Abstract
We developed an efficient CRISPR prime editing protocol and generated isogenic-induced pluripotent stem cell (iPSC) lines carrying heterozygous or homozygous alleles for putatively causal single nucleotide variants at six type 2 diabetes loci (ABCC8, MTNR1B, TCF7L2, HNF4A, CAMK1D, and GCK). Our two-step sequence-based approach to first identify transfected cell pools with the highest fraction of edited cells significantly reduced the downstream efforts to isolate single clones of edited cells. We found that prime editing can make targeted genetic changes in iPSC and optimization of system components and guide RNA designs that were critical to achieve acceptable efficiency. Systems utilizing PEmax, epegRNA modifications, and MLH1dn provided significant benefit, producing editing efficiencies of 36–73%. Editing success and pegRNA design optimization required for each variant differed depending on the sequence at the target site. With attention to design, prime editing is a promising approach to generate isogenic iPSC lines, enabling the study of specific genetic changes in a common genetic background.
Introduction
Advances in human molecular genetics and genomics have identified thousands of loci associated with common disease risk. As of April 2023, the NHGRI-EBI genome-wide association study (GWAS) catalog contained over 500,000 associated loci/regions, each of which typically includes multiple candidate variants.1 However, most variants are noncoding, making it challenging to identify molecular mechanisms underlying the associations. To address this challenge, experimental approaches have been used to measure molecular readouts, such as differential chromatin accessibility, transcription factor binding, and gene expression.2 These measures of variant effects often exhibit differences based on genetic background and cell type.
The availability of efficient and precise genome editing technologies such as the clustered regularly interspersed short palindromic repeat (CRISPR)-Cas systems potentially provides a straightforward approach to test functional effects of risk and nonrisk alleles of candidate variants, allowing exploration of molecular consequences in isogenic cells and maximizing the power to detect variant-specific effects.3–5 Furthermore, generating an isogenic series in pluripotent stem cells can enable study of cell-type-specific mechanisms.3–7
Multiple iterations of CRISPR technology have expanded the scope and precision of targeted genome editing, moving from targeted knockouts involving double-strand breaks to more precise approaches.8 Prime editing is an elegant strategy to mediate editing of virtually any single nucleotide substitution.9 As developed by Liu et al., prime editing has low off-target and bystander effects and avoids double-strand breaks that may have deleterious consequences to cell homeostasis.9–13
The 2nd generation prime editing complex consists of two components: the prime editor PE2, a Cas9 nickase (nCas9) domain fused to an engineered reverse transcriptase (RT) domain, and the prime editing guide pegRNA that both specifies the target site and encodes the sequence to be installed (Fig. 1A).9 CRISPR-based technology is dependent on the presence of a nearby protospacer adjacent motif (PAM). The PE2 nCas9 domain recognizes a GG dinucleotide PAM sequence and binds to the target genomic DNA as specified by the pegRNA spacer sequence. The nCas9 domain nicks the PAM-containing DNA strand and the pegRNA primer binding site (PBS) sequence captures the untethered 3′ end of the nicked DNA strand, where the RT domain begins DNA synthesis using the pegRNA's RT extension sequence (reverse transcriptase template [RTT]) as a template. The RTT sequence encodes the “alternate” nucleotide(s) to incorporate the desired edit into the final product.
FIG. 1.
Prime editing (PE3) components and experimental process. (A) The prime editor (nCas9-RT) is directed to the target site by the pegRNA. nCas9-RT nicks the genomic strand containing the PAM sequence, allowing the pegRNA PBS sequence to hybridize with the newly released 3′ end of genomic strand. DNA synthesis occurs (not shown) at this untethered 3′ end of the nicked DNA strand using the RTT as a template. The sgRNA is usually positioned 40–90 bp from the pegRNA-induced nick and will introduce a second nick to promote repair of that strand. (B) The prime editing process includes design of pegRNA and sgRNA oligos to target candidate SNVs, transfection of iPSCs with combinations of prime editing components, expansion of transfected cells, pooled sequencing of the region spanning across the edit (1st Screen), and selection of top performing pools for single cell isolation and expansion to isolation isogenic lines (Single Cell Cloning inset). Clonal lines are sequenced (2nd Screen) to identify those with the correctly installed edit. Selected and expanded clones are sequenced to confirm the edited base and stained with cell surface markers to confirm pluripotency. gRNA, Guide RNA; nCas9, Cas9 nickase; PAM, protospacer adjacent motif; PBS, primer binding site; RT, reverse transcriptase; RTT, reverse transcriptase template; sgRNA, single-guide RNA; SNV, single nucleotide variant.
The 3rd generation system PE3 includes an additional guide (single guide RNA: sgRNA) to nick the nonedited strand 40–90 bp from the pegRNA-induced nick, promoting selective repair of the nonedited strand.9 Subsequent prime editing systems discussed here include the optimized prime editor PEmax, a MLH1dn dominant negative mismatch repair protein to reduce bias toward repair of the installed edit, and an epegRNA, which incorporates a structural addition to stabilize the pegRNA.14,15
CRISPR prime editing is a powerful platform for making targeted diverse and specific genome sequence changes. In practice, however, the success and the amount of optimization required is dependent on the details of the protocol, the target sequence, and the cell type in which changes are intended to be made.9,12,16,17 Thus, much effort has been made to improve editing precision and to establish guidelines for optimal pegRNA design.9,12,18–21
In this work, we used prime editing to interrogate the functional relevance of candidate variants associated with risk for developing type 2 diabetes (T2D). We varied multiple parameters to optimize the editing system and targeted six putatively causal single nucleotide variants (SNVs) for editing in induced pluripotent stem cells (iPSCs) derived from genetically diverse human donors. At each site, we began with a line that was homozygous for either the reference or the nonreference allele, and then used prime editing to generate the heterozygote and alternative homozygote. We carried out editing at each target in three iPSC lines, allowing us to assess dosage effects of causal alleles while controlling for genetic background. We found that prime editing enabled the creation of isogenic series in iPSCs at appreciable efficiencies for the majority of variants we targeted. Here, we present a summary of our experience optimizing the prime editing protocol to achieve 36–73% editing at SNVs and a streamlined processing pipeline to generate edited clones over a 4–5-week period.
Materials and Methods
Selection of genomic targets for prime editing
Six SNVs associated with risk of T2D (MTNR1B rs10830963, ADCY5 rs11708067, TCF7L2 rs7903146, HNF4A rs1800961, CAMK1D rs11257655, and GCK rs878521) were selected for editing. We prioritized variants in the 99% credible sets of SNVs identified in a large-scale multiancestry T2D GWAS that have a high posterior probability of association (>0.95)22 and that are within 20 kb of a reported T2D effector gene (https://t2d.hugeamp.org/method.html?trait=t2d&dataset=egls accessed October 1, 2022).23 As a positive control for downstream functional testing of isogenic lines, we included a seventh SNV, ABCC8 rs137852671 (E1506K), which is associated with congenital hyperinsulinism (CHI).24
Cell lines for transfections
Cell lines were cultured at 37°C and 5% CO2 and counted with the NucleoCounter NC-300 (Chemometec). HEK293T (ATCC crl-11268) were cultured in Dulbecco's modified Eagle's medium (DMEM), high glucose, GlutaMAX™ Supplement HEPES (Transfection System [TFS], Thermo Fisher Scientific), 10% fetal bovine serum (Biotechne), and 1 mM sodium pyruvate (TFS). iPSC lines derived from human fibroblast cells of consented study participants in the Finland–United States Investigation of NIDDM (FUSION) study (NIH protocol OH95-HG-N030; approved by NHGRI IRB) were generated at the New York Stem Cell Foundation Research Institute (New York, NY) using the Stemgent, #00–0071 mRNA transfection iPSC reprogramming kit.25
Parental fibroblast lines and their corresponding dedifferentiated lines (iPSCs) were digitally karyotyped at 400 genomic loci (Nanostring nCounter Plex2 Assay) and SNP-genotyped (Fluidigm SNPtrace™ Panel) to confirm genomic integrity and cell line identity, respectively. iPSC lines were also assessed for pluripotency via Tra-1-60 surface marker staining and expression of a panel of 25 gene markers, which included those for pluripotency and early markers of differentiation. The iPSC lines were cultured in mTeSR™ plus (Stem Cell Technologies) supplemented with a 1:10 dilution of CloneR2 (Stem Cell Technologies) and a final concentration of 2 μg/mL Normocin™ (Invitrogen), hereafter referred to as “iPSC media/Normocin.”
gRNA design
pegRNA and sgRNA designs options were generated in PrimeDesign26 using either default settings or those suggested by Anzalone et al.9
Plasmid constructs and transfections into HEK293T
pegRNA and sgRNA target-specific recombinant plasmids were generated with oligonucleotides synthesized at Integrated DNA Technologies (Coralville, IA). Annealed oligos were cloned into expression vectors pU6-pegRNA-GG-acceptor (gift from D. Liu, Addgene #132777) and pBK1520 (gift from K Joung, Addgene #65777) by Golden Gate assembly.9,27,28
HEK293T cells were transfected with the prime editor PE2 plasmid pCMV-PE2-P2A (gift from D. Liu, Addgene #132775) and different combinations of plasmid constructs encoding target-specific pegRNAs and sgRNAs at a ratio described previously,9 using the Amaxa Nucleofector 2B (Lonza) program A-023. Briefly, 2400 ng of pCMV-PE2-P2A, 600 ng of pegRNA construct, and 249 ng of sgRNA construct were added to 500,000 cells in a final volume of 100 μL HSC1 solution (Human Stem Cell Nucleofector Kit 2; Lonza). After nucleofection, cells were transferred with media to one well of a 24-well plate (Corning). At 48 h after transfection, cells were dissociated for 5 min with 250 μL of 0.25% trypsin (TFS) then diluted with another 250 μL of media for downstream processing for pooled DNA sequencing and cell expansion/cryopreservation.
In vitro transcribed mRNA and synthetic gRNA
mRNA for prime editing components MLH1dn (pCMV-PE2-P2A-hMLH1dn; gift from David Liu, Addgene #174827) and prime editors PE2 (pPC1412: gift from D. Liu) and PEmax (pT7-PEmax for in vitro transcription [IVT]: gift from D. Liu, Addgene # 178113) were generated via IVT.15 Briefly, IVT was performed using the HiScribe T7 High-Yield RNA Kit (New England Biolabs) with replacement of uridine triphosphate with N1-Methylpseudouridine-5′-triphosphate (TriLink Biotechnologies) and co-transcriptional capping with CleanCap AG (TriLink Biotechnologies). The IVT product was purified with LiCl precipitation and resuspended in nuclease-free water.14 Synthetic pegRNA and sgRNA oligonucleotides (Alt-R™ Custom gRNA) were purchased from Integrated DNA Technologies (Coralville, IA) with 2′-O-methyl and phosphorothioate modifications.15
RNA delivery for all targets: transfection with RNA reagents
iPSCs and HEK293T cells were transfected with RNA reagents using the Neon™ TFS according to the manufacturer's protocol for the 10 μL kit. A total of 100,000 cells were electroporated (voltage: 1200, width: 20 ms and pulse: 1) with 1 μg of IVT prime editor (PE2 or PEmax), 90 pmol pegRNA, 60 pmol sgRNA, and in some combinations, 2 μg of MLH1dn.
Transfected IPSCs were transferred to one well of a Geltrex™ (TFS)-coated 24-well plates (Corning) in 500 μL of iPSC media/Normocin and incubated overnight. A half-volume media change and a full media change was made with mTeSR plus media with Normocin at 24 and 48 h after transfection, respectively. At 72–96 h after transfection, confluent cells were dissociated in 250 μL of StemPro™ Accutase™ Cell Dissociation Reagent (TFS) for 10 min at 37°C and resuspended in 250 μL of iPSC media/Normocin.
Transfected HEK293T samples were transferred to one well of a 24-well plate, cultured overnight in media, with a half volume media change at 24 h after transfection and a full media change at 48 h after transfection. At 72–96 h after transfection, confluent cells were dissociated with trypsin and resuspended in 250 μL of media.
Pooled next-generation sequencing of all cells within transfected sample to predict editing efficiency
To prepare for pool next-generation sequencing (NGS) sequencing, cell suspensions were counted, and DNA was extracted from at least 50,000 cells using DNeasy Blood and Tissue Kit (Qiagen). The remaining cells from each sample were pelleted and resuspended for cryopreservation in 200 μL of CryoStor® CS10 (Stem Cell Technologies).
Sequencing was performed following the 16S metagenomic Sequencing Library Preparation protocol (Illumina). Briefly, a pool of six forward and reverse polymerase chain reaction (PCR) primers Integrated DNA Technologies (Coralville, IA) were designed with Primer329 to amplify a 200 bp amplicon for each targeted SNV. Illumina adapters were added as well as degenerate bases to increase PCR specificity (Supplementary Table S3). Amplicons were generated and a small aliquot (250 ng) of each was run on a 1% agarose gel to confirm the presence of a 200 bp fragment. To enable multiplex sequencing, Nextera XT DNA Indexes (Illumina) were added using KAPA HiFi HotStart ReadyMix (Roche), purified product size was confirmed with the Bioanalyzer DNA 1000 chip (Agilent), amplicons were quantified, and 4 nM aliquots of each indexed sample were pooled for sequencing with the MiSeq Reagent Nano Kit v2 300 cycles (Illumina).
Sequencing reads were aligned to the GRCh37 reference genome using “bwa mem” (v0.7.17-r1194) with the -M option30 and filtered for properly paired primary aligned reads with mapping quality ≥30 with “samtools view” v1.9.31 The fraction of the nonreference allele at each position was determined using the filtered aligned reads with “samtools mpileup.” Option “-Q 30” was applied to include only bases with quality score ≥30. The average coverage at the target SNVs was ∼7000 reads and ranged from 2809 to 8000 reads. Editing efficiency was calculated by dividing the number of reads with the installed edited, by the total reads for each sample.
Single clone isolation of iPSCs for expansion
Cryopreserved transfected cell samples were thawed briefly in a 37°C water bath, diluted with 5 mLs iPSC media/Normocin, transferred to a 24-well plate, and cultured for 2–3 days. Cells were pelleted, resuspended in a fresh 250 μL aliquot of iPSC media/Normocin, and counted. An aliquot of cells was diluted 1:100 in iPSC media/Normocin and serially diluted to seed two cells/well across three 96-well (Corning) Geltrex-coated plates with a final volume of 100 μL. Cells were incubated for 2 days at 37°C in 10% CO2 (higher concentration CO2 for culturing cells at very low density), with a media change (mTeSR plus + Normocin) at day 3. At day 7, plates were supplemented with 50 μL of media and visually inspected to identify wells with single colonies. At day 10, wells with single colonies were dissociated with 50 μL of ReLeSR™ (Stem Cell Technologies) for 1 min at room temperature. The ReLeSR was removed, and the plate incubated for another 5 min at 37°C and 5% CO2, after which cells were resuspended in 120 μL of iPSC media/Normocin.
This cell suspension was aliquoted for downstream processing steps: (1) Sanger sequencing: 5 μL transferred to one 96-well PCR plate; (2) expansion of individual clones: 105 μL transferred to a Geltrex-coated 48-well plate (Corning), and (3) backup: 10 μL transferred to one 96-well PCR plate for cryopreservation at −20°C.
Sanger sequencing to screen for edited clones
Immediately after clonal expansion and dissociation, individual clones were sequenced (Fig. 1B—2nd Screen) to identify those with the installed edit. An 800 bp-region surrounding each SNV target was amplified (Kapa HiFi HotStart Ready Mix, Roche) using 5 μL of the dissociated cell suspension (Supplementary Table S3). PCR products were purified using QIAquick PCR purification kit (Qiagen) and Sanger-sequenced at ACGT, Inc. (Germantown, MD). Sequence data were analyzed with Sequencher v5.4.6 (Genecode). The timing of this analysis is critical to avoid overgrowth of cells in the 48-well plates that are cultured in parallel in anticipation of selecting edited clones for expansion and cryopreservation.
FIG. 2.
Editing efficiencies predicted by pool sequencing are validated by single colony sequencing. Editing efficiencies predicted by pooled sequencing (X-axis) and editing efficiencies observed from subsequent single colony sequencing (Y-axis) across four T2D-associated SNVs near ABCC8, HNF4A, MTNR1B, and TCF7L2. The two sets of values were compared using the Pearson r2 statistic, which quantifies the degree to which single clone editing efficiency is approximated by pooled sequencing. The single colony sequencing efficiencies represent the proportion of edited alleles from all clones selected from one pool (transfection) and sequenced individually. T2D, type 2 diabetes.
Comparison of editing efficiency predicted by pool NGS sequencing vs single colony Sanger sequencing
Editing efficiencies determined by NGS sequencing of cell pools were compared to those determined by Sanger sequencing of single clones isolated from the corresponding cell pools. The degree to which single clone editing efficiency is approximated by pooled sequencing was evaluated using the coefficient of determination (Pearson's r2) and calculated using R.
Expansion of clones with installed edits
Edited clones cultured in 48-well plates were expanded and dissociated with 125 μL of ReLeSR, and 100,000 cells were seeded into Geltrex-coated 6-well plates (Corning) in 2 mLs of iPSC media/Normocin. When wells were confluent (day ∼5), cells were dissociated with 1 mL ReLeSR and aliquoted for Sanger sequencing (final sequencing to confirm edited clonal line), cryopreservation, and cell staining with pluripotency markers.
Cell staining with pluripotency markers
Fifty thousand cells were seeded into two wells of a Geltrex-coated 8-chamber Nunc™ Lab-Tek™ II Chamber Slide™ (TFS) and incubated overnight at 37°C and 5% CO2. Cells were stained according to the ES Cell Characterization Kit protocol (Millipore) except the rinse buffer was substituted with 1 × tris-buffered saline with Tween-20 buffer (20 mM Tris-HCL, pH 7.4, 0.15 M NaCl, 0.05% Tween-20), and the blocking solution was substituted with 1% bovine serum albumin in phosphate-buffered saline (1 × PBS ph 7.4, 0.1% Tween-20). Antibodies directed to pluripotency markers SSEA4, OCT4, SOX2, and TRA1–81 (Supplementary Table S4) were used to stain cells.
pegRNA design parameter analyses and statistics
Statistical analyses were performed using R software version 4.2.3. To assess the effect of pegRNA design features on editing efficiency, we focused on experiments using the optimized prime editing system, including the PEmax prime editor, epegRNA, and MLH1dn. We calculated the average editing efficiency across multiple transfections using the same pegRNA. We fit a simple linear regression model for each pegRNA design feature using the “lm” function, with average editing efficiency as the outcome variable and pegRNA design feature as the dependent variable.
Determining PRIDICT scores for pegRNA designs
To determine the PRIDICT score20 for the top-performing pegRNA for each target, we input 100 nucleotides flanking each SNV and searched the output for the pegRNA sequence. PRIDICT scores for predictions in HEK293T cells were available at six of the seven targets using default parameters, which limit the “nick-to-edit” distance to 25 nucleotides or less.
Results
Selection of diabetes SNVs and iPSC lines for genome editing
Among the several hundred association signals for T2D,22 we selected six diabetes-associated GWAS SNVs and one SNV associated with CHI for experimental interrogation with prime editing in human iPSC lines: ABCC8 rs137852671, MTNR1B rs10830963, ADCY5 rs11708067, TCF7L2 rs7903146, HNF4A rs1800961, CAMK1D rs11257655, and GCK rs878521 (Supplementary Table S1). To assess allelic effects in the context of variable genetic backgrounds, we edited iPSC lines from three donor sources for each target that were homozygous for one allele and designed editing to produce the heterozygote and the alternate homozygote. We isolated clones with homozygous reference, heterozygous and homozygous alternate genotypes for each SNV to assess allelic dosage effects (Fig. 1B).
To account for potential off target effects, we isolated three independent biological replicates of each clone, resulting in nine (3 × 3) clones for each SNV per iPSC line. Of note, only 6/9 clones are edited clones as the homozygous reference clones are of the same genotype as the parent line. In total 9 × 3 lines = 27 isogenic iPSC clones were identified and processed for each target variant (Supplementary Fig. S1). We also isolated “nonedited” clones from transfections with a primer editor (nCas9-RT) but no gRNAs. The genotype at the target site of the control lines thus matches that of the parent line used for each transfection.
Establishing optimal delivery of prime editing machinery into human iPSC
Our first prime editing effort was directed toward the ABCC8 variant by transfecting plasmid-encoded PE2, pegRNA, and sgRNA components into HEK293T cells, a line with well-established success in prime editing.9,14 We tested several pegRNA sequences that varied by either PBS or RTT length (Table 1A), and used an sgRNA that targets a site 55 bp away from the pegRNA-induced nick, within the recommended range (40–90 bp).9 We assessed editing efficiency estimates of cell pools from each transfection on a per allele basis by NGS, where we scored efficiency by the number of sequence reads with the installed edit divided by the total reads of the transfected cell pool.
Table 1.
pegRNA optimization with the PE3 system (A) transfection into HEK293T: plasmid-encoded prime editing components delivered into HEK293T included ABCC8 pegRNAs with variable primer binding site (PBS) and reverse transcriptase template (RTT) lengths; the prime editor Cas9 nickase-reverse transcriptase (RT); a pegRNA that targets the site for editing; and a single guide RNA (sgRNA) to target a 2nd nick on the nonedited strand, labeled with the distance of the sgRNA from the pegRNA-induced nick. Protospacer adjacent motif (PAM)-to-edit indicates the distance from the PAM site to the target nucleotide(s) being edited. PBS (underlined); (B) Transfection into induced pluripotent stem cell (iPSC): in vitro transcribed or synthetic RNA components delivered into iPSCs included an ABCC8 pegRNA (PD10) and a new guide PD15
|
(A)
| |||||||||
|---|---|---|---|---|---|---|---|---|---|
| peg RNA design |
Nucleofection reagents
|
pegRNA design parameters
|
|
||||||
| nCas9-RT | sgRNA | pegRNA | PAM to edit (nt) | pegRNA (nt) | pegRNA extension sequence (5′->3′) | PBS (nt) | RTT (nt) | Pool NGS % Editing | |
| Constant RTT and variable PBS | PE2 PE2 PE2 PE2 PE2 PE2 PE2 |
sg55 sg55 sg55 sg55 sg55 sg55 sg55 |
PD5 | 1 | 27 | TGGAAGCCGTGGCCTtGTCCATGATGA | 9 | 18 18 18 18 18 18 18 |
0.3 |
| PD6 | 1 | 28 | TGGAAGCCGTGGCCTtGTCCATGATGAA | 10 | 0.2 | ||||
| PD7 | 1 | 29 | TGGAAGCCGTGGCCTtGTCCATGATGAAG | 11 | 0.2 | ||||
| PD8 | 1 | 30 | TGGAAGCCGTGGCCTtGTCCATGATGAAGA | 12 | 1.2 | ||||
| PD9 | 1 | 31 | TGGAAGCCGTGGCCTtGTCCATGATGAAGAT | 13 | 1.2 | ||||
| PD10a | 1 | 32 | TGGAAGCCGTGGCCTtGTCCATGATGAAGATG | 14 | 1.7 | ||||
| PD11 | 1 | 33 | TGGAAGCCGTGGCCTtGTCCATGATGAAGATGC | 15 | 1.4 | ||||
| Constant PBS and variable RTT | PE2 PE2 PE2 PE2 |
sg55 sg55 sg55 sg55 |
PD12 | 1 | 27 | GCCGTGGCCTtGTCCATGATGAAGATG | 14 14 14 14 |
13 | 1.4 |
| PD10a | 1 | 32 | TGGAAGCCGTGGCCTtGTCCATGATGAAGATG | 18 | 1.7 | ||||
| PD13 | 1 | 37 | GTCAATGGAAGCCGTGGCCTtGTCCATGATGAAGATG | 23 | 1.3 | ||||
| PD14 | 1 | 42 | GCCATGTCAATGGAAGCCGTGGCCTtGTCCATGATGAAGATG | 28 | 0.6 | ||||
| Cells only | None | None | None | 0.2 | |||||
|
(B)
| |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Test category |
Nucleofection reagents
|
pegRNA design parameters
|
|
||||||
| nCas9-RT | sgRNA | pegRNA | PAM to edit (nt) | pegRNA (nt) | pegRNA extension sequence (5′->3′) | PBS (nt) | RTT (nt) | Pool NGS % Editing | |
| All reagents |
PE2 PE2 PE2 PE2 |
sg55 |
PD15 |
6 |
33 |
GCCGTGGCCTtGTCCATGATGAAGATGCTGGTC |
14 |
19 |
15.2 |
| sg55 |
PD10 |
1 |
32 |
TGGAAGCCGTGGCCTtGTCCATGATGAAGATG |
14 |
18 |
29.9b |
||
| sg135 |
PD10 |
1 |
32 |
TGGAAGCCGTGGCCTtGTCCATGATGAAGATG |
14 |
18 |
2.7 |
||
| sg18 |
PD10 |
1 |
32 |
TGGAAGCCGTGGCCTtGTCCATGATGAAGATG |
14 |
18 |
0.4 |
||
| Cas9-RT only |
PE2 |
None |
None |
|
|
|
|
|
0.2b |
| Cas9-RT and sgRNA |
PE2 |
sg15 |
None |
1 |
32 |
TGGAAGCCGTGGCCTtGTCCATGATGAAGATG |
14 |
18 |
0.3 |
| pegRNA and sgRNA |
None |
sg55 |
PD10 |
1 |
32 |
TGGAAGCCGTGGCCTtGTCCATGATGAAGATG |
14 |
18 |
0.2 |
| Cells only | None | None | None | 0.2 | |||||
PD10 filled two optimization categories (PBS and RTT length) but was only transfected once and shown twice to clarify the effect of varying these sequences.
An average of two nucleofections; the best guide RNA combination is pegRNA PD10/sg55.
nCas9, Cas9 nickase; NGS, next-generation sequencing; PAM, protospacer adjacent motif; PBS, primer binding site; RT, reverse transcriptase; RTT, reverse transcriptase template; sgRNA, single-guide RNA.
Editing efficiencies for all pegRNAs tested were mostly undetectable except for one pool that showed 1.7% editing efficiency (pegRNA PD10: PBS = 14 nucleotides [nt], RTT = 18 nt) (Table 1A). Considering the low expression of the fluorescent reporter co-expressed with the prime editor (data not shown), we concluded that the editing components were not entering the cell and/or expressing or assembling effectively in cell nuclei and that further optimization using plasmid transfection would be inefficient.
Based on reports that RNA delivery was more efficient than plasmids in multiple cell types including hPSCs,12,14,32,33 we revised our delivery strategy to use in vitro transcribed (IVT) PE2 RNA and synthetically generated pegRNA and sgRNA molecules.12 We transfected an iPSC line with IVT PE2 RNA and synthetic oligos of the highest performing ABCC8 pegRNA (PD10) and sgRNA (sg55) identified from the HEK293T optimizations (Table 1B). We also tested a new pegRNA design (PD15) that relied on a PAM site further from the targeted SNV site than PD10 and included two additional sgRNAs to nick the unedited strand 18 and 135 bp away.
Transfection of RNA reagents into iPSCs resulted in much improved editing efficiency of 30% for the original PD10/sg55 combination (Table 1B). Neither changing the pegRNA to one that uses a more distant PAM site (PD15), nor changing the position of the second nick targeted by the sgRNA led to a better outcome. Compared to the sgRNA positioned 55 bp away, the additional sgRNAs that were closer (18 bp) or further (135 bp) away performed significantly worse with 0.4% and 2.7% editing efficiency, respectively.
In a follow-up experiment, we transfected both iPSCs and HEK293T cells with the identical series of RNA components targeting the ABCC8 SNV and observed a 1.8-fold improvement for editing in iPSCs than in HEK293T cells—51% versus 29% (Table 2). This trend of higher efficiencies with iPSC was also observed with another target (GCK: 63% in iPSCs vs 1% in HEK293T cells), while editing was similarly high (50%) in both cell lines for the target in HNF4A. This intriguing finding of higher prime editing in iPSCs with RNA delivery for some targets was previously reported by Surun et al.33 These results suggest that RNA delivery of CRISPR editing components can be more conducive to genome editing than transfection of plasmid-encoded components in iPSCs and that slight changes in pegRNA designs and sgRNA can have significant effects on editing efficiency.
Table 2.
RNA delivery to induced pluripotent stem cell and HEK293T
| PE components | Variant in gene |
Nucleofection reagents
|
Cell type
|
Pool NGS
|
|||
|---|---|---|---|---|---|---|---|
| nCas9-RT | MLH1dn | epegRNA | sgRNA | Line | % Editing | ||
| IVT Cas9-RT and synthetic gRNAs IVT Cas9-RT and synthetic gRNAs IVT Cas9-RT and synthetic gRNAs |
ABCC8 | PEMax PEMax PEMax |
Yes Yes Yes |
ABCC8_PD10 | ABCC8_sg55 | iPSC | 51 |
| HEK293T | 29 | ||||||
| HNF4A | HNF4A_PD5 | HNF4A_sg52 | iPSC | 50 | |||
| HEK293T | 51 | ||||||
| GCK | GCK_PD1 | GCK_sg73 | iPSC | 63 | |||
| HEK293T | 1 | ||||||
In vitro transcribed or synthetically synthesized RNA components delivered into iPSCs or HEK293T cells. The optimized prime editing system was used: PEmax, MLH1dn, and epegRNA.
iPSC, induced pluripotent stem cell; IVT, in vitro transcription; PE, prime editing.
Streamlined clone generation and pool sequencing
To determine the most effective prime editing system and the optimal pegRNA and sgRNA combination (or “gRNA combo”), we performed up to 10 transfections per target (and a maximum of 24 transfections for multiple targets) in a single experiment. We developed a streamlined approach that includes screening for edited cells at two different stages, (1) a primary NGS screen of pooled cells to identify transfected samples with the highest number of edited alleles7,9,32,34; and (2) a secondary Sanger sequencing screen of individual clones isolated from the best pool(s) (Fig. 1B) (see Methods). The inclusion of the primary NGS screen enabled the identification and selection of the best performing 1–2 of 10 transfected samples, reducing our downstream efforts for single cell isolation and individual clone Sanger sequencing (i.e., follow-up not needed for 9/10 samples).
Editing efficiency predicted by pool sequencing correlated well (r2 = 0.76, p = 4.89 × 10–11) with the editing efficiency of individual clones (Fig. 2). Two pooled sequencing results for the TCF7L2 (C to T) target predicted significantly lower efficiencies than observed when individual clones were sequenced, potentially due to the AT-rich sequence region surrounding this variant that had also made pegRNA design difficult. Notably, incorporating the edited allele (T) creates an uninterrupted run of 18 A/T that may have biased successful amplification of the “C” allele over the “T” allele in cell pools.
The effort required to generate the suites of 27 lines varied by target. ABCC8 and MTNR1B SNVs were transfected many times to serve as transfection controls across experiments, while other targets required testing more pegRNAs in up to 26 transfections (Fig. 3).
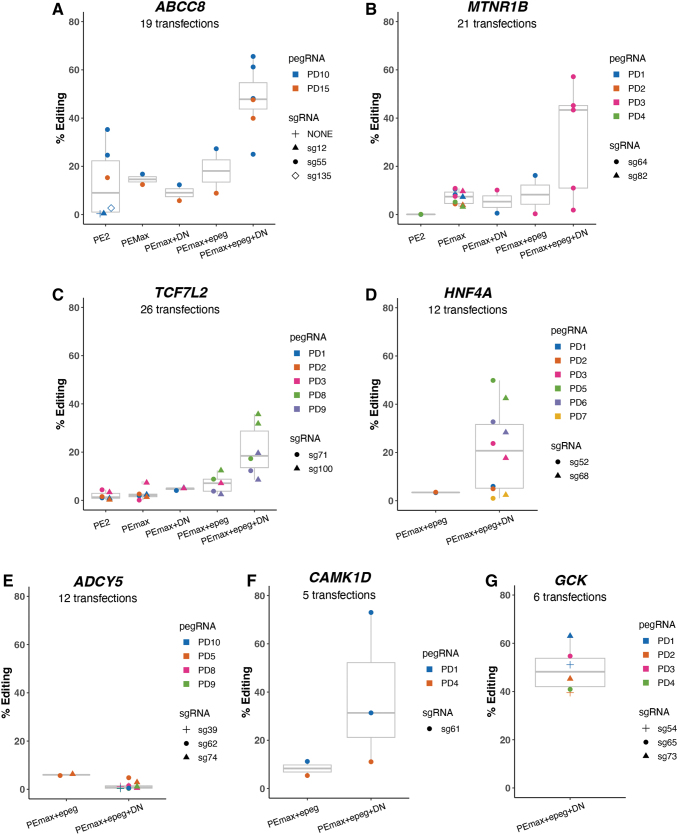
FIG. 3.
Editing efficiencies for different prime editing systems across seven targeted SNVs. (A–G) Different combinations of gRNA combinations (pegRNA and sgRNA) were transfected together with the prime editor (nCas9-RT). gRNA numbering is specific for each gene target. X-axis: prime editing complex systems represented by PE2 and PEmax in the presence or absence of modifications (epeg and DN). Y-axis: % editing efficiency as determined by read counts from sequencing of transfected cell pools. All SNVs except ADCY5 showed satisfactory efficiency. PD, pegRNAs; sg, sgRNAs, with distance from the pegRNA-induced nick represented by numeric values; epegRNA, modification on pegRNA; DN, MLH1dn; # rxns, number of transfections/data points in each plot.
Optimized prime editing systems increased editing efficiency
We tested multiple prime editing systems, starting with the PE2 editor and various gRNA combos, updating the systems by adopting components or modifications as the technology advanced to PE3 and PEmax.9,21 Targeted SNVs processed earlier in the project (ABCC8, MTNR1B, and TCF7L2) were attempted with several systems, while the last four targets (HNF4A, ADCY5, CAMK1D, and GCK) were processed with a more limited number of options (Fig. 3). Individual enhancements, such as the substitution of PEmax for PE2, the epegRNA for the pegRNA, and the addition of MLH1dn, had small and variable effectiveness that depended on the target (Fig. 3). In contrast, when we adopted all three enhancements, we ultimately achieved substantial editing efficiency of 36–73% in iPSCs for six of seven SNV targets.
pegRNA and sgRNA design combinations can result in variable editing efficiencies
Our results from the initial ABCC8 optimization efforts in HEK293T cells underscored the need to test multiple pegRNAs and sgRNAs, including their combined effects within each transfection. For each target, we tested multiple pegRNAs and paired each with one of several sgRNAs that were spaced at variable distances from the target site. Different pegRNAs paired with the same sgRNA exhibited variable editing success, as observed for ABCC8, TCF7L2, HNF4A, and CAMK1D (Fig. 3A, C, D, F). The same pegRNA paired with different sgRNAs also showed variable editing for some targets (ABCC8, TCF7L2, and HNF4A). sgRNAs positioned 50–75 bp from the site targeted for editing gave the best results (up to 50–73% depending on target), but an sgRNA 100 bp away still enabled significant editing for one of the more challenging targets (TCF7L2 sg100–36%). These results suggest that testing a greater number of pegRNA designs may be more productive than optimizing sgRNAs, as long as sgRNA are designed 40–100 nt from the edit.21
Optimizing pegRNA design
We incorporated pegRNA design recommendations as they became available.15,18 We empirically generated design data for a range of poor to good pegRNA for multiple target sequences and assessed the impact of four pegRNA design parameters on editing efficiency: (1) PAM-to-edit distance, (2) PBS length, (3) RTT length, and (4) RTT overhang, which is the nucleotide sequence installed after the edited nucleotide (Fig. 4A). Using the average editing efficiency across all transfections of a pegRNA as the outcome of interest, we fit a simple linear regression model for each pegRNA design feature (Methods). These analyses were restricted to experiments using the optimized prime editing system (with PEmax, epegRNA and MLH1dn), which included 56 transfections with 21 unique pegRNAs (Supplementary Table S2A).
FIG. 4.
pegRNA design parameters affected editing efficiency. (A) Examples of pegRNAs with a constant PBS length but variable RTT length and PAM-to-edit distance. PBS; RTT; PAM-to-edit, distance between PAM sequence and edit site; RTT overhang, nucleotides after the installed edit encoded by in the RTT; red tick mark, nucleotide to be installed. (B–E) Editing efficiency for each target, based on PAM-to-edit distance, PBS length, RTT length, or RTT overhang. Each dot represents the average % editing of all replicate transfections of each pegRNA. Error bars show standard deviation of percent editing across multiple transfections of each pegRNA. Only a single point is shown for pegRNAs where editing efficiency was only measured for a single transfection. p-Values were generated by fitting a simple linear model with average editing efficiency as the outcome variable. See methods for additional details.
The PAM sequence for the prime editors PE2 and PEmax is “NGG,” with N representing any one of the four bases. Thus, AT-rich target regions have fewer PAM sites, which limit pegRNA design options. We found the “PAM-to-edit distance” to be inversely correlated to editing efficiency, with higher editing for pegRNAs utilizing PAMs closer to the target site (p = 2.5 × 10–5, Fig. 4B, Supplementary Table S2B).
The pegRNA PBS is bound to the nicked genomic strand during editing, and target specificity and stability can be affected by the sequence and length of the PBS. Others have shown that editing is achievable using PBS lengths ranging from 8 to 15 nt.12,18 For six of seven targets, we observed >20% editing with at least one pegRNA that had a PBS length of 10–15 nt (Fig. 4C). However, length differences within this range did not have consistent effects (p = 0.35, Supplementary Table S2B). These findings suggest that a PBS length of 10 nt or more would likely provide sufficient thermodynamic stability to the PBS-genomic DNA duplex for appreciable and consistent DNA synthesis to incorporate the new nucleotide(s).
The RTT is immediately upstream of the PBS with its 3′ end positioned at the pegRNA-induced nick, while the 5′ end extends at least up to, and more commonly past, the site to be edited to incorporate the new nucleotide(s) (Fig. 1). Since the nick is made three nucleotides from the PAM site, the minimum length of the RTT is dependent on the PAM-to-edit distance. Not surprisingly given this correlation, our results also show higher editing efficiency with shorter RTT length (p = 2.0 × 10–4, Fig. 4D, Supplementary Table S2B). RTT lengths <20 nt were the most successful, but lengths as long as 27 nt also led to appreciable editing (>20%).
A longer RTT overhang after the installed edit may enhance flap resolution during DNA repair (D. Liu group, personal communication). We used RTT postedit lengths of 5–15 nt and observed efficient editing (20–73%) for all lengths, but high efficiencies of ≥40% were more consistently achieved with longer overhangs of >10 nt (p = 6.6 × 10–,3 Fig. 4E, Supplementary Table S2B). This result confirms reports of enhanced editing efficiencies with extended RTT overhangs.20,35
Frequency of indel errors
At each target SNV site, we sequenced an 800-bp region from hundreds of clones derived from several transfections for each of three target loci: MTNR1B (470 clones), HNF4A (295), and TCF7L2 (778). We observed variable indel rates across targets, with an overall rate of 3–9% depending on the target (Fig. 5A). These indel rates with the PE3 system are comparable to those previously observed in human immortalized cell lines and human embryonic stem cells.14,36 Indel rates also varied across transfections (experiments) for the same target, including those utilizing the identical gRNAs: MTNR1B (median 0.90%), HNF4A (7.32%), and TCF7L2 (8.48%) (Fig. 5B).
FIG. 5.
Insertion–deletion rates. (A) Overall insertion–deletion rate within 800 bp for all clones for one target. Numbers at the top of the bars represent the number of clones sequenced for each target. (B) Rate across all transfections for each target with each dot representing one transfection. Dot sizes are on a continuous scale with the size increasing as the number of clones screened increases. The total number of clones screened for each target corresponds to those in panel A (HNF4A = 295, MTNR1B = 470, TCF7L2 = 778).
Timeline: prime editing to edited clones
Our streamlined approach to manage the different stages of the editing process took 4–5 weeks from transfection of iPSCs with prime editing components to possession of validated pluripotent clones with the installed edit (Fig. 6). This timeframe required access to rapid-turnaround NGS capabilities during the primary screen to select the best-performing transfections. Fortunately, our process allowed for some leeway because an aliquot of each transfected sample of cells was cryopreserved and only thawed and expanded when pooled sequence data were available. This flexibility was also critical for partitioning the workload among multiple samples, as significant manual processing of clones is required after pegRNA and sgRNA combo selection in week 2.
FIG. 6.
Timeline: prime editing to edited clones. A general prime editing timeline illustrating a streamlined process that can be completed within 5 weeks. combo, combination of gRNAs (pegRNA and sgRNA); wp, well plate.
Downstream processing included substantial efforts with regards to cell culturing, single cell isolation, and expansion of desired clones. Spontaneous cellular differentiation and sample mix-ups were potential issues inherent to lengthy multistep processes. Thus, assessing the genotype identity and pluripotency of the final iPSC clones were critical to validate the integrity of the edited clones before differentiation (data not shown).
Discussion
We varied multiple experimental parameters in CRISPR prime editing to generate a genotype suite of 27 isogenic iPSC lines carrying heterozygous or homozygous T2D or CHI risk or nonrisk alleles at each of six disease-associated loci. In the process, we refined criteria regarding selection of prime editing system components and pegRNA design, and we optimized the logistics of screening hundreds of clones per target.
We observed low efficiency with plasmid-based delivery of prime editing components and independently confirmed that RNA delivery achieved better results in iPSC, as previously reported.14,32,33 This improved editing efficiency likely reflects better transfection efficiency of RNA compared to larger plasmids such as the PEmax editor and avoids need for a promoter that provides robust expression in iPSCs. RNA delivery may also reduce the potential for off-target effects by limiting prolonged expression of editing components. Furthermore, use of synthetic RNA components eliminated the substantial cloning effort needed to generate locus-specific pegRNA and sgRNA plasmids.
Disadvantages of RNA delivery include the high cost of synthetic oligos, potentially lower purity of full-length synthetic RNA molecules for long pegRNAs, and the absence of selectable markers to track successful transfection of prime editing components. To compensate for the lack of markers, we incorporated an initial pool sequencing analysis to prescreen transfected samples for a high proportion of edited cells. Several other studies have shown successful prime editing in iPSCs with RNA delivery, but efficiencies were low without incorporating successive rounds of re-editing.14,32,33 Our approach produced consistent and enhanced editing efficiencies of >20% in iPSCs with a single transfection.
Varying pegRNA design parameters also influenced editing success. Here, we observed better efficiency with a short distance between PAM and the edit site and a short RTT length while maintaining at least 5 and preferably 10 nucleotides after the edit site for the RTT overhang. Although shorter RTT length corresponds to shorter PAM-to-edit distance, the potentially lower quality/purity of long synthetic oligos may also contribute to the better performance of short RTTs. Lastly, the length of the PBS has been also a focus for pegRNA design, but we did not find significant differences with variable lengths once a minimum threshold of 10 nt was implemented to maintain stability of the RNA-DNA duplex. Notably, we observed differences only after we incorporated the optimized prime editing system.
The combination of PEmax, MLH1dn, epeg modification, and deliberate pegRNA design led to the highest editing efficiencies. When we scored our top pegRNAs with a recently developed deep learning-based prime editing guide prediction (PRIDICT) program,20 we found high scores for our targets in ABCC8 (71), MTNR1B (70), and CAMK1D (76), lower scores for GCK (53), HNF4A (48), and TCF7L2 (2.6), and no information for ADCY5 using PRIDICT default parameters. Thus, in most cases, the editing efficiencies predicted by PRIDICT correlated well with our experience.
The higher editing efficiency we observed with PE3 compared to PE2 underscores the value of the sgRNA.9 The window for positioning the sgRNA was broad, allowing for flexible design options. We positioned almost all sgRNA-induced nicks 3′ of the pegRNA-induced nick, that is, inward-facing PAMs,9,21 which worked well. Our overall indel rate with the PE3 system utilizing the PEmax nCas9-RT editor was 3–9% depending on the target sequence. This proportion of defective clones was easily accommodated by the high editing efficiencies.
While we achieved editing efficiencies of 36–73% for six targets, we were unable to achieve efficiencies above background for the SNV at ADCY5 (rs11708067) even after multiple transfections (12 with PEmax/epeg or PEmax/epeg/MLH1dn). This target had few “NGG” PAM options, leading to the longest PAM-to-edit distance (24 nt) and thus the longest RTT length of the seven SNVs (Fig. 4). We also were unsuccessful with a nCas-RT editor that recognized NGA” PAM sequences and transfected with pegRNAs targeting PAMs only 5–10 nt from the edit (10 transfections—data not shown), although this editor was not optimized as well as PEmax.
Editing efficiency is target-specific, suggesting a role for genomic architecture such as chromatin accessibility around the desired edit.37,38 ATAC-seq data from a pool of 12 iPSC lines showed that the ADCY5 variant was located in accessible chromatin (data not shown), suggesting that poor genome access was not the cause for the failure to edit at this site. Further work is needed to edit this variant and to improve predictions for sites that cannot be efficiently edited.
The addition of the sgRNA in the PE3 system enhanced editing efficiency, but it may also form DNA intermediates with nicks on both strands of the genomic DNA, leading to undesired insertions/deletions (indels) at the target site or nearby flanking region.9,21,36,39 Fiumara et al. recently showed that prime editing (with PE3) of beta-2 microglobulin in human hematopoietic stem/progenitor cell leads to an average of 4.5% indels nearby the nicking sites. Furthermore, they observed PE3-specific induction of the proapoptotic TP73 gene and suggested this may be a consequence of not rapidly resolved double-stranded break intermediates.39 Thus, the edited iPSC lines generated here and the beta-like cells differentiated in the next phase need to be assessed via whole genome sequencing or another method that allows broad interrogation of potential sites of double-stranded breaks.40,41 This comparison is needed to assess genome-wide off-target changes and ensures phenotypic changes (functional readouts) of edited and nonedited lines that are due to the targeted edits.
There are a few limitations to this study. We did not test an exhaustive matrix of all possible variations of multiple experimental parameters. The logistics of manual manipulation of potentially hundreds of clones for each optimization experiment and downstream processing limited the number of variations we could assess for each target. We generally ceased testing other options once achieving acceptable editing of a target, that is, efficiency is significantly high (in our case above 20%) such that we can reasonably accommodate screening for edited clones across multiple cell lines and in replicates for each genotype. Thus, our findings are based on empirical data that allow conclusions about general principles, but may miss important nuances, such as why very similar pegRNAs can achieve quite different efficiencies. Additionally, the editing efficiencies for the various prime editing system components and pegRNA designs were achieved under one transfection setting that may not be fully optimized to generate the best outcome for each of the prime editing variables tested.
There has been rapid evolution of the prime editing protein complex and/or accessory components,42,43 some of which we have adopted (i.e., PEmax and MLH1dn) and confirmed the value of these changes. We expect our results regarding pegRNA design to be transferable to the recent generation of prime editors but will have significant gains when used in combination with the enhanced efficiencies and capabilities of the evolved prime editor complexes.
Despite the incomplete knowledge of the complex nature of prime editing, we were able to obtain 27 edited iPSC lines for each of 6 SNVs. In the course of this work, we were able to advance from initial efforts, where achieving the desired edits often took months, to a pipeline where multiple edited iPSC clones can be achieved after 4–5 weeks. This strategy is easily adaptable by most molecular biology laboratories without the need for automation and opens a new window to understand the effect of genome variants on human health and disease pathogenesis.
Supplementary Material
Acknowledgments
We would like to thank the New York Stem Cell Foundation for the generation of iPSC lines and quality control assessments, the NIH NIAMS Genomic Technology Section for sequencing support, and David Liu for the kind gifts of the prime editing plasmids and the helpful advice from his group. Figures 1, 4A, 6, and Supplementary Figure S1 were created with BioRender.com.
Authors' Contributions
L.L.B., A.J.S., M.R.E., N.N., and F.S.C. conceived and designed research. L.L.B., A.J.S., E.C.M., A.L., E.W., E.S.L., and T.H. performed research. L.L.B., A.J.S., E.C.M., A.L., E.W., E.S.L., C.C.R., V.A.P., C.K., K.L.M., N.N., and F.S.C. analyzed and interpreted data. L.L.B., A.J.S., and F.S.C. supervised study; L.L.B., A.J.S., E.C.M., A.L., C.C.R., V.A.P., K.L.M., N.N., and F.S.C. wrote and edited article. All authors reviewed article.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This research was supported by the National Institutes of Health grant 1-ZIA-HG000024 (F.S.C.), T32 GM135128 (V.A.P.), and R01DK072193 (K.L.M.)
Supplementary Material
References
- 1. Buniello A, MacArthur JAL, Cerezo M, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 2019;47(D1):D1005–D1012; doi: 10.1093/nar/gky1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rao S, Yao Y, Bauer DE. Editing GWAS: Experimental approaches to dissect and exploit disease-associated genetic variation. Genome Med 2021;13(1):41; doi: 10.1186/s13073-021-00857-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Soldner F, Laganiere J, Cheng AW, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell 2011;146(2):318–331; doi: 10.1016/j.cell.2011.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hockemeyer D, Jaenisch R. Induced pluripotent stem cells meet genome editing. Cell Stem Cell 2016;18(5):573–586; doi: 10.1016/j.stem.2016.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nair AK, Traurig M, Sutherland JR, et al. Generation of isogenic hiPSCs with targeted edits at multiple intronic SNPs to study the effects of the type 2 diabetes associated KCNQ1 locus in American Indians. Cells 2022;11(9); doi: 10.3390/cells11091446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hotta A, Yamanaka S. From genomics to gene therapy: Induced pluripotent stem cells meet genome editing. Annu Rev Genet 2015;49:47–70; doi: 10.1146/annurev-genet-112414-054926 [DOI] [PubMed] [Google Scholar]
- 7. Li M, Zhong A, Wu Y, et al. Transient inhibition of p53 enhances prime editing and cytosine base-editing efficiencies in human pluripotent stem cells. Nat Commun 2022;13(1):6354; doi: 10.1038/s41467-022-34045-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Anzalone AV, Koblan LW, Liu DR. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol 2020;38(7):824–844; doi: 10.1038/s41587-020-0561-9 [DOI] [PubMed] [Google Scholar]
- 9. Anzalone AV, Randolph PB, Davis JR, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019;576(7785):149–157; doi: 10.1038/s41586-019-1711-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schene IF, Joore IP, Oka R, et al. Prime editing for functional repair in patient-derived disease models. Nat Commun 2020;11(1):5352; doi: 10.1038/s41467-020-19136-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gao P, Lyu Q, Ghanam AR, et al. Prime editing in mice reveals the essentiality of a single base in driving tissue-specific gene expression. Genome Biol 2021;22(1):83; doi: 10.1186/s13059-021-02304-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Doman JL, Sousa AA, Randolph PB, et al. Designing and executing prime editing experiments in mammalian cells. Nat Protoc 2022;17(11):2431–2468; doi: 10.1038/s41596-022-00724-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gao R, Fu ZC, Li X, et al. Genomic and transcriptomic analyses of prime editing guide RNA-independent off-target effects by prime editors. CRISPR J 2022;5(2):276–293; doi: 10.1089/crispr.2021.0080 [DOI] [PubMed] [Google Scholar]
- 14. Chen PJ, Hussmann JA, Yan J, et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell 2021;184(22):5635.e29–5652.e29; doi: 10.1016/j.cell.2021.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nelson JW, Randolph PB, Shen SP, et al. Engineered pegRNAs improve prime editing efficiency. Nat Biotechnol 2022;40(3):402–410; doi: 10.1038/s41587-021-01039-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Scholefield J, Harrison PT. Prime editing—An update on the field. Gene Ther 2021;28(7–8):396–401; doi: 10.1038/s41434-021-00263-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lu C, Kuang J, Shao T, et al. Prime editing: An all-rounder for genome editing. Int J Mol Sci 2022;23(17); doi: 10.3390/ijms23179862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim HK, Yu G, Park J, et al. Predicting the efficiency of prime editing guide RNAs in human cells. Nat Biotechnol 2021;39(2):198–206; doi: 10.1038/s41587-020-0677-y [DOI] [PubMed] [Google Scholar]
- 19. Li Y, Chen J, Tsai SQ, Cheng Y. Easy-Prime: A machine learning-based prime editor design tool. Genome Biol 2021;22(1):235; doi: 10.1186/s13059-021-02458-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mathis N, Allam A, Kissling L, et al. Predicting prime editing efficiency and product purity by deep learning. Nat Biotechnol 2023; doi: 10.1038/s41587-022-01613-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen PJ, Liu DR. Prime editing for precise and highly versatile genome manipulation. Nat Rev Genet 2022; doi: 10.1038/s41576-022-00541-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mahajan A, Spracklen CN, Zhang W, et al. Multi-ancestry genetic study of type 2 diabetes highlights the power of diverse populations for discovery and translation. Nat Genet 2022;54(5):560–572; doi: 10.1038/s41588-022-01058-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Costanzo MC, von Grotthuss M, Massung J, et al. The type 2 diabetes knowledge portal: An open access genetic resource dedicated to type 2 diabetes and related traits. Cell Metab 2023;35(4):695.e6–710.e6; doi: 10.1016/j.cmet.2023.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huopio H, Reimann F, Ashfield R, et al. Dominantly inherited hyperinsulinism caused by a mutation in the sulfonylurea receptor type 1. J Clin Invest 2000;106(7):897–906; doi: 10.1172/JCI9804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Paull D, Sevilla A, Zhou H, et al. Automated, high-throughput derivation, characterization and differentiation of induced pluripotent stem cells. Nat Methods 2015;12(9):885–892; doi: 10.1038/nmeth.3507 [DOI] [PubMed] [Google Scholar]
- 26. Hsu JY, Grunewald J, Szalay R, et al. PrimeDesign software for rapid and simplified design of prime editing guide RNAs. Nat Commun 2021;12(1):1034; doi: 10.1038/s41467-021-21337-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cong L, Zhang F. Genome engineering using CRISPR-Cas9 system. Methods Mol Biol 2015;1239:197–217; doi: 10.1007/978-1-4939-1862-1_10 [DOI] [PubMed] [Google Scholar]
- 28. Sakuma T, Sakamoto T, Yamamoto T. All-in-one CRISPR-Cas9/FokI-dCas9 vector-mediated multiplex genome engineering in cultured cells. Methods Mol Biol 2017;1498:41–56; doi: 10.1007/978-1-4939-6472-7_4 [DOI] [PubMed] [Google Scholar]
- 29. Untergasser A, Cutcutache I, Koressaar T, et al. Primer3—New capabilities and interfaces. Nucleic Acids Res 2012;40(15):e115; doi: 10.1093/nar/gks596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25(14):1754–1760; doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics 2009;25(16):2078–2079; doi: 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li H, Busquets O, Verma Y, et al. Highly efficient generation of isogenic pluripotent stem cell models using prime editing. Elife 2022;11; doi: 10.7554/eLife.79208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Surun D, Schneider A, Mircetic J, et al. Efficient generation and correction of mutations in human iPS cells utilizing mRNAs of CRISPR base editors and prime editors. Genes (Basel) 2020;11(5); doi: 10.3390/genes11050511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gaudelli NM, Komor AC, Rees HA, et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 2017;551(7681):464–471; doi: 10.1038/nature24644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Siegner SM, Karasu ME, Schroder MS, et al. PnB Designer: A web application to design prime and base editor guide RNAs for animals and plants. BMC Bioinformatics 2021;22(1):101; doi: 10.1186/s12859-021-04034-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Habib O, Habib G, Hwang GH, et al. Comprehensive analysis of prime editing outcomes in human embryonic stem cells. Nucleic Acids Res 2022;50(2):1187–1197; doi: 10.1093/nar/gkab1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Park SJ, Jeong TY, Shin SK, et al. Targeted mutagenesis in mouse cells and embryos using an enhanced prime editor. Genome Biol 2021;22(1):170; doi: 10.1186/s13059-021-02389-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu N, Zhou L, Lin G, et al. HDAC inhibitors improve CRISPR-Cas9 mediated prime editing and base editing. Mol Ther Nucleic Acids 2022;29:36–46; doi: 10.1016/j.omtn.2022.05.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fiumara M, Ferrari S, Omer-Javed A, et al. Genotoxic effects of base and prime editing in human hematopoietic stem cells. Nat Biotechnol 2023; doi: 10.1038/s41587-023-01915-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Atkins A, Chung CH, Allen AG, et al. Off-target analysis in gene editing and applications for clinical translation of CRISPR/Cas9 in HIV-1 therapy. Front Genome Ed 2021;3:673022; doi: 10.3389/fgeed.2021.673022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tao J, Bauer DE, Chiarle R. Assessing and advancing the safety of CRISPR-Cas tools: From DNA to RNA editing. Nat Commun 2023;14(1):212; doi: 10.1038/s41467-023-35886-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhao Z, Shang P, Mohanraju P, et al. Prime editing: Advances and therapeutic applications. Trends Biotechnol 2023;41(8):1000–1012; doi: 10.1016/j.tibtech.2023.03.004 [DOI] [PubMed] [Google Scholar]
- 43. Doman JL, Pandey S, Neugebauer ME, et al. Phage-assisted evolution and protein engineering yield compact, efficient prime editors. Cell 2023;186(18):3983.e26–4002.e26; doi: 10.1016/j.cell.2023.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.