26 proteins (Xu et al., 2003). To examine the involvement of these elements in BTB protein binding in Arabidopsis, we generated a mutant version of AtCUL3A in the H2 region (S48A/F49A) and tested it for interaction with At1g21780 using a yeast two-hybrid assay (Figure 5B). The results clearly demonstrate that the mutation in the H2 helix of AtCUL3A completely abolishes its ability to interact with At1g21780 (Figure 5D). We were also interested in defining the effect of RUB modification on the AtCUL3A–At1g21780 interaction. Therefore, we generated an AtCUL3A K676R mutation that abolished the RUB modification of AtCUL3A (Figure 5B). It is interesting that the AtCUL3A K676R mutant protein, which is not RUB-modified in yeast (Figure 5E, lane 2), is still able to interact with At1g21780 at a wild-type level (Figure 5D). This result indicates that the RUB modification of AtCUL3A is not required for and does not affect the AtCUL3A interaction with At1g21780 in the yeast assay. Previous structural analyses indicate that SKP1 and elongin C adopt α/β-structures similar to the BTB domain of human promyelocytic leukemia zinc finger (Schulman et al., 2000). In the case of SKP1, several residues located in β-strand 3 (S3) and in α-helix 5 (H5) contact CUL1 (Zheng et al., 2002). In addition, recent studies have shown that analogous residues in the MEL-26 BTB domain are important for the interaction between MEL-26 and CUL3 (Xu et al., 2003). Therefore, we generated mutations in analogous residues of the BTB domain (residues 151 to 259) of At1g21780 (Figure 5C), which are conserved in several Arabidopsis BTB proteins (see Supplemental Figure 2 online), and tested their ability to interact with AtCUL3A by yeast two-hybrid assay. Some mutations in the predicted S3 (I204R) or H5 (Y242A) abolished the interaction of the BTB protein with AtCUL3A (Figure 5D), whereas other mutations in S3 (D206A) or H5 (I244R) reduced the interaction by three times with respect to wild-type At1g21780 (Figure 5D). Clearly, these data suggest that analogous structural elements in the SKP1, MEL-26, and At1g21780 proteins may be used to interact with a common interface located in the N terminus of distinct cullin proteins. On the other hand, those same point mutations in the BTB domain did not significantly affect the self-interaction of the BTB protein (Figure 5D), suggesting that distinct interacting surfaces are responsible for its self-interaction and for its interaction with AtCUL3.
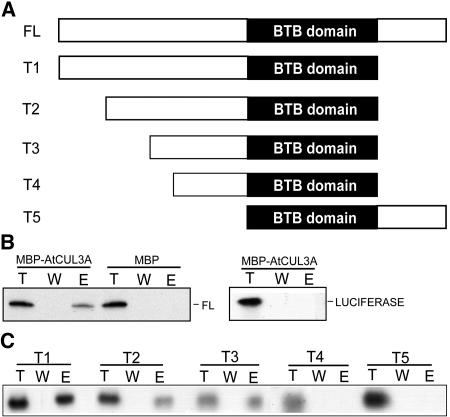
The BTB Domain of At1g21780 May Not Be Sufficient to Interact with AtCUL3A
As our yeast two-hybrid results suggested that several residues of the BTB domain of At1g21780 are critical for the interaction with AtCUL3 (Figure 5D), we further tested whether the BTB domain itself is sufficient to interact with AtCUL3A. Several truncated versions of At1g21780 (Figure 6A) were generated, and their ability to interact with AtCUL3A was analyzed using in vitro pull-down assays. To verify the specificity of this in vitro assay for interaction between AtCUL3A and At1g21780, two different negative controls were included. First, an in vitro translated full-length At1g21780 was used in the in vitro binding assay with MBP alone (Figure 6B, left). Second, an in vitro–translated Luciferase was used in the in vitro binding assay with MBP-CUL3A (Figure 6B, right). In both cases, no detectable interactions were observed, confirming the specificity of the results described below.
Figure 6.
The BTB Domain of At1g21780 Is Not Sufficient for the Interaction with AtCUL3A.
(A) Scheme of the truncations of At1g21780 used in the in vitro binding assay. FL, full-length At1g21780 protein.
(B) An in vitro pull-down assay was performed using purified MBP-AtCUL3A or MBP and in vitro–translated full-length At1g21780 (left) or purified MBP-AtCUL3A and Luciferase proteins (right). Lane T, total in vitro–translated protein; lane W, the last wash before elution; lane E, the protein eluted.
(C) An in vitro pull-down assay was performed using purified MBP-AtCUL3A and in vitro–translated truncated versions of At1g21780. T1 to T5 correspond to the truncated version of the At1g21780 protein represented in (A).
The deletion of 62 residues at the C-terminal region of At1g21780 (truncation T1, Figure 6C) did not affect the binding to MBP-AtCUL3A. Two other deletions of 50 and 100 residues at the N-terminal region of At1g21780, in addition to the 62 residues at the C terminus, also did not affect the binding to AtCUL3A (truncations T2 and T3, Figure 6C). Nevertheless, a third truncated version of At1g21780, lacking 124 residues at the N terminus and 62 residues at the C terminus but still containing the entire BTB region, did not bind to MBP-AtCUL3A (truncation T4, Figure 6C). Similarly, a truncated version of At1g21780, lacking 145 residues at the N terminus but containing the complete C-terminal region, did not bind to MBP-AtCUL3A (truncation T5, Figure 6C). The binding results obtained with truncations T4 and T5 suggest that the BTB domain of At1g21780 is not sufficient to confer a detectable interaction with AtCUL3A in our assay. Furthermore, comparison of the binding results for truncations T3 and T4 suggests that a region between residues 100 and 124 of At1g21780 is important for the interaction between this BTB protein and AtCUL3A.
The AtCUL3, RBX1, and BTB Proteins Likely Form E3 Ubiquitin Ligase Complexes in Vivo
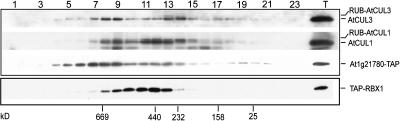
To test whether AtCUL3 forms the expected E3 ubiquitin ligase complex in vivo, we performed a gel filtration analysis of protein extracts obtained from tandem affinity purification (TAP)-AtRBX1 and At1g21780-TAP transgenic lines (see Methods). Protein blot analysis with anti-AtCUL3 antibodies showed that AtCUL3 is present in a broad range of size fractions, from the monomeric form to high molecular mass complex(es) (Figure 7, top gel, first row). The gel filtration profile of AtCUL3 is somewhat similar to that of AtCUL1 (Figure 7, top gel, second row), which forms well-characterized SCF complexes. Based on the fact that the AtCUL3A protein is predominantly expressed with respect to AtCUL3B in the same tissue (Figure 1D), we conclude that AtCUL3A can form high molecular mass complexes in vivo. The gel filtration profile of At1g21780 with a TAP tag (detected using anti-myc antibodies) is broad and overlaps with that of AtCUL3A (Figure 7, top gel, third row, fractions 8 to 14). Meanwhile, the gel filtration profile of TAP-RBX1 protein from TAP-AtRBX1 transgenic seedlings (Figure 7, bottom) overlaps well with the corresponding fractions of AtCUL3A and At1g21780-TAP proteins (fractions 8 to 14), in agreement with the idea that the three proteins can form a multisubunit complex in vivo.
Figure 7.
AtCUL3 Protein Fractionation Profile in Vivo.
Gel filtration fractions of protein extracts from 6-d-old light-grown At1g21780-TAP (top) or TAP-RBX1 (bottom) seedlings were subjected to protein blot analysis with anti-CUL3, anti-CUL1, and anti-myc antibodies. Lanes 1 through 23 contain the fractions from the gel filtration elution after the void volume was achieved. The numbers below the arrows indicate the peak positions of the molecular mass markers in kilodaltons. Lane T, total protein extract.
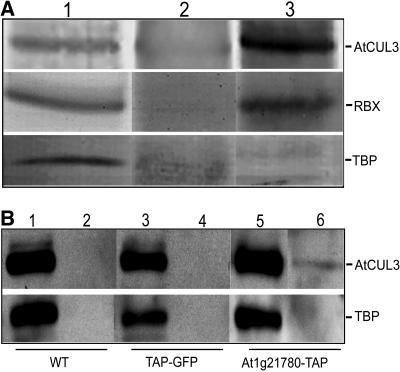
To test whether AtCUL3 interacts with RBX1 in vivo, coimmunoprecipitation experiments of the two proteins were performed. After immunoprecipitation of Arabidopsis seedling extracts with anti-AtCUL3 antibodies, the blot was probed with either anti-AtCUL3 or anti-RBX1 antibodies. As shown in Figure 8A, RBX1 coimmunoprecipitated well with AtCUL3 (lane 3), whereas the control TATA binding protein (TBP) did not (lane 3, bottom). This assay suggests that AtCUL3 associates specifically with RBX1 in vivo.
Figure 8.
AtCUL3 Is Specifically Associated with AtRBX1 and the BTB Protein.
(A) AtCUL3 associates with AtRBX1 in vivo. Lane 1, total protein extract; lane 2, extract after immunoprecipitation with the preimmune antibodies; lane 3, extract after immunoprecipitation with specific antibodies against AtCUL3. The blot was probed with antibodies against AtCUL3, AtRBX1, and TBP proteins (control for the specificity).
(B) The At1g21780 BTB protein coimmunoprecipitates with AtCUL3 in vivo. Protein extracts from 6-d-old wild-type (WT) or transgenic seedlings were immunoprecipitated with IgG-coupled Sepharose. Protein blot analysis was subsequently performed using anti-AtCUL3A and anti-TBP antibodies. Odd-numbered lanes represent total proteins, and even-numbered lanes represent immunoprecipitated products with IgG beads. Lanes 1 and 2, protein extract from wild-type seedlings; lanes 3 and 4, protein extracts from TAP-GFP line; lanes 5 and 6, protein extracts from the At1g21780-TAP transgenic line.
To verify the interaction between AtCUL3 and At1g21780, the protein extract from At1g21780-TAP transgenic plants was used for coimmunoprecipitation analysis. The results, shown in Figure 8B, demonstrate that the IgG beads can specifically pull down At1g21780-TAP and coimmunoprecipitate AtCUL3 but not the control protein TBP. The same beads cannot pull down AtCUL3 in either wild-type or TAP-green fluorescent protein (GFP) seedling extracts, suggesting that AtCUL3 specifically associates with the At1g21780 BTB protein in vivo.
Arabidopsis BTB Proteins Are a Diverse Group of Proteins with Distinct Expression Patterns
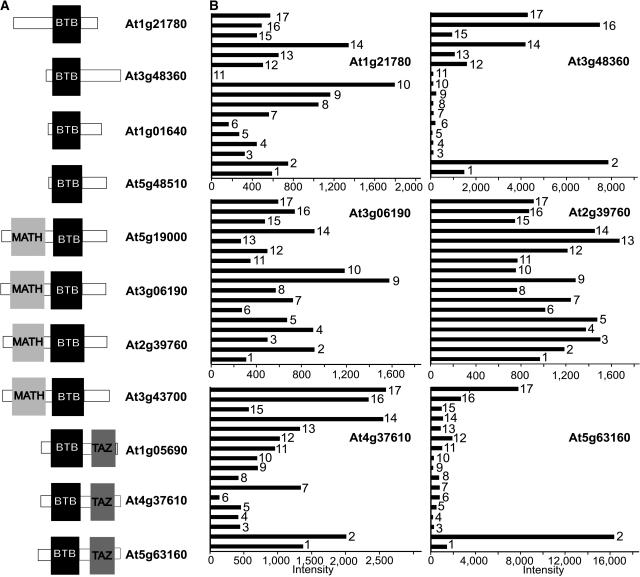
Careful analysis of the domain structure of the Arabidopsis BTB proteins using the PFAM domain database revealed a broad arrangement of different additional domains present in Arabidopsis BTB proteins. A comparison of the domain structures in the 11 cloned BTB proteins is shown in Figure 9A. In addition to the Meprin and TRAF-Homology (MATH) domain and the TAZ zinc finger domain shown in Figure 9A, ankyrin repeats, armadillo/β-catenin–like repeats, and BSD (for found in BTF2-like transcription factors, synapse-associated, and DOS2-like proteins) domains are present in other BTB proteins (data not shown).
Figure 9.
Representative BTB Protein Domain Structures and Organ-Specific Expression Profiles.
(A) Schematic comparison of the domain structures of selected Arabidopsis BTB proteins. The protein sequences of several Arabidopsis BTB proteins were analyzed using the PFAM domain database. The black boxes denote the BTB domains, the light gray boxes denote the MATH domains, and the dark gray boxes denote the TAZ domains.
(B) Expression profile of six selected BTB genes in different Arabidopsis tissues/organs derived from a genome-profiling study using a 70-mer oligonucleotide microarray (Ma et al., 2005). For each tissue/organ under consideration, the relative expression levels of the BTB genes are shown, with the normalized intensities represented by black bars. Bar 1, cauline leaf; bar 2, rosette leaf; bar 3, pistil, 1 d after pollination; bar 4, pistil, 1 d before pollination; bar 5, silique, 3 d after pollination; bar 6, silique, 8 d after pollination; bar 7, stem; bar 8, sepal; bar 9, stamen; bar 10, petal; bar 11, germinated seed; bar 12, root in dark; bar 13, root in white light; bar 14, hypocotyl in dark; bar 15, hypocotyl in white light; bar 16, cotyledon in dark; bar 17, cotyledon in white light.
To gain a quick glimpse into the possible function of the BTB proteins under study, the expression of the BTB genes at the mRNA level was examined by DNA microarray analysis (Figure 9B) (Ma et al., 2005). All of the BTB genes show distinct organ or tissue expression profiles and also are not identical to those of the AtCUL3 genes (Figure 3B). For example, the At1g21780 gene shows higher relative levels in petal, stamen, and hypocotyl of the dark-grown seedling.
DISCUSSION
In mice, a null mutation in cul3 results in early embryonic lethality (Singer et al., 1999), whereas depletion of CUL3 by RNA interference in C. elegans causes defects in early embryogenesis, resulting in abnormal microfilament and microtubule organization in spindle positioning and failed cytokinesis (Kurz et al., 2002). Therefore, CUL3 is important for early embryo development in animals. Here, we show that the Arabidopsis cul3 double mutant arrests at the transition between the globular and heart stages, demonstrating that AtCUL3A and AtCUL3B together play redundant functions that are essential for plant embryogenesis. In fact, although we were not able to see any obvious phenotype in either of the single cul3a or cul3b null mutants (Figure 1F), mutations in both the AtCUL3A and AtCUL3B genes result in the failure to produce viable double homozygous progeny. This observation establishes an essential role for AtCUL3 in plant development. DIC analyses of a large number of developing seeds indicate that the majority of the cul3 double mutant embryos were arrested between the late globular and transition stages (Figures 2M, 2O, and 2Q), even though we observed a minority of embryos arrested at a heart-like stage (Figure 2S). However, the embryo lethality in the double null mutant of AtCUL3 occurs considerably later than the embryo defect caused by a null mutation in AtCUL1, which impairs the very first cell division of the fertilized zygotes (Shen et al., 2002). Based both on the higher level of AtCUL3 protein in flowers and in light-grown seedlings (Figure 3A) and on the ubiquitous expression patterns of the two AtCUL3 genes, it is plausible that AtCUL3 plays important roles in other developmental processes beyond embryo development.
Our data collectively are consistent with the possibility that in Arabidopsis, AtCUL3 forms multisubunit E3 ubiquitin ligase complexes containing the common core component RBX1 and the substrate recognition subunit BTB protein(s). First, similar to AtCUL1, AtCUL3 is present in monomeric as well as in complexed forms in vivo. Second, AtCUL3, At1g21780, and RBX1 have similar gel filtration profiles. Third, AtCUL3 interacts directly with RBX1 in vitro and associates specifically with RBX1 in vivo. Two recent publications also independently demonstrated these interactions through in vitro binding assays or in yeast (Dieterle et al., 2005; Weber et al., 2005). Fourth, AtCUL3 is subjected to the familiar RUB modification in vivo, like the well-characterized AtCUL1 subunit of the SCF complex. Fifth, AtCUL3 directly interacts with 11 distinct Arabidopsis BTB proteins tested in vitro, and in one sample case, specific in vivo association of AtCUL3 and a BTB protein (At1g21780) was demonstrated. Because the Arabidopsis genome encodes a large number of BTB proteins, it is likely that the core components AtCUL3 and RBX1 associate with different individual BTB proteins, resulting in the formation of distinct combinations of CUL3-BTB-RBX E3 ubiquitin ligase complexes potentially involved in the recognition and degradation of multiple substrates. Based on their widespread expression profiles, these E3 ligases could be critical for a large number of events during development throughout the plant life cycle.
The Arabidopsis BTB proteins, as in other organisms, have broad diversity in their protein domain structure. Our initial analysis identified six subgroups based on additional shared domains or motifs: BTB proteins that contain a MATH domain (Uren and Vaux, 1996), BTB proteins that contain a BTB domain as the only recognizable domain (Ahmad et al., 1998), BTB proteins that contain an ankyrin repeat (Kohl et al., 2003), BTB proteins that contain a TAZ zinc finger domain (De Guzman et al., 2000), BTB proteins that contain an armadillo/β-catenin–like repeat (Huber et al., 1997), and BTB proteins that contain a BSD domain (Doerks et al., 2002).
One of the best-characterized BTB proteins is the C. elegans MEL-26 protein, which contains a MATH domain in the N-terminal region and a BTB domain in the C-terminal region. All six Arabidopsis BTB proteins that contain both domains are clustered in the same branch of the phylogenetic tree for Arabidopsis BTB proteins (Figure 4A). Based on the observation made in C. elegans, we can speculate that the MATH domain in those Arabidopsis BTB proteins could be responsible for mediating their interaction with the substrates. On the other hand, several other characterized BTB proteins, such as KIAA1309, KIAA1354, and KEAP1 from human and BTB1 and BTB2 from S. pombe, contain six kelch repeats and an ankyrin repeat, respectively (Furukawa et al., 2003; Geyer et al., 2003; Cullinan et al., 2004; Kobayashi et al., 2004). It was speculated that those kelch or ankyrin repeats might be responsible for recognizing the substrates. Thus, parts of the BTB protein other than the BTB domain, with or without known structural features, may be responsible for recognizing the substrates, whereas the BTB domain itself is responsible for the interaction with AtCUL3 and for the possible dimerization with other BTB-containing proteins. However, the results shown in Figure 6 suggest that, in the case of At1g21780, at least another upstream motif is required for the binding to AtCUL3A. The great variety of the modules present in BTB proteins, beyond the BTB domain itself, would confer a vast diversity on the substrates controlled by those AtCUL3-based E3 ubiquitin ligases (Dieterle et al., 2005; Weber et al., 2005).
It is interesting that some Arabidopsis BTB proteins do not seem to contain any other known modules or features (Figure 9A). At least three general possibilities exist to explain how BTB domain proteins recognize their substrates: the BTB protein interacts with the substrate using a small structural feature not yet recognized; the BTB protein forms a heterodimer with a BTB protein with known protein-interacting domains, such as MATH, ankyrin, TAZ zinc finger, armadillo, or BSD domains, to recognize the substrate; and the BTB protein is the substrate itself. In support of this last possibility, the human RhoBTB2 protein has been shown to be a substrate of the CUL3 E3 ubiquitin ligase (Wilkins et al., 2004). This is reminiscent of the ubiquitination and subsequent degradation of the F-box protein SKP2 by the CUL1-based SCF-type E3 ubiquitin ligase (Wirbelauer et al., 2000), except that SKP2 can form part of an SCF ligase that targets other substrates for ubiquitination.
In the literature, several BTB proteins have been characterized in plants: NPH3 and RPT2, signal transducers of the phototropic response (Motchoulski and Liscum, 1999; Sakai et al., 2000); NPR1, which is important in systemic acquired resistance (Cao et al., 1997); ETO1, which regulates ethylene production (Wang et al., 2004); AtBT1-5, a calmodulin binding protein (Du and Poovaiah, 2004); GMPOZ, which is involved in gibberellic acid–responsive gene expression in aleurone (Woodger et al., 2004); BOP1, which is required for leaf morphogenesis (Ha et al., 2004); and ARIA, a novel abscisic acid signaling component (Kim et al., 2004). However, to date, direct in vitro interaction with AtCUL3 has been reported only for ETO1 (Wang et al., 2004). The reported interaction between ETO1 and AtCUL3A by in vitro pull-down assay supports the notion that the ETO1-AtCUL3A-RBX complex could be involved in the regulation of ethylene biosynthesis (Wang et al., 2004). The facts that RUB2 is able to modify AtCUL3 (Figure 3C) and that silencing of RUB expression resulted in the overproduction of ethylene and a constitutive partial triple response (Bostick et al., 2004) support this idea.
The observation that AtCUL3 also forms similar E3 ubiquitin ligase complexes with RBX1 and BTB proteins sets the stage for addressing many exciting questions. Those questions include, but are not limited to, the following. What are the substrates recognized by each of the BTB proteins in the AtCUL3 E3 ligases? What are their physiological and developmental roles? How is the activity of this group of AtCUL3-based ubiquitin ligases regulated?
METHODS
Plant Materials and Growth Conditions
We obtained the T-DNA insertion line for Arabidopsis thaliana CUL3A from the Salk collection (SALK_050756; Columbia ecotype) (Alonso et al., 2003) and the T-DNA insertion line for AtCUL3B from screening the Wisconsin collection (Wassilewskija ecotype) (Sussman et al., 2000). Plants homozygous for the T-DNA insertion were identified by PCR-based genotyping. F1 plants from cul3a and cul3b plants were self-crossed. F2 plants were then genotyped by PCR, and the siliques from CUL3A/cul3a cul3b/cul3b plants were analyzed. 3HA-RUB1 and 3HA-RUB2 transgenic lines were described previously (Bostick et al., 2004). The wild-type Arabidopsis plants used in this study were of the Columbia-0 ecotype. To grow Arabidopsis seedlings, seeds were surface sterilized, put on MS plates (Invitrogen, Carlsbad, CA) containing 1% sucrose, and cold-treated at 4°C for 3 to 5 d before being placed in a continuous white light growth chamber (150 μmol·m−2·s−1) or in darkness. To obtain adult plants, 7- to 9-d-old light-grown seedlings were transferred to soil and grown in a standard long-day (16 h of light/8 h of dark) growth room.
For the microarray experiments, we used wild-type Arabidopsis tissues or organs as described below. The germinating seeds were collected after the seeds were planted on agar plates containing growth medium plus 1% sucrose and grown under continuous white light (150 μmol·m−2·s−1) for 48 h. The Arabidopsis seedlings used in this microarray study were 6 d old. The cotyledon, hypocotyl, and root were collected from the same seedling. Rosette leaves were collected from 3-week-old plants grown under continuous white light. The adult Arabidopsis plant was grown under continuous white light (250 μmol·m−2·s−1). Cauline leaves and stems were collected from 4-week-old plants. Floral organs were collected on the day of flowering. Siliques were collected 3 or 8 d after pollination. Suspension culture cells were prepared starting from seeds as described by Mathur and Koncz (1998). The cultured cells used for RNA isolation were collected at the logarithmic growth phase.
Whole-Mount Preparation of Ovules
Siliques at different developmental stages were harvested, dissected using a stereoscope, and fixed overnight in an ethanol/acetic acid (8:1) solution at 4°C. Fixed siliques were cleared in a derivative of Hoyer's solution (cloral hydrate/glycerol/water, 80:13:30 g) for 24 to 48 h at 4°C. Cleared ovules were further removed from their siliques in a drop of the same clearing solution, whole-mounted, and observed using a Zeiss Axiophot Microscope (Zeiss, Jena, Germany) equipped with Nomarsky optics (DIC). Digital images were captured using Axiovision 3.0 software (Zeiss).
Oligonucleotide Microarray
The 70-mer oligonucleotide set of the Arabidopsis genome was designed and synthesized by Qiagen/Operon (http://oligos.qiagen.com/arrays/omad.php) based on the Arabidopsis genome information available as of February 20, 2002. The Arabidopsis 26K 70-mer oligomer microarray contains 26,090 individual oligomers that represent 25,676 unique locus identifiers (genes) of Arabidopsis (Ma et al., 2005). The 70-mer sequences whose expression data are reported in this article are as follows: At1g26830 (AtCUL3A), 5′-TTCCGTTCTTACTACCTTGGGACACATACCGGTAGAAGATTGTCCTGGCAAACAAACATGGGCACAGCAG-3′; At1g69670 (AtCUL3B), 5′-AGGCAACTGAAATCCCCACCCCAGACCTAAAGCGTTGCTTGCAGTCAATGGCGTGTGTAAAAGGTAAAAA-3′; At1g21780, 5′-TTCAAAAGCATGTTCCACCACGACCTCATGGAAAAGGAATCATCCACAATCCACATAGACGACATGTCGA-3′; At3g48360, 5′-CGTAGTAGATAACAACAAAAAGTCAATGACTGCGGAAAAGTCAGAGCCTTGTAAGGCGTTTTCCACGTGT-3′; At3g06190, 5′-AAAACTCTGTGAAGGAATCAGCATAAATACGGTAGCAACCACCTTGGCTCTTGCAGAGCAGCATCATTGT-3′; At2g39760, 5′-AACTTGATGTTGATAATGTGGCAACAACACTTGCGTTGGCTGAACAGCACCAATTCTTACAGCTCAAAGC-3′; At4g37610, 5′-AAAGCTACGAGAGATTCATGGGATCGTATGTTCGACGAAGCTCATGGAGCTGATGTTTTGATTCATACTG-3′; and At5g63160, 5′-TCATCGAGAAGCCGAGGAAGATTCACGGCGGATCATCGAAGAAAGTTATTAAGATTCTCGGTGTTCCATG-3′.
RNA Isolation, Probe Labeling, and Hybridization in Microarray Analysis
The expression profiles of the 2 CUL3 genes and 11 BTB domain–containing genes in Arabidopsis were extracted from a recent whole-genome microarray-profiling study (Ma et al., 2005). Details of the materials and methods can be found in the original publication. In brief, total RNA was extracted from 17 Arabidopsis organs: light-grown cotyledon, dark-grown cotyledon, light-grown hypocotyl, dark-grown hypocotyl, rosette leaf, cauline leaf, stem, sepal, petal, stamen, pistil 1 d before pollination, pistil 1 d after pollination, silique 3 d after pollination, silique 8 d after pollination, germinated seed, light-grown root, and dark-grown root. Total RNA (50 μg) was first labeled with aminoallyl-dUTP (Sigma-Aldrich, St. Louis, MO) by direct incorporation of aminoallyl-dUTP during reverse transcription, as described previously (Ma et al., 2002). The purified probe was further labeled with fluorescent dye by conjugating aminoallyl-dUTP and monofunctional Cy-3 or Cy-5 (Amersham Biosciences, Piscataway, NJ). The dye-labeled probe was purified from the unincorporated dye molecules by washing three times through a Microcon YM-30 filter (Millipore, Bedford, MA). The purified labeled probes from each specific organ versus cultured cells as common control pairs or dark versus light organ pairs were combined to hybridize the microarray slide for 12 to 16 h at 42°C (Ma et al., 2002). Except for petal (two replicates) and pistil 1 d after pollination (four replicates), there were three biological replicates used for all organ types, with one quality data set from each replicate. In this genome-profiling study, all 17 organ sample probes were labeled with the same Cy-3 dye and were directly comparable after normalization (see below).
Microarray Data Processing and Normalization
Spot intensities on microarray slides after hybridization were scanned with an Axon GenePix 4000B scanner (Foster City, CA) set at the same voltage and quantified using Axon GenePix Pro 3.0 image-analysis software. The net intensities for each channel were measured using the GenePix Pro 3.0 median of intensity. Because the 70-mer oligomer microarray slides are highly uniform and reproducible from slide to slide, and all of the organ mRNAs were labeled with the same Cy-3 dye, a simple two-step normalization based on the median intensities of each microarray slide was performed for the entire data sets of 17 organs—namely, normalization of the replicates from the same organ type followed by normalization among all organs. For each specific gene (spot), the mean of the normalized intensities of the same spots from all replicates for a given organ was considered representative of the relative expression level of the gene in that organ. This is shown as bar graphs in Figures 3B and 9. It should be noted that because of the differences in the 70-mer oligonucleotide sequences and thus the hybridization efficiency for distinct genes, only the relative levels of each given transcript in different organs are comparable. Therefore, quantitative comparison of the relative mRNA levels of the different genes is not reliable.
Nucleic Acid Preparation and mRNA Gel Blot or RT-PCR Analysis
Total RNA was extracted from whole seedlings using the RNeasy Plant Mini Kit (Qiagen, Valencia, CA). Poly(A)+ RNA was enriched with Oligotex according to the manufacturer's protocol (Qiagen). RNA gel blot analysis was performed by standard procedures under stringent conditions with 32P-labeled probe. The AtCUL3A probe was a 167-bp DNA fragment obtained by PCR amplification from the 3′ untranslated region. The probe was radiolabeled using the Rediprime kit (Amersham Biosciences).
Cloning of Arabidopsis BTB cDNAs
To obtain the full-length open reading frame of AtBTB cDNAs, first-strand cDNA synthesis was performed on 1 μg of poly(A)+ RNA with oligo(dT) according to the instructions of the Superscript Preamplification System (Invitrogen). PCR amplifications were performed on one-tenth of the cDNA using Herculase DNA polymerase (Stratagene, La Jolla, CA) and specific primers. PCR products were purified by agarose gel electrophoresis, recombined with the pCR-Blunt II-TOPO plasmid (Invitrogen), and sequenced. The cDNA sequences are identical to those of the spliced products predicted by the genomic accessions.
In Vitro Binding Assays
The AtBTB cDNAs and truncated versions of At1g21780 cDNA were subcloned into the pTNT plasmid (Promega, Madison, WI) under the control of the T7 promoter. In vitro–translated AtBTBs were produced with the TNT-Quick Coupled transcription/translation system (Promega) using 35S-Met (Amersham Bioscience). The AtCUL3A coding region was subcloned downstream of the MBP coding sequence of plasmid pMAL2c (New England Biolabs, Beverly, MA). The fusion protein was produced in BL21 Codon Plus Escherichia coli (Stratagene) and purified using amylose–Sepharose 4B (New England Biolabs). The protein (5 μg) was incubated with in vitro–translated AtBTBs in 500 μL of binding buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, and 0.2% Nonidet P-40). After 45 min at room temperature, 20 μL of amylose resin was added to the reaction and incubated for an additional 45 min. After several washes with binding buffer, bound proteins were resuspended in Laemmli buffer and separated by SDS–PAGE. The gel was then exposed for fluorography.
Protein Blot Analysis and Antibodies
The N-terminal region of AtCUL3A (333 N-terminal residues) was expressed in the vector pET28a (Novagen, San Diego, CA) and the E. coli strain BL21(DE3)lysS (Novagen). The protein (insoluble fraction) was electrophoresed by SDS-PAGE and electroeluted. Rabbits were injected with the purified protein, and the antibodies were affinity-purified using the purified Cullin3A protein attached to a polyvinylidene difluoride membrane. For detection, the ECL Plus protein gel blotting detection system (Amersham Biosciences) was used.
For direct immunoblot analysis of plant extracts, Arabidopsis seedlings or tissues were homogenized in lysis buffer: 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 50 mM NaF, 10 mM Na3VO4, 60 mM β-glycerolphosphate, 0.1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, and 1× complete protease inhibitor (Roche, Basel, Switzerland). The extracts were centrifuged twice at 13,000g for 10 min each at 4°C, and the protein concentration in the supernatant was determined by Bradford assay (Bio-Rad, Hercules, CA). Protein samples were boiled in sample buffer, run on SDS-PAGE gels (10%), and blotted onto polyvinylidene difluoride membranes (Millipore). The blots were probed with different primary antibodies. Other primary antibodies used in this study include anti-AtCUL1 (Schwechheimer et al., 2001), anti-RPT5 (Kwok et al., 1999), anti-RPN6 (Kwok et al., 1999), anti-AtRBX1 (Schwechheimer et al., 2002), and anti-TBP (Schwechheimer et al., 2001).
In Vivo Coimmunoprecipitation Experiments
Coimmunoprecipitation experiments were performed as described previously (Yanagawa et al., 2004) with minor modifications. The lysis/binding/washing buffer was 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 50 mM NaF, 10 mM Na3VO4, 60 mM β-glycerolphosphate, 0.1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, and 1× complete protease inhibitor (Roche). After immunoprecipitation with IgG-coupled Sepharose (Amersham Biosciences), the proteins were eluted by cleavage of the IgG binding domain with 3C protease (Amersham Biosciences). The eluted proteins were concentrated using Strataclean resin (Stratagene).
Gel Filtration Chromatography
For gel filtration chromatography, 6-d-old seedlings were homogenized in lysis buffer (see above). The extract was centrifuged at 13,000g for 10 min at 4°C and subsequently filtered though a 0.2-μm syringe filter. Total soluble protein was fractionated through a 25-mL Superdex-6 HR fast protein liquid chromatography column (Amersham Biosciences) with the lysis buffer. After a 7-mL void volume was achieved, consecutive fractions of 0.5 mL each were collected. Proteins were concentrated using Strataclean resin (Stratagene).
Yeast Two-Hybrid Assay
The QuickChange II XL site-directed mutagenesis kit (Stratagene) was used to create AtCUL3A(K676R), AtCUL3A(S48A, F49A), and At1g21780 (I204R, D206A, Y242A, I244R) constructs. The full-length cDNA clones for AtCUL3A, AtCUL1, At1g21780, and the mutated versions of AtCUL3A and At1g21780 were cloned as translational fusions to the LexA DNA binding domain (in pEG202; Origene Technologies, Rockville, MD). At1g21780, ASK1, ASK2, and mutated versions of At1g21780 were cloned fused to the transcription activation domain (in pJG4-5; Origene Technologies). All of the LexA fusion constructs were transformed into yeast strain EGY48 (Invitrogen). The activation domain fusion constructs were transformed into yeast strain L40 (Invitrogen), which contains a β-galactosidase reporter gene. The transformants were selected and mated (Bendixen et al., 1994), and the resulting diploids were placed in medium selective for the diploids and containing X-Gal as a substrate for detection of the interaction. Selected pair-wise interactions were analyzed further by liquid assay as described previously (Serino et al., 1999).
At1g21780-TAP and RBX1-TAP Transgenic Plants
Full-length open reading frames of Arabidopsis At1g21780 and AtRBX1 were cloned via Gateway reactions (Invitrogen) into a plant binary vector with a C-terminal modified TAP tag fusion and a N-terminal modified TAP tag fusion, respectively (Rubio et al., 2005). The transgenes were introduced individually into wild-type Arabidopsis plants by the floral dip infiltration method using Agrobacterium tumefaciens strain GV3101 (pMP90). Transgenic plants were selected with gentamycin (200 μg/mL; Sigma-Aldrich).
BTB Protein Phylogenetic Analysis
Arabidopsis proteins containing a BTB domain were searched in the PFAM protein families and domains database (Bateman et al., 2004). Sequence alignment (∼110 amino acid residues of the BTB domain) of the BTB protein families was conducted using ClustalX software (version 1.83) (Thompson et al., 1997). The Gonnet protein weight matrices (Gonnet 40, 80, 120, 160, 250, and 350 matrices [Gonnet et al., 1992]) were selected, and the gap-opening and extension parameters were 10 and 0.2, respectively. Distance (mean standard distance) phylogenetic analysis was performed in PAUP 4.0 (score 3.41; Swofford, 2000). Bootstrap support (Felsenstein, 1985) for nodes was estimated using 1000 replicates under mean standard distance.
Sequence data for the proteins used in the phylogenetic analysis were taken from the EMBL/GenBank databases, with the following accession numbers (Arabidopsis Genome Initiative locus number with GenBank accession number in parentheses): At1g21780 (AAM65090), At5g19000 (AAM97116), At3g06190 (AAF30312), At2g39760 (NP_030522), At4g37610 (AAN15363), At1g01640 (AAF78396), At5g48510 (NP_199662), At3g43700 (CAB83071), At1g05690 (AAQ87006), At3g48360 (AAN31824), At5g63160 (AAR24650), C. elegans MEL-26 (NP_492449), AtCUL3A (CAC87120), AtCUL3B (AAG52544), and human CUL3 (NP_003581).
Supplementary Material
Acknowledgments
We are grateful to Vicente Rubio for advice on the TAP coimmunoprecipitation experiments and to Lena Hileman for help with the phylogenetic analysis. We also thank the Salk Institute Genomic Analyses Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion line. This research was supported by grants from the National Science Foundation (MCB-0077217 and NSF 2010 MCB-0115870) to X.W.D. P.F. was initially supported by a fellowship from the Pew Latin American Program in the Biomedical Sciences, and G.G. was initially supported by a fellowship from the Universitá degli Studi di Milano.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Xing Wang Deng (xingwang.deng@yale.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.031989.
References
- Ahmad, K.F., Engel, C.K., and Privé, G.G. (1998). Crystal structure of the BTB domain from PLZF. Proc. Natl. Acad. Sci. USA 95, 12123–12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Bardwell, V.J., and Treisman, R. (1994). The POZ domain: A conserved protein-protein interaction motif. Genes Dev. 8, 1664–1677. [DOI] [PubMed] [Google Scholar]
- Bateman, A., et al. (2004). The Pfam protein families database. Nucleic Acids Res. 32, D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendixen, C., Gangloff, S., and Rothstein, R. (1994). A yeast mating-selection scheme for detection of protein–protein interactions. Nucleic Acids Res. 22, 1778–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick, M., Lochhead, S., Honda, A., Palmer, S., and Callis, J. (2004). Related to Ubiquitin 1 and 2 are redundant and essential and regulate vegetative growth, auxin signaling, and ethylene production in Arabidopsis. Plant Cell 16, 2418–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Glazebrook, J., Clarke, J.D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. [DOI] [PubMed] [Google Scholar]
- Clark-Maguire, S., and Mains, P.E. (1994). mei-1, a gene required for meiotic spindle formation in Caenorhabditis elegans, is a member of a family of ATPases. Genetics 136, 533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullinan, S.B., Gordan, J.D., Jin, J., Harper, J.W., and Diehl, J.A. (2004). The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: Oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell. Biol. 24, 8477–8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Guzman, R.N., Liu, H.Y., Martinez-Yamout, M., Dyson, H.J., and Wright, P.E. (2000). Solution structure of the TAZ2 (CH3) domain of the transcriptional adaptor protein CBP. J. Mol. Biol. 303, 243–253. [DOI] [PubMed] [Google Scholar]
- del Pozo, J.C., and Estelle, M. (1999). The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proc. Natl. Acad. Sci. USA 96, 15342–15347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies, R.J. (2003). SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15, 435–467. [DOI] [PubMed] [Google Scholar]
- Dieterle, M., Thomann, A., Renou, J.-P., Parmentier, Y., Cognat, V., Lemonnier, G., Müller, R., Shen, W.-H., Kretsch, T., and Genschick, P. (2005). Molecular and functional characterization of Arabidopsis Cullin 3A. Plant J. 41, 386–399. [DOI] [PubMed] [Google Scholar]
- Doerks, T., Huber, S., Buchner, E., and Bork, P. (2002). BSD: A novel domain in transcription factors and synapse-associated proteins. Trends Biochem. Sci. 27, 168–170. [DOI] [PubMed] [Google Scholar]
- Du, L., and Poovaiah, B.W. (2004). A novel family of Ca2+/calmodulin-binding proteins involved in transcriptional regulation: Interaction with fsh/Ring3 class transcription activators. Plant Mol. Biol. 54, 549–569. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39, 783–791. [DOI] [PubMed] [Google Scholar]
- Furukawa, M., He, Y.J., Borchers, C., and Xiong, Y. (2003). Targeting of protein ubiquitination by BTB–Cullin 3–Roc1 ubiquitin ligases. Nat. Cell Biol. 5, 1001–1007. [DOI] [PubMed] [Google Scholar]
- Geyer, R., Wee, S., Anderson, S., Yates, J., and Wolf, D.A. (2003). BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol. Cell 12, 783–790. [DOI] [PubMed] [Google Scholar]
- Gonnet, G.H., Cohen, M.A., and Benner, S.A. (1992). Exhaustive matching of the entire protein sequence database. Science 256, 1443–1445. [DOI] [PubMed] [Google Scholar]
- Ha, C.M., Jun, J.H., Nam, H.G., and Fletcher, J.C. (2004). BLADE-ON-PETIOLE1 encodes a BTB/POZ domain protein required for leaf morphogenesis in Arabidopsis thaliana. Plant Cell Physiol. 45, 1361–1370. [DOI] [PubMed] [Google Scholar]
- Hellmann, H., and Estelle, M. (2002). Plant development: Regulation by protein degradation. Science 297, 793–797. [DOI] [PubMed] [Google Scholar]
- Hershko, A., and Ciechanover, A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Huber, A.H., Nelson, W.J., and Weis, W.I. (1997). Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell 90, 871–882. [DOI] [PubMed] [Google Scholar]
- Kawakami, T., Chiba, T., Suzuki, T., Iwai, K., Yamanaka, K., Minato, N., Suzuki, H., Shimbara, N., Hidaka, Y., Osaka, F., Omata, M., and Tanaka, K. (2001). NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 20, 4003–4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S., Choi, H., Ryu, H.-J., Park, J.H., Kim, M.D., and Kim, S.Y. (2004). ARIA, an Arabidopsis arm repeat protein interacting with a transcriptional regulator of abscisic acid-responsive gene expression, is a novel abscisic acid signaling component. Plant Physiol. 136, 3639–3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, A., Kang, M.I., Okawa, H., Ohtsuji, M., Zenke, Y., Chiba, T., and Yamamoto, I.K. (2004). Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 24, 7130–7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl, A., Binz, H.K., Forrer, P., Stumpp, M.T., Plückthun, A., and Grütter, M.G. (2003). Designed to be stable: Crystal structure of a consensus ankyrin repeat protein. Proc. Natl. Acad. Sci. USA 100, 1700–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz, T., Pintard, L., Willis, J.H., Hamill, D.R., Gonczy, P., Peter, M., and Bowerman, B. (2002). Cytoskeletal regulation by the Nedd8 ubiquitin-like protein modification pathway. Science 295, 1294–1298. [DOI] [PubMed] [Google Scholar]
- Kwok, S., Staub, J., and Deng, X.W. (1999). Characterization of two subunits of Arabidopsis 19S proteasome regulatory complex and its possible interaction with the COP9 complex. J. Mol. Biol. 285, 85–95. [DOI] [PubMed] [Google Scholar]
- Laux, T., Wurschum, T., and Breuninger, H. (2004). Genetic regulation of embryonic pattern formation. Plant Cell 16 (suppl.), S190–S202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L., Gao, Y., Qu, L., Chen, Z., Li, J., Zhao, H., and Deng, X.W. (2002). Genomic evidence for COP1 as a repressor of light-regulated gene expression and development in Arabidopsis. Plant Cell 14, 2383–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L., Sun, N., Liu, X., Jiao, Y., Zhao, H., and Deng, X.W. (2005). Organ-specific genome expression atlas during Arabidopsis development. Plant Physiol., in press. [DOI] [PMC free article] [PubMed]
- Mathur, J., and Koncz, C. (1998). Establishment and maintenance of cell suspension cultures. In Methods in Molecular Biology, Arabidopsis Protocols, J. Martinez-Zapater and J. Salinas, eds (New York: Humana Press), pp. 27–30. [DOI] [PubMed]
- Mistry, H., Wilson, B.A., Roberts, I.J.H., O'Kane, C.J., and Skeath, J.B. (2004). Cullin-3 regulates pattern formation, external sensory organ development and cell survival during Drosophila development. Mech. Dev. 121, 1495–1507. [DOI] [PubMed] [Google Scholar]
- Motchoulski, A., and Liscum, E. (1999). Arabidopsis NPH3: A NPH1 photoreceptor-interacting protein essential for phototropism. Science 286, 961–964. [DOI] [PubMed] [Google Scholar]
- Pintard, L., Kurz, T., Glaser, S., Willis, J.H., Peter, M., and Bowerman, B. (2003. a). Neddylation and deneddylation of CUL-3 is required to target MEI-1/Katanin for degradation at the meiosis-to-mitosis transition in C. elegans. Curr. Biol. 13, 911–921. [DOI] [PubMed] [Google Scholar]
- Pintard, L., Willems, A., and Peter, M. (2004). Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. EMBO J. 23, 1681–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintard, L., et al. (2003. b). The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 425, 311–316. [DOI] [PubMed] [Google Scholar]
- Risseeuw, E.P., Daskalchuk, T.E., Banks, T.W., Liu, E., Cotelesage, J., Hellmann, H., Estelle, M., Somers, D.E., and Crosby, W.L. (2003). Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. Plant J. 34, 753–767. [DOI] [PubMed] [Google Scholar]
- Rubio, V., Shen, Y., Saijo, Y., Liu, Y., Gusmaroli, G., Dinesh-Kumar, S.P., and Deng, X.W. (2005). An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 41, 767–778. [DOI] [PubMed] [Google Scholar]
- Sakai, T., Wada, T., Ishiguro, S., and Okada, K. (2000). RPT2. A signal transducer of the phototropic response in Arabidopsis. Plant Cell 12, 225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman, B.A., Carrano, A.C., Jeffrey, P.D., Bowen, Z., Kinnucan, E.R., Finnin, M.S., Elledge, S.J., Harper, J.W., Pagano, M., and Pavletich, N.P. (2000). Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408, 381–386. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., Serino, G., Callis, J., Crosby, W.L., Lyapina, S., Deshaies, R.J., Gray, W.M., Estelle, M., and Deng, X.W. (2001). Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science 292, 1379–1382. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., Serino, G., and Deng, X.W. (2002). The COP9 signalosome and AXR1 are required for multiple E3 ubiquitin ligase-mediated processes in Arabidopsis thaliana. Plant Cell 14, 2553–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serino, G., Tsuge, T., Kwok, S., Matsui, M., Wei, N., and Deng, X.W. (1999). Arabidopsis cop8 and fus4 mutations define the same gene that encodes subunit 4 of the COP9 signalosome. Plant Cell 11, 1967–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, W.H., Parmentier, Y., Hellmann, H., Lechner, E., Dong, A., Masson, J., Granier, F., Lepiniec, L., Estelle, M., and Genschik, P. (2002). Null mutation of AtCUL1 causes arrest in early embryogenesis in Arabidopsis. Mol. Biol. Cell 13, 1916–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer, J.D., Gurian-West, M., Clurman, B., and Roberts, J.M. (1999). Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev. 13, 2375–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins, C.E., Kaelin, W.G., and Pavletich, N.P. (1999). Structure of the VHL-ElonginC-ElonginB complex: Implications for VHL tumor suppressor function. Science 284, 455–461. [DOI] [PubMed] [Google Scholar]
- Sullivan, J.A., Shirasu, K., and Deng, X.W. (2003). The diverse roles of ubiquitin and the 26S proteasome in the life of plants. Nat. Rev. Genet. 4, 948–958. [DOI] [PubMed] [Google Scholar]
- Sussman, M.R., Amasino, R.M., Young, J.C., Krysan, P.J., and Austin-Phillips, S. (2000). The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol. 124, 1465–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D.L. (2000). PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods). (Sunderland, MA: Sinauer Associates).
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL X Windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren, A.G., and Vaux, D.L. (1996). TRAF proteins and meprins share a conserved domain. Trends Biochem. Sci. 21, 244–245. [PubMed] [Google Scholar]
- Wang, K.L., Yoshida, H., Lurin, C., and Ecker, J.R. (2004). Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428, 945–950. [DOI] [PubMed] [Google Scholar]
- Weber, H., Bernhardt, A., Dieterle, M., Hano, P., Mutlu, A., Estelle, M., Genschik, P., and Hellmann, H. (2005). Arabidopsis AtCUL3a and AtCUL3b form complexes with members of the BTB/POZ-MATH protein family. Plant Physiol. 137, 83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins, A., Ping, Q., and Carpenter, C.L. (2004). RhoBTB2 is a substrate of the mammalian Cul3 ubiquitin ligase complex. Genes Dev. 18, 856–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston, J.T., Chu, C., and Harper, J.W. (1999). Culprits in the degradation of cyclin E apprehended. Genes Dev. 13, 2751–2757. [DOI] [PubMed] [Google Scholar]
- Wirbelauer, C., Sutterluty, H., Blondel, M., Gstaiger, M., Peter, M., Reymond, F., and Krek, W. (2000). The F-box protein Skp2 is a ubiquitylation target of a Cul1-based core ubiquitin ligase complex: Evidence for a role of Cul1 in the suppression of Skp2 expression in quiescent fibroblasts. EMBO J. 19, 5362–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodger, F.J., Jacobsen, J.V., and Gubler, F. (2004). GMPOZ, a BTB/POZ domain nuclear protein, is a regulator of hormone responsive gene expression in barley aleurone. Plant Cell Physiol. 45, 945–950. [DOI] [PubMed] [Google Scholar]
- Wu, K., Chen, A., and Pan, Z.Q. (2000). Conjugation of Nedd8 to CUL1 enhances the ability of the ROC1–CUL1 complex to promote ubiquitin polymerization. J. Biol. Chem. 275, 32317–32324. [DOI] [PubMed] [Google Scholar]
- Xu, L., Wei, Y., Reboul, J., Vaglio, P., Shin, T.H., Vidal, M., Elledge, S.J., and Harper, J.W. (2003). BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 425, 316–321. [DOI] [PubMed] [Google Scholar]
- Yanagawa, Y., Sullivan, J.A., Komatsu, S., Gusmaroli, G., Suzuki, G., Yin, J., Ishibashi, T., Saijo, Y., Rubio, V., Kimura, S., Wang, J., and Deng, X.W. (2004). Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev. 18, 2172–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H., Peters, J.M., King, R.W., Page, A.M., Hieter, P., and Kirschner, M.W. (1998). Identification of a cullin homology region in a subunit of the anaphase-promoting complex. Science 279, 1219–1222. [DOI] [PubMed] [Google Scholar]
- Zheng, N., et al. (2002). Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416, 703–709. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.