Abstract
Histone acetylation is modulated through the action of histone acetyltransferases and deacetylases, which play key roles in the regulation of eukaryotic gene expression. Previously, we have identified a yeast histone deacetylase REDUCED POTASSIUM DEPENDENCY3 (RPD3) homolog, HISTONE DEACETYLASE19 (HDA19) (AtRPD3A), in Arabidopsis thaliana. Here, we report further study of the expression and function of HDA19. Analysis of Arabidopsis plants containing the HDA19:β-glucuronidase fusion gene revealed that HDA19 was expressed throughout the life of the plant and in most plant organs examined. In addition, the expression of HDA19 was induced by wounding, the pathogen Alternaria brassicicola, and the plant hormones jasmonic acid and ethylene. Using green fluorescent protein fusion, we demonstrated that HDA19 accumulated in the nuclei of Arabidopsis cells. Overexpression of HDA19 in 35S:HDA19 plants decreased histone acetylation levels, whereas downregulation of HDA19 in HDA19-RNA interference (RNAi) plants increased histone acetylation levels. In comparison with wild-type plants, 35S:HDA19 transgenic plants had increased expression of ETHYLENE RESPONSE FACTOR1 and were more resistant to the pathogen A. brassicicola. The expression of jasmonic acid and ethylene regulated PATHOGENESIS-RELATED genes, Basic Chitinase and β-1,3-Glucanase, was upregulated in 35S:HDA19 plants but downregulated in HDA19-RNAi plants. Our studies provide evidence that HDA19 may regulate gene expression involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis.
INTRODUCTION
Covalent modifications of the N termini of the core histones in nucleosomes, in particular acetylation, play important roles in gene regulation (Berger, 2002). Histone acetylation levels are determined by the action of histone acetyltransferases and histone deacetylases (HDAC). Acetylation of the histones is often associated with increased gene activity, whereas deacetylation of histones is correlated with transcriptional repression. Recent work has identified a series of histone acetyltransferase-coactivator proteins and HDAC-associated corepressor complexes in yeast (Saccharomyces cerevisiae) and mammalian cells, suggesting that the enzymatic modulation of histone acetylation is an integral component of transcription regulation (Khochbin et al., 2001; Berger, 2002). This includes plants where many essential biological processes are controlled by genes that are regulated by histone acetylation-mediated chromatin remodeling. For example, HDAC complexes are centrally involved in plant cellular functions, such as the cell cycle (Rossi et al., 2003). It has also been shown that histone acetyltransferases can be recruited through transcription factors, such as C-Repeat Binding Factor1, to cold-induced genes through multiprotein complexes similar to those found in other eukaryotes (Stockinger et al., 2001). The repression of a MADS box transcription factor, FLOWERING LOCUS C, which controls flowering time in Arabidopsis thaliana, is regulated by histone deacetylation (He et al., 2003; Ausin et al., 2004). Furthermore, H3 and H4 associated with the promoter of the pea (Pisum sativum) plastocyanin gene are hyperacetylated and sensitive to digestion by nucleases in transcriptionally active green shoots but not etiolated shoots and roots (Chua et al., 2003).
Eukaryotic HDACs can be grouped into three classes based on their primary homology to three yeast HDACs: REDUCED POTASSIUM DEPENDENCY3 (RPD3), HISTONE DEACETYLASE1 (HDA1), and SIRTUIN2 (Pandey et al., 2002; Yang and Seto, 2003). In addition, plants contain an uncommon class of HDACs, the HD2 class, which was identified in plants only (Lusser et al., 1997; Aravind and Koonin, 1998; Wu et al., 2000a, 2003; Dangl et al., 2001; Zhou et al., 2004). Studies on the mechanism of action of HDACs in plants are beginning to emerge. In maize (Zea mays), the RPD3-type HDACs were shown to be physically associated with the maize retinoblastoma-related protein, a key regulator of cell cycle progression (Rossi et al., 2003; Varotto et al., 2003). Several RPD3-type HDACs, HDA19 (also called AtRPD3A or AtHD1), HDA6 (AtRPD3B), HDA7, and HDA9 were identified in Arabidopsis (Wu et al., 2000b; Murfett et al., 2001; Tian and Chen, 2001; Pandey et al., 2002). Mutations in HDA6 affected transgene expression, DNA methylation, and regulation of rRNA genes (Murfett et al., 2001; Aufsatz et al., 2002; Probst et al., 2004). A range of developmental abnormalities, including delayed flowering, were observed in plants expressing an antisense HDA19 construct (Wu et al., 2000b; Tian and Chen, 2001) and a HDA19 T-DNA insertion mutant (Tian et al., 2003). The diversity of phenotypes displayed by different HDAC mutants suggests that different HDACs might have evolved a range of specialized functions in plant growth and development.
The involvement of HDAC in the plant defense response has previously been inferred from the interaction between maize and the pathogen Cochiobolus carbonum (Brosch et al., 1995), in which the host-selective HC toxin of the pathogen inhibited host HDAC activity. In maize, treatment of plants with HC toxin or infection by C. carbonum induced histone hyperacetylation (Ransom and Walton, 1997). It was suggested that HC toxin acts as a suppressor of elicitor-activated defense response through the inhibition of HDACs (Brosch et al., 1995; Ransom and Walton, 1997). The HDAC activity of the plant-pathogenic fungi C. carbonum and Alternaria brassicicola are both insensitive to HC toxin (Brosch et al., 2001; Baidyaroy et al., 2002). Recently, it was demonstrated that HDA6 can interact with CORONATINE-INSENSITIVE1 (COI1), an F-box protein that is involved in jasmonate (JA)-mediated plant defense responses, suggesting a possible role for HDA6 in plant–pathogen interaction (Devoto et al., 2002).
Previously, we have identified an Arabidopsis RPD3-type HDAC gene, HDA19, which appeared to be functionally involved in many developmental processes (Wu et al., 2000b; Tian and Chen, 2001; Tian et al., 2003). Here, we report evidence that HDA19 is involved in the ethylene, JA, and pathogen response. We demonstrated that HDA19 was induced by JA and ethylene. In addition, the expression of HDA19:β-glucuronidase (GUS) was induced by wounding, JA, ethylene, and the pathogen A. brassicicola. Overexpression of HDA19 in transgenic Arabidopsis plants resulted in increased resistance to the pathogen A. brassicicola, whereas silencing of HDA19 by RNA interference (RNAi) resulted in decreased resistance to this pathogen. The expression of JA and ethylene regulated PATHOGENESIS-RELATED (PR) genes was upregulated in 35S:HDA19 plants but downregulated in HDA19 RNAi plants compared with the wild type. The role of HDA19 in plant pathogen response is discussed.
RESULTS
The Effects of JA, Ethylene, Wounding, and a Pathogen on the Expression of HDA19:GUS
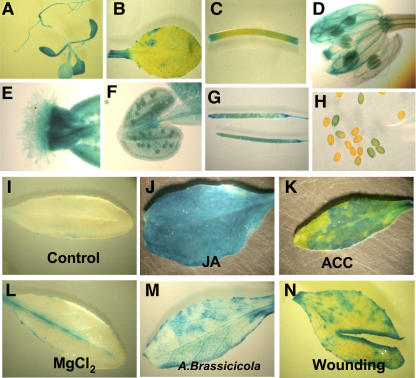
A 1.4-kb HDA19 promoter fragment was used to construct a GUS reporter gene fusion (HDA19:GUS) for stable transformation of Arabidopsis. In 2-week-old transgenic seedlings, high levels of GUS expression were observed in all parts of the seedlings (Figure 1A). In adult plants, GUS expression was detected in the mature leaves, stems, flowers, siliques, and seeds (Figures 1B to 1H). In developing or mature flowers, the GUS activity was detected in all parts of the flowers, including the sepal, style, and stamens (Figures 1D to 1F).
Figure 1.
GUS Activity in HDA19:GUS Plants.
GUS staining patterns of a seedling (A), leaf (B), stem (C), flower (D), stigma (E), anther (F), siliques (G), and seeds (H) from HDA19:GUS plants. GUS staining in HDA19:GUS leaves without treatment (I), treated with 1 mM methyl-jasmonate (JA) (J), 1 mM ACC (an ethylene precursor) (K), or 10 mM MgCl2 (L), infected with A. brassicicola for 24 h (M), and after cutting (N).
As shown in the Figure 1, the GUS reporter gene under the control of the HDA19 promoter was strongly induced by JA and 1-aminocyclopropane-1-carboxylic acid (ACC; an ethylene precursor). Furthermore, infections with the necrotrophic pathogen, A. brassicicola, induced GUS expression at the sites of infection by the pathogen (Figure 1M). In many of the tissues shown in Figure 1, GUS expression was induced by wounding of the tissues at the sites of excision. This was examined more carefully as shown in Figure 1N. GUS driven by the HDA19 promoter was shown to be wound inducible and limited to the cells at the actual wound site.
HDA19 Transcript Was Induced by JA and Ethylene
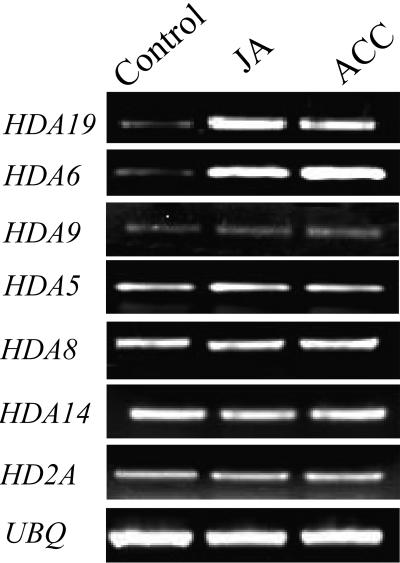
We further analyzed the expression of HDA19 by RT-PCR. As shown in Figure 2, HDA19 mRNA accumulated in the presence of JA and ACC. In addition to HDA19, three other homologs of yeast RPD3 (i.e., HDA6, HDA7, and HDA9) were also identified in the Arabidopsis genome (Pandey et al., 2002; Plant Chromatin Database, http://chromdb.org). We compared the expression of HDA19 with other RPD3-type HDAC genes in Arabidopsis. Like HDA19, the expression of HDA6 can also be induced by ethylene and JA (Figure 2). The expression of HDA9, however, cannot be induced. We could not detect the expression of HDA7 by RT-PCR. These data suggest that there is differential expression among RPD3-type HDAC genes in Arabidopsis. Furthermore, the expression of members of HDA1-type HDACs, HDA5, HDA8, and HDA14, as well as an HD2-type HDAC, HD2A, cannot be induced by ethylene and JA (Figure 2).
Figure 2.
RT-PCR Analysis of Expression of HDAC Genes.
Total RNA for RT-PCR analysis was isolated from leaf tissues of Arabidopsis plants treated without (control) or with 1 mM JA and 1 mM ACC (an ethylene precursor) for 6 h. Ubiquitin (UBQ) is shown as an internal control.
Targeting of HDA19 into the Nucleus
To investigate the cellular distribution of HDA19, we performed an in vivo targeting experiment using green fluorescent protein (GFP). HDA19-GFP gene fusion was created and introduced into Arabidopsis under the 35S promoter to achieve high levels of constitutive expression. To confirm that the fusion protein entered the nucleus, we monitored the fluorescence of GFP at the cellular level. Protoplasts were isolated from seedlings of transgenic Arabidopsis, and localization of the fusion protein was determined by fluorescence microscopy. As shown in Figure 3, the HDA19-GFP fusion protein was localized in the nucleus of the Arabidopsis cells.
Figure 3.
Subcellular Localization of HDA19.
Protoplasts were isolated from the leaves of 35S:GFP ([A] and [B]) and 35S:HDA19-GFP ([C] and [D]) transgenic Arabidopsis plants. GFP fluorescence was examined by fluorescence microscopy under UV light ([A] and [C]) and white light ([B] and [D]).
Altered HDA19 Expression Affected Histone Acetylation Levels
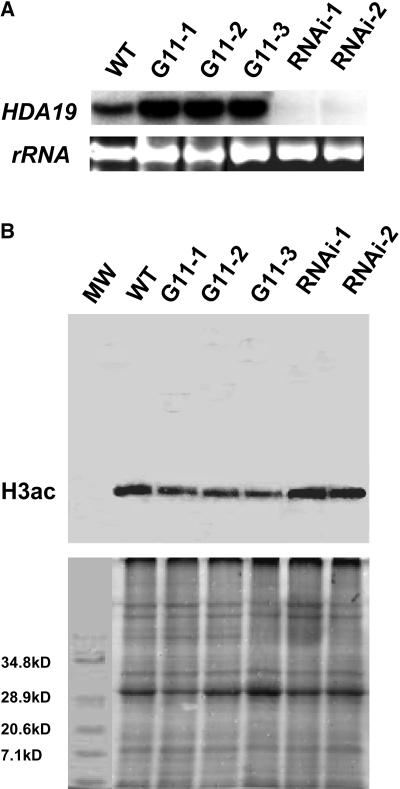
Transgenic Arabidopsis plants were developed that overexpressed HDA19 from the constitutive 35S promoter of Cauliflower mosaic virus. PCR and DNA gel blot analysis were performed to determine the integration of transgenes into the genome in transgenic plants (data not shown). The three 35S:HDA19 transgenic lines, G11-1, G11-2, and G11-3, showed significantly increased levels of HDA19 mRNA accumulation compared with wild-type plants (Figure 4A). Overexpression of HDA19 cDNA in Arabidopsis resulted in an assortment of pleiotropic developmental abnormalities, including aberrant cotyledons, abnormal leaves, delayed flowering, and reduced fertility (Figure 5).
Figure 4.
Levels of Tetra-Acetylated H3 in the 35S:HDA19 and HDA19-RNAi Lines.
(A) RNA gel blot analysis of HDA19 expression in wild-type, 35S:HDA19 (G11-1, G11-2, and G11-3), and HDA19-RNAi (RNAi-1 and RNAi-2) plants. Twenty micrograms of total RNA were probed with HDA19. Ethidium bromide–stained rRNA are shown to illustrate the gel loading.
(B) Protein gel blot analysis detecting tetra-acetylated H3 (H3ac) (top panel) using α-H3ac antibodies on protein extracts from wild-type, 35S:HDA19 (G11-1, G11-2, and G11-3), and HDA19-RNAi (RNAi-1 and RNAi-2) plants. Bottom panel, Coomassie blue staining shows equal protein loading. MW, molecular weight marker.
Figure 5.
Phenotypic Abnormalities in 35S:HDA19 Plants.
(A) to (C) The 35S:HDA19 transgenic seedlings from the T2 line showed aberrant cotyledons and lacked shoot and root development ([B] and [C]) compared with a wild-type seedling (A).
(D) to (F) The 35S:HDA19 transgenic plants from the T2 line with branching leaf (E) and narrow leaf (F) compared with a normal leaf (D).
(G) and (H) A 6-week-old 35S:HDA19 plant (H) from the T2 line showed delayed flowering when compared with a 6-week-old wild-type plant (G).
(I) and (J) A stem from a 35S:HDA19 transgenic plant of the T2 line with stunted siliques (I) and a wild-type stem with full silique elongation (J).
We further analyzed two HDA19-RNAi lines, RNAi-1 and RNAi-2, generated by expressing a transgene that encodes double-stranded HDA19 RNA (Plant Chromatin Database, http://chromdb.org). RNA gel blots revealed significantly reduced HDA19 mRNA levels in the HDA19-RNAi plants relative to nontransgenic control plants (Figure 4A). HDA19-RNAi plants were vigorous and displayed phenotypes similar to the HDA19 antisense plants and T-DNA insertion plants described previously (Wu et al., 2000b; Tian and Chen, 2001; Tian et al., 2003).
The levels of tetra-acetylated H3 in wild-type, 35S:HDA19, and HDA19-RNAi plants were analyzed by protein gel blot analysis. As shown in Figure 4B, there were obviously decreased levels of tetra-acetylated H3 in three 35S:HDA19 transgenic lines compared with the wild type. By comparison, the levels of tetra-acetylated H3 in two HDA19-RNAi lines were increased, suggesting that HDA19 transcript level affects histone acetylation levels globally. Our protein gel blot analysis indicated that the anti-tetra-acetylated H3 antibody was specific to the histone H3 because we were able to detect only one protein band with molecular mass of ∼17 kD corresponding to H3. Changed levels of tetra-acetylated H3 in 35S:HDA19 and HDA19-RNAi plants indicated that HDA19 has HDAC activity.
Overexpression HDA19-Induced PR Gene Expression and Enhanced Resistance to A. brassicicola
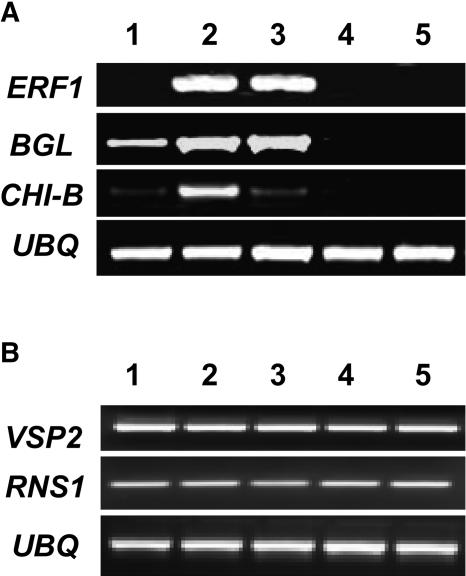
The finding that HDA19 can be induced by JA and ethylene prompted us to test whether changed HDA19 expression levels could affect JA and ethylene-regulated gene expression. It has been demonstrated that JA and ethylene coregulate the expression of a subset of PR genes, such as Basic Chitinase (CHI-B) and β-1,3-glucanase (BGL) (Lorenzo et al., 2003). RT-PCR analyses indicated that the expression of CHI-B and BGL was increased in the 35S:HDA19 plants (Figure 6A). By contrast, the expression of CHI-B and BGL was decreased in the HDA19-RNAi plants. These data indicate that there is a correlation between HDA19 expression and the expression of these PR genes. Recent studies indicate that the transcriptional factor ETHYLENE RESPONSE FACTOR1 (ERF1) acts downstream of the interaction between the ethylene and JA pathways and is a key element in the integration of both signals for the regulation of defense response genes (Lorenzo et al., 2003). RT-PCR analysis indicated that the expression of ERF1 was upregulated in the 35S:HDA19 plants (Figure 6A), suggesting that overexpression of HDA19 induced ERF1 expression.
Figure 6.
Gene Expression in the 35S:HDA19 and HDA19-RNAi Plants.
(A) RT-PCR analysis of ERF1, BGL, and CHI-B expression in wild-type (1), 35S:HDA19 (2 and 3), and HDA19-RNAi (4 and 5) transgenic lines.
(B) RT-PCR analysis of VSP2 and RNS1 expression in wild-type (1), 35S:HDA19 (2 and 3), and HDA19-RNAi (4 and 5) transgenic lines. Total RNA for RT-PCR analysis was isolated from leaf tissues of Arabidopsis plants. Ubiquitin (UBQ) is shown as an internal control.
We also analyzed the expression of the marker genes of the wound response, VSP2 (COI1 dependent) and RNS1 (COI1 independent) (Reymond et al., 2000). As shown in Figure 6B, no difference was found in the expression of VSP2 and RNS1 in the 35S:HDA19 and HDA19-RNAi plants compared with the wild type, suggesting that HDA19 does not affect the expression of these wound-inducible genes. It has been demonstrated that JA induces two groups of genes regulated by different mechanisms (Lorenzo et al., 2004). The first group, including PR genes, which are involved in defense responses against pathogens, is activated by ERF1; the second group, including VSP2, integrated by genes involved in JA-mediated systemic responses to wounding, is activated by another transcriptional factor, AtMYC2 (Lorenzo et al., 2004). Our analyses indicate that HDA19 can modulate the expression of the first group of genes involved in defense response but may not affect the second group of genes involved in wound response.
We further compared the susceptibility of wild-type, 35S:HDA19, and HDA19-RNAi transgenic plants to the necrotrophic pathogen A. brassicicola. Four-week-old plants grown in soil were challenged with a suspension containing 5 × 105 spores mL−1 of A. brassicicola, and the progress of the infection was followed. Compared with wild-type plants, 35S:HDA19 plants showed a clear reduction in the symptoms of infection, whereas HDA19-RNAi plants displayed an increase in symptoms (Figure 7). These results demonstrate that HDA19 is required for resistance to A. brassicicola and that overexpression of HDA19 is sufficient to increase resistance to this necrotrophic fungus.
Figure 7.
Resistance to A. brassicicola by HDA19 Overexpression.
Graphical representation of disease symptoms in wild-type, 35S:HDA19 (G11-1 and G11-2), and HDA19-RNAi (RNAi-1 and RNAi-2) transgenic lines. Disease severity index was calculated 48 and 72 h after inoculation with A. brassicicola. Disease severity index was calculated based on the degree of symptom severity as measured by leaf necrosis area: 0, no symptom; 1, 1 to 5% necrosis areas; 2, 5 to 10% necrosis areas; 3, 10 to 25% necrosis areas; 4, >25% necrosis areas. The leaf necrosis severity ratings were summed for 60 to 80 leaves of 15 to 25 plants per genotype and divided by the number of leaves rated times the maximum possible rating to given the final disease severity index. Asterisks mark values that are significantly different from the wild type (χ2 test, P < 0.05). The experiment was repeated three times with similar results.
DISCUSSION
Our study revealed that the HDA19 promoter produced GUS activity in all of the organs analyzed in the HDA19:GUS transgenic plants. This is in good agreement with the pattern of HDA19 transcript accumulation observed by RNA gel blot analysis (Wu et al., 2000b; Plant Chromatin Database, http://chromdb.org), suggesting that HDA19 is constitutively expressed in Arabidopsis. Furthermore, we have shown that HDA19:GUS expression was induced by JA, ethylene, wounding, and a pathogen. RT-PCR analysis also demonstrated that the expression of HDA19 could be induced by JA and ethylene. The induction of HDA19 by pathogen, JA, and ethylene implies that HDA19 may play a role in the plant defense response.
The finding that 35S:HDA19 plants were more resistant to A. brassicicola, whereas HDA19-RNAi plants were more sensitive to this pathogen than wild-type plants further supports the hypothesis that HDA19 is involved in the plant defense response. The induction of plant defense responses by pathogen infection involves the action of the plant hormones ethylene, JA, and salicylic acid (SA) (Wang et al., 2002). JA and ethylene were shown to synergistically induce defense genes such as CHI-B, BGL, and PDF1.2 (Penninckx et al., 1998; Lorenzo et al., 2003). Overexpression of HDA19 in Arabidopsis caused an increase in the steady state abundance of CHI-B and BGL transcripts, supporting a role for HDA19 in ethylene and JA-mediated defense response. SA regulates the expression of different sets of PR genes, such as PR1 (Gu et al., 2000). It has been suggested that SA may have an antagonistic effect on ethylene/JA signaling (Wang et al., 2002). Our study indicates that overexpression of HDA19 in Arabidopsis did not increase the SA-regulated PR gene expression (data not shown).
In Arabidopsis, defense responses under SA control are critical for resistance to the bacterial pathogen Pseudomonas syringae (Glazebrook, 2001). By contrast, JA and ethylene signaling is required for resistance to the fungal pathogene A. brassicicola (van Wees et al., 2003). Recent studies indicate that the transcriptional factor, ERF1, acts downstream of the junction between ethylene and JA pathways, and it is a key element in the integration of both signals for the regulation of defense response genes (Lorenzo et al., 2003). The expression of ERF1 was upregulated in 35S:HDA19 plants. Overexpression of ERF1 in Arabidopsis conferred resistance to necrotrophic fungi, such as Botrytis cinerea (Berrocal-Lobo et al., 2002). Similarly, we found that overexpression of HDA19 in Arabidopsis conferred increased resistance to the fungal pathogen A. brassicicola, suggesting that HDA19 might act upstream of ERF1 in the JA and ethylene signaling in plant defense.
A recent study indicated that another Arabidopsis RPD3-type HDAC, HDA6, could interact with COI1, an F-box protein, that was required for JA-mediated plant defense responses (Devoto et al., 2002). F-box proteins interact with SKP1 and cullin proteins to form E3 ubiquitin ligases known as the SCF complexes that selectively recruit regulatory proteins targeted for ubiquitination (Deshaies, 1999). Coimmunoprecipitation experiments confirmed the interaction in planta of COI1 with SKP1-like proteins and HDA6. It remains to be determined whether HDA19 can also interact with COI1. These results suggest that COI1 may form a functional E3-type ubiquitin ligase in plants to regulate expression of jasmonate responsive genes, possibly by targeted ubiquitination of an HDAC (Devoto et al., 2002). We find that the expression of HDA6 and HDA19 can be induced by JA. These observations suggest that HDA6 and probably HDA19 are subjected to transcriptional regulation as well as ubiquitination. Similarly, an Arabidopsis ABA-INSENSITIVE gene, ABI5, which encodes a basic Leu zipper transcription factor, was also found to be regulated by both transcription and ubiquitination (Lopez-Molina et al., 2003). The expression of ABI5 is induced by ABA, but this hormone also signals the inhibition of ABI5 protein degradation via the ubiquitin (Lopez-Molina et al., 2001). Further analysis is required to reveal whether HDA6 and HDA19 are regulated by ubiquitin-mediated proteolysis and whether JA can modulate this process. Our study indicates that among the members of HDAC gene families, only HDA6 and HDA19 can be induced by JA and ethylene. In addition, the transcript abundances of the JA-regulated PR genes were increased in the 35S:HDA19 transgenic plants compared with wild-type plants. These studies provide evidence that the RPD3-type HDACs, HDA6 and HDA19, may be involved in JA responsive pathways in pathogen response (Devoto et al., 2002).
HDACs are transcriptional repressors that reduce histone acetylation levels to create local regions of repressed chromatin. Deletion of RPD3 in yeast cells results in both upregulating and downregulating gene expression (Bernstein et al., 2000; Kurdistani et al., 2002). It was proposed that deacetylation of histone by RPD3 in certain cases may activate transcription by preventing binding of other repressor complexes in yeast (Bernstein et al., 2000). Our study indicated that overexpression of HDA19 in Arabidopsis resulted in upregulation of ERF1. Given the repressive nature of a HDA, ERF1 may not be directly regulated by HDA19. It is possible that HDA19 induces ERF1 expression by preventing binding of an unknown transcription repressor that regulates ERF1 expression directly. Chromatin immunoprecipitation experiments are needed to identify the direct target genes of HDA19 (Kurdistani et al., 2002). It is interesting to note that the expression of ERF1 and the PR genes, CHI-B and BGL, is also upregulated in the T-DNA insertion Arabidopsis mutants of GCN5 that encode a histone acetyltransferase (Vlachonasios et al., 2003). Mutation in GCN5 could result in decreased acetylation of core histone. We found that the tetra-acetylated H3 level was decreased in the 35S:HDA19 plants. These observations support the hypothesis that histone acetylation is involved in gene regulation in JA and ethylene signaling of pathogen response.
In addition to pathogen responses, overexpression of HDA19 resulted in pleitropic phenotypes, including aberrant cotyledons and leaves, delayed flowering, and reduced fertility, suggesting that HDA19 may be involved in many aspects of plant development (Wu et al., 2000b; Tian and Chen, 2001; Tian et al., 2003). We find that overexpression of HDA19 decreased the acetylation levels of histones, which may affect the expression of genes involved in a wide range of biological functions. Overexpression of RPD3-type HDAC in rice (Oryza sativa) led to an increased growth rate and altered architecture (Jang et al., 2003). Similarly, downregulation of HDA19 by antisense RNA (Wu et al., 2000b; Tian and Chen, 2001; Tian et al., 2003) and RNAi in Arabidopsis resulted in pleitropic phenotypes, including aberrant leaves, delayed flowering, and male sterility. These data indicate that the spatial control of RPD3-type HDAC expression is essential for overall plant development.
In summary, our study supports a previously postulated correlation between histone acetylation and plant pathogen response mechanisms (Brosch et al., 1995; Ransom and Walton, 1997). Increasing evidence suggests that the set of defenses activated in plants in response to different types of stress and developmental cues depends on the type of interaction occurring between the hormonal signaling pathways. Recently, the involvement of histone acetylation in the regulation of cold stress-regulated genes has been demonstrated (Stockinger et al., 2001; Vlachonasios et al., 2003; Kim et al., 2004). Histone acetylation and deacetylation may therefore play a key role in the regulation of plant responses to pathogens and environmental stress.
METHODS
Plant Material
Arabidopsis thaliana plants were grown in a growth chamber (16 h of light and 8 h of darkness at 23°C) after a 2- to 4-d vernalization period. For growth under sterile conditions, seeds were surface sterilized (15-min incubation in 5% [v/v] sodium hypochlorite and a three-time rinse in sterile distilled water) and sown on half-strength MS salts (Sigma-Aldrich, St. Louis, MO) supplemented with 1% sucrose, pH 5.7, and 0.8% (w/v) agar in Petri dishes.
Plasmid Construction
To generate the HDA19:GUS construct, a 1.4-kb promoter for HDA19 was PCR amplified using the primer pairs 5′-GGTAAAGCTTAAGATGGAAGCATGTGC-3′ and 5′-GTATCCATTACCTCTGCACGCACGCCGATC-3′. The resulting PCR product was then digested by HindIII and NcoI and subcloned into the pCAMBIA1300 binary vector (Cambia, Canberra, Australia). To generate the 35S:HDA19 construct, the GUS gene in the pBI221 was replaced with the HDA19 coding region. The resulting plasmid was digested with EcoRI and HindIII, and the fragment containing the 35S promoter and the HDA19 cDNA was then subcloned into the multicloning sites of pCAMBIA2300 binary vector. To generate the 35S:HDA19-GFP construct, HDA19 cDNA was PCR amplified and subcloned in frame in front of the GFP of the pCAMBIA1302 vector. DNA and protein sequence analysis was performed using BLAST searches (http://www.ncbi.nlm.nih.gov/BLAST/) and the Vector NTI Suite program (InforMax, Bethesda, MD).
Plant Transformation and Selection
Plant transformation plasmids were electroporated into Agrobacterium tumefaciens GV3101 as described (Shaw, 1995). The Agrobacterium-mediated transformation of Arabidopsis was performed as described (Clough and Bent, 1998). T1 seeds were harvested, dried at 25°C, and germinated on sterile medium containing 40 μg/mL of kanamycin or hygromycin to select the transformants. Surviving T1 plantlets were transferred to soil to set seeds (T2).
GUS Assays
For fluorometric GUS assays (Jefferson, 1988), 50 μL of the crude extract from Arabidopsis tissues was incubated at 37°C with 1 mM 4-methylumbelliferyl glucuronide in 0.3 mL of GUS assay buffer (50 mM NaPO4, pH 7.0, 10 mM EDTA, 0.1% [v/v] Triton X-100, and 10 mM β-mercaptoethanol). After 0, 0.5, 1, and 2 h of incubation, 0.1-mL aliquots were removed and added to 1.9 mL of 0.2 M Na2CO3 to end the reaction. GUS activity was expressed as picomoles of 4-methylumbelliferone per milligram of protein per hour. For histochemical GUS assay, Arabidopsis tissues were incubated in a 0.5-mg/mL solution of 5-bromo-4-chloro-indolyl-β-d-glucuronide in 100 mM sodium phosphate buffer, pH 7.0, and incubated at 37°C overnight, followed by washing with 70% ethanol to remove the chlorophyll.
GFP Localization
Protoplasts were isolated from Arabidopsis seedlings as described (Zhou et al., 2004). The fluorescence photographs of protoplasts were taken using an Olympus florescent microscope (Tokyo, Japan) fitted with fluorescein isothiocyanate filters (excitation filter, 450 to 490 nm; emission filter, 520 nm; and dichroic mirror, 510 nm).
DNA Gel Blot and RNA Gel Blot Analysis
Total genomic DNA from Arabidopsis was extracted using Plant DNAzol reagent (Invitrogen, Carlsbad, CA). For DNA gel blots, Arabidopsis genomic DNA was digested with restriction enzymes, separated by agarose gel electrophoresis, and transferred to nylon membranes (Sambrook and Russell, 2001). For RNA gel blots, total RNA was isolated from 100 to 200 mg of Arabidopsis leaf tissues using Trizol reagent as described by the manufacturer (Invitrogen). RNA gel blots were prepared by electrophoresis of 10- to 20-μg samples of total RNA through agarose gels in the presence of formaldehyde, followed by transfer to nylon membranes.
DNA gel blots and RNA gel blots were probed with 32P-labeled probes. Prehybridization and hybridization were performed at 65°C in 0.5 M Na2HPO4, pH 7.2, 7% SDS, and 1 mM EDTA (Strommer et al., 1993). Filters were washed once for 15 min in 2× SSC with 0.1% SDS at room temperature, then twice for 20 min in 0.1× SSC, 0.1% SDS at 65°C. The damp filters were autoradiographed at −80°C using two intensifying screens. Filters were stripped in 5 mM Tris-HCl, pH 7.5, 1 mM EDTA, and 0.05% SDS at 100°C for 2 min when reprobing was required.
RT-PCR Analysis
One microgram of total RNA was used for the first-strand cDNA synthesis after incubation at 65°C for 10 min. cDNA was synthesized in a volume of 20 μL that contained Moloney murine leukemia virus reverse transcriptase buffer (Promega, Madison, WI), 10 mM DTT, 1.5 μM poly(dT) primer, 0.5 mM dNTPs, and 2 units of Moloney murine leukemia virus reverse transcriptase at 37°C for 1 h. All PCR reactions were performed with 0.5 units of Taq polymerase (PGC Scientific, Gaithersburg, MD), the buffer provided by the supplier, 0.2 mM dNTPs, and a pair of primers (0.1 μM each) in a final volume of 20 μL. PCR reaction parameters differed for each gene: thermocycling conditions were 94°C for 2 min followed by 25 to 35 cycles of 94°C for 1 min, 55 to 68°C for 1 min, and 72°C for 2 min, with a final polymerization step at 72°C for 10 min. The gene-specific primer pairs used for the RT-PCR are as follows: HDA19, 5′-ACAAGATGCCGGAGCATGAA-3′ and 5′-TTTAGGAGGAAACGCCTGCT-3′; HDA6, 5′-TAGAGCCGGACAACAAACTC-3′ and 5′-TTCACGTCTGGCTCTGGGTT-3′; HDA7, 5′-GGTGATCCGTTTGGTACATT-3′ and 5′-TCTTCTCCATGTCCACTTCC-3′; HDA9, 5′-TTACAGGAGGTGGAGGATAC-3′ and 5′-CGTTATCGTTGTCTCCATCG-3′; ERF1, 5′-TTCTATCGGATCTTCTCCAG-3′ and 5′-CGGTGATCAAAGTCACTATC-3′; CHI-B, 5′-GAAGAGGACCAATGCAACTG-3′ and 5′-AGGCCGTTAACGAAGGATCT-3′; BGL, 5′-TGTTGGGAACAATTTGCCTTC-3′ and 5′-TCATGCCGATGTTACGGTTG-3′; VSP2, 5′-GTACTGGTTGTGGTTAGGGAC-3′ and 5′-GAAGGTACGTAGTAGAGTGG-3′; RNS1, 5′-ATGGTCTTTGGCCTAACTAC-3′ and 5′-GAATTCGATCTCAGCTCCAC-3′.
Protein Gel Blot Analysis
For nuclear isolation, 500 mg of Arabidopsis seedling tissues were homogenized in 1 mL of Honda buffer (2.5% Ficoll 400, 5% dextran T40, 0.4 M sucrose, 25 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 10 mM β-mercaptoethenol, 100 μg/mL of phenylmethylsulfonyl fluoride, 0.5 μg/mL of antipain, and 0.5 μg/mL of leupeptin) and filtered through a 62-μm nylon mesh (Weigel and Glazebrook, 2002). Then, 0.5% Triton X-100 was added to the extract, which was incubated for 15 min on ice and centrifuged at 1500g for 5 min. The pellet was washed with Honda buffer containing 0.1% Triton X-100, gently resuspended in 1 mL of Honda buffer, and centrifuged at 100g for 5 min to pellet starch and cell debris. The supernatant was transferred to a microcentrifuge tube and centrifuged at 1800g for 5 min to pellet the nuclear.
The nuclear extract was suspended in 200 μL of 5× SDS-PAGE loading buffer (0.2 M Tris-HCl, pH 6.8, 25% SDS, 25% glycerol, and 12.5% 2-mercaptoethanol). The protein samples were loaded on 15% polyacrylamide gel and blotted onto a nitrocellulose membrane. The membrane was blocked in PBS containing 3% dry milk for 60 min and then incubated with 0.01 to 0.05 μg/mL of antiacetyl-histone H3 (catalog no. 06-599; Upstate, Charlottesville, VA) for 2 h at room temperature. After washing, the primary antibody was detected with secondary anti-rabbit horseradish peroxidase-coupled antibody (Amersham, Buckinghamshire, UK) at room temperature for 45 min. Visualization was achieved using the ECL system (Amersham).
Plant Infection with Pathogens
Alternaria brassicicola was cultured on malt media at 22°C for 7 d. Mycelia were ground with a sterile plastic pestle in sterile water. The resulting suspension was transferred onto V8 media and incubated for 7 d with constant light. Spores were harvested by brushing the surface with a sterile paintbrush. Concentration of spores was determined and adjusted to 5 × 105 spores mL−1. Plants were inoculated by placing one droplet of 10 μL of suspension onto the surface of the sixth through eleventh true leaves. Inoculated plants were kept at 100% relative humidity at 24°C.
Acknowledgments
We thank Jian-Min Zhou for reading the manuscript. We are grateful to Jian-Min Zhou and Xinnian Dong for providing P. syringae pv tomato DC3000 and A. brassicicola strains. This work was supported by grants from the USDA (award number 2002-35301-12208) and the National Institute of Justice (Office of Justice Programs, award number 2001-RC-CX-K003) to K.W.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Keqiang Wu (kewu@mail.wvu.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.028514.
References
- Aravind, L., and Koonin, E.V. (1998). Second family of histone deacetylases. Science 280, 1167a. [Google Scholar]
- Aufsatz, W., Mette, M.F., Van Der Winden, J., Matzke, M., and Matzke, A.J. (2002). HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J. 21, 6832–6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausin, I., Alonso-Blanco, C., Jarillo, J.A., Ruiz-Garcia, L., and Martinez-Zapater, J.M. (2004). Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat. Genet. 36, 162–166. [DOI] [PubMed] [Google Scholar]
- Baidyaroy, D., Brosch, G., Graessle, S., Trojer, P., and Walton, J.D. (2002). Characterization of inhibitor-resistant histone deacetylase activity in plant-pathogenic fungi. Eukaryot. Cell 1, 538–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, S.L. (2002). Histone modification in transcriptional regulation. Curr. Opin. Genet. Dev. 12, 142–148. [DOI] [PubMed] [Google Scholar]
- Bernstein, B.E., Tong, J.K., and Schreiber, S.L. (2000). Genomewide studies of histone deacetylase function in yeast. Proc. Natl. Acad. Sci. USA 97, 13708–13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrocal-Lobo, M., Molina, A., and Solano, R. (2002). Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29, 23–32. [DOI] [PubMed] [Google Scholar]
- Brosch, G., Dangl, M., Graessle, S., Loidl, A., Trojer, P., Brandtner, E.M., Mair, K., Walton, J.D., Baidyaroy, D., and Loidl, P. (2001). An inhibitor-resistant histone deacetylase in the plant pathogenic fungus Cochliobolus carbonum. Biochemistry 40, 12855–12863. [DOI] [PubMed] [Google Scholar]
- Brosch, G., Ransom, R., Lechner, T., Walton, J.D., and Loidl, P. (1995). Inhibition of maize histone deacetylases by HC toxin, the host-selective toxin of Cochliobolus carbonum. Plant Cell 7, 1941–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua, Y.L., Watson, L.A., and Gray, J.C. (2003). The transcriptional enhancer of the pea plastocyanin gene associates with the nuclear matrix and regulates gene expression through histone acetylation. Plant Cell 15, 1468–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Dangl, M., Brosch, G., Haas, H., Loidl, P., and Lusser, A. (2001). Comparative analysis of HD2 type histone deacetylases in higher plants. Planta 213, 280–285. [DOI] [PubMed] [Google Scholar]
- Deshaies, R.J. (1999). SCF and cullin/ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15, 435–467. [DOI] [PubMed] [Google Scholar]
- Devoto, A., Nieto-Rostro, M., Xie, D., Ellis, C., Harmston, R., Patrick, E., Davis, J., Sherratt, L., Coleman, M., and Turner, J.G. (2002). COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 32, 457–466. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (2001). Genes controlling expression of defense responses in Arabidopsis—2001 status. Curr. Opin. Plant Biol. 4, 301–308. [DOI] [PubMed] [Google Scholar]
- Gu, Y.Q., Wildermuth, M.C., Chakravarthy, S., Loh, Y.T., Yang, C., He, X., Han, Y., and Martin, G.B. (2000). Tomato transcription factors pti4, pti5, and pti6 activate defense responses when expressed in Arabidopsis. Plant Cell 14, 817–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y., Michaels, S.D., and Amasino, R.M. (2003). Regulation of flowering time by histone acetylation in Arabidopsis. Science 302, 1751–1754. [DOI] [PubMed] [Google Scholar]
- Jang, I.C., Pahk, Y.M., Song, S.I., Kwon, H.J., Nahm, B.H., and Kim, J.K. (2003). Structure and expression of the rice class-I type histone deacetylase genes OsHDAC1-3: OsHDAC1 overexpression in transgenic plants leads to increased growth rate and altered architecture. Plant J. 33, 531–541. [DOI] [PubMed] [Google Scholar]
- Jefferson, R. (1988). Plant reporter genes: The GUS gene fusion system. In Genetic Engineering: Principles and Methods, Vol. 10, J.K. Setlow, ed (New York: Plenum Press), pp. 247–263.
- Khochbin, S., Verdel, A., Lemercier, C., and Seigneurin-Berny, S. (2001). Functional significance of histone deacetylase diversity. Curr. Opin. Genet. Dev. 419, 157–160. [DOI] [PubMed] [Google Scholar]
- Kim, H.J., Hyun, Y., Park, J.Y., Park, M.J., Park, M.K., Kim, M.D., Kim, H.J., Lee, M.H., Moon, J., Lee, I., and Kim, J. (2004). A genetic link between cold responses and flowering time through FVE in Arabidopsis thaliana. Nat. Genet. 36, 167–171. [DOI] [PubMed] [Google Scholar]
- Kurdistani, S.K., Robyr, D., Tavazoie, S., and Grunstein, M. (2002). Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat. Genet. 31, 248–254. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina, L., Mongrand, S., and Chua, N.H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 98, 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina, L., Mongrand, S., Kinoshita, N., and Chua, N.H. (2003). AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev. 17, 410–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo, O., Chico, J.M., Sanchez-Serrano, J.J., and Solano, R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16, 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo, O., Piqueras, R., Sanchez-Serrano, J.J., and Solano, R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15, 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusser, A., Brosch, G., Loidl, A., Haas, H., and Loidl, P. (1997). Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science 277, 88–91. [DOI] [PubMed] [Google Scholar]
- Murfett, J., Wang, X., Hagen, G., and Guilfoyle, T.J. (2001). Identification of Arabidopsis histone deacetylase HDA6 mutants that affect transgene expression. Plant Cell 13, 1047–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, R., Muller, A., Napoli, C.A., Selinger, D.A., Pikaard, C.S., Richards, E.J., Bender, J., Mount, D.W., and Jorgensen, R.A. (2002). Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30, 5036–5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I.A., Thomma, B.P., Buchala, A., Metraux, J.P., and Broekaert, W.F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10, 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst, A.V., Fagard, M., Proux, F., Mourrain, P., Boutet, S., Earley, K., Lawrence, R.J., Pikaard, C.S., Murfett, J., Furner, I., Vaucheret, H., and Scheid, O.M. (2004). Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell 16, 1021–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransom, R.F., and Walton, J.D. (1997). Histone hyperacetylation in maize in response to treatment with HC-toxin or infection by the filamentous fungus Cochliobolus carbonum. Plant Physiol. 115, 1021–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond, P., Weber, H., Damond, M., and Farmer, E.E. (2000). Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12, 707–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi, V., Locatelli, S., Lanzanova, C., Boniotti, M.B., Varotto, S., Pipal, A., Goralik-Schramel, M., Lusser, A., Gatz, C., Gutierrez, C., and Motto, M. (2003). A maize histone deacetylase and retinoblastoma-related protein physically interact and cooperate in repressing gene transcription. Plant Mol. Biol. 51, 401–413. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D.W. (2001). Molecular Cloning: A Laboratory Manual, 3rd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Shaw, C.H. (1995). Introduction of cloning plasmids into Agrobacterium tumefaciens. Methods Mol. Biol. 49, 33–37. [DOI] [PubMed] [Google Scholar]
- Stockinger, E.J., Mao, Y., Regier, M.K., Triezenberg, S.J., and Thomashow, M.F. (2001). Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res. 29, 1524–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strommer, J., Gregerson, R., and Vayda, M. (1993). Isolation and characterization of plant mRNA. In Methods in Plant Molecular Biology and Biotechnology, B.R. Glik and J.E. Thompson, eds (Boca Raton, FL: CRC Press), pp. 49–66.
- Tian, L., and Chen, Z.J. (2001). Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc. Natl. Acad. Sci. USA 98, 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, L., Wang, J., Fong, M.P., Chen, M., Cao, H., Gelvin, S.B., and Chen, Z.J. (2003). Genetic control of developmental changes induced by disruption of Arabidopsis histone deacetylase 1 (AtHD1) expression. Genetics 165, 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wees, S.C., Chang, H.S., Zhu, T., and Glazebrook, J. (2003). Characterization of the early response of Arabidopsis to Alternaria brassicicola infection using expression profiling. Plant Physiol. 132, 606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varotto, S., Locatelli, S., Canova, S., Pipal, A., Motto, M., and Rossi, V. (2003). Expression profile and cellular localization of maize Rpd3-type histone deacetylases during plant development. Plant Physiol. 133, 606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachonasios, K.E., Thomashow, M.F., and Triezenberg, S.J. (2003). Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell 15, 626–638.12615937 [Google Scholar]
- Wang, K.L., Li, H., and Ecker, J.R. (2002). Ethylene biosynthesis and signaling networks. Plant Cell 14 (suppl.), S131–S151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, D., and Glazebrook, J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Wu, K., Malik, K., Tian, L., Brown, D., and Miki, B. (2000. b). Functional analysis of a RPD3 histone deacetylase homolog in Arabidopsis thaliana. Plant Mol. Biol. 44, 167–176. [DOI] [PubMed] [Google Scholar]
- Wu, K., Tian, L., Malik, K., Brown, D., and Miki, B. (2000. a). Functional analysis of HD2 histone deacetylase homologues in Arabidopsis thaliana. Plant J. 22, 19–27. [DOI] [PubMed] [Google Scholar]
- Wu, K., Tian, L., Zhou, C., Brown, D., and Miki, B. (2003). Repression of gene expression by Arabidopsis HD2 histone deacetylases. Plant J. 34, 241–247. [DOI] [PubMed] [Google Scholar]
- Yang, X.J., and Seto, E. (2003). Collaborative spirit of histone deacetylases in regulating chromatin structure and gene expression. Curr. Opin. Genet. Dev. 13, 143–153. [DOI] [PubMed] [Google Scholar]
- Zhou, C., Labbe, H., Sridha, S., Wang, L., Tian, L., Latoszek-Green, M., Yang, Z., Brown, D., Miki, B., and Wu, K. (2004). Expression and function of HD2-type histone deacetylases in Arabidopsis development. Plant J. 38, 715–724. [DOI] [PubMed] [Google Scholar]