Abstract
The aim of the present study was to identify differentially expressed proteins in the lymph fluid of rabbits with breast cancer lymphatic metastasis compared with healthy rabbits and to analyze and verify these proteins using proteomics technologies. In the process of breast cancer metastasis, the composition of the lymph fluid will also change. Rabbits with breast cancer lymph node metastasis and normal rabbits were selected for analysis. Lymph fluid was extracted under the guidance of percutaneous contrast-enhanced ultrasound. Label-free quantitative proteomics was used to detect and compare differences between the rabbit cancer model and healthy rabbits and differential protein expression results were obtained. Bioinformatics analysis was performed using Kyoto Encyclopedia of Genes and Genomes and Gene Ontology analysis software, selecting the most significantly differentially expressed proteins. Finally, parallel reaction monitoring technology was applied for validation. A total of 547 significantly differentially expressed proteins were found in the present study, which included 371 upregulated proteins and 176 downregulated proteins. The aforementioned genes were mainly involved in various cellular and metabolic pathways, including upregulated proteins, such as biliverdin reductase A and isocitrate dehydrogenase 2 and downregulated proteins, such as pyridoxal kinase. The upregulated proteins protein disulfide-isomerase 3, protein kinase cAMP-dependent type I regulatory subunit α and ATP-binding cassette sub-family C member 4 participated in immune regulation, endocrine regulation and anti-tumor drug resistance regulation, respectively. Compared with healthy rabbits, rabbits with breast cancer metastasis differentially expressed of a number of different proteins in their lymph, which participate in the pathophysiological process of tumor occurrence and metastasis. Through further research, these differential proteins can be used as predictive indicators of breast cancer metastasis and new therapeutic targets.
Keywords: breast cancer, lymph node metastasis, contrast-enhanced ultrasonography, proteomics, differentially expressed protein
Introduction
Breast cancer is one of the most common cancers among women globally and its incidence rate is increasing annually (1). Breast cancer is the second leading cause of cancer deaths among women (2). Breast cancer is a metastatic cancer that initially spreads through the lymphatic system to various levels of lymph nodes and can then metastasize to distant organs through the bloodstream, such as the bones, liver, lungs and brain, which contributes to the difficulties in effectively treating this disease (3). Accurate metastasis prediction is related to the formulation of treatment plans, prognosis, survival rate and quality of life (4). However, breast cancer is a highly heterogeneous malignant disease with various functional phenotypes (5). Previous proteomic, genomic and transcriptomic studies have explored different cellular subtypes and the development of breast cancer lymphatic metastasis biomarkers (6,7). Therefore, finding protein biomarkers related to breast cancer is important for predicting the progression of the disease, implementing early drug or intervention treatment and thus reducing mortality caused by metastasis.
The process of lymphatic metastasis involves the lymphatic circulation and tumor-related lymphatic vessels provide a direct route to the lymph nodes, which enables primary tumors to transmit cytokine signals, gradually reshaping and hijacking lymph node function from afar (8). After a tumor develops, various aspects of the internal tumor environment will undergo complex biological changes (9). Lymph fluid, an essential component of the lymph node environment, is subject to these changes, with variations occurring in its component cytokines and antibodies; for example, programmed death ligand 1 and tumor-infiltrating lymphocytes (10). However, due to the difficulties associated with obtaining lymph fluid [due to individualized vessel routing and the technical difficulty of extraction via contrast-enhanced ultrasound (CEUS)], this issue has been under-researched to date. Percutaneous CEUS technology can provide support in this area (11), enabling observation of the lymphatic vessels entering and leaving the sentinel lymph nodes and assisting in the accurate extraction of lymph fluid. Therefore, in the present study, CEUS technology was used to precisely extract lymph fluid from rabbits with breast cancer metastasis and healthy rabbits.
Materials and methods
Establishment of the rabbit breast cancer model
Purebred New Zealand white rabbits (male, n=1; female, n=6; 6 months old), weighing 2.0–2.5 kg were purchased from the Experimental Animal Center of Ningxia Medical University. Maintenance conditions for the rabbits were as follows: Temperature, 18–29°C; relative humidity, 40–70%; noise, ≤60 dB; animal illumination, 100–001×; and food and water, sufficient food and water sources to meet the physiological needs of the rabbits. The VX2 rabbit mesenchymal squamous cell carcinoma tumor cell line (cat. no. RP-0097) was purchased from Shanghai Yinxi Biotechnology Co., Ltd. Cells were cultured in RPMI 1640 culture medium. The ratio was basic culture medium:serum (FBS) at 9:1, and the double antibody was added at 1% volume. Cells were cultivated in a 5% CO2 incubator at 37°C. The aforementioned reagents were purchased from Thermo Fisher Scientific, Inc. To establish the tumor model, 1 ml of a 1×107 cells/ml suspension of VX2 cells was injected into the lateral muscles of the hind legs of the male rabbit. After 2 weeks, a solid mass was formed. Animals were euthanized by an intravenous injection of pentobarbital sodium (100 mg/kg) into tumor bearing rabbits. Under sterile conditions, the solid tumor from the hind leg of the tumor bearing rabbit was removed, washed with physiological saline and placed in a glass dish containing 20 ml of RPMI 1640 solution. A fish-shaped piece of tissue (~5×5×5 mm in size) with vigorous growth at the edge of the tumor was cut into tissue blocks with a diameter of 0.5–1.0 mm using ophthalmic scissors. The tumor blocks were mixed well and placed with 10 ml RPMI1640 solution into a 20 ml syringe to make a tissue block suspension. Tissues were placed on ice until used. Three female rabbits were held in a supine position using a fixator and administered inhalant isoflurane anesthesia (induction concentration, 2%; maintenance concentration, 1.5%) for 2 min. Once the corneal reflex weakened, muscles relaxed or breathing slowed down, the injection was performed. A 10 ml volume of tissue block suspension was injected under the breast pad of the second nipple on the left side of the rabbit chest wall. After 2 weeks, the rabbits were observed to have a 100% tumor formation rate and 1 tumor per animal. Humane endpoints were as follows: During the experiment, if any uncontrollable pain or other conditions were found in the animal, euthanasia would be carried out promptly. During the experiment, the size of the tumor was observed and if the tumor volume exceeded 10% of the experimental rabbit's body weight, the experiment would be terminated in a timely manner. The in vivo experiments performed in the present study were approved by the Medical Research Ethics Review Committee of Ningxia Medical University General Hospital (approval no. KYLL-2022-0173).
Precision extraction of lymph fluid
A total of three healthy rabbits and three rabbits with lymph node metastasis of breast cancer were analyzed. Ultrasound contrast agent (Sonazoid; GE Healthcare) was injected subcutaneously at the edge of the tumor in the breast cancer model rabbits. The first (group) lymph node traced from the injection point along the lymphatic vessel was the sentinel lymph node (SLN). CEUS can clearly display the input lymphatic vessels of the SLN, which may reflect the state of the lymph fluid after tumor microenvironment formation (11,12). CEUS can used to dynamically observe the progression of lymph node metastasis. Lymph fluid from the rabbit's SLN at 8 weeks after tumor implantation was used to ensure that the metastatic lymph node was malignant. Accurate lymph fluid extraction from normal rabbits under CEUS guidance was performed as a control. This process occurred once every 2 days and lasted for 2 weeks, with 10 µl of lymph extracted each time. During the extraction process, animals were anaesthetized through the inhalation of isoflurane (induction concentration, 2%; maintenance concentration, 1.5%) for 3 min. After the experiment was completed, all rabbits were euthanized by intravenous injection of pentobarbital sodium (100 mg/kg). The criteria for determining death were: No breathing or pulse in the rabbit, no heartbeat lasting for more than 5 min when auscultating or touching the chest with a stethoscope, rabbit corneal reflex disappeared, pupil dilation occurred and nerve reflexes ceased. At the end of the experiment, the maximum tumor diameter of both female and male rabbits did not exceed 4 cm.
Reagents and equipment
Ammonium bicarbonate buffer (NH4HCO3; pH 8.0), dithiothreitol (DTT), iodoacetamide (IAA) and sodium carbonate were purchased from MilliporeSigma. Urea, SDS and the BCA protein assay kit were purchased from Bio-Rad Laboratories, Inc. Trypsin was purchased from Promega Corporation and the Q Exactive™ Plus mass spectrometer and EASY-nLC™ 1200 were purchased from Thermo Fisher Scientific, Inc.
Label-free quantification (LFQ) sample preparation
After freeze-drying the lymph fluid, each sample was mixed with 100–200 µl SDT lysis buffer [4% sodium dodecyl sulfate, 100 mM dithiothreitol, 100 mM Tris-HCl (pH 8.0); Bio-Rad Laboratories, Inc.] based on the volume of lymph fluid obtained (2:1 ratio). Lysate was homogenized and transferred to an Eppendorf® tube. The lysate was then incubated in a boiling water bath (100°C) for 3 min, sonicated for 2 min (50 W, 60°C for 2 sec), centrifuged at 16,000 × g and 4°C for 20 min and the supernatant was collected. The BCA method was used for protein quantification. A total of 100 µg protein was taken from each sample for filter-aided sample preparation digestion. The following steps were performed: DTT (1 M to a final concentration of 100 mM) was added to each sample followed by incubation in a boiling water bath (100°C) for 5 min, after which the samples were cooled to room temperature. A 200 µl volume of urea buffer (8 M urea; 150 mM Tris-HCl; pH 8.0) was added to each sample and mixed before transferring to a 10 kDa ultrafiltration centrifuge tube and centrifuged at 12,000 × g for 15 min at 4°C. UA buffer (200 µl) was added to samples and centrifuged at 12,000 × g for 15 min at 4°C before the filtrate was discarded. A 100 µl volume of IAA (50 mM IAA in UA) was added and samples were shaken at 600 rpm for 1 min. Next, samples were kept in the dark at room temperature for 30 min, then centrifuged at 12,000 × g for 10 min at 4°C. UA buffer (100 µl) was added and samples were centrifuged at 12,000 × g for 10 min at 4°C and repeated twice. A 100-µl volume of NH4HCO3 buffer was added and samples were centrifuged at 14,000 × g for 10 min at 4°C and repeated twice. A 40 µl volume of trypsin buffer (6 µg trypsin in 40 µl NH4HCO3 buffer) was added to each sample before shaking at 600 rpm for 1 min and incubating at 37°C for 16–18 h. Samples were moved to a new collection tube and centrifuged at 12,000 × g for 10 min at 4°C. The filtrate was collected and an appropriate volume (100 µl) of 0.1% trifluoroacetic acid (TFA) solution was added. Then, the digested peptides were desalted using the C18 Cartridge (Bio-Rad Laboratories, Inc.) and freeze-dried under vacuum. After drying, the digested peptides were reconstituted with 0.1% TFA. The peptide concentration was measured and samples were prepared for liquid chromatography with tandem mass spectrometry (LC-MS/MS) analysis.
LC-MS/MS analysis
Chromatographic separation of peptide samples was performed using a nanoflow Easy nLC 1200 chromatography system (Thermo Fisher Scientific, Inc.). The buffer solutions were prepared as follows: Buffer A was a 0.1% formic acid aqueous solution and buffer B was a formic acid (0.1%), acetonitrile (80%) and water solution. The chromatographic column was equilibrated with 95% buffer A solution. The sample was injected into a trap column (100 µm; 20 mm; 5 µm; C18; Dr. Maisch HPLC GmbH) and separated by a gradient through the chromatographic analysis column (75 µm; 150 mm; 3 µm; C18; Dr. Maisch HPLC GmbH) at a flow rate of 300 nl/min. The liquid phase separation gradient was as follows: 0–2 min, linear gradient of buffer B solution from 5–8%; 2–90 min, linear gradient of buffer B solution from 8–23%; 90–100 min, linear gradient of buffer B solution from 23–40%; 100–108 min, linear gradient of buffer B solution from 40–100%; 108–120 min, buffer B solution was maintained at 100%. After peptide separation, a Q-Exactive HF-X mass spectrometer (Thermo Fisher Scientific, Inc.) was used for data-dependent acquisition mass spectrometry analysis. The analysis time was 120 min, the detection mode was positive ionization, the parent ion scan range was 300–1,800 m/z, the primary mass spectrometry resolution was 60,000 at m/z 200, the AGC target was 3×106 and primary mass spectrometry maximum ion time (IT) was 50 msec. Peptide secondary mass spectrometry was collected according to the following methods: After each full scan, 20 secondary mass spectra (MS2 scans) of the highest intensity parent ions were collected, with a secondary mass spectrometry resolution of 15,000 at m/z 200, an AGC target of 1×105, secondary mass spectrometry maximum IT of 50 msec, MS2 activation type of HCD, an isolation window of 1.6 m/z and a normalized collision energy of 28.
Database retrieval
The resulting LC-MS/MS raw files were imported into the Proteome Discoverer software (version 2.4; Thermo Fisher Scientific, Inc.) and the search engine Sequest HT was used for database retrieval. The database used for searching was uniprot-Oryctolagus cuniculus (Rabbit) [9986]-43526-20211222.fasta, which was sourced from the Uniprot protein database (https://www.uniprot.org/taxonomy/9986), with a protein entry of 43,526 and a download date of 22.12.2021.
Parallel reaction monitoring (PRM) sample preparation
Urea (8 M) was added to 200-µl samples, which were then sonicated in an ice bath and centrifuged at 4°C and 16,000 × g for 20 min to collect the supernatant. BCA quantification was performed on the supernatant and 15 µg of each sample was run on a 1% agarose gel. DTT (final concentration 10 mM) was added to 200 µg of each sample and incubated at 37°C for 1 h. IAA was added to a final concentration of 50 mM and samples were incubated in darkness for 30 min at 25°C. Trypsin was added to each sample (1:50) and incubated at 37°C overnight. After quantitative desalting, samples were mixed in equal volumes for testing.
LC-PRM/MS analysis
A total of 2 µg of peptide from each sample was used for LC-PRM/MS analysis. After sample loading, chromatographic separation was performed using a nanoflow Easy nLC1200 chromatography system (Thermo Fisher Scientific, Inc.). The liquid phase separation gradient was as follows: 0–5 min, linear gradient of buffer B solution from 2–5%; 5–45 min, the linear gradient of buffer B solution from 5–23%; 45–50 min, linear gradient of buffer B solution from 23–40%; 50–52 min, linear gradient of buffer B solution from 40–100%; and 52–60 min, buffer B solution was maintained at 100%. The Q Exactive HF-X mass spectrometer (Thermo Fisher Scientific, Inc.) was used for targeted PRM mass spectrometry analysis. The original PRM raw data files of the mass spectra obtained were analyzed using Skyline software (version 4.1) (13).
Bioinformatics analysis
Proteins with fold-change (FC) >1.5 or <0.667 and P<0.05 were considered as significantly differentially expressed proteins. Bioinformatics data were analyzed using Perseus (14), Microsoft Excel (Microsoft Corporation) and R (version 4.0.3; RStudio, Inc.) statistical computing software. Sequence annotations were extracted from UniProtKB/Swiss-Prot, Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) databases. GO and KEGG enrichment analyses were performed using Fisher's exact test and false discovery rate adjustment for multiple testing. GO terms were divided into three categories: Biological processes (BP), molecular functions (MF) and cellular components (CC). Enriched GO and KEGG pathways were classed as statistically significant if P<0.05.
Statistical analysis
Data were analyzed using SPSS software (version 20.0; IBM Corp.). Measurement data were presented as the mean ± standard deviation of three replicates for each group and the unpaired t-test was used for inter-group comparisons. Count data were presented as percentages and inter-group differences were compared using the χ2 test. P<0.05 was considered to indicate a statistically significant difference.
Results
Observation of breast cancer lymph node metastasis under CEUS guidance
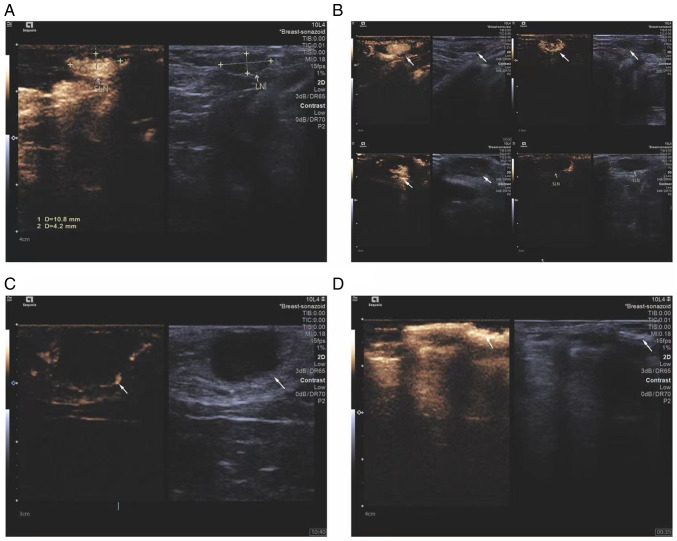
CEUS was used to observe the changes in sentinel lymph nodes during the metastasis process of the breast cancer lymphatic metastasis group of rabbits. Percutaneous superficial ultrasound with Sonozoid demonstrated four contrast patterns in sentinel lymph nodes during breast cancer progression (Fig. 1): i) Uniform enhancement; ii) non-uniform enhancement; iii) peripheral ring enhancement with no internal enhancement; and iv) complete absence of enhancement in the lymph node with the presence of surrounding lymphatic vessels bypassing. To ensure that the extracted lymph originated from metastatic lymph nodes, the status of rabbit lymph nodes using CEUS was observed and ultrasound-guided lymph extraction was performed when there was no enhancement under contrast-enhanced ultrasound. During the experiment, the volume, maximum diameter and minimum diameter of the rabbit tumor were measured and recorded (Fig. 2) (Table I).
Figure 1.
Ultrasound imaging patterns of sentinel lymph nodes. (A) Homogeneous enhancement, (B) heterogeneous enhancement, (C) peripheral ring enhancement with no internal enhancement and (D) no enhancement with lymphatic vessel bypass around the lymph node. Respective features are indicated with arrows.
Figure 2.
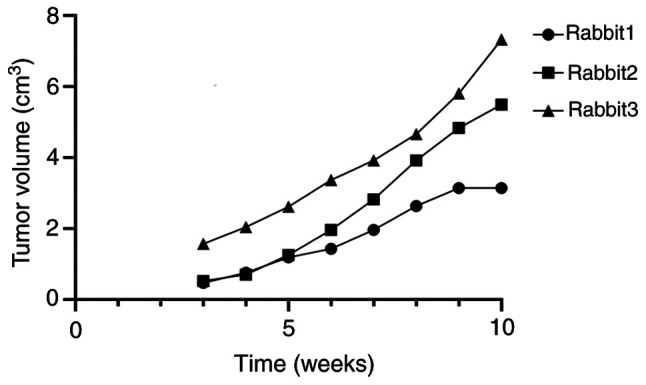
Tumor volume in rabbits over the course of the experiment.
Table I.
Maximum diameter of tumors observed in rabbits at the end of the experiment.
| Animal | Long diameter of tumor, cm | Short diameter of tumor, cm |
|---|---|---|
| Male rabbit | 2.0 | 1.5 |
| Female rabbit 1 | 3.0 | 2.0 |
| Female rabbit 2 | 3.5 | 3.0 |
| Female rabbit 3 | 4.0 | 3.0 |
Analysis of lymphatic fluid differentially expressed proteins
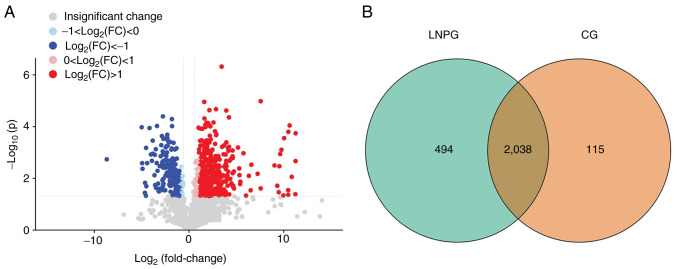
A total of 2,647 proteins were detected in six samples using the LFQ method. Using a 1.2-fold increase or decrease in protein expression as the criterion for significant changes, 547 differentially expressed proteins were identified. Compared with the normal group (CG), the metastasis group (LNPG) demonstrated 371 upregulated proteins and 176 downregulated proteins (Fig. 3).
Figure 3.
Differentially expressed proteins in LNPG and CG groups of rabbits. (A) Volcano plot of differentially expressed proteins and a (B) Venn diagram of differentially expressed proteins in the LNPG and CG groups. CG, normal group; LNPG, metastasis group; FC, fold-change.
Differentially expressed protein GO functional enrichment analysis
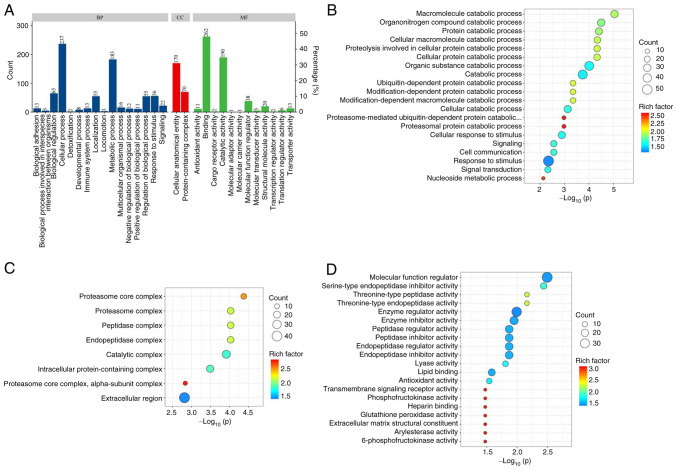
GO functional enrichment analysis was conducted on the differentially expressed proteins in the lymphatic fluid of breast cancer rabbits and normal rabbits (Fig. 4A). The main BPs included ‘macromolecule catabolic process’, ‘organonitrogen compound catabolic process’ and ‘protein catabolic process’ (Fig. 4B). The main CCs included ‘proteasome core complex’, ‘proteasome complex’, ‘peptidase complex’, ‘endopeptidase complex’ and ‘catalytic complex’ (Fig. 4C). The main MFs included ‘molecular function modulator’, ‘serine-type endopeptidase inhibitor activity’ and ‘threonine-type peptidase activity’ (Fig. 4D).
Figure 4.
GO enrichment analysis of the differentially expressed proteins in LNPG and CG groups of rabbits. (A) GO term annotation, (B) top 20 BP term enrichment bubble chart, (C) top 20 CC term enrichment bubble chart and (D) top 20 MF term enrichment bubble chart. GO, gene ontology; LNPG, metastasis group; CG, normal group; BP, biological processes; CC, cellular components; MF, molecular functions.
Differentially expressed protein KEGG enrichment analysis
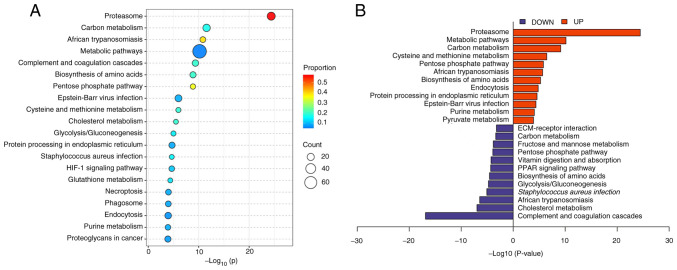
KEGG analysis was used to subject the differentially expressed proteins to pathway enrichment analysis. It was demonstrated that the differentially expressed proteins were involved in 20 main pathways (Fig. 5A), which included the proteasome pathway, complement and coagulation cascades pathway and pentose phosphate pathway. The upregulated proteins were mainly involved in pathways such as the proteasome pathway, carbon metabolism pathway and cysteine and methionine metabolism pathway. The downregulated proteins were mainly involved in pathways such as the complement and coagulation cascade pathway, cholesterol metabolism pathway and Staphylococcus aureus infection pathway (Fig. 5B).
Figure 5.
KEGG enrichment analysis of differentially expressed proteins in LNPG and CG groups of rabbits. (A) Top 20 KEGG pathway bubble chart and (B) top 12 KEGG pathway enrichment butterfly chart. KEGG, Kyoto Encyclopedia of Genes and Genomes; LNPG, metastasis group; CG, normal group.
PRM validation
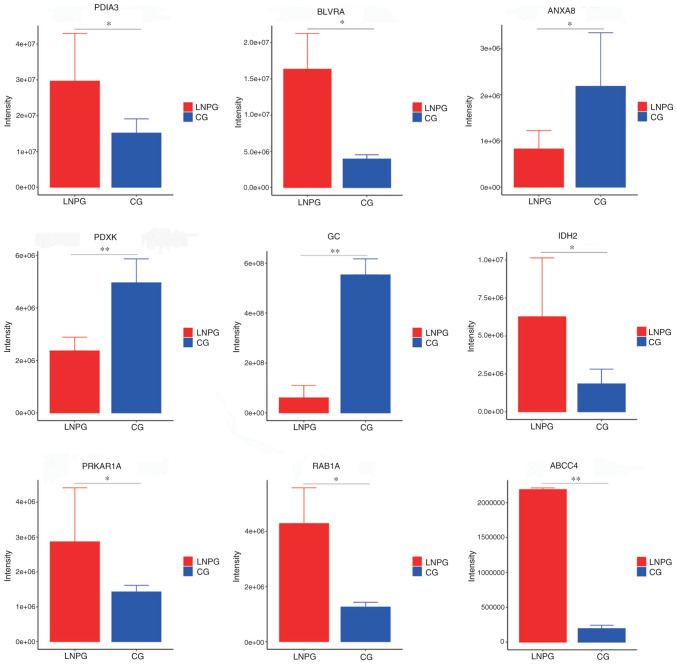
PRM was conducted on the differentially expressed proteins in the lymphatic fluid of the metastasis group and healthy group of rabbits. A total of 10 differentially expressed proteins were demonstrated to have consistent trends with the results from the LFQ proteomics technology assay (Table II). The upregulated proteins included protein disulfide-isomerase 3 (PDIA3), biliverdin reductase A, isocitrate dehydrogenase 2, protein kinase cAMP-dependent type I regulatory subunit α (PRKAR1A), Ras-related protein Rab-1A, ATP-binding cassette sub-family C member 4 (ABCC4) and microtubule-associated protein 4, while the downregulated proteins included annexin A8, pyridoxal kinase and envelope glycoprotein C (Fig. 6).
Table II.
Label-free quantification proteomic validation of differentially expressed proteins in the normal and metastasis groups.
| Gene | Protein | NCBI accession no. | Peptide sequence | Score | Fold-change | P-value |
|---|---|---|---|---|---|---|
| PDIA3 | Protein disulfide-isomerase | B7NZF1 | ELSDFISYLQR | 187.53 | 0.62 | <0.05 |
| BLVRA | Biliverdin reductase A | G1SRZ6 | FGFPAFSGISR | 21.76 | 1.88 | <0.05 |
| IDH2 | Isocitrate dehydrogenase (NADP) | G1SZF7 | LNEHFLNTTDFLDTIK | 130.9 | 2.35 | <0.05 |
| PRKAR1A | Protein kinase cAMP-dependent type I regulatory subunit alpha | G1TDN4 | NVLFSHLDDNER | 48.67 | 6.97 | <0.05 |
| RAB1A | RAB1A, member RAS oncogene family | G1TCS8 | QWLQEIDR | 61.72 | 1.78 | <0.05 |
| ABCC4 | ATP binding cassette subfamily C member 4 | A0A5F9CTH3 | SFAELIASLR | 59.42 | 0.97 | <0.05 |
| ANXA8 | Annexin | G1T6W4 | GAGTLDGTLIR | 141.5 | 7.97 | <0.01 |
| PDXK | Pyridoxal kinase | G1UZB5 | GQVLTSDELHELHELYEGLR | 21.46 | 4.49 | <0.05 |
| GC | Gc-globulin | G1SU8Z | HLSLLTTLSNR | 628.2 | 2.35 | <0.05 |
Figure 6.
Parallel reaction monitoring validation results for differentially expressed proteins. LNPG, metastasis group; CG, normal group. *P<0.05, **P<0.01 vs. CG group.
Discussion
Lymphatic metastasis is a major factor affecting the prognosis of various cancers, including breast cancer (15). Although cancer cells spread in a number of different ways, the structure of the lymphatic system makes it the prime site of cancer metastasis (16). Despite the current understanding of the pathophysiology of breast cancer, the molecular mechanisms, especially those related to lymphatic metastasis, are still unclear. This may be due to the difficulty in obtaining lymphatic fluid. Identifying differentially expressed proteins and signaling pathways involved in breast cancer metastasis is critical to understanding its mechanism and exploring biomarkers related to metastasis. In the present study, CEUS technology was used to extract lymphatic fluid for comparative analysis, quantitative evaluation and functional identification of LFQ proteomics and validated differentially expressed proteins with PRM technology.
In the present study, using LC-MS/MS analysis, proteins in the lymph fluid were quantified and bioinformatics analysis was performed on differentially expressed proteins to identify protein biomarkers for breast cancer lymphatic metastasis and explore the function of differentially expressed proteins and the signaling pathways involved. The present study laid the foundation for further exploration of predictive indicators and therapeutic targets for breast cancer metastasis in the future. As breast cancer develops, the tumor microenvironment inevitably changes, as does the body's metabolic environment. For example, amino acids are essential nutrients for all living cells and are vital for the proliferation and maintenance of tumor cells. Since tumor cells grow faster compared with normal cells, tumor cells have a higher demand for amino acids (17). Previous studies have reported that some amino acid metabolic pathways, such as glutamine, serine, glycine and proline pathways, are altered in breast cancer, which suggests that amino acid transport may be crucial for the proliferation and progression of breast cancer (18–20). At the same time, mechanisms of hypoxia-adaptive metabolic responses (21), including increased glycolysis and decreased tricarboxylic acid cycle, serve to reduce the production of mitochondrial reactive oxygen species (22).
The present study identified upregulated proteins PDIA3, PRKAR1A and ABCC4 in the lymphatic fluid of metastatic breast cancer rabbits using LC-MS/MS. PDIA3, also known as ERp57, is a 58 kDa glucose-regulated protein that also acts as a chaperone, modifying and folding proteins, and has redox functions (23,24). PDIA3 serves a role in the quality control of newly synthesized glycoproteins, participates in the assembly of major histocompatibility complex class I molecules and regulates immune responses and immunogenic cell death (25). Despite, to the best of our knowledge, no reports of a direct link between PDIA3 and breast cancer development to date, PDIA3 has previously been reported to be upregulated in various cancers and is involved in cancer initiation, progression and chemosensitivity, which suggests its potential as a cancer biomarker and therapeutic target (26–28). PRKAR1A is a gene that directs the synthesis of protein kinase A (PKA) regulatory subunits and is one of the critical components of the PKA tetramer (29). It is the primary mediator of cAMP function in various mammalian cellular processes, including cell differentiation, proliferation and apoptosis (30). PRKAR1A inactivating mutations can lead to Carney syndrome, which is characterized by cardiac myxoma and multiple endocrine tumors (31). Therefore, PRKAR1A mutations may serve as a predictor of breast cancer metastasis. ABCC4 is the fourth member of the ATP-binding cassette protein C subfamily, also known as multidrug resistance-associated protein 4 (MRP4). ABCC4 was discovered due to its role in mediating drug resistance in various tumor types (32). High expression levels of ABCC4 have been reported in drug-resistant tumors, including neuroblastoma, prostate cancer, pancreatic cancer and acute myeloid leukemia, where MRP4 expression is associated with poor prognosis (33–35). Thus, the increased drug resistance in breast cancer may be linked to the high expression levels of ABCC4.
However, the technical difficulty in accurately extracting lymphatic fluid using CEUS limits its application to animal research. Additionally, proteomics technology has high economic costs, making it challenging to promote in clinical practice.
In conclusion, the present study employed CEUS technology for lymphatic fluid extraction, as well as proteomic and mass spectrometry analysis techniques to investigate differentially expressed proteins and related functions in metastatic breast cancer rabbits. The up-regulated or down-regulated proteins and multiple enrichment pathways identified in the present study were related to the pathophysiological process of the occurrence and development of breast cancer. The data provided by the present study may provide new ideas for future follow-up research on new metastasis targets of breast cancer.
Acknowledgments
The authors would like to thank Shanghai Bioprofile (Shanghai, China) for providing technical support by performing the proteomic testing and analysis.
Funding Statement
The present study was supported by the National Science Foundation of China (grant no. 82260351).
Availability of data and materials
The datasets used in the present study can be found in online repositories. The LC-MS/MS data was uploaded to iProX (accession no. IPX0007648000).
Authors' contributions
JX, XZ, GY, WS, WW and CM designed the research concept. WW and CM coordinated the study. Lymph extraction was performed by JX, XZ, GY and WS. XZ conducted the statistical analysis. JX is responsible for the language editing of this manuscript and writing the main manuscript. JX, XZ, GY, WS, WW and CM participated in the planning and supervision of the work, as well as in the interpretation of data. JX and XZ confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
All experimental animals in the present study were provided by the Experimental Animal Center of Ningxia Medical University and all experimental procedures met the review standards of the Medical Research Ethics Review Committee of Ningxia Medical University General Hospital (approval no. KYLL-2022-0173).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17–48. doi: 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 2.Sun YS, Zhao Z, Yang ZN, Xu F, Lu HJ, Zhu ZY, Shi W, Jiang J, Yao PP, Zhu HP. Risk factors and preventions of breast cancer. Int J Biol Sci. 2017;13:1387–1397. doi: 10.7150/ijbs.21635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park M, Kim D, Ko S, Kim A, Mo K, Yoon H. Breast cancer metastasis: Mechanisms and therapeutic implications. Int J Mol Sci. 2022;23:6806. doi: 10.3390/ijms23126806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gouri A, Benarba B, Dekaken A, Aoures H, Benharkat S. Prediction of late recurrence and distant metastasis in early-stage breast cancer: Overview of current and emerging biomarkers. Curr Drug Targets. 2020;21:1008–1025. doi: 10.2174/1389450121666200312105908. [DOI] [PubMed] [Google Scholar]
- 5.Yeo SK, Guan JL. Breast cancer: Multiple subtypes within a tumor? Trends Cancer. 2017;3:753–760. doi: 10.1016/j.trecan.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mani DR, Krug K, Zhang B, Satpathy S, Clauser KR, Ding L, Ellis M, Gillette MA, Carr SA. Cancer proteogenomics: Current impact and future prospects. Nat Rev Cancer. 2022;22:298–313. doi: 10.1038/s41568-022-00446-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Kong X, Wang Z, Xuan L. Recent advances of transcriptomics and proteomics in triple-negative breast cancer prognosis assessment. J Cell Mol Med. 2022;26:1351–1362. doi: 10.1111/jcmm.17124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.du Bois H, Heim TA, Lund AW. Tumor-draining lymph nodes: At the crossroads of metastasis and immunity. Sci Immunol. 2021;6:eabg3551. doi: 10.1126/sciimmunol.abg3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arneth B. Tumor microenvironment. Medicina (Kaunas) 2019;56:15. doi: 10.3390/medicina56010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rizzo A, Ricci AD. Biomarkers for breast cancer immunotherapy: PD-L1, TILs, and beyond. Expert Opin Investig Drugs. 2022;31:549–555. doi: 10.1080/13543784.2022.2008354. [DOI] [PubMed] [Google Scholar]
- 11.Cui Q, Dai L, Li J, Xue J. Accuracy of CEUS-guided sentinel lymph node biopsy in early-stage breast cancer: A study review and meta-analysis. World J Surg Oncol. 2020;18:112. doi: 10.1186/s12957-020-01890-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karaman S, Detmar M. Mechanisms of lymphatic metastasis. J Clin Invest. 2014;124:922–928. doi: 10.1172/JCI71606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 15.Das S, Skobe M. Lymphatic vessel activation in cancer. Ann N Y Acad Sci. 2008;113:235–241. doi: 10.1196/annals.1413.021. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z, Wang R, Zhou J, Zheng Y, Dong Y, Luo T, Wang X, Zhan W. Ultrasound lymphatic imaging for the diagnosis of metastatic central lymph nodes in papillary thyroid cancer. Eur Radiol. 2021;31:8458–8467. doi: 10.1007/s00330-021-07958-y. [DOI] [PubMed] [Google Scholar]
- 17.Cha YJ, Kim ES, Koo JS. Amino acid transporters and glutamine metabolism in breast cancer. Int J Mol Sci. 2018;19:907. doi: 10.3390/ijms19030907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El Ansari R, McIntyre A, Craze ML, Ellis IO, Rakha EA, Green AR. Altered glutamine metabolism in breast cancer; subtype dependencies and alternative adaptations. Histopathology. 2018;72:183–190. doi: 10.1111/his.13334. [DOI] [PubMed] [Google Scholar]
- 19.Kim S, Kim DH, Jung WH, Koo JS. Expression of glutamine metabolism-related proteins according to molecular subtype of breast cancer. Endocr Relat Cancer. 2013;20:339–348. doi: 10.1530/ERC-12-0398. [DOI] [PubMed] [Google Scholar]
- 20.Craze ML, Cheung H, Jewa N, Coimbra NDM, Soria D, El-Ansari R, Aleskandarany MA, Wai Cheng K, Diez-Rodriguez M, Nolan CC, et al. MYC regulation of glutamine-proline regulatory axis is key in luminal B breast cancer. Br J Cancer. 2018;118:258–265. doi: 10.1038/bjc.2017.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Semenza GL. Hypoxia-inducible factors: Coupling glucose metabolism and redox regulation with induction of the breast cancer stem cell phenotype. EMBO J. 2017;36:252–259. doi: 10.15252/embj.201695204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Heer EC, Jalving M, Harris AL. HIFs, angiogenesis, and metabolism: Elusive enemies in breast cancer. J Clin Invest. 2020;130:5074–5087. doi: 10.1172/JCI137552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chichiarelli S, Altieri F, Paglia G, Rubini E, Minacori M, Eufemi M. ERp57/PDIA3: New insight. Cell Mol Biol Lett. 2022;27:12. doi: 10.1186/s11658-022-00315-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hettinghouse A, Liu R, Liu CJ. Multifunctional molecule ERp57: From cancer to neurodegenerative diseases. Pharmacol Ther. 2018;181:34–48. doi: 10.1016/j.pharmthera.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song D, Liu H, Wu J, Gao X, Hao J, Fan D. Insights into the role of ERp57 in cancer. J Cancer. 2021;12:2456–2464. doi: 10.7150/jca.48707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park S, Park JH, Jung HJ, Jang JH, Ahn S, Kim Y, Suh PG, Chae S, Yoon JH, Ryu SH, Hwang D. A secretome profile indicative of oleate-induced proliferation of HepG2 hepatocellular carcinoma cells. Exp Mol Med. 2018;50:1–14. doi: 10.1038/s12276-018-0069-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y, Lin MC, Wang H, Chan CY, Jiang L, Ngai SM, Yu J, He ML, Shaw PC, Yew DT, et al. Proteomic analysis of EZH2 downstream target proteins in hepatocellular carcinoma. Proteomics. 2007;7:3097–3104. doi: 10.1002/pmic.200700019. [DOI] [PubMed] [Google Scholar]
- 28.Li S, Zhao X, Chang S, Li Y, Guo M, Guan Y. ERp57-small interfering RNA silencing can enhance the sensitivity of drug-resistant human ovarian cancer cells to paclitaxel. Int J Oncol. 2019;54:249–260. doi: 10.3892/ijo.2018.4628. [DOI] [PubMed] [Google Scholar]
- 29.Czarnecka AM, Synoradzki K, Firlej W, Bartnik E, Sobczuk P, Fiedorowicz M, Grieb P, Rutkowski P. Molecular Biology of Osteosarcoma. Cancers (Basel) 2020;12:2130. doi: 10.3390/cancers12082130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pitsava G, Stratakis CA, Faucz FR. PRKAR1 and thyroid tumors. Cancers (Basel) 2021;13:3834. doi: 10.3390/cancers13153834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirschner LS. PRKAR1A and the evolution of pituitary tumors. Mol Cell Endocrinol. 2010;326:3–7. doi: 10.1016/j.mce.2010.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kochel TJ, Fulton AM. Multiple drug resistance-associated protein 4 (MRP4), prostaglandin transporter (PGT), and 15-hydroxyprostaglandin dehydrogenase (15-PGDH) as determinants of PGE2 levels in cancer. Prostaglandins Other Lipid Mediat. 2015:116–117. 99–103. doi: 10.1016/j.prostaglandins.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huynh T, Norris MD, Haber M, Henderson MJ. ABCC4/MRP4: A MYCN-regulated transporter and potential therapeutic target in neuroblastoma. Front Oncol. 2012;2:178. doi: 10.3389/fonc.2012.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rasmuson A, Kock A, Fuskevåg OM, Kruspig B, Simón-Santamaría J, Gogvadze V, Johnsen JI, Kogner P, Sveinbjörnsson B. Autocrine prostaglandin E2 signaling promotes tumor cell survival and proliferation in childhood neuroblastoma. PLoS One. 2012;7:e29331. doi: 10.1371/journal.pone.0029331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montani M, Hermanns T, Müntener M, Wild P, Sulser T, Kristiansen G. Multidrug resistance protein 4 (MRP4) expression in prostate cancer is associated with androgen signaling and decreases with tumor progression. Virchows Arch. 2013;462:437–443. doi: 10.1007/s00428-013-1390-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used in the present study can be found in online repositories. The LC-MS/MS data was uploaded to iProX (accession no. IPX0007648000).