Abstract
Cellular senescence, a process that arrests the cell cycle, is a cellular response mechanism for various stresses and is implicated in aging and various age-related diseases. However, the understanding of senescence in living organisms is insufficient, largely due to the scarcity of sensitive tools for the detection of cellular senescence in vivo. Herein, we describe the development of a self-immobilizing near-infrared (NIR) fluorogenic probe that can be activated by senescence-associated β-galactosidase (SA-β-Gal), the most widely used senescence marker. The NIR signal is turned on only in the presence of SA-β-Gal, and the fluorescence signal is retained to the site of activation via in situ labeling, significantly enhancing the sensitivity of the probe. We demonstrate its efficient non-invasive imaging of senescence in mice xenograft models.
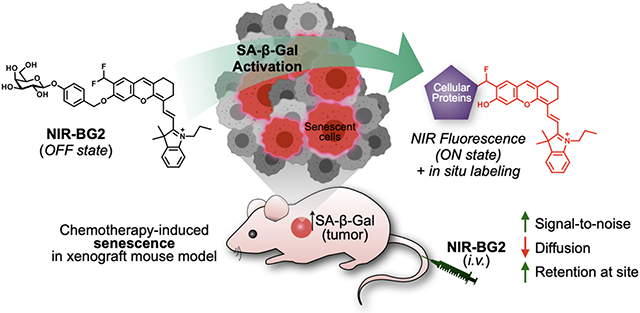
Graphical Abstract
INTRODUCTION
Cellular senescence, a non-proliferative but viable cellular state with prolonged and mostly irreversible cell-cycle arrest, not only acts as an endogenous tumor suppression mechanism, but also is a cellular response to various stresses, including telomere shortening, DNA damage, chromatin perturbation, and oncogenes activation1-4. Mounting evidence has linked cellular senescence with aging and numerous age-related diseases, such as cardiovascular disease, diabetes, neurodegenerative disorders, and fibrosis at various vital organs3-4. Therapeutic removal of senescent cells (also known as senotherapy) has drawn drastically increasing attention in recent years5. Monitoring the status of cellular senescence in living subjects allows the study of senescence in real-time without the need to terminate the experiments, enabling long-term study of age-related disease progression, and evaluation of treatment responses of both anticancer therapies and senolytic therapies6. However, the non-invasive detection of senescence in vivo has been very challenging, due to the lack of sensitive probes. Senescence is regularly characterized by the overexpression of cell cycle inhibitors, such as p16 and p212, 7, altered secretome (known as senescence associated secretory phenotype)8, and deregulated metabolism5. Among these features, senescence-associated β-galactosidase (SA-β-Gal)9, which is derived from the increased lysosomal content of senescent cells7, 10, has been the most widely used marker for senescence detection, and the detection of SA-β-Gal is mostly achieved with a colorimetric assay using 5-bromo-4-chloro-3-indoyl β-D-galactopyranoside (X-gal) as a chromogenic substrate11. However, this approach is limited to cells and fresh tissue sections10. Fluorescent probes12-16 developed for β-Gal detection in lacZ(+) (gene encoding β-Gal) cells can potentially be used for senescence detection, and some have been applied for the detection of SA-β-Gal in vitro17-21. However, these probes lack the capability of visualizing cellular senescence in living animals due to short-wavelength excitation/emission of fluorophores or poor sensitivity9.
NIR fluorescent probes offer high penetration depth, minimal photodamage to tissues, decreased background autofluorescence, and have been applied in noninvasive detection and imaging of biological targets in vivo22-24. We have previously developed a fluorogenic near-infrared (NIR) molecular probe NIR-BG and applied it in the imaging of drug-induced cellular senescence in human xenograft tumor models25. To obtain a sensitive NIR probe for senescence, we envisioned that a self-immobilizing group that could be activated by the target enzyme can further enhance the imaging efficiency by labeling the surrounding proteins in situ to retain the probe molecules to the site of activation while decreasing the rapid diffusion of the freed small molecules26 (Figure 1). In this paper, we demonstrated the first self-immobilizing turn-on NIR fluorescent probe, NIR-BG2, for enhanced real-time imaging of drug-induced senescence in vivo.
Figure 1.
Non-invasive imaging of chemotherapy-induced senescence in vivo using probes NIR-BG1 or NIR-BG2. Both probes are at their fluorescence OFF state without the target enzyme SA-β-Gal. Upon activation by SA-β-Gal, both probes become fluorescent. NIR-BG2 is converted to an intermediate with electrophilic quinone methide, which reacts with endogenous cellular proteins in situ, increasing the detection sensitivity.
RESULTS AND DISCUSSION
We therefore designed and chemically synthesized probe NIR-BG2 consisting of four moieties: a β-Gal substrate, a NIR fluorophore reporter, a self-immolative linker and a self-immobilizing moiety. Quinone methide chemistry has been successfully used in the design of covalent inhibitors27-28, immobilization of coumarin or rhodol tag29-35, and photo-controlled chemical cross-linking of proteins36-37. We incorporated a difluoromethyl group in the NIR fluorescent probe in which the hemicyanine skeleton is utilized as a NIR chromophore38 and a β-galactose residue is utilized as an enzyme recognizable trigger39, so that the electrophilic quinone methide species can be released upon activation by the SA-β-Gal, then trapped by the target enzymes or the nearby proteins to form a covalent linkage resulting in retained NIR signals. It remains challenging to develop NIR probes that can be turned on while possessing the feature to self-immobilize, as there is still a lack of efficient chemistry to install self-immobilizing functional groups under conditions that are compatible with NIR probes. In addition, even if the self-immobilizing fluorescent probe can be synthesized, the introduction of a self-immobilizing group may destabilize the fluorescent probe. For example, the QCy7-based probes40-41 bearing a self-immobilizing group was found to readily decompose42, such as -CHF2/-CH2F at the ortho position of an optically tunable hydroxyl group in NIR fluorophore. In our study, (E)-2-(2-(6-hydroxy-2,3-dihydro-1H-xanthen-4-yl)vinyl)-3,3-dimethyl-1-propyl-3H-indol-1-ium (HXPI) was employed as the NIR fluorophore due to its high quantum yield, photostability43, and membrane permeability38, 44-48. More importantly, HXPI was compatible with the chemistry for the incorporation of a quinone methide-based self-immobilizing group, and the final probe NIR-BG2 was very stable (Figure 1). To demonstrate the self-immobilizing characteristics, a control probe NIR-BG1 (similar to the first-generation probe NIR-BG), without the self-immobilizing properties, was also synthesized as well (Figure S1). The compounds were characterized by mass spectrometry, 1H, and 13C NMR in the supporting information. There are previous reports of monofluoromethylated probe having higher labeling efficiency than their difluoromethylated analogue30, 34. We also tested a monofluoromethylated analogue of this probe and to our surprise this analogue hydrolyzes quickly at room temperature (Figure S2E). In comparison NIR-BG2 showed much better stability and thus is more suitable for cell and in vivo studies (Fig. S2A-D).
We first investigated the spectroscopic properties of these two probes in PBS buffer with or without β-Gal. Probe NIR-BG1 (5 μM) exhibited typical absorption maximum of caged HXPI at 601 nm and 650 nm, and the absorption maximum for probe NIR-BG2 appeared at 596 nm and 644 nm (Figure 2). Upon the treatment with β-Gal, a remarkable bathochromic shift for both probes was observed. As expected, prior to β-Gal treatment, both probes NIR-BG2 and NIR-BG1 were almost nonfluorescent because the hydroxyl group of HXPI was caged with a β-Gal substrate, suppressing the intramolecular charge transfer (ICT) process that would lead to fluorescence. However, upon addition of β-Gal, probe NIR-BG1 produced a dramatic fluorescence enhancement (100-fold) over the background at 699 nm which can be attributed to the enzyme-triggered cleavage of the glycosylic bond to liberate the free hydroxyl group of NIR chromophore as a strong electron donor in the D-π-A system , thereby recovering ICT process and lighting up the fluorescence39. Whereas, NIR-BG2 exhibited a small fluorescence response (16-fold) at 709 nm to β-Gal due to the formation of formyl group partially quenching the fluorescence30, 32. To confirm the enzymatic hydrolysis mechanism, HPLC equipped with PDA detector and ESI mass spectrometry were utilized to analyze the enzymatic hydrolysis products of β-Gal treated probes. As shown in Figure S3, a new peak was observed at 15.84 min, corresponding to the NIR chromophore, in HPLC trace after incubation of 5 μM NIR-BG1 (15.23 min) with β-Gal (2 U) for 30 minutes. For the reaction of NIR-BG2 with β-Gal, the corresponding peak was found at 16.51 min. Also, the UV-vis spectrum of these peaks recorded by the PDA detector is in alignment with that of the corresponding NIR chromophores. Furthermore, these peaks were subjected to ESI mass analysis and a signal was observed at m/z 412.2284 [M]+ for NIR-BG1, 440.2295 [M]+ for NIR-BG2, respectively. We also investigated the effect of pH on the UV absorbance and fluorescent intensity of the probes. UV absorbance of both NIR-BG1 and NIR-BG2 showed blue-shift at acidic pH and red-shift at basic pH and the emission intensity of the activated probes are slightly decreased at acidic pH (Figure S4). While LacZ β-galactosidase has optimal activity near neutral pH, SA-β-gal is a lysosomal enzyme with optimal activity at around pH = 4. Thus, we also tested the activation of NIR-BG1 and NIR-BG2 by human β-galactosidase-1/GLB1 at pH = 3.5, which showed similar fast activation of the probes within 15 min (Figure S5). These results unambiguously confirmed the release of the NIR chromophore, resulting in a fluorescent response.
Figure 2.
Photophysical properties of the NIR probes. UV/Vis absorption and fluorescence spectra of probes NIR-BG1 (A, B) and NIR-BG2 (C, D) (5 μM) before (blue line) and after (black line) incubation with β-Gal (2 U) in PBS (pH 7.4) buffer for 5 min at 37 °C.
We then conducted the fluorescence-titration experiments with NIR-BG1 or NIR-BG2 and various concentrations of pure β-Gal (0.005–0.2 U/mL). The fluorescence intensity dramatically increased in the presence of a high concentration of β-Gal in both cases (Figure S6). Also, the fluorescence intensity against concentrations of β-Gal from0.006 to 0.2 U/mL exhibited a good relationship at 700 nm for NIR-BG1, and 708 nm for NIR-BG2, respectively. The regression equation is calculated as F700 nm = 324.5[β-Gal] +1.671 for NIR-BG1 and F708 nm = 554.9[β-Gal] + 0.452 for NIR-BG2 with a linear coefficient of 0.9957 and 0.9972 respectively. The kinetic parameters of the Michaelis constant (Km), the turnover number (kcat), and the catalytic efficiency constant (kcat/Km), were subsequently studied by monitoring the fluorescent intensity change at various concentrations of probes with β-Gal (0.1 U/mL) to obtain the enzymatic hydrolysis reaction rate (Figure S7). Then the kinetic parameters were determined by plotting the Lineweaver-Burk equation: 1/V0 = Km/kcat[E0][S] + 1/kcat[E0], where [E0] is the concentration of β-Gal. Thus, the kinetic parameters Km, kcat and kcat / Km were calculated to be 2.0 μM, 6.4 s−1, and 3.2 μM−1•s−1 for NIR-BG1; 9.3 μM, 14.6 s−1, and 1.6 μM−1•s−1 for NIR-BG2.
To validate the self-immobilization of probe NIR-BG2 to β-Gal upon activation, the fluorescence western blot analysis was carried out as it enables the quantification of β-Gal and probe concentration under different fluorescence channels to observe the colocalization of NIR-BG2 and β-Gal (via dye-labeled antibody) to verify the activation and self-immobilization of the probe. Also, a quantitative analysis could be performed to estimate the relationship between the β-Gal concentration and the probe activation and binding. The results showed that the bright NIR-BG2 fluorescence signal exactly colocalized with the signal from the anti-β-Gal antibody while only baseline signal was observed for the control probe NIR-BG1 (Figure 3 and Figure S8), which demonstrated the NIR-BG2 could be activated and linked to the β-Gal enzyme, while NIR-BG1 lacking the self-immobilizing handle could not. The quantification data indicated the linear correlation between the concentration of β-Gal enzyme and the fluorescence intensity of NIR-BG2, which demonstrated the ability of NIR-BG2 for quantitatively imaging the cellular senescence by measuring the concentration of β-Gal enzyme.
Figure 3.
Fluorescent western blot of NIR-BG1 and NIR-BG2 (5 μM) incubated with different concentrations of recombinant β-Gal for 4 hours. (A) Fluorescence western blot imaging. (B) Fluorescence intensity of the probes. (C) Fluorescence intensity from anti-β-Gal antibody.
To get a quantitative measure of the cellular uptake and activation of NIR-BG1 and NIR-BG2 in therapy-induced senescence in human cancer cells, flow cytometry analysis was performed. It showed significantly higher fluorescence intensity in senescent HeLa cells compared to normal HeLa cells (Figure 4A-4B and Figure S10). Cellular senescence in HeLa cells was induced by treatment with camptothecin (CPT) at 7.5 nM for 7 days as confirmed by X-gal staining (Figure. S9). In a parallel experiment, we also confirmed the uptake and activation of the two probes using CT26.CL25 cells and CT26.WT cells, with or without β-Gal encoding lacZ gene; both probes had remarkably higher fluorescence turn-on signal in CT26.CL25 cells than in the control CT26.WT cells (Figure S11-12). Importantly, the fluorescence signal from NIR-BG2 was significantly higher than that from NIR-BG1 in both CT26.CL25 cells (expressing endogenous β-Gal) and senescent HeLa cells (induced expression of SA-β-Gal), demonstrating the continuous activation and accumulation of NIR-BG2 due to its self-immobilizing activity. The viability of the cells was unaffected after incubation with NIR-BG1 or NIR-BG2 (Figure S13). These results further suggested the potential of using NIR-BG2 to monitor cellular senescence.
Figure 4.
(A) Flow cytometric analyses of untreated and CPT-treated HeLa cells incubated with NIR-BG1 or NIR-BG2 for 4 h. (B) Quantitative analysis of HeLa cells with or without CPT treatment incubated with NIR-BG1 or NIR-BG2 at 10 min, 1 h and 4 h. (λex/λem = 642 nm/675±25 nm) Fluorescent microscope images of (C) untreated and (D) CPT-treated HeLa cells incubated with 5 μM NIR-BG1 or NIR-BG2 for 2 hours, followed by washout for another 4 hours. (λex/λem = 395 nm/460 nm for DAPI, 470 nm/535 nm for β-Gal and 740 nm/767 nm for probe). Scale bar: 10 μm.
To investigate the imaging efficiency and the difference between NIR-BG2 and NIR-BG1 for senescence detection, fluorescence microscopy imaging of HeLa cells with or without chemotherapy-induced senescence was performed (Figure 4C-D). It is worth to note that the CPT treated HeLa cells were significantly enlarged in size compared to normal HeLa cells, indicating their morphological change upon senescence induction. Although both NIR-BG1 and NIR-BG2 produced remarkably higher fluorescence in CPT treated cells, only NIR-BG2 showed retained fluorescence signals in senescent HeLa cells after an extra probe washout step, and the fluorescence signal of NIR-BG2 was highly colocalized with β-Gal immunostaining (Figure 4C-D). The dynamic clearance of probes in CT26.CL25 cells showed that NIR-BG1 could be cleared out within 24 h while NIR-BG2 could accumulate in the cells for more than 24 hours (Figure S12A). The photostability of NIR-BG2 was measured by monitoring its fluorescent intensity in CT26.CL25 cells with continuous excitation. The fluorescent intensity showed about only 15 percent decrease after 30 min (Figure S14). We also tested NIR-BG1 and NIR-BG2 in doxorubicin-induced senescence in MDA-MB-231 cells, CPT-induced senescence in MCF7 cells and peroxide induced senescence in IMR-90 cells. The retention of NIR-BG2 was similarly higher in the senescent MDA-MB-231 cells and MCF7 cells than their untreated control cells; while NIR-BG1 did not provide contrast in the senescent cells after prolonged incubation and washing steps (Figure S15-17). These results suggested retained fluorescence in senescent cells as a result of the attachment of the activated NIR-BG2 to the SA-β-Gal and the surrounding proteins. Collectively, probe NIR-BG2 could be activated by SA-β-Gal to turn on its NIR fluorescence signal, meanwhile its self-immobilizing chemistry further enhanced the imaging efficiency and prolonged the probe retention at site of activation, suggesting its suitability for enhanced in vivo imaging of cellular senescence.
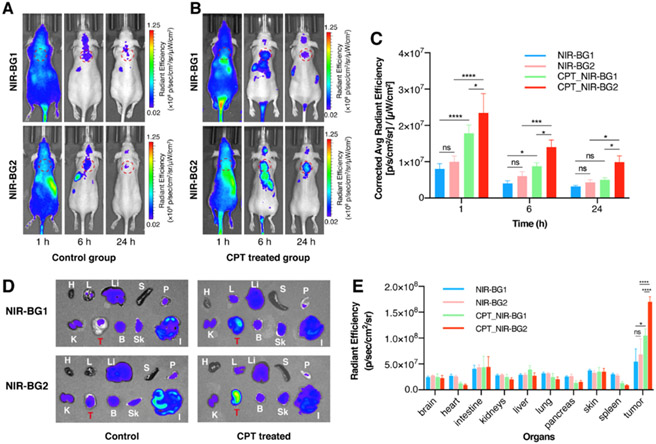
In vivo fluorescence imaging of HeLa xenografts was performed to evaluate the capability of NIR-BG2 to visualize chemotherapy-induced senescence in human tumor xenograft models. HeLa tumor-bearing mice were treated with chemotherapy to induce cellular senescence following a previously reported method by our group25. The CPT-treated mice showed significantly increased fluorescence intensity in the tumor compared to the control group receiving saline (Figure 5A-B). The quantification of the optical imaging showed that the fluorescence intensity at 24 h from the tumors with or without drug treatment was (1.04 ± 0.15) × 107 and (0.53 ± 0.13) × 107 (NIR-BG2 vs. NIR-BG1) (Figure 5C). Importantly, NIR-BG2 showed significantly higher fluorescence signal than NIR-BG1 in drug treated tumors due to in situ labelling of senescent cells. The ex vivo imaging (Figure 5D) and quantitative (Figure 5E) analysis of the tumors and organs further confirmed the in vivo imaging results. X-gal staining (Figure 6A) and fluorescence imaging of the tumor slices (Figure 6B) validated the probes were retained in tumor tissue receiving chemotherapy, suggesting NIR-BG2 superior to NIR-BG1 for detecting cellular senescence in vivo, due to prolonged signal retention.
Figure 5.
Live animal imaging of (A) saline-treated (control) and (B) CPT-treated HeLa xenografts mice after tail vein injection of NIR-BG1 (top) or NIR-BG2 (bottom) at 1 h (left), 6 h (middle) and 24 h (right). (C) The quantitative analysis of the in vivo fluorescence imaging at 1 h, 6 h and 24 h post injection. Ex vivo imaging (D) and quantification (E) of tumors and major organs of the control and CPT treated animals. (H = heart, L = lung, Li = liver, S = spleen, P = pancreas, K = kidneys, T = tumor, B = brain, Sk = skin, I = intestine) (λex/λem = 675 nm/720 ± 20 nm) (* p<0.05, *** p<0.0005, **** p<0.0001).
Figure 6.
Pathological staining of tumor sections of saline-treated (control) and CPT-treated HeLa xenografts mice. (A) Conventional X-Gal (blue) staining for SA-β-Gal activity and eosin (pink) staining of HeLa tumor slides without (control) or with CPT treatment. (B) Fluorescent imaging of tumor slice sections receiving i.v. injected NIR-BG1 (top) or NIR-BG2 (bottom) (λex/λem = 675 nm/720 ± 20 nm). Scale bar: 50 μm.
CONCLUSIONS
In summary, we have developed a self-immobilizing NIR probe for the imaging of cellular senescence in living animals. This self-immobilizing NIR probe was efficiently activated by SA-β-Gal to produce intense fluorescence signals both in vitro and in vivo. Compared with the control probe NIR-BG1 lacking the immobilizing group, probe NIR-BG2 showed stronger NIR fluorescence after activation in the senescent cells and longer retention inside the senescent cells. Importantly, NIR-BG2 showed significant prolonged retention in animal models induced with senescence, offering a wider time window to allow the clearance of background fluorescence signals. We anticipate that the combined advantage of the NIR optical probe and long-term tracking makes our self-immobilizing probe useful for investigating the senescence in living organisms.
EXPERIMENTAL SECTION
General materials and instruments.
Solvents and chemicals were purchased from commercial sources and used directly without further purification. Flash chromatography was performed manually with Agela Technologies Flash Silica (40-60 μm, 60 Angstroms). HPLC was performed on a Dionex UltiMate 3000 with a pump and an in-line Diode Array Detector (DAD-3000). A reverse-phase C18 (Teledyne, 5 μm, 10 x 250 mm) column was used for analysis and semi-preparation. All compounds are >95% pure by HPLC analysis. Deuterated solvents were purchased from Sigma-Aldrich and Merck Millipore. 1H NMR spectra were recorded using either a Bruker Avance III 300, Bruker Avance 500, or a Bruker Avance Neo 600, and chemical shifts were reported in ppm with either TMS or deuterated solvents as internal standards (1H: δ 0.00 for TMS, δ 7.26 for CDCl3, δ 3.31 for CD3OD respectively). The following are used for multiplicities: s, singlet; d, doublet; t, triplet; m, multiplet; and dd, doublet of doublet.) 13C NMR spectra were recorded at 75.4 or 125.7 MHz, and chemical shifts were reported in ppm with deuterated solvents as internal standards (13C: δ 77.16 for CDCl3, δ 49.0 for CD3OD respectively). High resolution mass spectrometry was recorded on Waters LCT Premier Mass Spectrometer. UV-vis spectra were collected on a Shimadzu UV-2700 spectrophotometer. Absorption spectra were taken on a Shimadzu UV-1800 UV-VIS Spectrophotometer. Fluorescence spectra were recorded on Edinburgh FLS980 fluorescence spectrometer and Shimadzu RF-5301pc spectrophotometer. β-Galactosidase (E. coli.) (catalog. P5269) was purchased from Abnova. 4’,6-diamidino-2-phenylindole (DAPI) was purchased from Biotium, CA, USA. Bovine serum albumin (BSA), Fetal Bovine Serum (FBS), and PBS were obtained from VWR, PA, USA. Dulbecco's Modified Eagle's medium (DMEM) and Eagle's minimal essential medium (EMEM) were from Corning Inc, USA. β-Galactosidase (E. coli) was purchased from Abnova (catalog #P5269). Goat anti-rabbit IgG H&L-AF488 (Catalog #ab150077) was purchased from Abcam (USA). Anti-β-Galactosidase antibody (Catalog # A-11132), and goat anti-rabbit IgG H&L Secondary Antibody, HRP (Catalog # 65-6120), ActinGreen 488 ReadyProbes (Catalog # R37110) and ProLong Gold Antifade Mountant were purchased from Invitrogen, USA. Radioimmunoprecipitation assay (RIPA) cell lysis buffer was from Enzo Life Sciences, NY, USA. HeLa (human cervical cancer cell line), CT26.WT (wild type mouse colon fibroblast carcinoma cells), CT26.CL25 (lacZ+ CT26 cell, engineered cells that highly express β-gal), MCF7 and IMR-90 cell lines were purchased from American Type Culture Collection (ATCC), VA, USA. Mini-PROTEAN® TGX™ Precast Gels and polyvinylidene fluoride (PVDF) membrane were purchased from Bio-Rad, USA. Dimethyl sulfoxide (DMSO) was from Sigma-Aldrich, USA.
Chemical Synthesis.
(E)-2-(2-(7-formyl-6-hydroxy-2,3-dihydro-1H-xanthen-4-yl)vinyl)-3,3-dimethyl-1-propyl-3H-indol-1-ium (2b) was prepared following a literature procedure.49 1H NMR (500 MHz, CDCl3) δ 10.04 (s, 1H), 8.66 (d, J = 15.1 Hz, 1H), 7.65 (s, 1H), 7.54 – 7.50 (m, 2H), 7.48 – 7.45 (m, 1H), 7.38 (d, J = 7.8 Hz, 1H), 7.08 (s, 1H), 6.98 (s, 1H), 6.59 (d, J = 15.1 Hz, 1H), 4.36 (t, J = 7.3 Hz, 2H), 2.72 (t, J = 6.5 Hz, 2H), 2.68 (t, J = 6.5 Hz, 2H), 1.99 – 1.92 (m, 4H), 1.81 (s, 6H), 1.07 (t, J = 7.5 Hz, 3H). 13C NMR (126 MHz, CDCl3) δ 193.55, 179.00, 165.14, 159.63, 157.99, 146.53, 142.37, 141.35, 131.81, 131.35, 129.49, 128.29, 128.10, 122.81, 119.81, 115.86, 115.35, 113.20, 106.01, 104.16, 51.34, 47.43, 29.24, 28.14, 23.97, 21.62, 20.30, 11.49. HRMS (ESI) Calcd. For C29H30NO3+ [M]+: 440.2220; found: 440.2215.
3,3-dimethyl-1-propyl-2-((E)-2-(6-((4-(((2S,3R,4S,5S,6R)-3,4,5-triacetoxy-6-(acetoxymethyl)tetrahydro-2H-pyran-2-yl)oxy)benzyl)oxy)-2,3-dihydro-1H-xanthen-4-yl)vinyl)-3H-indol-1-ium (3a).
To a solution of compound 2a (27.0 mg, 0.05 mmol), K2CO3 (8 mg, 0.06 mmol) and NaI (14.9 mg, 0.1 mmol) in 2 mL of DMF, 1 (31 mg, 0.06 mmol) was added. Then the reaction mixture was stirred at room temperature in the dark for 3 days. HPLC was used to monitor reaction process. Once the reaction was completed. The reaction mixture was purified by HPLC to afford 3a (18.0 mg, 37%). 1H NMR (500 MHz, CDCl3) δ 8.66 (d, J = 14.8 Hz, 1H), 7.51-7.47 (m, 2H), 7.43–7.37 (m, 4H), 7.30 (d, J = 7.9 Hz, 1H), 7.24 (s, 1H), 7.06 (d, J = 8.6 Hz, 2H), 6.97-7.96 (m, 2H), 6.35 (d, J = 14.8 Hz, 1H), 5.51 (dd, J = 10.4, 8.0 Hz, 1H), 5.47 (d, J = 3.2 Hz, 1H), 5.14- 5.12 (m, 3H), 5.09 (d, J = 7.9 Hz, 1H), 4.26 – 4.20 (m, 3H), 4.16 (dd, J = 11.2, 6.4 Hz, 1H), 4.09 (dd, J = J = 6.6 Hz, 1H), 2.75 (t, J = 5.5 Hz, 2H), 2.65 (t, J = 5.9 Hz, 2H), 2.19 (s, 3H), 2.08 (s, 3H), 2.06 (s, 3H) 2.02 (s, 3H), 1.96 – 1.93 (m, 4H), 1.79 (s, 6H), 1.07 (t, J = 7.4 Hz, 3H). 13C NMR (126 MHz, CDCl3) δ 177.42, 170.62, 170.39, 170.31, 169.64, 162.34, 162.19, 157.19, 154.59, 145.96, 141.76, 141.55, 134.30, 130.60, 129.63, 129.33, 128.97, 127.54, 127.48, 122.72, 117.28, 116.13, 114.92, 113.83, 112.44, 103.36, 102.07, 99.65, 71.17, 70.92, 70.53, 68.72, 66.97, 61.42, 50.73, 46.84, 29.85, 29.24, 28.30, 24.10, 21.29, 20.87, 20.80, 20.73, 20.32, 11.53. HRMS (ESI) Calcd. For C49H54NO12+ [M]+: 848.3641; found: 848.3608.
2-((E)-2-(7-formyl-6-((4-(((2S,3R,4S,5S,6R)-3,4,5-triacetoxy-6-(acetoxymethyl)tetrahydro-2H-pyran-2-yl)oxy)benzyl)oxy)-2,3-dihydro-1H-xanthen-4-yl)vinyl)-3,3-dimethyl-1-propyl-3H-indol-1-ium (3b) was synthesized in 32% yield with a similar procedure for 3a. 1H NMR (500 MHz, CDCl3) δ 10.41 (s, 1H), 8.70 (d, J = 15.0 Hz, 1H), 7.84 (s, 1H), 7.58 (d, J = 7.2 Hz, 1H), 7.52 – 7.47 (m, 2H), 7.45 (d, J = 8.3 Hz, 2H), 7.36 (d, J = 7.7 Hz, 1H), 7.15 (s, 1H), 7.07 (s, 1H), 7.06 (d, J = 8.6 Hz, 2H), 6.49 (d, J = 15.0 Hz, 1H), 5.50 (dd, J = 10.3, 8.1 Hz, 1H), 5.47 (d, J = 3.2 Hz, 1H), 5.29 (s, 2H), 5.13 (dd, J = 10.5, 3.4 Hz, 1H), 5.10 (d, J = 7.9 Hz, 1H), 4.31 (t, J = 7.3 Hz, 2H), 4.23 (dd, J = 11.2, 6.9 Hz, 1H), 4.16 (dd, J = 11.2, 6.3 Hz, 1H), 4.10 (t, J = 6.6 Hz, 1H), 2.73 (t, J = 5.8 Hz, 2H), 2.66 (t, J = 5.8 Hz, 2H), 2.18 (s, 3H), 2.08 (s, 3H), 2.06 (s, 3H), 2.02 (s, 3H), 1.99 – 1.93 (m, 4H), 1.82 (s, 6H), 1.08 (t, J = 7.4 Hz, 3H). 13C NMR (126 MHz, CDCl3) δ 187.90, 179.08, 170.58, 170.38, 170.26, 169.63, 163.73, 160.06, 158.09, 157.22, 146.62, 142.45, 141.23, 131.75, 130.16, 129.52, 129.39, 128.62, 128.40, 127.87, 123.06, 117.30, 115.69, 115.43, 112.96, 105.48, 101.00, 99.64, 71.30, 71.23, 70.96, 68.74, 67.01, 61.46, 51.40, 47.32, 29.39, 27.94, 24.19, 21.54, 20.88, 20.79, 20.78, 20.73, 20.24, 11.55. HRMS (ESI) Calcd. For C50H54NO13+ [M]+: 876.3590; found: 876.3686.
3,3-dimethyl-1-propyl-2-((E)-2-(6-((4-(((2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl)oxy)benzyl)oxy)-2,3-dihydro-1H-xanthen-4-yl)vinyl)-3H-indol-1-ium (NIR-BG1).
To a solution of 3a (18 mg, 0.0187 mmol) in methanol (1 mL), NaOMe (0.43 μL) was added. The reaction was monitored with HPLC. After starting material was consumed, the crude product was purified with HPLC to afford NIR-BG1 in 68% yield. 1H NMR (500 MHz, CD3OD) δ 8.77 (d, J = 15.0 Hz, 1H), 7.68 (d, J = 7.5 Hz, 1H), 7.55-7.52 (m, 2H), 7.49–7.42 (m, 4H), 7.40 (s, 1H), 7.18-7.15 (m, 2H), 7.09 (d, J = 2.2 Hz, 1H), 7.04 (dd, J = 8.6, 2.4 Hz, 1H), 6.52 (d, J = 14.9 Hz, 1H), 5.21 (s, 2H), 4.88 (d, J = 8.0 Hz, 1H), 4.33 (t, J = 7.5 Hz, 2H), 3.90 (d, J = 3.0 Hz, 1H), 3.82-3.73 (m, 3H), 3.69 (dd, J = 6.8, 5.7 Hz, 1H), 3.58 (dd, J = 9.7, 3.4 Hz, 1H), 2.79 (t, J = 6.0 Hz, 2H), 2.72 (t, J = 6.0 Hz, 2H), 1.99-1.92 (m, 4H), 1.83 (s, 6H), 1.08 (t, J = 7.5 Hz, 3H). 13C NMR (126 MHz, CD3OD) δ 179.29, 163.79, 163.13, 159.30, 155.82, 147.08, 143.50, 143.06, 135.17, 131.44, 130.29, 130.21, 130.06, 128.61, 128.40, 123.84, 117.94, 117.29, 115.64, 115.40, 113.92, 104.75, 102.91, 102.68, 77.06, 74.85, 72.24, 71.55, 70.20, 62.44, 52.02, 47.49, 30.05, 28.38, 25.03, 22.25, 21.65, 11.58. HRMS (ESI) Calcd. For C41H46NO8+ [M]+: 680.3218; found: 680.3215.
2-((E)-2-(7-(difluoromethyl)-6-((4-(((2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl)oxy)benzyl)oxy)-2,3-dihydro-1H-xanthen-4-yl)vinyl)-3,3-dimethyl-1-propyl-3H-indol-1-ium (NIR-BG2).
To a solution of 3b (5.4 mg, 5.3 umol) in dichloromethane (1 mL), DAST (23 uL, 0.16 mmol) was added. The reaction was stirred at room temperature and monitored with HPLC. Once the starting material was consumed, the reaction was quenched with methanol for 1 h to consume remaining DAST. Next, the solvents were removed under reduced pressure, and the residue was dissolved in methanol. NaOMe was added to this mixture until the reaction was completed. Then the mixture was purified with HPLC to afford NIR-BG2 in 70% yield over 2 steps. 1H NMR (500 MHz, CD3OD) δ 8.77 (d, J = 15.0 Hz, 1H), 7.73–7.69 (m, 2H), 7.59 (d, J = 15.0 Hz, 1H), 7.56 (t, J = 7.6 Hz, 1H), 7.51 (t, J = 7.4 Hz, 1H), 7.46 (d, J = 8.5 Hz, 2H), 7.34 (s, 1H), 7.21– 7.18 (m, 3H), 7.00 (t, JH-F = 55.3 Hz, 1H), 6.60 (d, J = 15.0 Hz, 1H), 5.33 (s, 2H), 4.88 (d, J = 8.0 Hz, 1H), 4.38 (t, J = 7.4 Hz, 2H), 3.89 (d, J = 3.5 Hz, 1H), 3.82–3.72 (m, 3H), 3.69–3.67 (m, 1H), 3.56 (dd, J = 9.7, 3.3 Hz, 1H), 2.78 (t, J = 5.5 Hz, 2H), 2.72 (t, J = 6.0 Hz, 2H), 1.99-1.93 (m, 4H), 1.86 (s, 6H), 1.09 (t, J = 7.5 Hz, 3H). 13C NMR (126 MHz, CD3OD) δ 180.14, 161.97, 160.69, 159.43, 156.82, 147.40, 143.79, 142.92, 133.44, 130.98, 130.31, 130.18, 129.66, 128.92, 126.71, 123.94, 118.11, 116.72, 115.88, 114.35, 112.55 (JC-F = 236.4 Hz), 106.06, 102.95, 101.84, 101.40, 77.10, 74.86, 72.25, 72.14, 70.22, 62.47, 52.37, 47.79, 30.11, 28.26, 28.24, 25.01, 22.40, 21.54, 11.56. HRMS (ESI) Calcd. For C42H46F2NO8+ [M]+: 730.3186; found: 730.3162.
HPLC analysis of NIR-BG1 and NIR-BG2 activation by β-galactosidase.
β-Gal (5 unit, 50 μL) was added to the NIR-BG probe solution (30 μL, 1 mM) and the mixture was incubated for 10 minutes at 37 °C. The resulting mixture was then quenched with HCl (1 M, 120 uL). The supernatant was obtained by centrifugation at 10,000 rpm for 20 min and then injected into HPLC for analysis. HPLC analysis was performed under the following conditions - mobile phase A: water with 0.1% TFA; B: acetonitrile with 0.1% TFA; 0-10 min: gradient elution, 2-95% B; 10-20min: isocratic elution, 95% B. The reaction was monitored using UV-vis absorbance at 600 nm. The peak was collected and was subjected to mass spectrometry analysis, respectively. The results further confirmed the observed peaks in HPLC trace are the enzymatic hydrolysis products.
UV-Vis and fluorescence spectra.
To a solution of 20 μL of 50 μM probes NIR-BG1 or NIR-BG2 and 160 μL PBS (pH = 7.4) was added 20 μL 0.1U/μL β-Galactosidase to obtain 5 μM probes with 2 U β-Galactosidase solution. After incubation at 37 °C for 5 min, the reaction solution was transferred to quartz cuvettes to measure absorbance or fluorescence. Absorbance spectrum scan range from 350 nm to 800 nm (1 nm increment). fluorescence spectrum setting for NIR-BG1: λex = 679 nm, Slit Width 5 nm. fluorescence spectrum setting for NIR-BG2: λex = 675 nm, Slit Width 5 nm. Emission was record from 690 nm to 800 nm, Slit Width 5 nm. Absorption spectra were recordes on Shimadzu UV-2700 UV-VIS Spectrophotometer. Fluorescence spectra were recorded on Shimadzu RF-5301pc spectrophotometer.
Time-dependent fluorescence intensity increment using β-Galactosidase (0.1 U/mL) with different concentrations of probes.
To a solution of 4, 6.7, 10, 13.3, 20 μL of 50 μM probe NIR-BG1 or NIR-BG2 and 176, 173.3, 170, 166.7, 160 μL PBS (pH = 7.4) buffers was added 20 μL β-Galactosidase (1 U/mL) in quartz cuvettes. Then final concentration of NIR-BG1 or NIR-BG2 is 1.0, 1.67, 2.5, 3.34, 5 μM and β-Galactosidase is 0.1 U/mL. Then the fluorescence intensity was recorded on Shimadzu RF-5301pc spectrophotometer at each timepoint. fluorescence spectrum setting for NIR-BG1: λex/λem = 679/700 nm, Slit Width 5 nm/ Slit Width 5 nm. Fluorescence spectrum setting for NIR-BG2: λex/λem = 675/708 nm, Slit Width 5 nm/ Slit Width 10 nm.
Time-dependent fluorescence intensity with various amounts of β-Galactosidase.
To a solution of 20 μL of 50 μM probe NIR-BG1 or NIR-BG2 and 160 μL PBS (pH = 7.4) buffer was added 20 μL β-Galactosidase with various concentrations (0.05-2.0 U/mL) in quartz cuvettes. Then final concentration of β-Galactosidase is from 0.005-0.2 U/mL. Then the fluorescence intensity was recorded on Shimadzu RF-5301pc spectrophotometer at each timepoint. fluorescence spectrum setting for NIR-BG1: λex/λem = 679/700 nm, Slit Width 5 nm/ Slit Width 5 nm. Fluorescence spectrum setting for NIR-BG2: λex/λem = 675/708 nm, Slit Width 5 nm/ Slit Width 10 nm.
Cells culture and induction of cellular senescence.
Cells were cultured at 37 °C in DMEM supplemented with 10% FBS and 1% penicillin under 5% CO2 and 95% humidity. To induce cellular senescence in HeLa cells, HeLa cells were cultured with freshly prepared culture medium containing 7.5 nM camptothecin (CPT) for 7 days. To induce cellular senescence in MDA-MB-231 cells, MDA-MB-231 cells were cultured with freshly prepared culture medium containing 20 nM doxorubicin for 4 days. To induce cellular senescence in MCF7 cells, MCF7 cells were cultured with freshly prepared culture medium containing 20 nM CPT for 4 days. To induce cellular senescence in IMR-90 cells, IMR-90 cells were cultured with freshly prepared culture medium containing 20 μM H2O2 for 2 h and was replaced with fresh medium. After 3 days, the cells were treated with freshly prepared culture medium containing 20 μM H2O2 for 2 h and was replaced with fresh medium for another 3 days.
Cell viability assays.
Cell growth and cytotoxicity were evaluated by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay. Cells were seeded in a 96-well plate at a density of 1 × 104 cells/well, then cells were incubated with 0, 0.5, 1, 2, 5 and 10 μM probe for 4 hours at 37 °C. Afer removal of the old medium, MTT reagent (3 mg/mL) was added, and cells were incubated for 3 hours at 37 °C. Upon the formation of visible purple crystals (formazan), medium was aspirated and dimethyl sulfoxide (DMSO, 100 μL) was added to dissolve the formazan for 20 min. Absorbance was read at 570 nm and cell viability was calculated based on untreated wells.
Fluorescence Western blot analysis.
To verify the binding affinity of activated NIR-BG1 and NIR-BG2, different concentration of the β-gal enzyme was incubated with 5 μM probes (in 1×PBS containing 5% DMSO) for 4 hours at 37 °C. Then samples were separated using 4-20% Mini-PROTEAN® TGX™ Precast Gels. After being transferred onto the PVDF membrane, β-gal was stained with anti-β-Galactosidase antibody (1:5000) and goat anti-chicken IgY H&L-AF568 (1:5000). Then the PVDF membrane was imaged using ChemiDoc MP (Bio-Rad, USA) to observe the colocalization of β-gal bands and probe signals.
Immunofluorescence cell staining.
HeLa cells were seeded on glass coverslips (0.13-0.16 mm thickness) at a density of 3 × 104 cells/well (total 8 wells, 4 wells for inducing senescence and 4 wells for control) and cultured overnight. On the next day, cells were treated with CPT (20 nM) or PBS for 4 days. After treatment, all cells were incubated with 5 μM probe (NIR-BG1 or NIR-BG2) for 2 hours, Then the culture medium was changed to probe-free MEM to allow cells to wash out the probes for 0 and 4 hours. Cells were then fixed and subsequently stained with actin (ActinGreen 488 ready probes reagent 2 drops/mL) for 15 min and DAPI for 5 min. Fluorescence microscopy imaging was performed to observe and compare the residual of probes in cells. Then, all coverslips were mounted on the slides with ProLong Gold Antifade Mounting media and were acquired for fluorescent microscope imaging (Nikon Ti2, Japan). CT26.WT and CT26.CL25 cells were also stained and image with same procedure just without treatment to confirm the uptake and binding of the probes. Dynamic clearance of the probes in the β-gal overexpressed CT26.CL25 cells was also performed to estimate the pharmaceutical kinetics of the probes in cells. CT26.CL25 cells were incubated with probes (5 μM) for 2 hours. Then the culture medium was changed to probe-free DMEM to allow cells to wash out the probes for 0, 1, 4, and 24 hours. Cells were then fixed and subsequently stained with β-actin and DAPI. Fluorescence microscopy imaging was performed to observe and compare the residual of probes in cells.
Flow cytometry.
HeLa cells were seeded in a 24-well plate at a density of 3 × 105 cells/well with or without CPT treatment (20 nM) for 7 days. CT26.WT and CT26.CL25 cells were seeded in a 24-well plate at a density of 4 × 105 cells/well and cultured at 37 °C overnight. Then cells were incubated with 5 μM of probes for 10 min, 1 hour, and 4 hours at 37 °C. After being washed 3 times with PBS, cells were digested with 0.25% trypsin and resuspended in 200 μL PBS buffer for flow cytometry analysis (Accuri C6 Plus, Becton Dickinson and Company, USA).
In vivo imaging.
In vivo imaging was performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of the University of New Mexico and followed the National Institutes of Health guidelines for animal care. Twenty thymic female nude mice (from Harland Laboratories) were injected subcutaneously with 2 × 106 HeLa cells to establish a tumor xenograft model. When tumor size reaches 100 mm3, mice were randomly divided into 4 groups for treatment. Two groups were administrated with CPT by gavage (2 mg/kg, every two days for 4 times, total dose was 8 mg/kg) and two groups were given saline for control. At day 10, mice were injected with probes (10 nmol, 100 μL) through tail vein. The fluorescence images were acquired at 1 h, 6 h, and 24 h post-injection using an IVIS Spectrum optical imaging system (PerkinElmer, USA) with a 680 nm excitation and 720 nm emission filter set. All mice were euthanized after the last imaging for tumors and major organs collection, ex vivo imaging. After a fast imaging, all tumors were immersed in optimal cutting temperature (OCT) compound and frozen on dry ice immediately for later immunohistochemistry experiments.
Immunohistochemistry Staining.
The frozen tumors were sectioned into 4 μm slices and were fixed with 4% formaldehyde for 10 min. The slides were then stained with Eosin and X-gal according to our previously reported method25.
Statistical analysis.
Values are reported as the mean ± standard deviation unless otherwise noted. Student’s t-test and two-way analysis of variance (ANOVA) were used to determine the statistical significance with probability values less than 0.05 (p < 0.05). All statistical calculations were performed using Prism 7.0 (GraphPad Software).
Supplementary Material
ACKNOWLEDGMENT
We thank Prof. Wei Wang (UNM) for the use of the fluorescence spectrometer, UNM Fluorescence Microscopy Shared Resource for use of the confocal microscopes, and Dr. Mara Steinkamp at the UNM Cancer Center Animal Models Shared Resource for assistance in the animal care and studies.
Funding Sources
The work was supported by research grants to Prof. L. Cui from the University of New Mexico, National Institute of General Medical Sciences of National Institutes of Health (Maximizing Investigators' Research Award for Early Stage Investigators, R35GM124963), Department of Defense (Career Development Award, Peer Re-viewed Cancer Research Program, Congressionally Directed Medical Research Programs, W81XWH-17-1-0529), the UNM Comprehensive Cancer Center, and the National Cancer Institute of the United States (P30CA118100).
Footnotes
Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
Conflict of Interest
The University of New Mexico has filed a patent application for the molecules described here.
REFERENCES
- 1.Campisi J; d'Adda di Fagagna F Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol 2007, 8, 729–740. [DOI] [PubMed] [Google Scholar]
- 2.Munoz-Espin D; Serrano M Cellular senescence: from physiology to pathology. Nat. Rev. Mol. Cell Biol 2014, 15, 482–496. [DOI] [PubMed] [Google Scholar]
- 3.Childs BG; Durik M; Baker DJ; van Deursen JM Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat. Med 2015, 21, 1424–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Deursen JM The role of senescent cells in ageing. Nature 2014, 509, 439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorgoulis V; Adams PD; Alimonti A; Bennett DC; Bischof O; Bishop C; Campisi J; Collado M; Evangelou K; Ferbeyre G; Gil J; Hara E; Krizhanovsky V; Jurk D; Maier AB; Narita M; Niedernhofer L; Passos JF; Robbins PD; Schmitt CA; Sedivy J; Vougas K; von Zglinicki T; Zhou D; Serrano M; Demaria M Cellular senescence: defining a path forward. Cell 2019, 179, 813–827. [DOI] [PubMed] [Google Scholar]
- 6.Chang J; Wang Y; Shao L; Laberge RM; Demaria M; Campisi J; Janakiraman K; Sharpless NE; Ding S; Feng W; Luo Y; Wang X; Aykin-Burns N; Krager K; Ponnappan U; Hauer-Jensen M; Meng A; Zhou D Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat. Med 2016, 22, 78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collado M; Serrano M The power and the promise of oncogene-induced senescence markers. Nat. Rev. Cancer 2006, 6, 472–476. [DOI] [PubMed] [Google Scholar]
- 8.Kuilman T; Peeper DS Senescence-messaging secretome: SMS-ing cellular stress. Nat. Rev. Cancer 2009, 9, 81–94. [DOI] [PubMed] [Google Scholar]
- 9.Lozano-Torres B; Estepa-Fernández A; Rovira M; Orzáez M; Serrano M; Martínez-Máñez R; Sancenón F The chemistry of senescence. Nat. Rev. Chem 2019, 3, 426–441. [Google Scholar]
- 10.Dimri GP; Lee X; Basile G; Acosta M; Scott G; Roskelley C; Medrano EE; Linskens M; Rubelj I; Pereira-Smith O A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 1995, 92, 9363–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Debacq-Chainiaux F; Erusalimsky JD; Campisi J; Toussaint O Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat. Protoc 2009, 4, 1798–1806. [DOI] [PubMed] [Google Scholar]
- 12.Urano Y; Kamiya M; Kanda K; Ueno T; Hirose K; Nagano T Evolution of fluorescein as a platform for finely tunable fluorescence probes. J. Am. Chem. Soc 2005, 127, 4888–4894. [DOI] [PubMed] [Google Scholar]
- 13.Kamiya M; Asanuma D; Kuranaga E; Takeishi A; Sakabe M; Miura M; Nagano T; Urano Y beta-Galactosidase fluorescence probe with improved cellular accumulation based on a spirocyclized rhodol scaffold. J. Am. Chem. Soc 2011, 133, 12960–12963. [DOI] [PubMed] [Google Scholar]
- 14.Sakabe M; Asanuma D; Kamiya M; Iwatate RJ; Hanaoka K; Terai T; Nagano T; Urano Y Rational design of highly sensitive fluorescence probes for protease and glycosidase based on precisely controlled spirocyclization. J. Am. Chem. Soc 2013, 135, 409–414. [DOI] [PubMed] [Google Scholar]
- 15.Egawa T; Koide Y; Hanaoka K; Komatsu T; Terai T; Nagano T Development of a fluorescein analogue, TokyoMagenta, as a novel scaffold for fluorescence probes in red region. Chem. Commun 2011, 47, 4162–4164. [DOI] [PubMed] [Google Scholar]
- 16.Gu K; Xu Y; Li H; Guo Z; Zhu S; Zhu S; Shi P; James TD; Tian H; Zhu WH Real-time tracking and in vivo visualization of beta-galactosidase activity in colorectal tumor with a ratiometric near-infrared fluorescent probe. J. Am. Chem. Soc 2016, 138, 5334–5340. [DOI] [PubMed] [Google Scholar]
- 17.Safir Filho M; Dao P; Gesson M; Martin AR; Benhida R Development of highly sensitive fluorescent probes for the detection of beta-galactosidase activity - application to the real-time monitoring of senescence in live cells. Analyst 2018, 143, 2680–2688. [DOI] [PubMed] [Google Scholar]
- 18.Chen X; Ma X; Zhang Y; Gao G; Liu J; Zhang X; Wang M; Hou S Ratiometric fluorescent probes with a self-immolative spacer for real-time detection of beta-galactosidase and imaging in living cells. Anal. Chim. Acta 2018, 1033, 193–198. [DOI] [PubMed] [Google Scholar]
- 19.Lozano-Torres B; Galiana I; Rovira M; Garrido E; Chaib S; Bernardos A; Munoz-Espin D; Serrano M; Martinez-Manez R; Sancenon F An OFF-ON two-photon fluorescent probe for tracking cell senescence in vivo. J. Am. Chem. Soc 2017, 139, 8808–8811. [DOI] [PubMed] [Google Scholar]
- 20.Gao Y; Hu Y; Liu Q; Li X; Li X; Kim CY; James TD; Li J; Chen X; Guo Y Two-dimensional design strategy to construct smart fluorescent probes for the precise tracking of senescence. Angew. Chem. Int. Ed 2021, 60, 10756–10765. [DOI] [PubMed] [Google Scholar]
- 21.Li X; Qiu W; Li J; Chen X; Hu Y; Gao Y; Shi D; Li X; Lin H; Hu Z; Dong G; Sheng C; Jiang B; Xia C; Kim C-Y; Guo Y; Li J First-generation species-selective chemical probes for fluorescence imaging of human senescence-associated β-galactosidase. Chem. Sci 2020, 11, 7292–7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan L; Lin W; Zheng K; He L; Huang W Far-red to near infrared analyte-responsive fluorescent probes based on organic fluorophore platforms for fluorescence imaging. Chem. Soc. Rev 2013, 42, 622–661. [DOI] [PubMed] [Google Scholar]
- 23.Owens EA; Henary M; El Fakhri G; Choi HS Tissue-specific near-infrared fluorescence imaging. Acc. Chem. Res 2016, 49, 1731–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo Z; Park S; Yoon J; Shin I Recent progress in the development of near-infrared fluorescent probes for bioimaging applications. Chem. Soc. Rev 2014, 43, 16–29. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y; Liu J; Ma X; Cui C; Deenik PR; Henderson PKP; Sigler AL; Cui L Real-time imaging of senescence in tumors with DNA damage. Sci. Rep 2019, 9, 2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z; Chen S; Lam JW; Qin W; Kwok RT; Xie N; Hu Q; Tang BZ Long-term fluorescent cellular tracing by the aggregates of AIE bioconjugates. J. Am. Chem. Soc 2013, 135, 8238–8245. [DOI] [PubMed] [Google Scholar]
- 27.Halazy S; Berges V; Ehrhard A; Danzin C Ortho- and para-(difluoromethyl)aryl-β-d-glucosides: A new class of enzyme-activated irreversible inhibitors of β-glucosidases. Bioorg. Chem 1990, 18, 330–344. [Google Scholar]
- 28.Myers JK; Widlanski TS Mechanism-based inactivation of prostatic acid phosphatase. Science 1993, 262, 1451–1453. [DOI] [PubMed] [Google Scholar]
- 29.Kwan DH; Chen HM; Ratananikom K; Hancock SM; Watanabe Y; Kongsaeree PT; Samuels AL; Withers SG Self-immobilizing fluorogenic imaging agents of enzyme activity. Angew. Chem. Int. Ed 2011, 50, 300–303. [DOI] [PubMed] [Google Scholar]
- 30.Doura T; Kamiya M; Obata F; Yamaguchi Y; Hiyama TY; Matsuda T; Fukamizu A; Noda M; Miura M; Urano Y Detection of LacZ-positive cells in living tissue with single-cell resolution. Angew. Chem. Int. Ed 2016, 55, 9620–9624. [DOI] [PubMed] [Google Scholar]
- 31.Gao Z; Thompson AJ; Paulson JC; Withers SG Proximity ligation-based fluorogenic imaging agents for neuraminidases. Angew. Chem. Int. Ed 2018, 57, 13538–13541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mao W; Xia L; Wang Y; Xie H A self-immobilizing and fluorogenic probe for beta-lactamase detection. Chem. Asian J 2016, 11, 3493–3497. [DOI] [PubMed] [Google Scholar]
- 33.Jiang J; Tan Q; Zhao S; Song H; Hu L; Xie H Late-stage difluoromethylation leading to a self-immobilizing fluorogenic probe for the visualization of enzyme activities in live cells. Chem. Commun 2019, 55, 15000–15003. [DOI] [PubMed] [Google Scholar]
- 34.Ito H; Kawamata Y; Kamiya M; Tsuda-Sakurai K; Tanaka S; Ueno T; Komatsu T; Hanaoka K; Okabe S; Miura M; Urano Y Red-shifted fluorogenic substrate for detection of lacZ-positive cells in living tissue with single-cell resolution. Angew. Chem. Int. Ed 2018, 57, 15702–15706. [DOI] [PubMed] [Google Scholar]
- 35.Chiba M; Kamiya M; Tsuda-Sakurai K; Fujisawa Y; Kosakamoto H; Kojima R; Miura M; Urano Y Activatable photosensitizer for targeted ablation of lacZ-positive cells with single-cell resolution. ACS Cent. Sci 2019, 5, 1676–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J; Li S; Aslam NA; Zheng F; Yang B; Cheng R; Wang N; Rozovsky S; Wang PG; Wang Q; Wang L Genetically encoding photocaged quinone methide to multitarget protein residues covalently in vivo. J. Am. Chem. Soc 2019, 141, 9458–9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J; Cai L; Sun W; Cheng R; Wang N; Jin L; Rozovsky S; Seiple IB; Wang L Photocaged quinone methide crosslinkers for light-controlled chemical crosslinking of protein-protein and protein-DNA complexes. Angew. Chem. Int. Ed 2019, 58, 18839–18843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan L; Lin W; Zhao S; Gao W; Chen B; He L; Zhu S A unique approach to development of near-infrared fluorescent sensors for in vivo imaging. J. Am. Chem. Soc 2012, 134, 13510–13523. [DOI] [PubMed] [Google Scholar]
- 39.Burke HM; Gunnlaugsson T; Scanlan EM Recent advances in the development of synthetic chemical probes for glycosidase enzymes. Chem. Commun 2015, 51, 10576–10588. [DOI] [PubMed] [Google Scholar]
- 40.Karton-Lifshin N; Segal E; Omer L; Portnoy M; Satchi-Fainaro R; Shabat D A unique paradigm for a turn-ON near-infrared cyanine-based probe: noninvasive intravital optical imaging of hydrogen peroxide. J. Am. Chem. Soc 2011, 133, 10960–10965. [DOI] [PubMed] [Google Scholar]
- 41.Redy-Keisar O; Kisin-Finfer E; Ferber S; Satchi-Fainaro R; Shabat D Synthesis and use of QCy7-derived modular probes for the detection and imaging of biologically relevant analytes. Nat. Protoc 2014, 9, 27–36. [DOI] [PubMed] [Google Scholar]
- 42.Unpublisded results.
- 43.Fang Y; Chen W; Shi W; Li H; Xian M; Ma H A near-infrared fluorescence off-on probe for sensitive imaging of hydrogen polysulfides in living cells and mice in vivo. Chem. Commun 2017, 53, 8759–8762. [DOI] [PubMed] [Google Scholar]
- 44.Wu X; Li L; Shi W; Gong Q; Ma H Near-infrared fluorescent probe with new recognition moiety for specific detection of tyrosinase activity: design, synthesis, and application in living cells and zebrafish. Angew. Chem. Int. Ed 2016, 55, 14728–14732. [DOI] [PubMed] [Google Scholar]
- 45.Ning J; Liu T; Dong P; Wang W; Ge G; Wang B; Yu Z; Shi L; Tian X; Huo X; Feng L; Wang C; Sun C; Cui J; James TD; Ma X Molecular design strategy to construct the near-infrared fluorescent probe for selectively sensing human cytochrome P450 2J2. J. Am. Chem. Soc 2019, 141, 1126–1134. [DOI] [PubMed] [Google Scholar]
- 46.Wrobel AT; Johnstone TC; Deliz Liang A; Lippard SJ; Rivera-Fuentes P A fast and selective near-infrared fluorescent sensor for multicolor imaging of biological nitroxyl (HNO). J. Am. Chem. Soc 2014, 136, 4697–4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan Y; Zhang L; Man KH; Peltier R; Chen G; Zhang H; Zhou L; Wang F; Ho D; Yao SQ; Hu Y; Sun H Reaction-based off-on near-infrared fluorescent probe for imaging alkaline phosphatase activity in living cells and mice. ACS Appl. Mater. Interfaces 2017, 9, 6796–6803. [DOI] [PubMed] [Google Scholar]
- 48.Li L; Li Z; Shi W; Li X; Ma H Sensitive and selective near-infrared fluorescent off-on probe and its application to imaging different levels of beta-lactamase in Staphylococcus aureus. Anal. Chem 2014, 86, 6115–6120. [DOI] [PubMed] [Google Scholar]
- 49.Li Z; He XY; Wang Z; Yang RH; Shi W; Ma HM, in vivo imaging and detection of nitroreductase in zebrafish by a new near-infrared fluorescence off-on probe. Biosensors & Bioelectronics 2015, 63, 112–116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.