Abstract
The Arabidopsis thaliana NONEXPRESSER OF PR GENES1 (NPR1, also known as NIM1) protein is an essential positive regulator of salicylic acid (SA)-induced PATHOGENESIS-RELATED (PR) gene expression and systemic acquired resistance (SAR). PR gene activity is regulated at the level of redox-dependent nuclear transport of NPR1. NPR1 interacts with members of the TGA family of transcription factors that are known to bind to SA-responsive elements in the PR-1 promoter. In an attempt to identify proteins involved in SA-mediated signal transduction, we previously described the isolation of three novel genes encoding distinct albeit structurally related proteins designated NIMIN1 (for NIM1-INTERACTING1), NIMIN2, and NIMIN3 that interact with NPR1 in the yeast two-hybrid system. Here, we show that NIMIN1 and NPR1 can be copurified from plant extracts, providing biochemical evidence for their interaction. We provide functional evidence for this interaction by describing transgenic plants constitutively expressing high amounts of NIMIN1. These plants show reduced SA-mediated PR gene induction and a compromised SAR, thus mimicking the described phenotype conferred by npr1. Moreover, they showed reduced RESISTANCE gene–mediated protection. These effects were dependent on the ability of NIMIN1 to interact with NPR1. Mutant plants with a T-DNA insertion in NIMIN1 as well as transgenic plants with reduced NIMIN1 mRNA levels showed hyperactivation of PR-1 gene expression after SA treatment but no effect on the disease resistance phenotype. Our results strongly suggest that NIMIN1 negatively regulates distinct functions of NPR1, providing a mechanism to modulate specific features of SAR.
INTRODUCTION
Nutrients synthesized by plants attract all kinds of organisms. To avoid being exploited, plants have evolved sophisticated mechanisms to protect themselves against pathogens. These include both preformed and pathogen-inducible physical and chemical barriers. RESISTANCE (R) gene–mediated protection, for example, is an induced response. A so-called avirulence (Avr) gene product of the pathogen is recognized directly or indirectly by a plant R gene product (for recent reviews, see Belkhadir et al., 2004; Innes, 2004). R gene–mediated recognition triggers a diverse set of cellular responses. Signaling molecules such as reactive oxygen species, nitric oxide, jasmonic acid (JA), ethylene, and salicylic acid (SA) are produced, and defense-related genes (e.g., the PATHOGENESIS-RELATED [PR] genes) are transcriptionally activated (Lamb and Dixon, 1997). This often coincides with the formation of necrotic lesions at the site of infection, which is termed the hypersensitive response. The pathogen is contained in these lesions and thus unable to spread throughout the plant (Dangl et al., 1996). Ultimately, the plant develops a kind of long-lasting, systemic immunity to subsequent challenge by a wide range of normally virulent pathogens. This immunity is called systemic acquired resistance (SAR) (Uknes et al., 1992; Ryals et al., 1994). SAR is preceded by an accumulation of SA, which is observed not only locally during the hypersensitive response but subsequently also in distal uninfected tissue. This leads to the concerted induction of a subset of PR genes that code for proteins such as β-glucanases and chitinases (Van Loon and Van Kammen, 1970; Ward et al., 1991). PR genes can also be induced by exogenous application of SA (White, 1979). They are commonly used as marker genes for SAR. SA is a necessary and sufficient signal for SAR, because Arabidopsis thaliana mutants defective in SA biosynthesis are strongly impaired in SAR (Wildermuth et al., 2001).
A plethora of genetic screens has been conducted to identify proteins involved in the signal transduction leading to SAR. Several groups have identified mutants in the NONEXPRESSER OF PR GENES1 (NPR1, also known as NIM1 and SAI1) locus, which are impaired in SA-induced PR gene expression and SAR (Cao et al., 1994; Delaney et al., 1995; Glazebrook et al., 1996; Shah et al., 1997). The analysis of npr1 mutants and the fact that lines overexpressing NPR1 show enhanced disease resistance led to the conclusion that NPR1 is a key positive regulator of SAR (Cao et al., 1997, 1998). The question of how NPR1 acts as a mediator between the SA signal and the activation of PR gene expression was investigated intensively during the last years. It was recently demonstrated that PR gene activity is regulated at the level of redox-dependent nuclear localization of NPR1 (Kinkema et al., 2000; Mou et al., 2003). Before SAR induction, NPR1 resides in a homooligomeric complex in the cytoplasm. After induction of SAR, SA accumulation triggers a redox change that brings about monomerization of NPR1, presumably as a result of the reduction of intermolecular disulfide bridges. In its monomeric form, NPR1 is transported to the nucleus and activates PR gene expression. The mechanism by which NPR1 regulates PR gene activity is of great interest. Analysis of the NPR1 primary structure predicted neither a DNA binding domain nor a transcriptional activation domain (Cao et al., 1997). Thus, it is unlikely that NPR1 acts as a transcription factor itself. On the other hand, two putative protein–protein interaction domains, an ankyrin repeat domain and a BTB/POZ (for BROAD-COMPLEX, TRAMTRACK, AND BRIC-A-BRAC/POXVIRUS, ZINC FINGER) domain, were identified in NPR1 (Cao et al., 1997; Aravind and Koonin, 1999).
Using the yeast two-hybrid system with NPR1 as a bait, two classes of interacting proteins were identified. Several groups discovered members of the basic domain/leucine zipper family of transcription factors, the TGA factors that were demonstrated to bind to SA-responsive elements of the PR-1 promoter in vitro (Strompen et al., 1998; Zhang et al., 1999; Després et al., 2000; Niggeweg et al., 2000; Zhou et al., 2000). The in vivo interaction between TGA2 and NPR1 was shown to be SA-dependent (Subramaniam et al., 2001). Moreover, NPR1- and SA-dependent recruitment of a TGA factor to its target sequence of the PR-1 promoter was reported (Fan and Dong, 2002; Johnson et al., 2003). In an attempt to identify proteins involved in SA signal transduction, we previously described the isolation of three novel genes encoding distinct albeit structurally related proteins designated NIMIN1 (for NIM1-INTERACTING1), NIMIN2, and NIMIN3 that interact with NPR1 (also known as NIM1) in the yeast two-hybrid system (Weigel et al., 2001). The NIMIN proteins fail to bind a mutant NPR1 that abolishes SAR in Arabidopsis. In addition, their mRNAs are transiently expressed after SA induction. These data, together with the finding that NIMIN proteins, NPR1, and TGA factors form a ternary complex in yeast, suggest that they are involved in the signal transduction pathway leading to SAR.
Here, we show that NPR1 interacts with NIMIN1 in plant extracts and that a ternary complex of NIMIN1, NPR1, and TGA factor can bind to an SA-responsive promoter element in the yeast one-hybrid system. Analysis of plants overexpressing NIMIN1 revealed that SA-mediated PR gene induction is repressed and that these plants are impaired in SAR and R gene–mediated resistance. We demonstrate that this effect is contingent on the ability of NIMIN1 to interact with NPR1. In a complementary approach, we identified a NIMIN1 knockout mutant and created double-stranded RNA interference (dsRNAi) transgenic lines. Both strategies yielded plants showing enhanced PR-1 gene induction after SA treatment. However, as this hyperactivation of gene expression did not coincide with enhanced resistance, endogenous NIMIN1 seems to regulate only distinct functions of NPR1.
RESULTS
NPR1 Interacts with NIMIN1 but Not with a Mutant Derivative, NIMIN1-2, in Plant Extracts
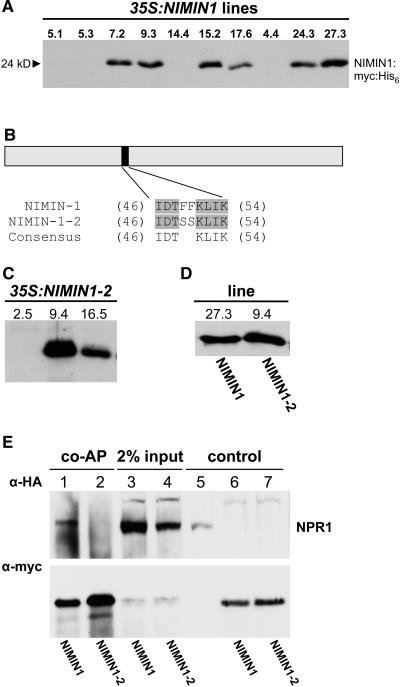
In an attempt to identify proteins involved in SA signal transduction, we previously described the isolation of three novel genes encoding distinct albeit structurally related proteins, NIMIN1, NIMIN2, and NIMIN3, that interact with NPR1 in the yeast two-hybrid system. NIMIN2 and NIMIN3 were also shown to bind NPR1 in vitro (Weigel et al., 2001). To analyze whether this interaction occurs in planta, we created transgenic lines expressing NIMIN1 constitutively. To facilitate the detection of NIMIN1, a sequence encoding a c-myc:hexahistidine (His6) tag was added to the C terminus of the protein. Yeast three-hybrid analysis confirmed that the NIMIN1:myc:His6 fusion still forms a ternary complex with NPR1 and TGA factors (data not shown), as described previously (Weigel et al., 2001). The NIMIN1:myc:His6 gene fusion under the control of the Cauliflower mosaic virus 35S promoter was transformed via Agrobacterium tumefaciens–mediated gene transfer into wild-type plants (Arabidopsis ecotype Columbia [Col-0]). Thirty-two transformants were selected and 10 T0 lines were brought to homozygosity (35S:NIMIN1 lines), as inferred from segregation analysis. As shown by the protein gel blot in Figure 1A, NIMIN1:myc:His6 accumulates to high levels in lines 7.2, 9.3, 15.2, 24.3, and 27.3, to intermediate levels in line 17.6, and is hardly detectable in lines 5.1, 5.3, 14.4, and 4.4. The protein migrates at a position corresponding to 24 kD, which is close to its predicted molecular mass of 21 kD. The lines with strong NIMIN1:myc:His6 expression display subtle morphological changes: the petioles tend to be longer and the laminae are a bit shorter than in wild-type plants, but rosette size is similar to that of wild-type plants. They also appear to be earlier flowering than wild-type plants (data not shown).
Figure 1.
NPR1 Interacts with NIMIN1 but Not with a Mutant Derivative, NIMIN1-2, in Plant Extracts.
(A) Characterization of plants overexpressing NIMIN1:myc:His6 (35S:NIMIN1 lines). Protein gel blot analysis of the NIMIN1:myc:His6 protein in 35S:NIMIN1 transformants. Expression levels of 10 homozygous lines are shown. NIMIN1:myc:His6 was enriched from denaturing extracts using nickel affinity chromatography and detected using an antibody against the c-myc tag. The position and molecular mass of NIMIN1:myc:His6 are indicated by an arrowhead at right.
(B) Top, scheme of NIMIN1 and its NPR1 interaction domain. Bottom, alignment of the amino acid sequences of wild-type and mutant NIMIN1.
(C) Characterization of plants overexpressing a mutant derivative of NIMIN1 designated NIMIN1-2 that does not interact with NPR1 (35S:NIMIN1-2 lines). Protein gel blot analysis of the NIMIN1-2:myc:His6 protein in 35S:NIMIN1-2 transformants. Expression levels of three homozygous lines are shown. NIMIN1-2:myc:His6 was detected using an antibody against the c-myc tag.
(D) NIMIN1:myc:His6 and NIMIN1-2:myc:His6 are expressed at similar levels in transgenic lines. Protein gel blot analysis of 35S:NIMIN1 and 35S:NIMIN1-2 transgenic lines. Native protein extracts were prepared from lines strongly expressing the wild type (line 27.3) and the mutant allele (line 9.4) of NIMIN1. Total protein (50 μg) was loaded in each lane.
(E) Coaffinity purification (co-AP) of HA3:NPR1 with NIMIN1:myc:His6. HA3:NPR1 was pulled down with NIMIN1:myc:His6 (lane 1) but not with NIMIN1-2:myc:His6 (lane 2) in protein extracts from transgenic plants. Top, Detection of HA3:NPR1 using an HA tag antibody. Bottom, detection of NIMIN1:myc:His6 or NIMIN1-2:myc:His6 using a c-myc tag antibody. Total extract (2.5 mg) from plants expressing either HA3:NPR1 and NIMIN1:myc:His6 (lane 3) or HA3:NPR1 and NIMIN1-2:myc:His6 (lane 4) was used for coaffinity purification with Ni-NTA. Lanes 5 to 7 show extracts from plants expressing HA3:NPR1, NIMIN1:myc:His6, or NIMIN1-2:myc:His6 alone.
As a control for the in planta interaction assay, we created transgenic lines expressing a NIMIN1 mutant that does not interact with NPR1. In our previous study, we had mapped the NPR1 interaction domain of NIMIN1 to amino acids 49 to 54 (Weigel et al., 2001). To identify crucial residues within the six–amino acid region that are important for the interaction with NPR1, a rational site-directed mutagenesis approach was taken based on a multiple alignment of the region of three NIMIN proteins (NIMIN1, NIMIN1b, and NIMIN2) (Weigel et al., 2001) and G8-1, a NIMIN-like protein from tobacco (Nicotiana tabacum) (Horvath et al., 1998). Only two consecutive Phe residues are conserved in all four proteins (Phe-49 and Phe-50 in NIMIN1). We have chosen to exchange these large hydrophobic residues by small, slightly polar Ser residues (Betts and Russell, 2003) to disrupt the NPR1 interaction domain. The NIMIN1 mutant was designated NIMIN1-2 (Figure 1B). Using the yeast two-hybrid system, we confirmed that NIMIN1-2 does not interact with NPR1 (data not shown).
To create transgenic lines expressing NIMIN1-2, the cDNA was fused as the wild-type cDNA to a c-myc:His6 tag and was expressed under the control of the Cauliflower mosaic virus 35S promoter. Several lines were brought to homozygosity (35S:NIMIN1-2 lines), as inferred from segregation analysis. As shown by the protein gel blot in Figure 1C, NIMIN1-2:myc:His6 accumulates to a high level in line 9.4, to an intermediate level in line 16.5, and is hardly detectable in line 2.5. The subtle growth phenotype of 35S:NIMIN1 lines was also observed with 35S:NIMIN1-2 lines.
To test for interaction between NIMIN1 and NPR1 in planta, we crossed 35S:NIMIN1 line 27.3 and 35S:NIMIN1-2 line 9.4 to homozygous plants expressing a triple hemagglutinin epitope (HA3)-tagged NPR1 (35S:NPR1) in the npr1-1 background. 35S:NIMIN1-2 line 9.4 was chosen as a control because it exhibits similar levels of transgene expression as 35S:NIMIN1 line 27.3 (Figure 1D). The resulting F1 generation was analyzed for complex formation between either NIMIN1 and NPR1 or NIMIN1-2 and NPR1 using coaffinity purification. Because NIMIN1 is localized to the nucleus (Weigel et al., 2001), nuclear localization of NPR1 was induced by SA treatment of F1 plants 8 h before harvesting. Total protein extracts of plants expressing similar levels of either NIMIN1:myc:His6 and HA3:NPR1 or NIMIN1-2:myc:His6 and HA3:NPR1 were prepared and incubated with nickel–nitrilotriacetic acid agarose (Ni-NTA) as described previously (Fan and Dong, 2002). His6-tagged NIMIN1 and NIMIN1-2, together with corresponding interaction partners, were bound to the matrix and were eluted after washing away nonspecific interactors. Protein gel blot analysis confirmed that similar amounts of NIMIN1 and NIMIN1-2, and similar amounts of NPR1, were present in the input extracts of both transgenic lines (Figure 1E, lanes 3 and 4, bottom and top panels), and similar amounts of either NIMIN1 or NIMIN1-2 were purified from these extracts (Figure 1E, lanes 1 and 2, bottom panel). As shown in Figure 1E, NPR1 was copurified with NIMIN1 (lane 1, top panel) but not with NIMIN1-2 (lane 2, top panel). This finding clearly demonstrates that interaction between NIMIN1 and NPR1 occurs in plant extracts and that interaction between NIMIN1-2 and NPR1 is disrupted not only in yeast but also in plant extracts.
A Ternary Complex of NIMIN1, NPR1, and TGA Factor Binds to the activation sequence-1 Element in Yeast
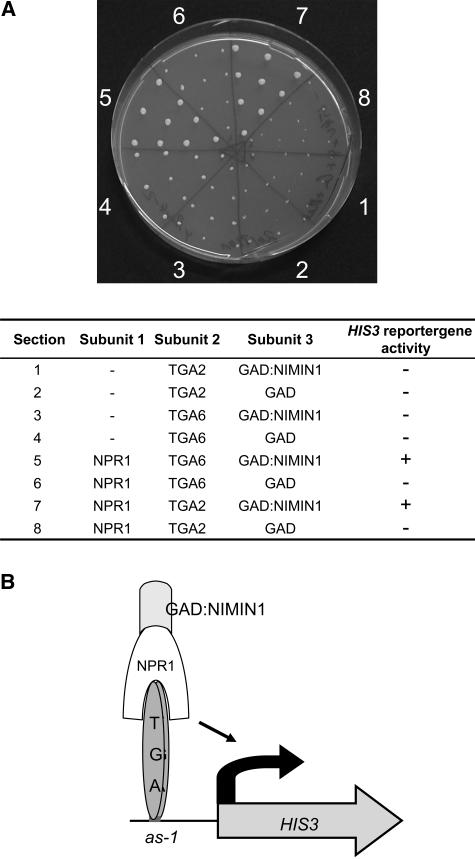
Next, we addressed the question of whether NIMIN1 has the potential to modulate PR gene expression. Yeast three-hybrid data suggest that NIMIN1 may be involved in PR gene regulation as part of a ternary complex with NPR1 and TGA factors. In vitro experiments failed to show that NPR1 interacts with a TGA factor bound to an activation sequence-1 (as-1)–like cis element of the PR-1 gene promoter (Després et al., 2000). To examine whether NPR1 can interact with DNA-bound TGA factors in an in vivo situation and to test whether NIMIN1 may be part of this complex, we used the yeast one-hybrid system to modulate PR gene expression. A yeast strain was created carrying the HIS3 reporter gene driven by a minimal promoter with three tandemly repeated as-1 DNA binding sites. NIMIN1 was fused to the GAL4 transcription activation domain (GAD:NIMIN1) because none of the subunits of the putative complex exhibits transcriptional activation in yeast (R. Weigel, unpublished data). If GAD:NIMIN1 is part of a complex together with NPR1 and as-1–bound TGA factor, the reporter gene is activated and the yeast cells can grow on medium lacking His. As shown in Figure 2A, only in the presence of all three subunits of the ternary complex between TGA2 and TGA6, respectively, NPR1, and GAD:NIMIN1 was reporter gene activation observed and were yeast cells able to grow on medium lacking His (sections 5 and 7). These data demonstrate the interaction between NPR1 and DNA-bound TGA factor together with GAD:NIMIN1 in an in vivo system (Figure 2B). The data indicate that an assembly of the ternary complex on the PR-1 promoter is possible.
Figure 2.
A Ternary Complex of NIMIN1, NPR1, and TGA Factor Binds the as-1 Promoter Element in the Yeast One-Hybrid System.
(A) Cells of the yeast strain yTSH1 carrying a 3× as-1:HIS3 reporter gene construct were cotransformed with the respective plasmids encoding interaction partners. Transformants were grown on selective medium lacking Leu, Trp, and uracil (data not shown). Expression of NPR1 and TGA factors was under the control of the Met-25 promoter. GAD:NIMIN1 was expressed under the control of the CUP1 promoter. To test for protein–DNA interaction, similar amounts of cells were transferred on medium lacking Leu, Trp, uracil, and His in the absence of Met. The proteins expressed in yeast cells that were spotted on the different sectors of the plate are listed in the table. Cells expressing only one or two subunits of the ternary DNA binding complex were transferred to sectors 1 to 4, 6, and 8. They served as negative controls. Only in the presence of all three subunits of the ternary complex between TGA2 and TGA6, NPR1, and GAD:NIMIN1 was reporter gene activation observed and were yeast cells able to grow on medium lacking His (sections 5 and 7).
(B) Scheme of the ternary complex formed by GAD:NIMIN1, NPR1, and TGA factors bound to an as-1 element of the HIS3 reporter gene construct in yTSH1 cells.
Constitutive Expression of NIMIN1 but Not of NIMIN1-2 Leads to Repression of PR Gene Expression after SA Induction
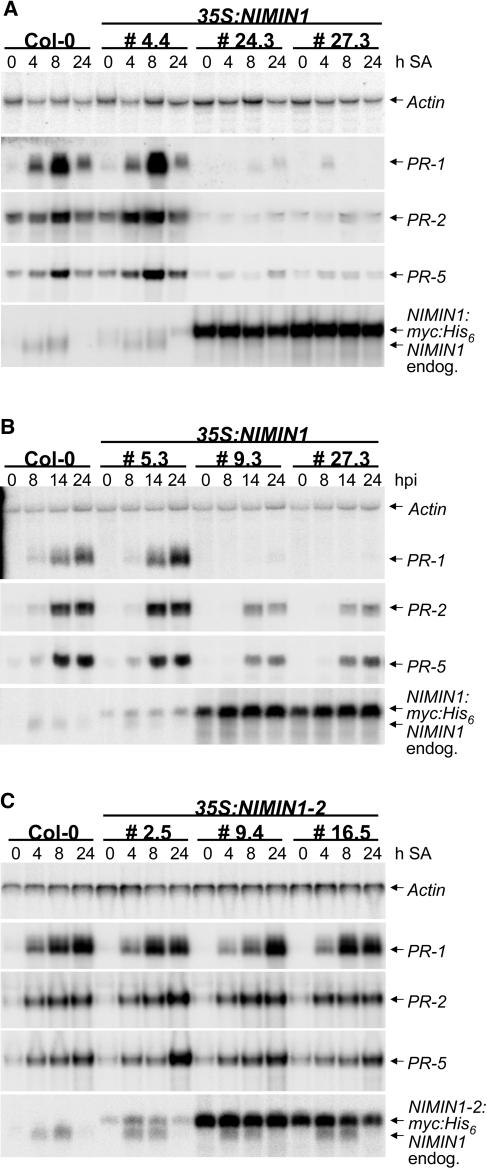
SA and benzo(1,2,3)thiadiazole-7-carbothioic acid inducibility of NIMIN1, and the interaction between NIMIN1, NPR1, and TGA factors in yeast, prompted us to test whether the expression of SAR marker genes is altered in 35S:NIMIN1 lines. We examined the expression of PR genes in a time-course experiment 0, 4, 8, and 24 h after SA treatment. As shown by the RNA gel blot in Figure 3A, SA-induced expression of PR-1, PR-2, and PR-5 was strongly reduced in lines with high levels of NIMIN1:myc:His6 expression (similar results were obtained with all lines showing NIMIN1:myc:His6 expression presented in Figure 1A). Interestingly, induction of the endogenous NIMIN1 transcript was also repressed. This reduction was not observed in lines with low levels of NIMIN1:myc:His6. To corroborate our findings, we tested whether PR genes are also repressed in 35S:NIMIN1 lines using an independent inducing stimulus. 35S:NIMIN1 lines were challenged with the avirulent bacterial strain Pseudomonas syringae pv maculicola ES4326 avrRpt2 (Psm 4326 avrRpt2) (Dong et al., 1991; Whalen et al., 1991), and PR gene expression was examined 0, 8, 12, and 24 h after infection. As shown in Figure 3B, Psm 4326 avrRpt2–induced expression of SAR genes was reduced in lines with high levels of NIMIN1:myc:His6, whereas PR-2 and PR-5 were less strongly repressed than PR-1. Again, induction of the endogenous NIMIN1 transcript was also repressed. In lines with low levels of NIMIN1:myc:His6, PR genes were induced comparably with the wild type. These results indicate that NIMIN1 may be a negative regulator of PR gene expression.
Figure 3.
PR Gene Expression in Plants Overexpressing NIMIN1 and NIMIN1-2.
(A) RNA gel blot analysis of PR gene expression in wild-type and 35S:NIMIN1 lines in response to treatment with SA. 35S:NIMIN1 line 4.4 expresses NIMIN1:myc:His6 to low levels, whereas lines 24.3 and 27.3 express NIMIN1:myc:His6 to high levels. Total RNA was extracted from 3-week-old plants kept under short-day conditions (21/19°C, 8-h-light/16-h-dark cycle, and 60% relative humidity) at 0, 4, 8, and 24 h after SA treatment. Detection of ACTIN mRNA served as a loading control.
(B) Expression of PR genes in wild-type and 35S:NIMIN1 lines in response to bacterial infection. 35S:NIMIN1 line 5.3 expresses NIMIN1:myc:His6 to low levels, whereas lines 9.3 and 27.3 express NIMIN1:myc:His6 to high levels. Total RNA was extracted from 3-week-old plants at 0, 8, 14, and 24 h after Psm 4326 avrRpt2 infection. Detection of ACTIN mRNA served as a loading control.
(C) Expression of PR genes in 35S:NIMIN1-2 lines in response to treatment with SA. 35S:NIMIN1-2 line 2.5 expresses NIMIN1-2:myc:His6 to low levels, whereas lines 9.4 and 16.5 express NIMIN1-2:myc:His6 to high levels. Total RNA was extracted from 3-week-old plants at 0, 4, 8, and 24 h after SA treatment. Detection of ACTIN mRNA served as a loading control.
To test whether PR gene repression is contingent on the ability of NIMIN1 to interact with NPR1, we investigated PR gene expression in 35S:NIMIN1-2 lines in a time-course experiment 0, 4, 8, and 24 h after SA treatment. As shown by RNA gel blot analysis in Figure 3C, PR gene expression in 35S:NIMIN1-2 plants did not differ from that in wild-type plants, although NIMIN1-2 was as highly expressed as NIMIN1 (Figure 1D). Induction of the endogenous NIMIN1 transcript also was not affected. This result shows that PR gene and NIMIN1 repression in 35S:NIMIN1 lines depends on the ability of NIMIN1 to interact with NPR1. Constitutive expression of NIMIN1:myc:His6 in the wild-type background may antagonize NPR1 activity and lead to a phenotype like that conferred by npr1.
SAR Is Abolished in 35S:NIMIN1 Lines
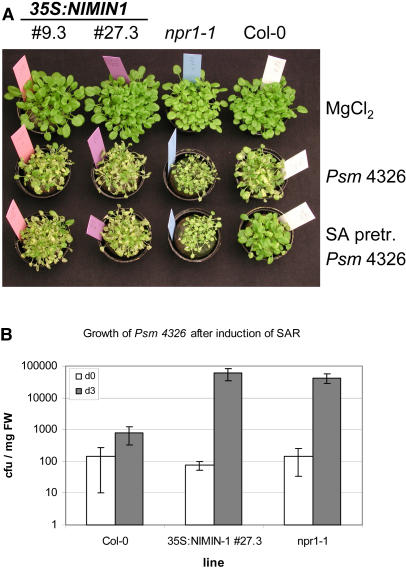
To investigate whether 35S:NIMIN1 lines have additional npr1-like characteristics, we tested whether the loss of SA-induced PR gene expression would coincide with impaired SAR-mediated protection against infection with a virulent pathogen. Two-week-old plants (35S:NIMIN1 lines 9.3 and 27.3, npr1-1, and Col-0) were inoculated with P. syringae pv maculicola ES4326 (Psm 4326; OD600 = 0.2) (Dong et al., 1991; Whalen et al., 1991) with or without pretreatment with 1 mM SA or were mock inoculated (Figure 4A). Without pretreatment with SA, all plant lines tested were infected by the virulent bacterial strain and showed severe disease symptoms. Pretreatment with SA protected wild-type plants as a result of the establishment of SAR. In npr1-1 plants, however, SAR was abolished. They showed strong disease symptoms when infected with a virulent bacterial strain even after SAR-inducing treatment. Similarly, 35S:NIMIN1 lines 9.4 and 27.3 showed strong disease symptoms after SA pretreatment, indicating that the repression of SA-inducible PR gene expression coincides with an impaired SAR response. This is a second common feature of 35S:NIMIN1 lines and npr1 mutants. To confirm that the macroscopic disease symptoms reflect the bacterial growth inside the plant, we measured the bacterial titer in SA-pretreated wild-type, 35S:NIMIN1 line 27.3, and npr1-1 plants at 0 and 3 d after infection (DAI). In wild-type plants, the Psm 4326 titer increased only ∼10-fold after 3 DAI. By contrast, 35S:NIMIN1 line 27.3 showed an increase of approximately three orders of magnitude, similar to the situation in npr1-1 plants. This is in accordance with the observed macroscopic disease symptoms and with published data (Cao et al., 1994).
Figure 4.
Response of 35S:NIMIN1, npr1-1, and Wild-Type Lines to Psm 4326 Infection after SA Pretreatment.
(A) Macroscopic symptoms of 35S:NIMIN1 lines 9.3 and 27.3, npr1-1, and Col-0 plants 4 d after inoculation with Psm 4326. Plants were mock inoculated (top) or infected with Psm 4326 at an OD600 of 0.2 with (bottom) or without (middle) pretreatment with SA.
(B) Growth of Psm 4326 in wild-type, 35S:NIMIN1 line 27.3, and npr1-1 plants after pretreatment with SA. Plants were treated with SA before Psm 4326 infection at an OD600 of 0.002. Samples were taken at 0 and 3 DAI. Three to four samples were taken for each genotype and time point. Data points represent means of three independent experiments ± sd. At 3 DAI, significant differences were observed between in planta Psm 4326 growth in Col-0 plants and 35S:NIMIN1 plants (Student's t test; P < 0.002) and npr1-1 plants (Student's t test; P < 0.003). cfu, colony-forming units.
35S:NIMIN1 but Not 35S:NIMIN1-2 Lines Are Susceptible to an Avirulent Bacterial Strain
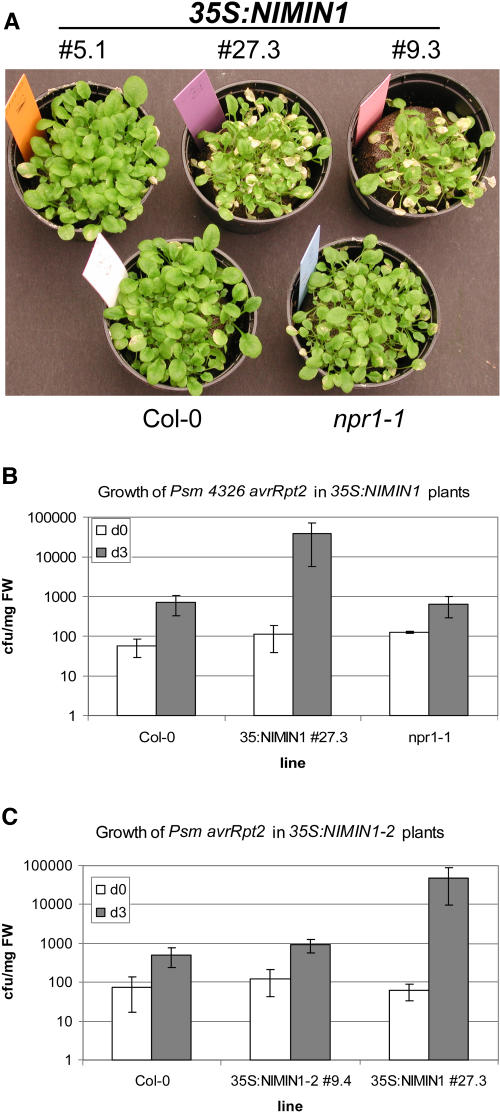
To examine whether 35S:NIMIN1 lines are impaired not only in SAR but also in R gene–mediated resistance, we infected 35S:NIMIN1 lines 5.1, 27.3, and 9.3 with the Psm 4326 avrRpt2 (Dong et al., 1991; Whalen et al., 1991). To observe macroscopic symptoms, plants were dipped in a bacterial suspension (OD600 = 0.2) and the progress of infection was followed over 5 d. Wild-type and npr1-1 plants hardly showed any macroscopic signs of infection, as depicted in Figure 5A. 35S:NIMIN1 line 5.1 expressing low levels of NIMIN1:myc:His6 behaved like a wild-type plant. The Resistance to Pseudomonas syringae2 gene confers resistance against Psm avrRpt2 in these lines. By contrast, 35S:NIMIN1 lines 27.3 and 9.3, with high NIMIN1:myc:His6 expression, showed a significant number of necrotic leaves compared with wild-type plants, indicating a higher susceptibility even to an avirulent bacterial pathogen. To confirm that the macroscopic disease symptoms reflect the bacterial growth inside the plant, we measured the bacterial growth in wild-type, 35S:NIMIN1 line 27.3, and npr1-1 plants at 0 and 3 DAI. As shown in Figure 5B, wild-type and npr1-1 plants did not support the growth of Psm 4326 avrRpt2, as described in the literature (Cao et al., 1994; Clarke et al., 2000). The bacterial titer increased only ∼10-fold. By contrast, 35S:NIMIN1 27.3 plants were less well protected. At 3 DAI, these plants showed an ∼85-fold higher bacterial titer compared with wild-type plants. These results are in accordance with the observed macroscopic disease symptoms and indicate that overexpression of NIMIN1:myc:His6 also affects R gene–mediated resistance and does not merely lead to a npr1 phenocopy.
Figure 5.
Response of 35S:NIMIN1, 35S:NIMIN1-2, npr1-1, and Wild-Type Plants to Psm 4326 avrRpt2 Infection.
(A) Macroscopic symptoms of 35S:NIMIN1 lines 5.1, 9.3, and 27.3, Col-0, and npr1-1 plants 5 d after inoculation with Psm 4326 avrRpt2. Plants were infected with Psm 4326 avrRpt2 at an OD600 of 0.2. In 35S:NIMIN1 line 5.1, NIMIN1:myc:His6 is not detectable (Figure 1A).
(B) Growth of Psm 4326 avrRpt2 in wild-type, 35S:NIMIN1 line 27.3, and npr1-1 plants. Plants were infected with Psm 4326 avrRpt2 at an OD600 of 0.002. Samples were taken at 0 and 3 DAI. Three to four samples were taken for each genotype and time point. Data points represent means of three independent experiments ± sd. At 3 DAI, significant differences were observed between in planta Psm 4326 avrRpt2 growth in Col-0 (Student's t test; P < 0.005) and npr1-1 plants (Student's t test; P < 0.005) compared with 35S:NIMIN1 plants. cfu, colony-forming units.
(C) Bacterial growth in 35S:NIMIN1-2 plants compared with wild-type and 35S:NIMIN1 plants. Col-0, 35S:NIMIN1-2 line 9.4, and 35S:NIMIN1 line 27.3 plants were infected with Psm 4326 avrRpt2 at an OD600 of 0.002. Samples were taken at 0 and 3 DAI. Three to four samples were taken for each genotype and time point. Data points represent means of three independent experiments ± sd. At 3 DAI, significant differences were observed between in planta Psm 4326 avrRpt2 growth in Col-0 compared with 35S:NIMIN1 plants (Student's t test; P < 0.03) and 35S:NIMIN1-2 plants (Student's t test; P < 0.03). cfu, colony-forming units.
To test whether enhanced susceptibility in 35S:NIMIN1 lines is contingent on the ability of NIMIN1 to interact with NPR1, we investigated the susceptibility of 35S:NIMIN1-2 line 9.4 to Psm 4326 avrRpt2 compared with wild-type plants and 35S:NIMIN1 line 27.3. Bacterial growth was measured in a titration experiment at 0 and 3 DAI. The data in Figure 5C show that 35S:NIMIN1-2 line 9.4 was as well protected against the avirulent Psm 4326 strain as the wild-type plants. In both lines, bacterial titers increased ∼10-fold at 3 DAI. These results strongly suggest that PR gene repression and enhanced susceptibility in 35S:NIMIN1 lines are attributable to the specific interaction between endogenous NPR1 and transgenic NIMIN1 and not to interference of NIMIN1 with a yet unknown factor.
A NIMIN1 Knockout Line and NIMIN1 dsRNAi Lines Show Enhanced PR-1 Gene Induction after SA Treatment
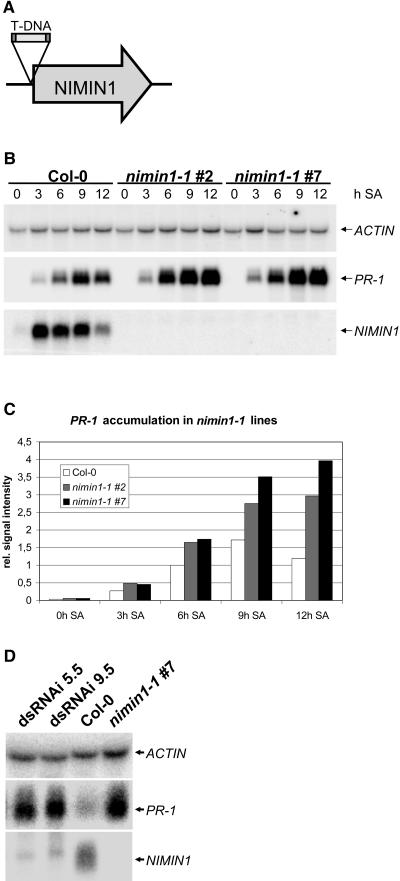
Using a complementary approach to verify that NIMIN1 is a negative regulator of NPR1 activity, a T-DNA insertion line for NIMIN1, designated nimin1-1 (SALK_086460), was obtained from the Nottingham Arabidopsis Stock Centre (Nottingham, UK). The exact position of the T-DNA insertion was determined by PCR and subsequent sequencing of the T-DNA flanking regions. The T-DNA insertion is 4 bp upstream of the start ATG, as depicted schematically in Figure 6A. Based on PCR and segregation analyses, we obtained progeny homozygous for the T-DNA insertion (nimin1-1 lines 2 and 7). RNA gel blot analysis revealed that the nimin1-1 mutant is a real knockout, because no NIMIN1 mRNA was detected after SA treatment, even after extensive exposure of the RNA gel blot, whereas in wild-type plants the transcript had already accumulated strongly at 3 h after SA treatment (Figure 6B). To examine PR-1 mRNA accumulation in nimin1-1 knockout lines, the same blot was incubated with a PR-1 probe. As shown in Figure 6B, PR-1 mRNA accumulation was stronger in nimin1-1 lines than in wild-type plants. Quantification of the signals shown in Figure 6B revealed that the difference in PR-1 signal strength was strongest at 12 h after SA induction (Figure 6C). Compared with Col-0 plants, nimin1-1 line 2 showed a 2.5-fold increase and nimin1-1 line 7 showed a 3.3-fold increase in PR-1 mRNA levels at 12 h after SA induction. Stronger PR-1 induction in nimin1-1 lines was reproducible in several experiments with RNAs from independently grown and induced plants. In a time-course experiment with plants harvested at later time points, differences were still present at 24 h after SA induction but were no longer observed after 36 or 48 h (data not shown), indicating that the amplitude but not the shutting down of PR-1 gene expression is affected in nimin1-1. Basal PR-1 expression was not notably different in nimin1-1 lines compared with wild-type plants.
Figure 6.
Characterization of a NIMIN1 Knockout Line nimin1-1 and NIMIN1 dsRNAi Lines with Respect to PR-1 Gene Expression.
(A) Scheme of the nimin1-1 locus. nimin1-1 carries a T-DNA insertion 4 bp upstream of the start codon of NIMIN1.
(B) RNA gel blot analysis of PR-1 and NIMIN1 gene expression in wild-type and nimin1-1 lines 2 and 7. Total RNA was extracted from 3-week-old plants at 0, 3, 6, 9, and 12 h after SA treatment. Detection of ACTIN mRNA served as a loading control.
(C) Quantification of PR-1 mRNA accumulation based on the data shown in (B). The graph depicts levels of PR-1 mRNA normalized to that of ACTIN.
(D) RNA gel blot analysis of PR-1 and NIMIN1 gene expression in individuals of dsRNAi lines 5.5 and 9.5, a wild-type plant, and nimin1-1 line 7. Total RNA was extracted from 2-week-old plants at 8 h after SA treatment. Detection of ACTIN mRNA served as a loading control.
To verify that the mutant phenotype is specifically attributed to the T-DNA insertion in NIMIN1 and not to unrelated T-DNA insertions or point mutations that are known to be generated as part of the transformation process (McElver et al., 2001), we generated dsRNAi transgenic lines to silence the NIMIN1 gene. A second NIMIN1 T-DNA insertion line was not available from the Nottingham Arabidopsis Stock Centre. First, we tested NIMIN1 mRNA levels in NIMIN1 dsRNAi lines compared with wild-type and nimin1-1 lines at 8 h after SA induction (all plant lines were grown and treated in parallel). RNA gel blot analysis revealed that NIMIN1 expression was strongly reduced in 63 of 74 individuals of nine independent NIMIN1 dsRNAi lines. Figure 6D shows representative results for two individuals of two independent T2 lines. Next, PR-1 gene expression was tested using the same blots. Compared with the wild type, PR-1 mRNA levels were higher in NIMIN1 dsRNAi lines and similar to the level of a nimin1-1 line (Figure 6D). These results strongly support the hypothesis that NIMIN1 counteracts NPR1 activity and modulates PR gene expression. To test whether stronger PR-1 gene expression in nimin1-1 plants coincides with enhanced resistance against a bacterial pathogen, the mutant was subjected to infection with both Psm 4326 avrRpt2 and Psm 4326 with and without SAR-inducing treatment. Using our experimental setup, no significant changes in the defense responses of nimin1-1 plants were observed (data not shown), indicating that hyperactivation of PR-1 gene expression does not coincide with enhanced resistance.
DISCUSSION
In addition to members of the TGA family of basic domain/leucine zipper transcription factors, NIMIN proteins have been found to interact with NPR1 in several independent two-hybrid screens (Weigel et al., 2001). Compared with the TGA factors, their mechanism of action is much less obvious. Before we started to analyze the biological function of the NIMIN proteins, we verified the interaction between NPR1 and NIMIN1 in plant extracts. NIMIN1 was chosen because it exhibits the strongest affinity with NPR1 compared with other NIMIN proteins (Weigel et al., 2001). We created transgenic plants expressing NIMIN1 under the control of the 35S promoter in the wild-type background (35S:NIMIN1 plants). As a control, transgenic plants were created expressing a mutant designated NIMIN1-2 (35S:NIMIN1-2 plants) that is unable to interact with NPR1 in the yeast two-hybrid system. NIMIN1-2 carries two amino acid exchanges (F49S and F50S) in the NPR1 interaction domain (Weigel et al., 2001). To facilitate the purification and detection of the proteins, a c-myc:His6 tag was fused to the C termini, resulting in the expression of NIMIN1:myc:His6 and NIMIN1-2:myc:His6 fusion proteins. Homozygous plants expressing comparable amounts of the transgene were crossed with homozygous plants expressing a HA-tagged NPR1 under the control of the 35S promoter (35S:NPR1) in the npr1-1 background. Using nickel affinity chromatography, we were able to copurify NPR1 from SA-treated F1 plants expressing HA3:NPR1 and NIMIN1:myc:His6 but not from F1 plants expressing HA3:NPR1 and NIMIN1-2:myc:His6, verifying our yeast data and demonstrating NIMIN1–NPR1 interaction in plant extracts.
The Role of NIMIN1 in Gene Expression
Transient expression of NIMIN mRNAs after SA treatment and the formation of a ternary complex between NIMIN proteins, NPR1, and TGA factors suggested a function of the NIMIN proteins in SA-mediated PR gene regulation. Therefore, we tested whether altered NIMIN1 expression would affect PR gene expression. NIMIN1 knockout mutant nimin1-1 and NIMIN1 dsRNAi lines showed enhanced PR-1 accumulation after SA treatment, clearly demonstrating a negative effect of NIMIN1 on PR-1 expression. Accordingly, PR gene activation was strongly suppressed in 35S:NIMIN1 plants after SA treatment or infection with an avirulent bacterial strain. This effect was not observed in plants constitutively expressing NIMIN1-2, thus demonstrating that NIMIN1 exerts its negative effect through its interaction with NPR1. Consistently, 35S:NIMIN1 plants mimic the known npr1 phenotype. On the other hand, nimin1-1 lines show a phenotype similar to that of 35S:NPR1 plants, at least with regard to PR-1 expression (Cao et al., 1998): both plant lines are characterized by an approximately threefold higher PR-1 expression after SA induction. Thus, higher amounts of active NPR1 can be made available, either by enhancing NPR1 protein levels or by reducing the amount of the negative regulator NIMIN1. This positive effect on gene expression can only be exerted in the presence of SA, as NPR1 is located in the cytosol in the uninduced state (Kinkema et al., 2000). This inactive condition is independent of the relative amounts of NPR1 and NIMIN1, as shown by unaltered PR-1 expression in the uninduced state in nimin-1 and 35S:NPR1 plants. The difference in PR-1 expression between wild-type and nimin1-1 lines was observed until 24 h after induction. At later time points, the difference was negligible, probably because of the lack of NIMIN1 in wild-type plants during this late state, when NIMIN1 transcription is shut down. Thus, NIMIN1 is involved in neither turning on nor turning off PR-1 expression. Rather, it modulates its amplitude. This function seems to be specific for NIMIN1, because it cannot be replaced by the other three members of the NIMIN family, albeit it is possible that in the absence of NIMIN1 other NIMIN proteins may partially take over its function. Interestingly, transcript levels of the endogenous NIMIN1 gene were reduced in 35S:NIMIN1 but not in 35S:NIMIN1-2 plants, suggesting that NIMIN1 regulates its own expression through its interaction with NPR1. Differential expression analysis using microarrays and RNA of induced wild-type and knockout plants will provide a suitable tool to identify additional genes regulated by NIMIN1.
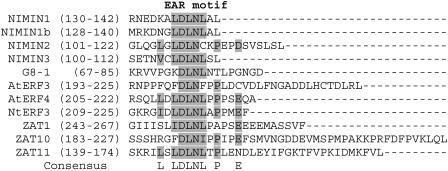
The mechanism by which NIMIN1 represses PR gene expression after SA induction remains to be analyzed. The fact that NIMIN1 was found to bind simultaneously with NPR1 and TGA factors to a PR-1 promoter element in an in vivo situation suggests that it acts as a transcriptional repressor. Two types of transcriptional repressors have been described: passive repressors, which directly bind to DNA, inhibiting access of transcriptional activators; and active repressors, which repress transcription through a distinct repression domain (Hanna-Rose and Hansen, 1996). The primary structure of NIMIN1 shows no homology with known DNA binding domains, suggesting that NIMIN1 may act as an active repressor after being recruited to the DNA/TGA/NPR1 complex. Interestingly, the previously described C-terminal LXL motif of NIMIN1 reveals striking similarity to the so-called ethylene-responsive element binding factor (ERF)–associated amphipathic repression motif [EAR; L/FDLNL/F(x)P] identified in ERF and zinc finger transcription factors (Figure 7) (Ohta et al., 2001). Recently, an EAR motif was also identified in Auxin/Indole-3-Acetic Acid repressors (Tiwari et al., 2004). These repressors are unable to bind to DNA themselves but are recruited to the DNA through their interaction with ARF transcriptional activators, which are rendered inactive through the action of the EAR motif. Similarly, NIMIN1 might bind to the NPR1/TGA factor complex, repressing or reducing PR gene expression (Figure 8). The assembly of a ternary complex of NIMIN1, NPR1, and TGA factors on an SA-responsive promoter element in the yeast one-hybrid system supports this view. All four Arabidopsis NIMIN proteins contain this putative EAR domain (Figure 7). Their low overall sequence similarity might enable them to interact with different additional proteins, facilitating their recruitment to different promoters. Through this mechanism, distinct subsets of NPR1-dependent genes might be regulated by different NIMIN proteins.
Figure 7.
The EAR Motif Is Conserved in the C-Terminal Regions of NIMIN, Class II ERF, and TFIIIA-Type Zinc Finger Proteins.
Comparison of the amino acid sequences of the C-terminal regions of NIMIN, class II ERF, and TFIIIA-type zinc finger proteins from Arabidopsis and tobacco. Numbers in parentheses indicate the positions of amino acid sequences. The EAR motif [L/FDLNL/F(x)P] and other conserved amino acids are shaded. G8-1, NIMIN2 homolog of tobacco encoded by a previously described cDNA (Horvath et al., 1998).
Figure 8.
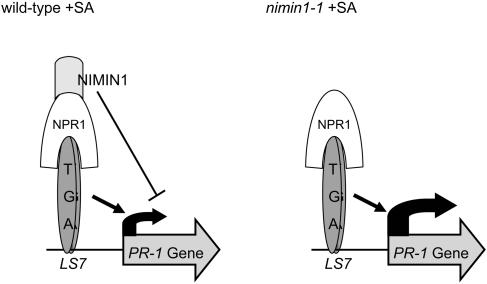
Proposed Model for the Regulatory Mechanism of SA-Mediated PR-1 Gene Expression.
In wild-type plants, NPR1 forms a ternary complex with NIMIN1 and TGA factors upon SAR induction that binds to a positive regulatory cis element of the PR-1 promoter, termed LS7 (Lebel et al., 1998). This leads to PR-1 gene induction. NIMIN1 decreases transcriptional activation, possibly through its EAR motif, which results in fine-tuning of PR-1 gene expression. In the nimin1-1 mutant, NIMIN1 is absent from the transactivating protein complex upon SAR induction, which leads to enhanced PR-1 gene expression caused by the lack of the repressing function of NIMIN1.
The Role of NIMIN1 in Disease Susceptibility
Transgenic plants encoding 35S:NIMIN1 were severely compromised with respect to mounting SAR after SA treatment. Thus, 35S:NIMIN1 plants mimicked the phenotype conferred by npr1, as already observed for PR-1 gene expression. However, enhanced inducible PR-1 induction in nimin1-1 lines did not coincide with enhanced resistance. In this regard, these plants differ from the 35S:NPR1 plants, which are more resistant to the virulent bacterial strain Psm 4326 (Cao et al., 1998). This finding is consistent with the above-mentioned hypothesis that NIMIN1 negatively regulates only a subset of NPR1-dependent genes. The enhanced expression of this subset in nimin1-1 plants may not be sufficient for enhanced resistance against Psm 4326. Alternatively, the closest relative of NIMIN1, designated NIMIN1b (Weigel et al., 2001), which shares 44% identity and 64% similarity, may partially take over its function in nimin1-1 plants.
The impaired defense observed in 35S:NIMIN1 plants is presumably attributable to high amounts of the NPR1/NIMIN1 complex, leading to severely reduced amounts of active NPR1. It can further be speculated that high levels of NIMIN1 compete with other members of the NIMIN family. Thus, additional subsets of NPR1-dependent genes may be downregulated in 35S:NIMIN1 plants, finally leading to a compromised SAR. T-DNA insertion lines are available for NIMIN1b, NIMIN2, and NIMIN3. They are currently used to generate double and triple knockouts to test whether the lack of multiple members of the NIMIN family can lead to enhanced disease resistance.
In contrast with their npr1-like features, 35S:NIMIN1 plants are impaired in R gene–mediated resistance against Psm 4326 avrRpt2. This is not found in the npr1-1 mutant (Cao et al., 1994; Clarke et al., 2000), an observation that has led to the conclusion that NPR1 plays only a minor role in local defense responses against avirulent pathogens (Clarke et al., 2000). This apparent contradiction can be resolved by assuming that 35S:NIMIN1 plants mimic an npr1 allele that is even stronger than the ones described to date. Alternatively, NIMIN1 but not NIMIN1-2 may interact with NPR1-like proteins present in Arabidopsis that possibly play a role in R gene–mediated resistance. It also can be speculated that for some functions the lack of npr1 does not have the same consequences as the presence of an NPR1/NIMIN1 complex, which might actively inhibit R gene–mediated resistance, providing a negative feedback control device. Either way, our data clearly indicate that NPR1 or NPR1-like proteins play a role in R gene–mediated resistance in Arabidopsis. Therefore, the dominant negative effect of constitutively expressed NIMIN1 on NPR1 has revealed a yet unknown function of NPR1. Similar observations were made in Solanaceae: NPR1-like proteins were shown to be essential for N gene–mediated resistance against Tobacco mosaic virus in tobacco (Liu et al., 2002) and for Pto-mediated resistance against Pseudomonas syringae pv tomato DC3000 avrPto in tomato (Ekengren et al., 2003).
In conclusion, we have shown that NIMIN1 can counteract NPR1-activated gene expression. However, only a subset of NPR1-dependent genes may be hyperactivated in the absence of NIMIN1, which is not sufficient to increase plant resistance. As mentioned above, simultaneous knockout of several NIMIN genes might lead to the same phenotype as revealed by 35S:NPR1 plants. Negative regulators of NPR1, such as NIMIN1, might ensure that plant resources are not allocated to the defense response to an extent that would adversely affect other fitness-relevant processes, such as growth and reproduction (Heil and Baldwin, 2002). Single PR proteins may account for ∼1% of the total soluble protein in infected tobacco plants (Antoniw and Pierpoint, 1978), indicating that large quantities of resources can be allocated to resistance traits. SA- and JA-dependent signal transduction pathways mutually inhibit each other (Kunkel and Brooks, 2002), also suggesting the necessity for shutting down certain defense responses to avoid exhaustion of resources. As a matter of fact, NPR1 is involved in the suppression of JA-dependent genes in the presence of SA (Spoel et al., 2003). In the absence of NIMIN proteins, hyperactivated NPR1 may lead to enhanced repression of JA-mediated protection, rendering plants even more susceptible to many necrotrophic pathogens (Heil and Baldwin, 2002). Thus, NIMIN proteins might be indirectly involved in the distribution of resources to either SA/JA-mediated defense responses or processes relevant for growth and reproduction. Given that plants are exposed to a complex environment, different NIMIN proteins might adjust the amplitude of activation of subsets of defense responses depending on the prevailing biotic and abiotic conditions.
METHODS
DNA Constructs
To generate constructs for expression in Arabidopsis thaliana, NIMIN1 was cloned as a BamHI fragment into the BglII-cleaved vector pTrcHis2c (Invitrogen, Karlsruhe, Germany), creating an in frame fusion with a c-myc and the His6 epitope tag. The NIMIN1:myc:His6 sequence was cut out as a BamHI/PmeI fragment and ligated to BamHI/SmaI-cut pUC19. NIMIN1:myc:His6 was then cut out as a BamHI/SacI fragment and cloned into the BamHI/SacI-cut vector pUC19/35S:β-glucuronidase (Beilmann et al., 1991), resulting in the plasmid pUC19/35S:NIMIN1:myc:His6. From there, the 35S:NIMIN1:myc:His6 cassette was cloned as an EcoRI fragment into the EcoRI-opened vector pBIN19/35S:β-glucuronidase, resulting in the plasmid pBIN19/35S:NIMIN1:myc:His6.
To create the NIMIN1-2 allele, the N terminus of NIMIN1 was amplified by PCR using oligonucleotides pUC19bck (5′-TGTGGAATTGTGAGCGGA-3′) and N1-M1 (5′-GGAAGCTTAGAGGACGTATCAATCTTC-3′) and pUC19/NIMIN1:myc:His6 as a template. Amplification with N1-M1 results in a DNA fragment coding for two amino acid exchanges (F49S and F50S) in the NPR1 interaction domain of NIMIN1 (Weigel et al., 2001). The amplified PCR fragment was cut with HindIII and then cloned into the HindIII-cut vector pUC19/NIMIN1:myc:His6. pBIN19/35S:NIMIN1-2:myc:His6 was cloned as described for pBIN19/35S:NIMIN1:myc:His6. To create a binary vector for the expression of a tagged version of NPR1 under the control of the 35S promoter, Gateway technology (Invitrogen) was used. The full-length cDNA of NPR1 was amplified using primers 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCCATGGACACCACCATTGATGG-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTCACCGACGACGATGAGAGAG-3′. The generated PCR product was recombined into pDONR207 (Invitrogen). The resulting plasmid pDONR207/NPR1 was further recombined into pAlligator2 (Bensmihen et al., 2004). To generate NIMIN1 dsRNAi lines, the coding sequence was amplified using primers 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTATATGTATCCTAAACAATTTAG-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCCAATGCAAGATTAAGATC-3′. The amplification product was recombined into the donor vector pDONR207 using the Gateway system (Invitrogen). A second recombination step with a Gateway-compatible derivative of pFGC5491 (Kerschen et al., 2004) yielded the binary construct pFGC5941/NIMIN1, containing a chalcone synthase intron designed to produce double-stranded RNA in plants. NIMIN1 was recombined upstream and downstream of the chalcone synthase intron in both the antisense and sense orientations. All DNA constructs were verified by dideoxynucleotide sequencing.
To create the 3× as-1 yeast reporter gene construct, two complementary oligonucleotides [5′-AATTC(TGACGTAAGGGATGACGCATTACC)3T-3′ and 5′-CTAGA(GGTAATGCGTCATCCCTTACGTCA)3G-3′] were aligned, creating EcoRI-compatible ends at the 5′ end and XbaI-compatible ends at the 3′ end. The resulting double-stranded DNA fragment was cloned into the plasmid pHISi-1 (BD Biosciences, Heidelberg, Germany). Integration, selection, and analysis of the resulting yeast strain yTSH1 (a derivative of YM4271) were performed as described by the manufacturer (BD Biosciences). To express the TGA factors in yeast, their corresponding cDNAs were cloned as BamHI fragments into the BglII site of a modified pBridge plasmid (Weigel et al., 2001), resulting in the plasmids pGBD-/TGA2 and pGBD-/TGA6, respectively (TRP1-selectable marker). To express NPR1 in yeast, the Met-25 expression cassette of pGBD-/NPR1 (Weigel et al., 2001) was cloned as a StuI fragment into the PvuII-cleaved pGAD424 plasmid (BD Bioscience), resulting in the plasmid pGAD-/NPR1 (LEU2-selectable marker). The GAD:NIMIN1 fusion was cloned as described in Weigel et al. (2001) for the GAL4BD fusion, resulting in the plasmid pGAD424/NIMIN1. To express the fusion in yeast under the control of the CUP1 promoter, pGAD424/NIMIN1 was partially digested with HindIII. The appropriate fragment was cloned into the HindIII-cleaved pCU426 plasmid (Labbe and Thiele, 1999), resulting in the plasmid pCU426/GAD:NIMIN1 (URA3-selectable marker).
Plant Growth Conditions and Transformation
Arabidopsis plants (ecotype Col-0) were grown in soil under controlled environmental conditions (21/19°C, 16-h-light/8-h-dark cycle, and 60% relative humidity unless stated otherwise). All seeds were vernalized at 4°C for 2 d before placement in a growth environment. For pathogen infection, 10 to 20 plants were grown on the mesh of Jiffy-7 peat pots. To maintain high humidity, plant trays were covered with a lid throughout the entire growth period.
Binary plasmids were electroporated into Agrobacterium tumefaciens strain GV3101 (pMP90). The resulting agrobacteria were used to transform Col-0 plants using a floral dipping method (Clough and Bent, 1998). Transgenic plants containing pBIN19 constructs were selected on MS agar plates containing 50 μg/mL kanamycin. To generate homozygous transgenic lines, T0 plants were selfed and the progeny of T1 plants were analyzed on MS plates containing 50 μg/mL kanamycin. If 100% of 50 to 100 T2 seedlings were resistant to kanamycin, the T1 mother plant was scored as homozygous. To generate plants expressing HA3:NPR1 under the control of the 35S promoter, npr1-1 plants were transformed with pAlligator/NPR1. Transgenic seeds containing pAlligator/NPR1 were selected using the seed-specific green fluorescent protein (GFP) marker. If all of the seeds of a T1 silique were GFP-positive, the plant was scored as homozygous. To create plants expressing HA3:NPR1 and NIMIN1:myc:His6 or NIMIN1-2:myc:His6, 35S:NIMIN1 line 27.3 or 35S:NIMIN1-2 line 9.4, respectively, was crossed with a 35S:NPR1 line (pollen donor). GFP-positive F1 seed was used to grow plants for the coaffinity purification experiment. Transgenic plants containing pFGC5941/NIMIN1 constructs were selected by spraying a solution of Basta (glufosinate) herbicide according to the manufacturer's instructions (AgrEvo, Duesseldorf, Germany).
Protein Gel Blot Analysis
Total denaturing protein extracts were made from Arabidopsis plants by grinding plants in liquid nitrogen and adding two volumes of denaturing urea buffer (100 mM NaH2PO4, 10 mM Tris-Cl, and 8 M urea, pH 8.0). The extracts were cleared of cell debris by centrifugation at 16,000g at 4°C. Extracts contained equal amounts of proteins, as confirmed by Coomassie Brilliant Blue G 250 staining of a protein gel. His6-tagged proteins were enriched by affinity chromatography using Ni-NTA as described by the manufacturer (Qiagen, Hilden, Germany). Extracts were separated and transferred to nitrocellulose membranes according to Sambrook et al. (1989).
Subsequently, the blots were probed with an antibody against the c-myc tag (New England Biolabs, Frankfurt, Germany) and the HA tag (Santa Cruz Biotechnology, Heidelberg, Germany), according to each manufacturer's conditions. The antibody-bound proteins were detected using the ECL Plus system (Amersham Biosciences, Freiburg, Germany). Native protein extracts were prepared as described by Fan and Dong (2002).
Copurification of NPR1 and NIMIN1
Copurification of NPR1 and NIMIN1 was done essentially as described by Fan and Dong (2002). The procedure was scaled down by a factor of 10.
Yeast One-Hybrid System
The yeast assay for DNA binding of the ternary complex between NIMIN1, NPR1, and TGA factor was performed essentially as described by Weigel et al. (2001).
RNA Gel Blot Analysis
Total RNA was extracted from 3-week-old plants, and 10 μg of each sample was used for RNA gel blot analysis as described previously (Weigel et al., 2001). Each time-course experiment was done in duplicate. Blots were stripped and reprobed two to three times. Fragments used to generate probes specific to PR-1, PR-2, PR-5, ACTIN2, and NIMIN1 were amplified by PCR using primer combinations as described previously (Uknes et al., 1992; An et al., 1996; Weigel et al., 2001). RNA gel blot analysis was performed as described (Weigel et al., 2001).
Infection with Psm 4326 after Pretreatment with 1 mM SA
To generate macroscopic symptoms, 3-week-old plants of each genotype were used in three different treatments: one group was sprayed with 1 mM SA at 2 and 1 d before dipping in a bacterial suspension of Psm 4326 (OD600 = 0.2; in 10 mM MgCl2 and 0.01% Silwett L-77; Lehle Seeds, Round Rock, TX). The second group was dipped in bacterial suspension without pretreatment with SA, and the third group was dipped in 10 mM MgCl2 and 0.01% Silwett L-77. Plants were photographed at 4 DAI.
To assay the growth of bacteria in the plants, a combination of two methods was used (Tornero and Dangl, 2001; Katagiri et al., 2002). Plants were sprayed with 1 mM SA at 2 and 1 d before dipping in a bacterial suspension of Psm 4326 (OD600 = 0.002; in 10 mM MgCl2 and 0.01% Silwett L-77). One hour after inoculation, the samples for day 0 were taken. Nine leaves were excised per genotype, and leaf discs were made from these samples using a hole puncher. Three sets of three discs per genotype were added to a preweighed 2-mL tube containing 1 mL of 10 mM MgCl2 and 0.2% Silwett L-77. The tubes were then shaken on a vortex mixer set to 4 at room temperature for 10 min. After this time, 10 and 20 μL of each tube was spread onto a Petri dish of King's B medium (King et al., 1954) containing the appropriate antibiotics. For day 3, 12 leaves were excised per genotype and four sets of three leaf discs were used for the extraction of bacteria as described above. Extract (100 μL) was added to a microwell plate, and serial 10-fold dilutions were prepared using a multichannel pipette. The bacteria were spotted in 5-μL samples onto a 150-mm Petri plate of King's B medium containing the appropriate antibiotics using a multichannel pipette. Each plate was replicated once. The plates were incubated for 2 d at 28°C. Dilutions that gave 1 to 60 colonies were counted. Each data point represents the mean and standard deviation of three independent experiments.
Infection with Psm 4326 avrRpt2 was done essentially as described above but without pretreatment with 1 mM SA.
Insertional Mutant Plant Isolation
A mutant carrying a T-DNA insertion in the NIMIN1 gene was identified in an insertional mutant population (ecotype Col-0) (Sessions et al., 2002). Genomic DNAs were isolated using the method described by Weigel et al. (2001). The T-DNA insertion event in NIMIN1 was confirmed by PCR and sequencing of the right and left border PCR products. Homozygous mutant plants for nimin1-1 were identified by segregation analysis using the resistance marker kanamycin and RNA gel blot analysis. PCR analysis of plants carrying a homozygous insertion consistently yielded a single band using the combination of gene-specific and T-DNA border primers; however, as a result of the large insert size, no band was found using the two gene-specific primers.
Acknowledgments
We thank D. Thiele (pCU426), F. Parcy (pAlligator2), S. Smeekens (pFGC5941, Gateway derivative), T. Siemsen (yTSH1), and the Nottingham Arabidopsis Stock Centre (SALK_086460) for providing materials used in this study as well as Wolfgang Droege-Laser and Corinna Thurow for carefully reading the manuscript. This work was supported by Grant WE 2716/1-2 from the Deutsche Forschungsgemeinschaft.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Ralf R. Weigel (rweigel@gwdg.de).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.027441.
References
- An, Y.Q., McDowell, J.M., Huang, S., McKinney, E.C., Chambliss, S., and Meagher, R.B. (1996). Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 10, 107–121. [DOI] [PubMed] [Google Scholar]
- Antoniw, J.F., and Pierpoint, W.S. (1978). Purification of a tobacco leaf protein associated with resistance to virus infection. Biochem. Soc. Trans. 6, 248–250. [DOI] [PubMed] [Google Scholar]
- Aravind, L., and Koonin, E.V. (1999). Fold prediction and evolutionary analysis of the POZ domain: Structural and evolutionary relationship with the potassium channel tetramerization domain. J. Mol. Biol. 285, 1353–1361. [DOI] [PubMed] [Google Scholar]
- Beilmann, A., Pfitzner, A.J., Goodman, H.M., and Pfitzner, U.M. (1991). Functional analysis of the pathogenesis-related 1a protein gene minimal promoter region: Comparison of reporter gene expression in transient and in stable transfections. Eur. J. Biochem. 196, 415–421. [DOI] [PubMed] [Google Scholar]
- Belkhadir, Y., Subramaniam, R., and Dangl, J.L. (2004). Plant disease resistance protein signaling: NBS-LRR proteins and their partners. Curr. Opin. Plant Biol. 7, 391–399. [DOI] [PubMed] [Google Scholar]
- Bensmihen, S., To, A., Lambert, G., Kroj, T., Giraudat, J., and Parcy, F. (2004). Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS Lett. 561, 127–131. [DOI] [PubMed] [Google Scholar]
- Betts, M.J., and Russell, R.B. (2003). Amino acid properties and consequences of substitutions. In Bioinformatics for Geneticists, M.R. Barnes and I.C. Gray, eds (Chichester, UK: Wiley), pp. 289–316.
- Cao, H., Bowling, S.A., Gordon, A.S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Glazebrook, J., Clarke, J.D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. [DOI] [PubMed] [Google Scholar]
- Cao, H., Li, X., and Dong, X. (1998). Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc. Natl. Acad. Sci. USA 95, 6531–6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, J.D., Volko, S.M., Ledford, H., Ausubel, F.M., and Dong, X. (2000). Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell 12, 2175–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., Dietrich, R.A., and Richberg, M.H. (1996). Death don't have no mercy: Cell death programs in plant–microbe interactions. Plant Cell 8, 1793–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney, T.P., Friedrich, L., and Ryals, J.A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA 92, 6602–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després, C., DeLong, C., Glaze, S., Liu, E., and Fobert, P.R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12, 279–290. [PMC free article] [PubMed] [Google Scholar]
- Dong, X., Mindrinos, M., Davis, K.R., and Ausubel, F.M. (1991). Induction of Arabidopsis defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. Plant Cell 3, 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekengren, S.K., Liu, Y., Schiff, M., Dinesh-Kumar, S.P., and Martin, G.B. (2003). Two MAPK cascades, NPR1, and TGA transcription factors play a role in Pto-mediated disease resistance in tomato. Plant J. 36, 905–917. [DOI] [PubMed] [Google Scholar]
- Fan, W., and Dong, X. (2002). In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. Plant Cell 14, 1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J., Rogers, E.E., and Ausubel, F.M. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143, 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna-Rose, W., and Hansen, U. (1996). Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 12, 229–234. [DOI] [PubMed] [Google Scholar]
- Heil, M., and Baldwin, I.T. (2002). Fitness costs of induced resistance: Emerging experimental support for a slippery concept. Trends Plant Sci. 7, 61–67. [DOI] [PubMed] [Google Scholar]
- Horvath, D.M., Huang, D.J., and Chua, N.H. (1998). Four classes of salicylate-induced tobacco genes. Mol. Plant-Microbe Interact. 11, 895–905. [DOI] [PubMed] [Google Scholar]
- Innes, R.W. (2004). Guarding the goods: New insights into the central alarm system of plants. Plant Physiol. 135, 695–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, C., Boden, E., and Arias, J. (2003). Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell 15, 1846–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri, F., Thilmony, R., and He, S.Y. (2002). The Arabidopsis thaliana-Pseudomonas syringae interaction. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/10.1199/tab.0039, http://www.aspb.org/publications/arabidopsis/. [DOI] [PMC free article] [PubMed]
- Kerschen, A., Napoli, C.A., Jorgensen, R.A., and Muller, A.E. (2004). Effectiveness of RNA interference in transgenic plants. FEBS Lett. 566, 223–228. [DOI] [PubMed] [Google Scholar]
- King, E.O., Ward, M.K., and Raney, D.E. (1954). Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44, 301–307. [PubMed] [Google Scholar]
- Kinkema, M., Fan, W., and Dong, X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12, 2339–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel, B.N., and Brooks, D.M. (2002). Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5, 325–331. [DOI] [PubMed] [Google Scholar]
- Labbe, S., and Thiele, D.J. (1999). Copper ion inducible and repressible promoter systems in yeast. Methods Enzymol. 306, 145–153. [DOI] [PubMed] [Google Scholar]
- Lamb, C., and Dixon, R.A. (1997). The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. 48, 251–275. [DOI] [PubMed] [Google Scholar]
- Lebel, E., Heifetz, P., Thorne, L., Uknes, S., Ryals, J., and Ward, E. (1998). Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J. 16, 223–233. [DOI] [PubMed] [Google Scholar]
- Liu, Y.L., Schiff, M., Marathe, R., and Dinesh-Kumar, S.P. (2002). Tobacco RAR1, EDS1 and NPR1/NIM1-like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30, 415–429. [DOI] [PubMed] [Google Scholar]
- McElver, J., et al. (2001). Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159, 1751–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou, Z., Fan, W.H., and Dong, X.N. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113, 935–944. [DOI] [PubMed] [Google Scholar]
- Niggeweg, R., Thurow, C., Weigel, R., Pfitzner, U., and Gatz, C. (2000). Tobacco TGA factors differ with respect to interaction with NPR1, activation potential and DNA-binding properties. Plant Mol. Biol. 42, 775–788. [DOI] [PubMed] [Google Scholar]
- Ohta, M., Matsui, K., Hiratsu, K., Shinshi, H., and Ohme-Takagi, M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13, 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals, J.A., Uknes, S., and Ward, E. (1994). Systemic acquired resistance. Plant Physiol. 104, 1109–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sessions, A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14, 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, J., Tsui, F., and Klessig, D.F. (1997). Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant-Microbe Interact. 10, 69–78. [DOI] [PubMed] [Google Scholar]
- Spoel, S.H., et al. (2003). NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15, 760–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strompen, G., Gruner, R., and Pfitzner, U.M. (1998). An as-1-like motif controls the level of expression of the gene for the pathogenesis-related protein 1a from tobacco. Plant Mol. Biol. 37, 871–883. [DOI] [PubMed] [Google Scholar]
- Subramaniam, R., Desveaux, D., Spickler, C., Michnick, S.W., and Brisson, N. (2001). Direct visualization of protein interactions in plant cells. Nat. Biotechnol. 19, 769–772. [DOI] [PubMed] [Google Scholar]
- Tiwari, S.B., Hagen, G., and Guilfoyle, T.J. (2004). Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16, 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero, P., and Dangl, J.L. (2001). A high-throughput method for quantifying growth of phytopathogenic bacteria in Arabidopsis thaliana. Plant J. 28, 475–481. [DOI] [PubMed] [Google Scholar]
- Uknes, S., Mauch-Mani, B., Moyer, M., Potter, S., Williams, S., Dincher, S., Chandler, D., Slusarenko, A., Ward, E., and Ryals, J. (1992). Acquired resistance in Arabidopsis. Plant Cell 4, 645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loon, L.C., and Van Kammen, A. (1970). Polyacrylamide disc electrophoresis of the soluble proteins from Nicotiana tabacum var. “Samsun” and “Samsun NN.” II. Changes in protein constitution after infection with tobacco mosaic virus. Virology 40, 199–211. [DOI] [PubMed] [Google Scholar]
- Ward, E.R., Uknes, S.J., Williams, S.C., Dincher, S.S., Wiederhold, D.L., Alexander, D.C., Ahl-Goy, P., Metraux, J.P., and Ryals, J.A. (1991). Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3, 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, R.R., Bauscher, C., Pfitzner, A.J., and Pfitzner, U.M. (2001). NIMIN-1, NIMIN-2 and NIMIN-3, members of a novel family of proteins from Arabidopsis that interact with NPR1/NIM1, a key regulator of systemic acquired resistance in plants. Plant Mol. Biol. 46, 143–160. [DOI] [PubMed] [Google Scholar]
- Whalen, M.C., Innes, R.W., Bent, A.F., and Staskawicz, B.J. (1991). Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3, 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, R.F. (1979). Acetylsalicylic acid (aspirin) induces resistance to tobacco mosaic virus in tobacco. Virology 99, 410–412. [DOI] [PubMed] [Google Scholar]
- Wildermuth, M.C., Dewdney, J., Wu, G., and Ausubel, F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Fan, W., Kinkema, M., Li, X., and Dong, X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. USA 96, 6523–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J.M., Trifa, Y., Silva, H., Pontier, D., Lam, E., Shah, J., and Klessig, D.F. (2000). NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol. Plant-Microbe Interact. 13, 191–202. [DOI] [PubMed] [Google Scholar]