Abstract
Background
Lung cancer is the leading cause of cancer-related death worldwide. The sex differences in the occurrence and fatality rates of non-small cell lung cancer (NSCLC), along with its association with estrogen dependence, suggest that estrogen receptors (ERs) contribute to the development of NSCLC. However, the influence of G protein-coupled estrogen receptor (GPER1) on NSCLC remains to be determined. Escape from ferroptosis is one of the hallmarks of tumor discovered in recent years. In this context, the present study evaluated whether GPER1 promotes NSCLC progression by preventing ferroptosis, and the underlying mechanism through which GPER1 protects against ferroptosis was also explored.
Methods
The effects of GPER1 on the cytotoxicity of H2O2, the ferroptosis inducer RSL3, and Erastin were assessed using the CCK8 assay and plate cloning. Lipid peroxidation levels were measured based on the levels of MDA and BODIPY™581/591C11. GPER1 overexpression and knockdown were performed and G1 was used, and the expression of SCD1 and PI3K/AKT/mTOR signaling factors was measured. Immunofluorescence analysis and immunohistochemistry were performed on paired specimens to measure the correlation between the expression of GPER1 and SCD1 in NSCLC tissues. The effect of GPER1 on the cytotoxicity of cisplatin was measured in vitro using the CCK8 assay and in vivo using xenograft tumor models.
Results
GPER1 and G1 alleviated the cytotoxicity of H2O2, reduced sensitivity to RSL3, and impaired lipid peroxidation in NSCLC tissues. In addition, GPER1 and G1 promoted the protein and mRNA expression of SCD1 and the activation of PI3K/AKT/mTOR signaling. GPER1 and SCD1 expression were elevated and positively correlated in NSCLC tissues, and high GPER1 expression predicted a poor prognosis. GPER1 knockdown enhanced the antitumor activity of cisplatin in vitro and in vivo.
Conclusion
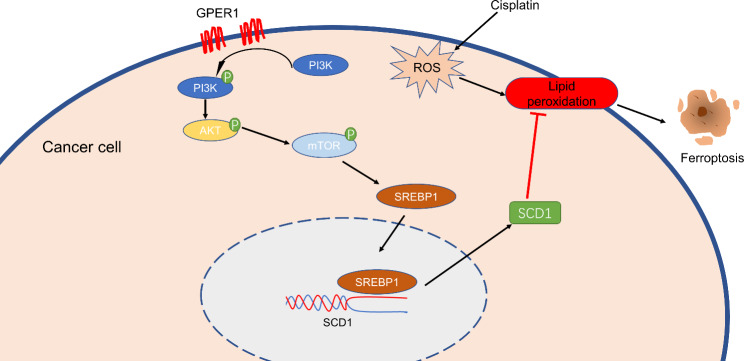
GPER1 prevents ferroptosis in NSCLC by promoting the activation of PI3K/AKT/mTOR signaling, thereby inducing SCD1 expression. Therefore, treatments targeting GPER1 combined with cisplatin would exhibit better antitumor effects.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s10020-023-00763-x.
Keywords: NSCLC, GPER1, Ferroptosis, Cisplatin and SCD1
Introduction
Lung cancer is the primary cause of cancer-related deaths worldwide in men and women (Sung et al. 2021). Non-small cell lung cancer (NSCLC) accounts for 85% of all cases of lung cancer (Zhang et al., 2021). Researchers have been tirelessly searching for novel therapeutic targets to combat NSCLC. Recently, research has presented compelling evidence linking estrogen to NSCLC. A study conducted on 6500 breast cancer survivors revealed that women who received antiestrogen therapy exhibited a lower mortality rate due to lung cancer (Bouchardy et al. 2011). A study conducted with over 16,000 postmenopausal women who were treated with a placebo or daily hormone replacement therapy (HRT) for 5 years revealed that those in the HRT group exhibited a higher incidence of lung cancer morbidity and mortality (Chlebowski et al. 2009). Furthermore, prospective investigations have demonstrated that the risk of HRT increasing the incidence and mortality of lung cancer was time-dependent (Slatore et al. 2010). These findings suggest that estrogens play a role in tumor etiology and progression to a certain extent.
Estrogen is known to play a crucial role in regulating numerous physiological processes under normal conditions. Consequently, disruptions in the estrogen signaling pathway could lead to a variety of diseases, including reproductive system-related conditions such as endometriosis, bone-related abnormalities such as osteoporosis, and tumor-related conditions such as lung and breast cancer. In addition, estrogen abnormalities may contribute to the development of diseases related to the cardiovascular and digestive systems (Chen et al. 2022; Iorga et al. 2017). Estrogens function by binding to their receptors and activating them. These receptors include the classical ERα and ERβ, as well as the membrane-bound G protein-coupled estrogen receptor (GPER1) (Barton et al. 2018; Luo and Liu, 2020). Over the past two decades, extensive research has been conducted on ERα, ERβ, and estrogen signaling in relation to lung adenocarcinoma, which has a higher incidence in women (Garon et al. 2013; Marquez-Garban et al. 2007; Toh et al. 2010). However, research focused on GPER1 in lung cancer has been scarce.
The GPER1 protein is known to be biologically active and highly expressed in a range of solid tumors, particularly those with sex-based differences in their incidence rates, such as breast, endometrial, thyroid, and colon cancer (Hernandez-Silva et al. 2020; Hsu et al. 2019; Jacenik et al. 2019; Xu et al. 2019). Recently, research revealed that GPER1 expression was elevated in NSCLC compared to normal lung tissue (Jala et al. 2012), and higher expression correlated with poor postoperative prognosis in NSCLC patients (Li et al. 2022). Studies have indicated that the activation of EGFR via GPER1 leads to the phosphorylation of ERK1/2, which promotes the expression of matrix metalloproteinase-2 (MMP-2) and matrix metalloproteinase-9 (MMP-9). This, in turn, promotes NSCLC invasion and metastasis (Zhang et al. 2014). Recent studies have demonstrated that GPER1 interacts with E2 or fulvestrant, which are antagonists of ERα and ERβ, respectively, to activate and promote LUAD cell proliferation in vitro and in vivo (Liu et al. 2015, 2019; Shen et al. 2021). Further research is necessary to elucidate the mechanism through which GPER1 promotes NSCLC progression. In this context, the present study aimed to investigate the specific mechanism through which GPER1 contributes to NSCLC progression.
Ferroptosis is a type of nonapoptotic cell death that relies on iron and may be detected based on lipid peroxidation (Dixon et al. 2012). Recent studies have suggested that inducing ferroptosis in tumor cells is an effective strategy to terminate tumor growth. In addition, inhibiting lipid peroxidation or reducing ferroptosis through pharmaceutical or genetic means could effectively prevent ferroptosis (Jiang et al. 2021; Zhang et al. 2022). Stearoyl CoA desaturase 1 (SCD1) is a crucial enzyme that catalyzes the conversion of saturated fatty acids (SFAs) into monounsaturated fatty acids (MUFAs). SCD1 is highly expressed in various tumors and reportedly promotes tumor progression. Furthermore, recent research revealed that SCD1 protects against tumor ferroptosis (Chen et al. 2021; Magtanong et al. 2019). In the context of pancreatic cancer, FBW7 plays a crucial role in regulating lipid peroxidation and promoting ferroptosis by suppressing SCD1 expression. Mechanistically, this is accomplished by inhibiting nuclear receptor subfamily 4 group A member 1 (NR4A1). This suppression of SCD1 ultimately leads to the promotion of ferroptosis in pancreatic cancer cells (Ye et al. 2021). SCD1 knockdown in the NSCLC cell line A549 led to a noteworthy decrease in the cell growth rate, along with an increase in intracellular iron levels and lipid peroxidation. These findings indicate that SCD1 plays a crucial role in regulating ferroptosis (Jiang et al. 2017). Tumor cells with PI3K hyperactive mutations are highly resistant to ferroptosis. However, when the PI3K/AKT/mTOR signaling axis is inhibited, cancer cells become sensitive to ferroptosis. Mechanistic studies have demonstrated that this signaling axis protects cancer cells from ferroptosis by producing MUFAs via SREBP1/SCD1 (Yi et al. 2020). Previous investigations by our research group have revealed that estrogen exerts antioxidant effects on tumors while promoting the expression of SCD1 (Belkaid et al. 2015; Chen et al. 2013; Kong et al. 2014). This finding led to the hypothesis that estrogen could regulate tumor ferroptosis. To date, no study has investigated the potential association between GPER1 and SCD1. In the present study, it was hypothesized that GPER1 is linked to ferroptosis, and SCD1 plays a role in this process.
Materials and methods
Patients and samples
The set of samples used in the present study were 96 formalin-fixed, paraffin-embedded tissue samples from patients with primary NSCLC, who had been diagnosed at the Department of Thoracic Surgery of Union Hospital, Tongji Medical College, Huazhong University of Science and Technology (Wuhan, China), between the years 2014 and 2018. None of these patients had used hormone therapy, radiation, or chemotherapy prior to the surgery. Patients’ clinical details were noted, and at least two pathologists verified the diagnosis. At the Union Hospital, the pTNM stage and tumor differentiation grade were also noted. The respective tissue samples were obtained with informed consent from all patients. The baseline characteristics of the study participants are presented in Table 1.
Table 1.
Clinical characteristics of patients
| Adenocarcinoma | Squamous | All | |
|---|---|---|---|
| All | 52(54.2%) | 44(45.8%) | 96(100%) |
| Gender | |||
| Male | 33(63.5%) | 40(90.9%) | 73(76.0%) |
| Female | 19(36.5%) | 4(9.1%) | 23(24.0%) |
| Age(years) | |||
| ≤ 60 | 34(65.4%) | 28(63.6%) | 62(64.6%) |
| >60 | 18(34.6%) | 16(36.4%) | 34(35.4%) |
| Tumor differentiation grade | |||
| Poor | 12(23.2%) | 15(34.1%) | 27(28.1%) |
| Poor ~ moderate | 19(36.5%) | 12(27.3%) | 31(32.3%) |
| Moderate | 18(34.6%) | 11(25%) | 29(30.2%) |
| Well /Moderate-well | 3(57.7%) | 6(13.6%) | 9(9.4%) |
| Pathological tumor (T) status | |||
| T1 ~ T2 | 37(71.1%) | 28(63.6%) | 65(67.7%) |
| T3 ~ T4 | 15(28.9%) | 16(36.4%) | 31(32.3%) |
| Pathological node (N) status | |||
| N0 ~ N1 | 15(28.9%) | 23(52.3%) | 38(39.6%) |
| N2 ~ N3 | 37(71.1%) | 21(47.7%) | 58(61.4%) |
| Clinical stage | |||
| I ~ II | 9(17.3%) | 18(40.9%) | 27(28.1%) |
| III | 43(82.7%) | 26(59.1%) | 69(71.9%) |
Reagents and antibodies
G-1 (HY-107,216), RSL3 (HY-100,218 A), Erastin (HY-15,763), A939572 (HY-50,709), NSC781406 (HY-100,470), and Fatostatin (HY-14,452) were purchased from Med-Chem-Express. All these reagents were dissolved in DMSO, which was obtained from Servicebio (GC203005). The primary antibodies used in the present study included GPER1 (ABclonal, A10217), SCD1 (ABclonal, A16429), SREBP1 (Servicebio, GB113804), PI3K (ZENBIO, R22768), Phospho-PI3K (Tyr467/Tyr199) (ZENBIO, 310,164), Phospho-mTOR (Ser2448) (ZENBIO, 381,557), AKT (ABmart, T55561), Phospho-AKT (Ser473) (ABmart, T40067), GAPDH (ABclonal, AC033), mTOR (ABmart, T55306), GPX4 (ZENBIO, R381958), and NRF2 (ABmart, T55136). The second antibodies, horseradish peroxidase (HRP) goat anti-rabbit IgG (ABclonal, AS014) and (Servicebio, G1213), HRP goat anti-mouse IgG (ABclonal, AS003) were applied. A dilution ratio of 1:1000 was used in western blotting and 1:100 in immunohistochemistry and immunofluorescence analyses.
Cell culture
The NSCLC cell lines A549 (CCL-185, ATCC) and H1299 (CRL-5803, ATCC) were obtained from ATCC. All cell lines were cultured in the RPMI-1640 medium (Gibico) supplemented with 10% fetal bovine serum (FBS; Gibico) and antibiotics (100 units/mL penicillin and 100 μg/mL streptomycin) using standard procedures. The cell culture was conducted in an incubator at 37 °C with 5% CO2 and 95% air atmosphere.
Transfection
The shRNA lentiviral and GPER1 plasmid lentiviral with puromycin resistance vectors were obtained from Genechem (Shanghai, China). The cells were seeded in the wells of 6-well plates and then treated with puromycin for a minimum of 5 days after being infected with the lentiviral for 72 h. afterward, the transfection efficiency was verified through qRT-PCR and western blotting. The sequence for shGPER1 was 5′- CGG CTT TGT GGG CAA CAT C-3′, while 5′-TTC TCC GAA CGT GTC ACG T-3′ was used as the negative control.
RNA extraction and qRT-PCR
TreliefTM-RNAprep-FastPure Tissue & Cell Kit (TSP413, TSINGKE) was employed for RNA extraction. The reverse transcription process was performed using the ABScript II cDNA First-Strand Synthesis Kit (RK20400, ABclonal). The subsequent qRT-PCR was conducted using the SYBR Green mix (G3326, Servicebio). The primers for the qPCR were obtained from TSINGKE, and the primer sequences are presented in Table 2.
Table 2.
Primer sequences used for qRT-PCR
| Name | Sequence (5’ ~ 3’) | Length |
|---|---|---|
| GPX4 (Forward Primer) | GAGGCAAGACCGAAGTAAACTAC | 23 |
| GPX4 (Reverse Primer) | CCGAACTGGTTACACGGGAA | 20 |
| NRF2 (Forward Primer) | TCAGCGACGGAAAGAGTATGA | 21 |
| NRF2 (Reverse Primer) | CCACTGGTTTCTGACTGGATGT | 22 |
| SLC7A11 (Forward Primer) | TCTCCAAAGGAGGTTACCTGC | 21 |
| SLC7A11 (Reverse Primer) | AGACTCCCCTCAGTAAAGTGAC | 22 |
| FTH1 (Forward Primer) | CCCCCATTTGTGTGACTTCAT | 21 |
| FTH1 (Reverse Primer) | GCCCGAGGCTTAGCTTTCATT | 21 |
| SCD1 (Forward Primer) | TCTAGCTCCTATACCACCACCA | 22 |
| SCD1 (Reverse Primer) | TCGTCTCCAACTTATCTCCTCC | 22 |
| CBS (Forward Primer) | GGCCAAGTGTGAGTTCTTCAA | 21 |
| CBS (Reverse Primer) | GGCTCGATAATCGTGTCCCC | 20 |
| FTMT (Forward Primer) | TGGAGTGTGCTCTACTCTTGG | 21 |
| FTMT (Reverse Primer) | ACGTGGTCACCTAGTTCTTTGA | 22 |
| MTOR (Forward Primer) | ATGCTTGGAACCGGACCTG | 19 |
| MTOR (Reverse Primer) | TCTTGACTCATCTCTCGGAGTT | 22 |
| GPER1 (Forward Primer) | CACCAGCAGTACGTGATCGG | 20 |
| GPER1 (Reverse Primer) | CATCTTCTCGCGGAAGCTGAT | 21 |
| GAPDH (Forward Primer) | GGAGCGAGATCCCTCCAAAAT | 21 |
| GAPDH (Reverse Primer) | GGCTGTTGTCATACTTCTCATGG | 23 |
Western blotting
The cells were fully lysed using the ice-cold RIPA lysis buffer [1% Nonidet P-40, 50 mM Tris-HCl (pH 7.4), 150 mM sodium chloride, and 0.5% sodium deoxycholate] mixed with 1% protease inhibitor and phosphatase inhibitor. The cell lysis was performed at 4 °C for 40 min, followed by centrifugation at 12,000 rpm/min for 25 min. The supernatants were harvested and subjected to protein concentration measurement using the BCA assay, followed by adding the loading buffer, mixing, and separating the proteins in the mixture on a 10% SDS-PAGE gel. The proteins separated on the gel were transferred to PVDF membranes, then the membrans had been blocked with 5% non-fat milk for 1 h at RT. Next, the membranes with proteins were incubated overnight with the primary antibody solution at 4 °C. After thorough washing followed by incubation with the HRP-conjugated secondary antibody solution for 1 h at RT, the protein bands were visualized and imaged using the ECL detection system (Bio-Rad Laboratories).
Immunohistochemistry
Tissues that were formalin-fixed, paraffin-embedded, and excised into 4-μm slices were examined by immunohistochemistry (IHC). After microwave heat-based antigen retrieval, the tissue sections were incubated overnight with primary antibodies and then with the secondary antibody. Next, the tissue sections were allowed to react with diaminobenzidine to develop color. Hematoxylin was added to the slides as a counterstain. In a double-blind process, the expression levels of GPER1 and SCD1 in the samples on the slides were determined, and the scoring system was logically modified as previously reported (Huang et al. 2018).
Tissue immunofluorescence
Polychrome immunofluorescence Kit (RC0086, Recordbio) was used according to the manufacturer’s instructions. After antigen retrieval, the tissue sections were sequentially incubated with primary antibodies overnight and then the secondary antibodies. Next, the fluorescence color development reaction was conducted. Each experiment was performed in triplicate. Finally, DAPI was added to stain the nuclei. Images were acquired using a scanner from Servicebio.
CCK8 assay
Cells were seeded in the wells of 96-well plates (same cell density in each well). The medium containing the drug was refreshed the next day, and the cell culture was continued at 37 °C for the next 48 h. Afterward, the cells were incubated for 30 min in a fresh medium containing 10% CCK8 reagent. The absorbance in each well was measured at 450 nm with a microplate scanner.
Plate cloning formation assay
A total of 1000 cells were seeded uniformly across the wells of 6-well plates, and the drug-containing medium was refreshed first on the third day and then every four days. The experiments were terminated when the appropriate cell mass was reached in 15th days. Then, the cells were fixed in 4% paraformaldehyde for ten minutes, followed by 15 min of staining using the crystal violet dye solution. After a thorough wash, images were acquired using a scanner.
Cell viability/cytotoxicity assay
Cells were seeded in the wells of 12-well plates (same cell density in each well) and the medium containing the drug was added the next day, and the cell culture was continued at 37 °C for the next 48 h. After removing the medium, an appropriate volume of Calcein AM/PI assay solution (C2015S, Beyotime) was added to plate and the cell was incubated at 37℃ in the dark for 20 min. The fluorescent microscope was used for observing and recording the staining effect.
Lipid peroxidation test
Lipid Peroxidation MDA Assay Kit (S0131M, Beyotime) was used according to the manufacturer’s instructions. The protein solutions were prepared using the lysate, and the BCA assay was performed. At 100 °C and in an acidic atmosphere, MDA and TBA combine to generate a red MDA-TBA adduct, and the resulting absorbance was measured at 532 nm using a microplate reader. The initial MDA content in the samples was expressed in protein content per weight.
BODIPY™ 581/591 C11 (D3861, Thermo Fisher) was used in the lipid peroxidation test. First, BODIPY™ 581/591 C11 was diluted to 10 μM. Next, 1 mL of the staining solution was added to each well of the 6-well plate, sufficient to cover the cells in the wells, following which the plate was placed in an incubator at 37 °C for 1 h. Subsequently, the cells were collected into flow tubes after washing away the excess staining solution using PBS. Afterward, the cells were resuspended with PBS containing 5% FBS. A flow cytometric analysis was performed, and the corresponding signals were measured at the FITC and PE channels.
Tumor formation assay
A549 cells (density = 4 × 106 cells) were infected with shGPER1 or negative control shRNA and then injected subcutaneously into 6-week-old nude mice. The growth status of these nude mice and the change in the size of the transplanted tumor were observed daily. In addition, the size change in the subcutaneous tumor of the nude mice was measured every 4 days beginning from the 4th day. Cisplatin treatment (2 mg/kg, i.p., q4d) was administered 5 times. The mice were euthanized at 28 days, and the tumors were retrieved for measurements after paraffin embedding and IHC staining.
FerrDb database
FerrDb (www.zhounan.org/ferrdb) (Zhou and Bao, 2020) is the world’s first database dedicated to the study of the association between ferroptosis regulators and diseases. FerrDb comprises seven independent datasets. In the present study, the “suppressor” dataset of FerrDb was searched for genes that prevent ferroptosis, and the top 8 genes were selected for validation.
Statistical analysis
The result data were analyzed using GraphPad Prism 9.03 statistical software. Unless otherwise stated, each experiment was conducted in triplicate, with comparable results. The unpaired two-tailed Student’s t-test and one-way analysis of variance were performed for inter-group comparisons. All results are expressed as mean ± SD (standard deviation). A p-value of < 0.05 was considered statistically significant. The relationship between GPER1 expression level and the prognosis of NSCLC patients was analyzed using the online database KMplotter (http://www.kmplot.com/lung) (Gyorffy et al. 2013).
Results
GPER1 promoted anti-oxidative stress and anti-lipid peroxidation in NSCLC cells
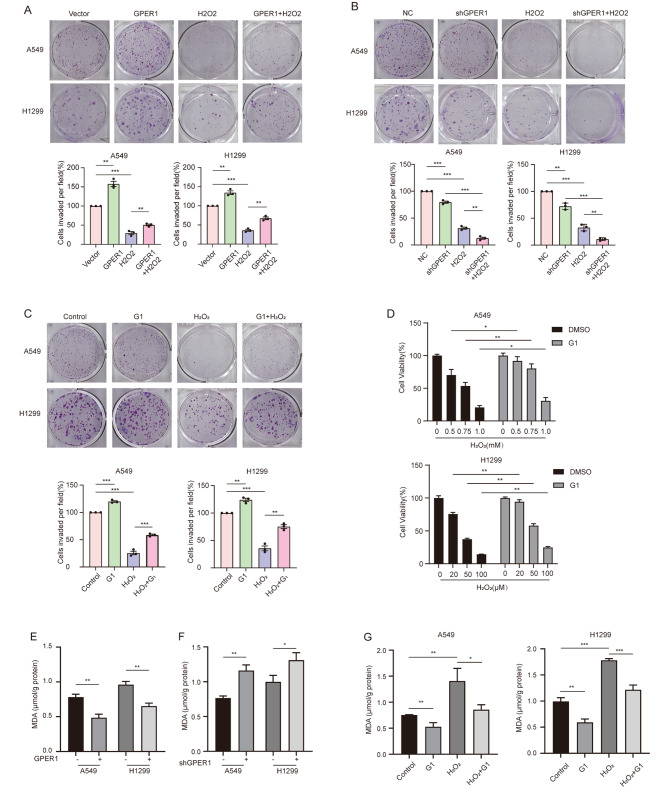
Anti-oxidative stress typically mediates tumor survival and proliferation. To explore the role of GPER1 in regulating oxidative stress, hydrogen peroxide (H2O2) was used to induce intracellular lethal oxidative stress in NSCLC cells, followed by the evaluation of whether GPER1 would exert an impact on cell viability. GPER1 overexpression improved the viability of A549 cells treated with 0.5, 0.75, and 1.0 mM H2O2 and H1299 cells treated with 20, 50, and 100 μM H2O2 (Figure S1A). In addition, GPER1 overexpression abrogated the reduction in cell proliferation in response to treatment with H2O2 (Fig. 1A). Conversely, GPER1 knockdown exacerbated the cytotoxicity of H2O2 and reduced the viability of the A549 and H1299 cell lines (Figure S1B). GPER1 knockdown significantly aggravated the damage to cell proliferation caused by H2O2 (Fig. 1B). Similar results were observed when the GPER1 agonist G1 was used (Fig. 1C and D). Moreover, GPER1 knockdown increased MDA levels, while GPER1 overexpression inhibited lipid peroxidation in the A549 and H1299 cell lines (Fig. 1E and F). G1 decreased the levels of MDA, a marker of lipid peroxidation. G1 also blocked the increase in the levels of MDA in response to treatment with H2O2 (Fig. 1G).
Fig. 1.
GPER1 promoted anti-oxidative stress and anti-lipid peroxidation in NSCLC cells. (A) The proliferation of A549 (250 μM) and H1299 (10 μM) cells with GPER1 overexpression and vectors that were treated with H2O2 was determined using the plate cloning formation assay. (B) The proliferation of GPER1-knockdown and negative control A549 (250 μM) and H1299 (10 μM) cells treated with H2O2 was determined using the plate cloning formation assay. (C) The proliferation of A549 and H1299 cells (as stated earlier) treated with G1 and H2O2 was determined using the plate cloning formation assay. (D) Cell viability was determined using the CCK8 assay after 48 h of G1 (1 μM) and H2O2 treatments in A549 and H1299 cells. (E, F) MDA levels in A549 and H1299 cells with GPER1 overexpression or knockdown. (G) MDA levels in A549 and H1299 cells after treatment with G1, H2O2, or both for 48 h. The results are presented as the mean ± SD. n = 3; *P < 0.05, **P < 0.01, ***P < 0.001
GPER1 protected NSCLC cells from ferroptosis
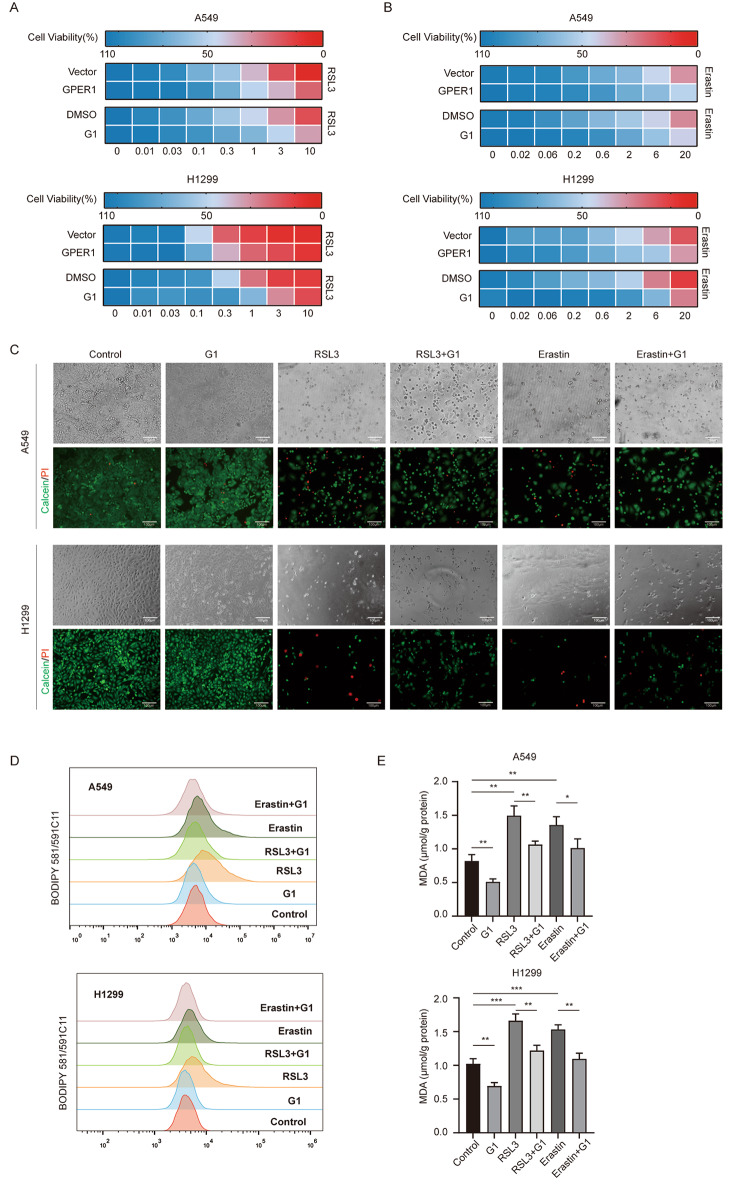
Lipid peroxidation is the most significant indicator of ferroptosis, and GPER1 reportedly reduces lipid peroxidation. Therefore, it was hypothesized that GPER1 could inhibit ferroptosis. To confirm this hypothesis, the effect of GPER1 on the cytotoxicity of ferroptosis inducers (RSL3 and Erastin) was evaluated in the present study. GPER1 overexpression and G1 treatment inhibited the cytotoxicity of RSL3 and Erastin on the A549 and H1299 cell lines (Fig. 2A and B, Figure S1C), and RSL3 exhibited more robust cytotoxicity. G1 also protected the cells from ferroptosis (Fig. 2C). Moreover, changes in lipid peroxidation levels were examined under the same conditions, and the results demonstrated that G1 significantly attenuated RSL3- and Erastin-induced lipid peroxidation (Fig. 2D and E). These results demonstrated that GPER1 protected NSCLC cells from ferroptosis.
Fig. 2.
GPER1 protected NSCLC cells from ferroptosis. (A, B) Heatmap showing that GPER1 reduced sensitivity to the ferroptosis inducers RSL3 and Erastin. Darker blue indicates higher cell viability, and darker red indicates worse viability. The number at the bottom of the heatmap indicates the concentration of G1. (C) Representative phase-contrast images and viability staining (green indicates live cells, and red indicates dead cells) of A549 and H1299 cells treated with G1, RSL3, or Erastin. (D) The fluorescence of BODIPY 581/591C11 in A549 and H1299 cells treated with G1, RSL3, or Erastin, was determined using flow cytometry. (E) MDA levels in A549 and H1299 cells treated with G1, RSL3, or Erastin. The results are presented as the mean ± SD. n = 3; *P < 0.05, **P < 0.01, ***P < 0.001
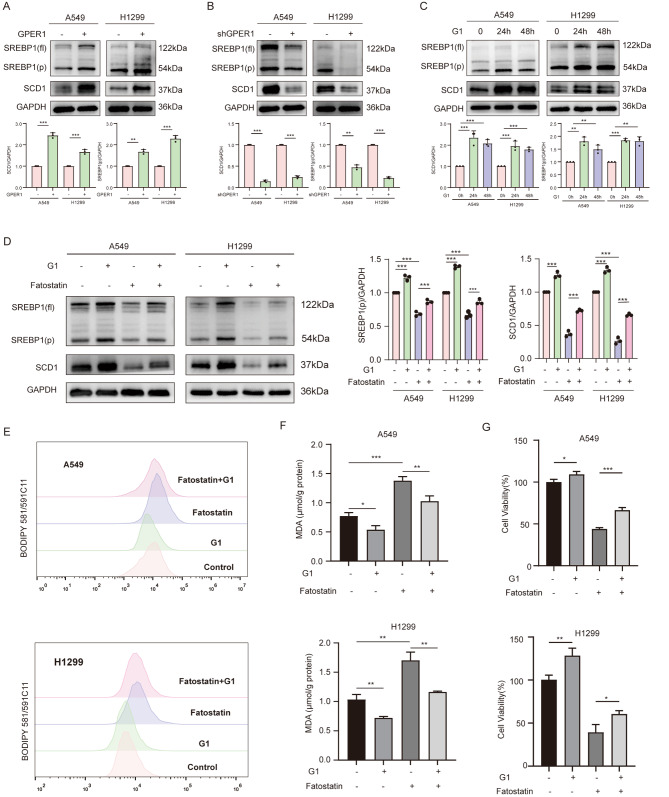
GPER1 prevented ferroptosis by upregulating SCD1
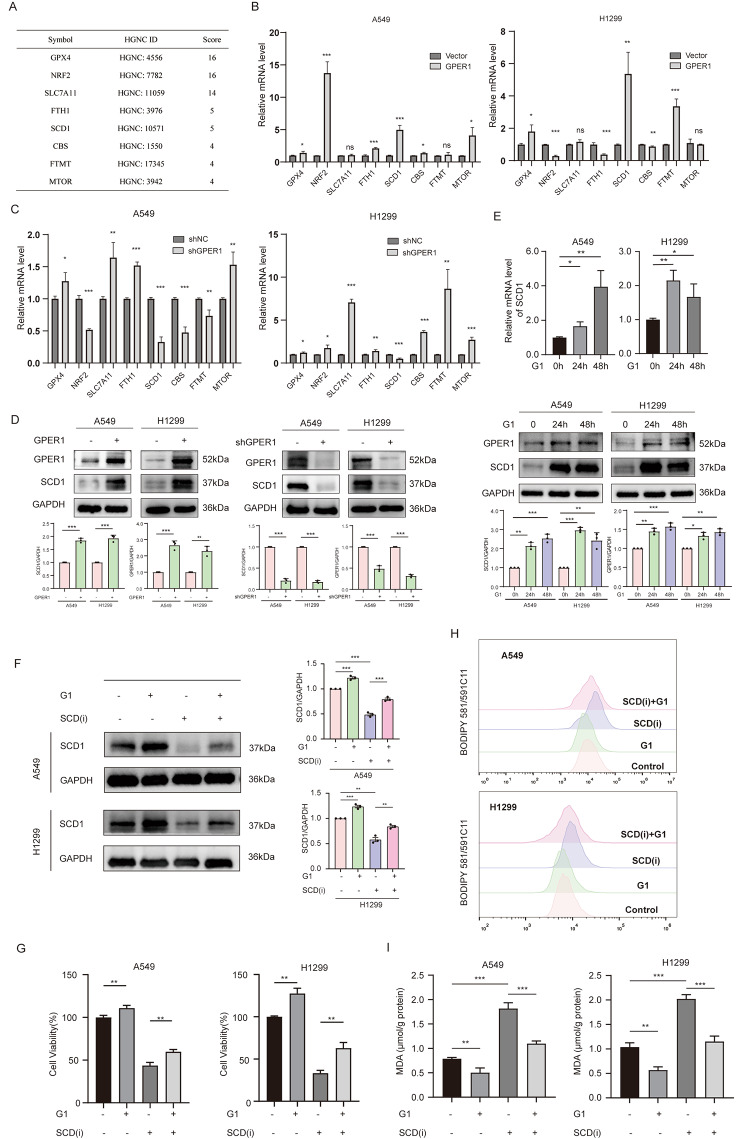
To determine how GPER1 inhibits ferroptosis, the FerrDb database was searched, and the top 8 genes in the ferroptosis suppressor gene module were selected for further analysis (Fig. 3A). The fold changes in mRNAs related to these top 8 genes in the A549 and H1299 cell lines in response to GPER1 overexpression and knockdown were assessed. When GPER1 was overexpressed, the mRNA expression levels of GPX4, NRF2, fth1, and SCD1 were significantly increased in A549 cells, while GPX4, SCD1, and FTMT expression was significantly increased in H1299 cells. In addition, when GPER1 was knocked down, only the expression of SCD1 was decreased in A549 and H1299 cells. These results suggest that GPER1 positively regulates SCD1 expression (Fig. 3B and C). SCD1 protein levels also exhibited the same trend (Fig. 3D). Moreover, G1 treatment for 24 and 48 h increased the mRNA and protein levels of SCD1 in A549 and H1299 cells (Fig. 3E). Therefore, it was hypothesized that GPER1 could prevent ferroptosis by promoting the expression of SCD1. Accordingly, the SCD1-specific inhibitor A939572 was used to block the SCD1 protein, followed by analysis. The results demonstrated that G1 could prevent the reduction in SCD1 protein expression caused by A939572 (Fig. 3F). G1 also promoted cell viability and inhibited the cytotoxicity of A939572 (Fig. 3G). Similarly, A939572 increased intracellular lipid peroxidation levels, while G1 inhibited lipid peroxidation caused by A939572 (Fig. 3H and I). Collectively, these results suggest that GPER1 upregulates SCD1 expression to prevent ferroptosis in NSCLC cells.
Fig. 3.
GPER1 prevented ferroptosis by upregulating SCD1. (A) The top 8 ferroptosis suppressors were searched in the FerrDb database. (B) The mRNA levels of the 8 ferroptosis suppressors after GPER1 overexpression in A549 and H1299 cells. (C) The mRNA levels of the 8 ferroptosis suppressors after GPER1 knockdown in A549 and H1299 cells. (D) The protein levels of SCD1 in GPER1-overexpressing or knockdown cells. (E) The mRNA and protein levels of SCD1 in A549 and H1299 cells treated with G1 for 24 and 48 h. (F) GPER1 and SCD1 levels were determined in A549 and H1299 cells that were treated with G1, A939572, or both for 48 h. (G) Cell viability was evaluated using the CCK8 assay. (H) The fluorescence of BODIPY 581/591C11 was determined using flow cytometry. (I) MDA levels in A549 and H1299 cells that were treated with G1, A939572, or both for 48 h. The results are presented as the mean ± SD. n = 3; *P < 0.05, **P < 0.01, ***P < 0.001
GPER1 promoted SCD1 transcription via SREBP1
As a membrane receptor, GPER1 is not directly involved in the transcription of any target genes. Therefore, it was hypothesized that GPER1 promotes SCD1 transcription through the activation of transcription factors that affect SCD1. Sterol regulatory element binding protein 1 (SREBP1) is a classic transcription factor that affects SCD1. Whether SREBP1 was involved in the regulation of SCD1 via GPER1 was investigated. The expression of full-length SREBP1(fl.) and its activated form SREBP1(p) were evaluated in the A549 and H1299 cell lines after GPER1 overexpression and knockdown. SREBP1(p) was significantly increased in GPER1-overexpressing cell lines and reduced in response to GPER1 knockdown (Fig. 4A and B). G1 treatment also increased the levels of SREBP1(p) in A549 and H1299 cells (Fig. 4C). Furthermore, fatostatin, an effective inhibitor of SREBP1 activation, was used to repress SREBP1(p) and inhibited the expression of SCD1. G1 significantly prevented the decrease in the levels of SREBP1(p), resulting in SCD1 induction by fatostatin (Fig. 4D). Fatostatin significantly increased intracellular lipid peroxidation levels, while G1 antagonized lipid peroxidation caused by fatostatin in A549 and H1299 cells (Fig. 4E and F), and the same held true for cell viability (Fig. 4G).
Fig. 4.
GPER1 promoted SCD1 transcription via SREBP1. (A, B) The protein levels of SREBP1(p) in GPER1-overexpressing or knockdown cells. (C) The protein levels of SREBP1 (p) in A549 and H1299 cells treated with G1 for 24 and 48 h. (D) SREBP1(p) levels were determined in A549 and H1299 cells that were treated with G1, fatostatin, or both for 48 h. (E) The fluorescence of BODIPY 581/591C11 in A549 and H1299 cells that were treated with G1, fatostatin, or both for 48 h was analyzed using flow cytometry. (F) MDA levels in A549 and H1299 cells that were treated with G1, fatostatin, or both for 48 h. (G) The viability of A549 and H1299 cells that were treated with G1, fatostatin, or both for 48 h was evaluated using a CCK-8 assay. The results are presented as the mean ± SD. n = 3; *P < 0.05, **P < 0.01, ***P < 0.001
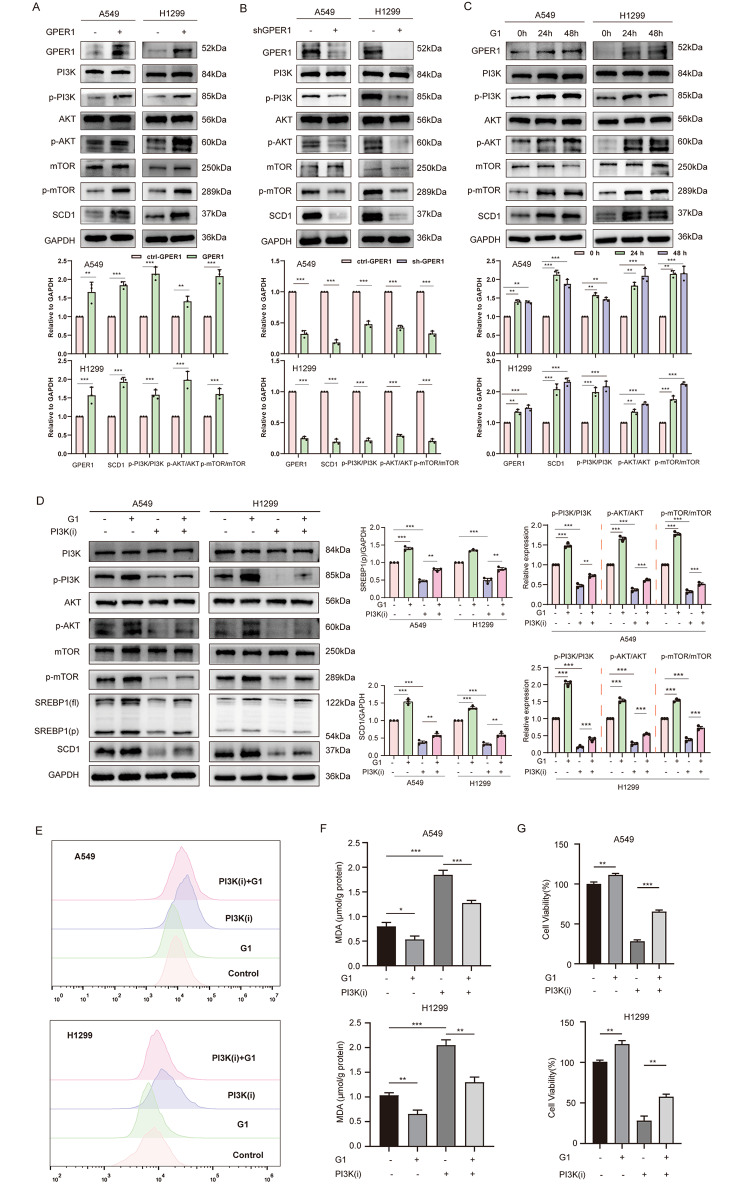
GPER1 promoted SREBP1-SCD1 via PI3K/AKT/mTOR signaling
GPER1 promotes tumor progression mainly by interacting with carcinogenic signaling pathways, such as the EGFR/ERK1/2 pathway, the PI3K/AKT/mTOR signaling pathway, and the MAPK/ERK signaling pathway. It was further revealed that excessive activation of the PI3K/AKT/mTOR signaling pathway could enhance ferroptosis resistance in lung cancer cells, and this was achieved by inducing the expression of SREBP1/SCD1. Therefore, it was hypothesized that GPER1 promoted SREBP1/SCD1 through the activation of PI3K/AKT/mTOR signaling. To verify this hypothesis, experiments were conducted, and GPER1 overexpression promoted activation of the PI3K/AKT/mTOR signaling pathway, while GPER1 knockdown reduced the expression of the PI3K/AKT/mTOR signaling pathway (Fig. 5A and B). G1 also significantly increased the expression of the PI3K/AKT/mTOR signaling pathway (Fig. 5C). NSC781406, a PI3K activity inhibitor, was used to block PI3K/AKT/mTOR signaling. The levels of SREBP1(p) and SCD1 were significantly reduced by NSC781406, and G1 prevented the decrease in the protein levels of SREBP1(p) and SCD1 caused by NSC781406 (Fig. 5D). NSC781406 significantly increased intracellular lipid peroxidation levels, while G1 antagonized lipid peroxidation caused by NSC781406 in A549 and H1299 cells (Fig. 5E and F); the same held true for cell viability (Fig. 5G). These results collectively indicated that GPER1 promoted SCD1 transcription by activating PI3K/AKT/mTOR signaling.
Fig. 5.
GPER1 promoted SREBP1-SCD1 via PI3K/AKT/mTOR signaling. (A, B) The activation of PI3K/AKT/mTOR signaling (the protein levels of p-PI3K, p-AKT, and p-mTOR) in the GPER1-overexpressing or knockdown cells. (C) The activation of PI3K/AKT/mTOR signaling (the protein levels of p-PI3K, p-AKT, and p-mTOR) in A549 and H1299 cells treated with G1 for 24 and 48 h. (D) The levels of SREBP1 (fl.), SREBP1 (p), and SCD1 and the activation of PI3K/AKT/mTOR signaling (the protein levels of p-PI3K, p-AKT, and p-mTOR) were determined in A549 and H1299 cells that were treated with G1, PI3K(i) (NSC781406), or both for 48 h. (E) The fluorescence of BODIPY 581/591C11 in A549 and H1299 cells that were treated with G1, PI3K(i) (NSC781406), or both for 48 h, as analyzed using flow cytometry. (F) MDA levels in the A549 and H1299 cells that were treated with G1, PI3K(i) (NSC781406), or both for 48 h. (G) The viability of A549 and H1299 cells that were treated with G1, PI3K(i) (NSC781406), or both for 48 h, determined using the CCK8 assay. The results are presented as the mean ± SD. n = 3; *P < 0.05, **P < 0.01, ***P < 0.001
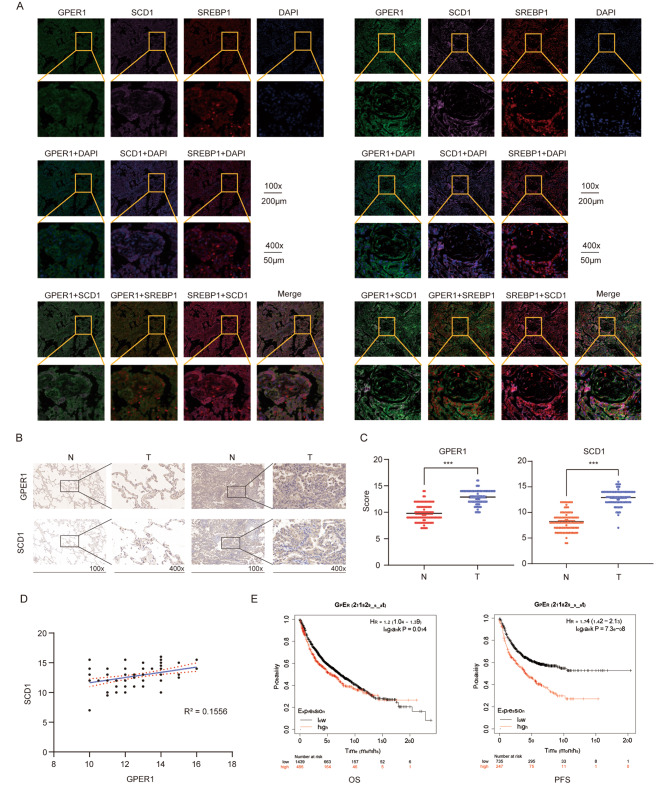
GPER1 correlated with SCD1 in NSCLC tissues
Previous studies have demonstrated that GPER1 is upregulated in lung cancer tissues. In the present study, the relationship among GPER1, SCD1, and SREBP1 in NSCLC tissues was investigated. A tissue immunofluorescence assay was performed, and GPER1, SCD1, and SREBP1 were labeled with different fluorescence signals. The results indicated that when GPER1 levels were high, SCD1 and SREBP1 were abundantly expressed (Fig. 6A). The IHC staining results indicated that the expression levels of GPER1 and SCD1 in tumor tissues were significantly higher than those in corresponding normal tissues (Fig. 6B and C). Furthermore, the correlation between GPER1 and SCD1 expression was determined. Correlation analysis showed a positive correlation between GPER1 and SCD1 expression in NSCLC tissues (Fig. 6D). Next, the association between the clinicopathological characteristics of NSCLC and GPER1 expression was investigated. High GPER1 expression was substantially associated with poor overall survival (OS) and progression-free survival (PFS) in NSCLC, as revealed by the analysis of Kaplan‒Meier survival curves and the log-rank test (Fig. 6E).
Fig. 6.
GPER1 correlated with SCD1 in NSCLC tissues. (A) An immunofluorescence assay was performed on GPER1 (Green, TYR-520), SCD1 (Purple, TYR-690), SREBP1 (Red, TYR-570) and nucleus (Blue, PADI) in the NSCLC tissue with different levels of GPER1 expression (the right panel shows higher levels and the left one shows lower levels). (B, C) The expression of GPER1 and SCD1 in tumor tissue and normal tissue were detected using immunohistochemistry. (D) The relationship between GPER1 and SCD1 expression in NSCLC tissue; R squared: 0.1556. (E) Overall survival (OS) and progression-free survival (PFS) of NSCLC cells with GPER1 expression. *P < 0.05, **P < 0.01, ***P < 0.001
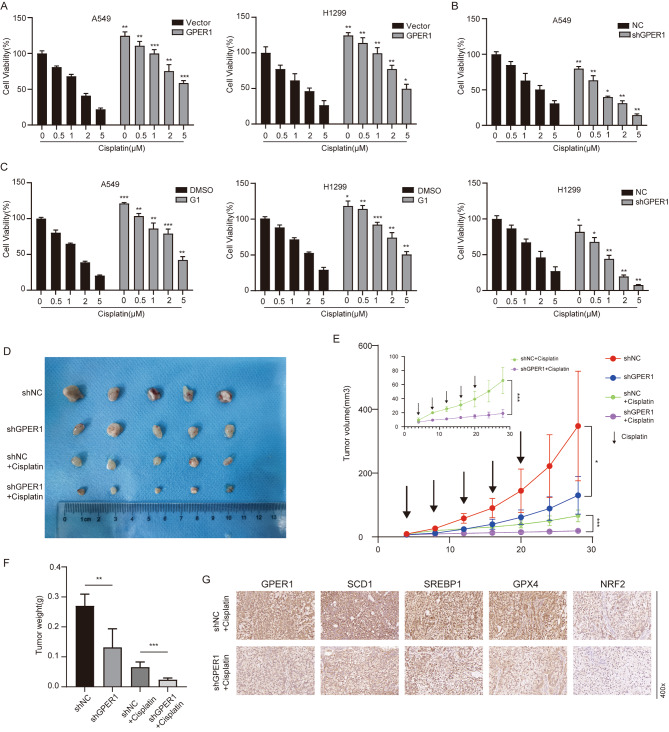
GPER1 deficiency potentiated the cytotoxic effect of cisplatin by inducing ferroptosis
Cisplatin is currently considered indispensable in the comprehensive treatment regimen for NSCLC. One of the cytotoxic mechanisms of cisplatin involves inducing intracellular oxidative stress. Studies have also demonstrated that cisplatin induces ferroptosis, and combination therapy with Erastin exerts a significant synergistic antitumor effect. Since GPER1 was shown to prevent ferroptosis in the present study, it was expected that the combination of GPER1 deficiency and cisplatin could enhance the antitumor activity of GPER1. To confirm this hypothesis, experiments were conducted, and GPER1 overexpression impaired the cytotoxic effects of cisplatin, while GPER1 knockdown enhanced the cytotoxic effects of cisplatin on A549 and H1299 cells (Fig. 7A and B). Combination treatment with G1 and cisplatin increased the viability of A549 and H1299 cells (Fig. 7C). Intriguingly, a subcutaneous graft model was established using A549 cells with GPER1 knockdown and negative control cells. GPER1 knockdown slowed tumor growth, resulting in reduced tumor size and weight (Fig. 7D, E and F). Moreover, GPER1 knockdown and cisplatin treatment exerted significant antitumor effects, and the tumor was in a state of growth arrest (Fig. 7D, E and F). Next, IHC staining of the subcutaneous graft was performed, and the results revealed that the expression levels of GPER1, SCD1, and SREBP1 were low in the GPER1 knockdown plus cisplatin treatment group. Correspondingly, GPX4 and NRF2, which are markers of ferroptosis, were also low (Fig. 7G). These results suggested that GPER1 deficiency enhanced the antitumor activity of cisplatin by promoting ferroptosis.
Fig. 7.
GPER1 deficiency potentiated the cytotoxic effect of cisplatin by inducing ferroptosis. (A, B) The viability of in A549 and H1299 cells with GPER1 overexpression or knockdown was determined using the CCK8 assay after treatment with cisplatin (0.5 μM, 1 μM, 2 μM, and 5 μM). (C) The viability of A549 and H1299 cells with or without G1 treatment was determined using the CCK8 assay after treatment with cisplatin (0.5 μM, 1 μM, 2 μM, and 5 μM). (D) Images of subcutaneous tumors from the indicated groups (n = 5 per group). (E) Tumor volumes in xenograft mice were measured at the indicated times. (F) Tumor weights in xenograft mice. (G) GPER1, SCD1, SREBP1, GPX4, and NRF2 expression in subcutaneous tumors from the indicated groups was determined using immunohistochemistry. The results are presented as the mean ± SD. n = 3; *P < 0.05, **P < 0.01, ***P < 0.001
Discussion
Substantial evidence indicates that estrogen is strongly linked to NSCLC. Specifically, estrogen triggers protumor effects by activating ERs. Initially, ERβ was believed to be the primary subtype in NSCLC, although antiestrogen therapy led to limited clinical efficacy due to the inhibition of ERβ signaling (Garon et al. 2018; Stabile et al. 2011). Consequently, GPER1, which is the most recently identified ER, has emerged as a target of therapeutic interest. In the present study, GPER1 was shown antagonize oxidative stress and reactive oxygen species. Additionally, it was demonstrated that inhibiting lipid peroxidation could maintain cell viability. Specifically, these findings suggested that GPER1 protected against iron death in NSCLC. Previous studies have shown that the GPER1 agonist G1 reduces ROS production in human umbilical vein endothelial cells stimulated by H2O2 and increases cell survival, while the GPER1 antagonist G15 further increases ROS production (Kong et al. 2014). Kaempferol increases the protein expression of GPER1, which, in turn, ameliorates atherosclerosis by decreasing intracellular ROS generation via the PI3K/AKT/Nrf2 pathway (Feng et al. 2021). In breast cancer, estrogen has been reported to inhibit ROS generation by inducing the expression of antioxidant genes (Cook et al. 2014; Tian et al. 2016). These findings and the hypothesis proposed in the present study were unexpected and were confirmed in NSCLC. Overall, the present study demonstrated that GPER1 in NSCLC promoted cell viability by inhibiting oxidation.
Ferroptosis may be regulated by a variety of factors and regulates cancer progression. A study investigated the role of RBMS1 in regulating ferroptosis in lung cancer through translational control of SLC7A11 (Zhang et al., 2021). However, few studies have examined estrogen and ERs in the regulation of ferroptosis. A study reported that ERα could prevent ferroptosis in breast cancer cells that had been exposed to ionizing radiation, and this was achieved through the NEDD4L/CD71 pathway (Liu et al. 2022). The present study, to the best of the author’s knowledge, is the first to report that GPER1 protects against ferroptosis in NSCLC.
SCD1 plays a crucial role in regulating iron death and can advance tumor growth by inhibiting ferroptosis. Recent studies have identified concurrent mutations in STK11 and KEAP1 that appear to protect against ferroptosis and promote SCD1 dependence in lung cancer (Wohlhieter et al. 2020). Tumor resistance to ferroptosis, which is driven by SCD1 in cancer cells or by fatty acid binding protein-4 (FABP4) in the tumor microenvironment, promotes tumor recurrence (Luis et al. 2021). The oncogenic activation of PI3K/AKT/mTOR signaling suppresses ferroptosis via SREBP1/SCD1-mediated lipogenesis (Yi et al. 2020). It has been reported that 17β-estradiol induces SCD1 expression in estrogen receptor-positive breast cancer cells, which suggests an interaction between ERs and SCD1 (Belkaid et al. 2015). Since SCD1 is downstream of PI3K/AKT/mTOR signaling, GPER1 must be involved in activation of the MAPK, PI3K/AKT, and Notch1 signaling pathways and promote NSCLC progression, indicating that GPER1 has a regulatory effect on SCD1 expression (Liu et al. 2015; Shen et al. 2021; Yi et al. 2020). In the present study, it was demonstrated that GPER1 was required to prevent ferroptosis in NSCLC, and it increased SREBP1 expression rather than stimulated its processing through PI3K/ATK/mTOR signaling, thereby promoting SCD1 transcription.
Cisplatin is indispensable in tumor therapy. It has been reported that GPER1-mediated epithelial-mesenchymal transition induces cisplatin resistance in gastric cancer cells (Wang et al. 2020). Moreover, cisplatin can induce ferroptosis in A549 cells, and combination therapy with cisplatin and erastin exerts a significant synergistic antitumor effect (Guo et al. 2018). Our study also demonstrated that GPER1 could reduce cisplatin cytotoxicity and that treatments targeting GPER1 combined with cisplatin could effectively enhance the antitumor activity of cisplatin by enhancing ferroptosis. In conclusion, the present study provides novel insights into the role of GPER1 in NSCLC and a novel strategy for the treatment of NSCLC.
Conclusion
The present study aimed to enhance our comprehension of estrogen signaling in the progression of NSCLC. Specifically, the findings suggest that GPER1 may be a promising target or biomarker for the treatment of NSCLC. We propose that GPER1 can prevent ferroptosis in NSCLC, which is achieved by promoting SCD1 transcription via PI3K/AKT/mTOR signaling. Furthermore, combining treatments that inhibit GPER1 with cisplatin is expected to enhance cisplatin’s antitumor activity by promoting ferroptosis in NSCLC. The present study provides a novel approach for treating NSCLC by targeting GPER1.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary Material 1: (A) The viability of in A549 and H1299 cells overexpressing GPER1 and the vector was determined using the CCK8 assay after 48 h of treatment with H2O2. (B) The viability of GPER1 knockdown and negative control A549 and H1299 cells was determined using the CCK8 assay after 48 h of treatment with H2O2. (C) The dose-viability curve revealed that GPER1 reduced sensitivity to the ferroptosis inducers RSL3 and Erastin. (D) GPER1, p-PI3K, p-AKT and p-mTOR expression in subcutaneous tumors in the GPER1-knockdown and control groups were determined using immunohistochemistry. The results are presented as the mean ± SD. n = 3; *P < 0.05, **P < 0.01, ***P < 0.001
Acknowledgements
Not applicable.
List of abbreviations
- ACSL4 Acyl
CoA synthetase long-chain family member 4
- DMSO
Dimethyl Sulfoxide
- ER
Estrogen recptor
- FABP4
Fatty acid binding protein-4
- GPER1
G-protein-coupled estrogen receptor 1
- GPX4
Glutathione peroxidase 4
- H2O2
Hydrogen peroxide
- HRP
Horseradish Peroxidase
- IHC
Immunohistochemistry
- NRF2
Nuclear factor-erythroid-2-related factor 2
- NSCLC
Non-small cell lung cancer
- PBS
Phosphate Buffer Saline
- RNA
Ribonucleic Acid
- ROS
Reactive oxygen species
- SCD1
Stearoyl CoA desaturase 1
- SREBP1
Sterol-regulatory-element binding protein 1
- WB
Western Blotting
Authors’ contributions
JPC, RZ, PYM and YDL designed this study. JPC, RZ, and YWW performed the experiments. RZ, PYM, HX, MXD, SWH, and WL analyzed the data. JPC, RZ, and YWW wrote the paper. All authors approved the final version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 82072593), and the Department of Science and Technology of Hubei Province (No. 2020BCB027).
Data Availability
The datasets and data used in this study can be obtained from the official website or corresponding author.
Declarations
Ethics approval and consent to participate
The study was approved by the Ethical Review Board of Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, and written informed consent was obtained from all subjects in accordance with the Declaration of Helsinki.
Consent
We declare that we have obtained written informed consent to publish the details from the affected individual.
Conflict of interest
The authors report no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jiaping Chen, Rong Zhao and Yangwei Wang contributed equally to this work.
Contributor Information
Peiyuan Mei, Email: m13260630812@163.com.
Yongde Liao, Email: liaoyongde@hust.edu.cn.
References
- Barton M, Filardo EJ, Lolait SJ, Thomas P, Maggiolini M, Prossnitz ER. Twenty years of the G protein-coupled estrogen receptor GPER: historical and personal perspectives. J Steroid Biochem Mol Biol. 2018;176:4–15. doi: 10.1016/j.jsbmb.2017.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid A, Duguay SR, Ouellette RJ, Surette ME. 17β-estradiol induces stearoyl-CoA desaturase-1 expression in estrogen receptor-positive Breast cancer cells. BMC Cancer. 2015;15:440. doi: 10.1186/s12885-015-1452-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchardy C, Benhamou S, Schaffar R, Verkooijen HM, Fioretta G, Schubert H, et al. Lung cancer mortality risk among Breast cancer patients treated with anti-estrogens. Cancer. 2011;117:1288–95. doi: 10.1002/cncr.25638. [DOI] [PubMed] [Google Scholar]
- Chen CS, Tseng YT, Hsu YY, Lo YC. Nrf2-Keap1 antioxidant defense and cell survival signaling are upregulated by 17beta-estradiol in homocysteine-treated dopaminergic SH-SY5Y cells. Neuroendocrinology. 2013;97:232–41. doi: 10.1159/000342692. [DOI] [PubMed] [Google Scholar]
- Chen X, Li J, Kang R, Klionsky DJ, Tang D. Ferroptosis: machinery and regulation. Autophagy. 2021;17:2054–81. doi: 10.1080/15548627.2020.1810918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Li B, Ou-Yang L. Role of estrogen receptors in health and Disease. Front Endocrinol (Lausanne) 2022;13:839005. doi: 10.3389/fendo.2022.839005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlebowski RT, Schwartz AG, Wakelee H, Anderson GL, Stefanick ML, Manson JE, et al. Oestrogen plus progestin and Lung cancer in postmenopausal women (women’s Health Initiative trial): a post-hoc analysis of a randomised controlled trial. Lancet. 2009;374:1243–51. doi: 10.1016/S0140-6736(09)61526-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook KL, Clarke PA, Parmar J, Hu R, Schwartz-Roberts JL, Abu-Asab M, et al. Knockdown of estrogen receptor-alpha induces autophagy and inhibits antiestrogen-mediated unfolded protein response activation, promoting ROS-induced Breast cancer cell death. FASEB J. 2014;28:3891–905. doi: 10.1096/fj.13-247353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–72. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Wang C, Yue, Jin, Meng Q, Wu J, et al. Kaempferol-induced GPER upregulation attenuates Atherosclerosis via the PI3K/AKT/Nrf2 pathway. Pharm Biol. 2021;59:1106–16. doi: 10.1080/13880209.2021.1961823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garon EB, Pietras RJ, Finn RS, Kamranpour N, Pitts S, Marquez-Garban DC, et al. Antiestrogen fulvestrant enhances the antiproliferative effects of epidermal growth factor receptor inhibitors in human non-small-cell Lung cancer. J Thorac Oncol. 2013;8:270–8. doi: 10.1097/JTO.0b013e31827d525c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garon EB, Siegfried JM, Stabile LP, Young PA, Marquez-Garban DC, Park DJ, et al. Randomized phase II study of fulvestrant and erlotinib compared with erlotinib alone in patients with advanced or metastatic non-small cell Lung cancer. Lung Cancer. 2018;123:91–8. doi: 10.1016/j.lungcan.2018.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Xu B, Han Q, Zhou H, Xia Y, Gong C, et al. Ferroptosis: a Novel Anti-tumor Action for Cisplatin. Cancer Res Treat. 2018;50:445–60. doi: 10.4143/crt.2016.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell Lung cancer. PLoS ONE. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Silva CD, Villegas-Pineda JC, Pereira-Suarez AL. Expression and role of the G Protein-Coupled Estrogen Receptor (GPR30/GPER) in the Development and Immune Response in Female Reproductive cancers. Front Endocrinol (Lausanne) 2020;11:544. doi: 10.3389/fendo.2020.00544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu LH, Chu NM, Lin YF, Kao SH. G-protein coupled estrogen receptor in breast cancer. Int J Mol Sci. 2019;20. 10.3390/ijms20020306 [DOI] [PMC free article] [PubMed]
- Huang Q, Zhang Z, Liao Y, Liu C, Fan S, Wei X, et al. 17beta-estradiol upregulates IL6 expression through the ERbeta pathway to promote lung adenocarcinoma progression. J Exp Clin Cancer Res. 2018;37:133. doi: 10.1186/s13046-018-0804-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorga A, Cunningham CM, Moazeni S, Ruffenach G, Umar S, Eghbali M. The protective role of estrogen and estrogen receptors in Cardiovascular Disease and the controversial use of estrogen therapy. Biol Sex Differ. 2017;8:33. doi: 10.1186/s13293-017-0152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacenik D, Beswick EJ, Krajewska WM, Prossnitz ER. G protein-coupled estrogen receptor in colon function, immune regulation and carcinogenesis. World J Gastroenterol. 2019;25:4092–104. doi: 10.3748/wjg.v25.i30.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jala VR, Radde BN, Haribabu B, Klinge CM. Enhanced expression of G-protein coupled estrogen receptor (GPER/GPR30) in Lung cancer. BMC Cancer. 2012;12:624. doi: 10.1186/1471-2407-12-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Mao C, Yang R, Yan B, Shi Y, Liu X, et al. EGLN1/c-Myc Induced lymphoid-specific helicase inhibits ferroptosis through lipid metabolic gene expression changes. Theranostics. 2017;7:3293–305. doi: 10.7150/thno.19988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Stockwell BR, Conrad M. Ferroptosis: mechanisms, biology and role in Disease. Nat Rev Mol Cell Biol. 2021;22:266–82. doi: 10.1038/s41580-020-00324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong BS, Cho YH, Lee EJ. G protein-coupled estrogen receptor-1 is involved in the protective effect of protocatechuic aldehyde against endothelial dysfunction. PLoS ONE. 2014;9:e113242. doi: 10.1371/journal.pone.0113242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZH, Liu C, Liu QH, Wang J, Wang Y, Wang YF, et al. Cytoplasmic expression of G protein-coupled estrogen receptor 1 correlates with poor postoperative prognosis in non-small cell Lung cancer. J Thorac Dis. 2022;14:1466–77. doi: 10.21037/jtd-22-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Liao Y, Fan S, Tang H, Jiang Z, Zhou B, et al. G protein-coupled estrogen receptor (GPER) mediates NSCLC progression induced by 17beta-estradiol (E2) and selective agonist G1. Med Oncol. 2015;32:104. doi: 10.1007/s12032-015-0558-2. [DOI] [PubMed] [Google Scholar]
- Liu C, Liao Y, Fan S, Fu X, Xiong J, Zhou S, et al. G-Protein-coupled estrogen receptor antagonist G15 decreases Estrogen-Induced Development of Non-small Cell Lung Cancer. Oncol Res. 2019;27:283–92. doi: 10.3727/096504017X15035795904677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhang C, Qu S, Liu R, Chen H, Liang Z, et al. ESR1 inhibits ionizing radiation-induced ferroptosis in Breast cancer cells via the NEDD4L/CD71 pathway. Arch Biochem Biophys. 2022;725:109299. doi: 10.1016/j.abb.2022.109299. [DOI] [PubMed] [Google Scholar]
- Luis G, Godfroid A, Nishiumi S, Cimino J, Blacher S, Maquoi E, et al. Tumor resistance to ferroptosis driven by Stearoyl-CoA Desaturase-1 (SCD1) in cancer cells and fatty acid biding Protein-4 (FABP4) in Tumor microenvironment promote Tumor recurrence. Redox Biol. 2021;43:102006. doi: 10.1016/j.redox.2021.102006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Liu D, Does GPER really function as a G protein-coupled estrogen receptor in vivo? Front Endocrinol (Lausanne) 2020;11:148. doi: 10.3389/fendo.2020.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magtanong L, Ko PJ, To M, Cao JY, Forcina GC, Tarangelo A, et al. Exogenous monounsaturated fatty acids promote a ferroptosis-resistant cell state. Cell Chem Biol. 2019;26:420–32. doi: 10.1016/j.chembiol.2018.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez-Garban DC, Chen HW, Fishbein MC, Goodglick L, Pietras RJ. Estrogen receptor signaling pathways in human non-small cell Lung cancer. Steroids. 2007;72:135–43. doi: 10.1016/j.steroids.2006.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Li C, Zhou L, Huang JA. G protein-coupled oestrogen receptor promotes cell growth of non-small cell Lung cancer cells via YAP1/QKI/circNOTCH1/m6A methylated NOTCH1 signalling. J Cell Mol Med. 2021;25:284–96. doi: 10.1111/jcmm.15997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatore CG, Chien JW, Au DH, Satia JA, White E. Lung cancer and hormone replacement therapy: association in the vitamins and lifestyle study. J Clin Oncol. 2010;28:1540–6. doi: 10.1200/JCO.2009.25.9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabile LP, Dacic S, Land SR, Lenzner DE, Dhir R, Acquafondata M, et al. Combined analysis of estrogen receptor beta-1 and progesterone receptor expression identifies Lung cancer patients with poor outcome. Clin Cancer Res. 2011;17:154–64. doi: 10.1158/1078-0432.CCR-10-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and Mortality Worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- Tian H, Gao Z, Wang G, Li H, Zheng J. Estrogen potentiates reactive oxygen species (ROS) tolerance to initiate carcinogenesis and promote cancer malignant transformation. Tumour Biol. 2016;37:141–50. doi: 10.1007/s13277-015-4370-6. [DOI] [PubMed] [Google Scholar]
- Toh CK, Ahmad B, Soong R, Chuah KL, Tan SH, Hee SW, et al. Correlation between epidermal growth factor receptor mutations and expression of female hormone receptors in East-Asian lung adenocarcinomas. J Thorac Oncol. 2010;5:17–22. doi: 10.1097/JTO.0b013e3181c0a602. [DOI] [PubMed] [Google Scholar]
- Wang X, Xu Z, Sun J, Lv H, Wang Y, Ni Y, et al. Cisplatin resistance in gastric cancer cells is involved with GPR30-mediated epithelial-mesenchymal transition. J Cell Mol Med. 2020;24:3625–33. doi: 10.1111/jcmm.15055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlhieter CA, Richards AL, Uddin F, Hulton CH, Quintanal-Villalonga A, Martin A, et al. Concurrent mutations in STK11 and KEAP1 promote Ferroptosis Protection and SCD1 dependence in Lung Cancer. Cell Rep. 2020;33:108444. doi: 10.1016/j.celrep.2020.108444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Yu S, Dong D, Lee LG, Protein-Coupled Estrogen receptor: a potential therapeutic target in Cancer. Front Endocrinol (Lausanne) 2019;10:725. doi: 10.3389/fendo.2019.00725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z, Zhuo Q, Hu Q, Xu X, Mengqi L, Zhang Z, et al. FBW7-NRA41-SCD1 axis synchronously regulates apoptosis and ferroptosis in Pancreatic cancer cells. Redox Biol. 2021;38:101807. doi: 10.1016/j.redox.2020.101807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J, Zhu J, Wu J, Thompson CB, Jiang X. Oncogenic activation of PI3K-AKT-mTOR signaling suppresses ferroptosis via SREBP-mediated lipogenesis. Proc Natl Acad Sci U S A. 2020;117:31189–97. doi: 10.1073/pnas.2017152117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang KS, Chen HQ, Chen YS, Qiu KF, Zheng XB, Li GC, et al. Bisphenol A stimulates human Lung cancer cell migration via upregulation of matrix metalloproteinases by GPER/EGFR/ERK1/2 signal pathway. Biomed Pharmacother. 2014;68:1037–43. doi: 10.1016/j.biopha.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Zhang C, Liu X, Jin S, Chen Y, Guo R. Ferroptosis in cancer therapy: a novel approach to reversing drug resistance. Mol Cancer. 2022;21:47. doi: 10.1186/s12943-022-01530-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Sun Y, Bai L, Zhi L, Yang Y, Zhao Q et al. RBMS1 regulates lung cancer ferroptosis through translational control of SLC7A11 2021;131. 10.1172/JCI152067 [DOI] [PMC free article] [PubMed]
- Zhou N, Bao J. FerrDb: a manually curated resource for regulators and markers of ferroptosis and ferroptosis-disease associations. Database (Oxford) 2020;2020. 10.1093/database/baaa021 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: (A) The viability of in A549 and H1299 cells overexpressing GPER1 and the vector was determined using the CCK8 assay after 48 h of treatment with H2O2. (B) The viability of GPER1 knockdown and negative control A549 and H1299 cells was determined using the CCK8 assay after 48 h of treatment with H2O2. (C) The dose-viability curve revealed that GPER1 reduced sensitivity to the ferroptosis inducers RSL3 and Erastin. (D) GPER1, p-PI3K, p-AKT and p-mTOR expression in subcutaneous tumors in the GPER1-knockdown and control groups were determined using immunohistochemistry. The results are presented as the mean ± SD. n = 3; *P < 0.05, **P < 0.01, ***P < 0.001
Data Availability Statement
The datasets and data used in this study can be obtained from the official website or corresponding author.