Abstract
Two peroxins of the AAA family, PpPex1p and PpPex6p, are required for peroxisome biogenesis in the yeast Pichia pastoris. Cells from the corresponding deletion strains (PpΔpex1 and PpΔpex6) contain only small vesicular remnants of peroxisomes, the bulk of peroxisomal matrix proteins is mislocalized to the cytosol, and these cells cannot grow in peroxisome-requiring media (J. A. Heyman, E. Monosov, and S. Subramani, J. Cell Biol. 127:1259–1273, 1994; A. P. Spong and S. Subramani, J. Cell Biol. 123:535–548, 1993). We demonstrate that PpPex1p and PpPex6p interact in an ATP-dependent manner. Genetically, the interaction was observed in a suppressor screen with a strain harboring a temperature-sensitive allele of PpPEX1 and in the yeast two-hybrid system. Biochemially, these proteins were coimmunoprecipitated with antibodies raised against either of the proteins, but only in the presence of ATP. The protein complex formed under these conditions was 320 to 400 kDa in size, consistent with the formation of a heterodimeric PpPex1p-PpPex6p complex. Subcellular fractionation revealed PpPex1p and PpPex6p to be predominantly associated with membranous subcellular structures distinct from peroxisomes. Based on their behavior in subcellular fractionation experiments including flotation gradients and on the fact that these structures are also present in a PpΔpex3 strain in which no morphologically detectable peroxisomal remnants have been observed, we propose that these structures are small vesicles. The identification of vesicle-associated peroxins is novel and implies a role for these vesicles in peroxisome biogenesis. We discuss the possible role of the ATP-dependent interaction between PpPex1p and PpPex6p in regulating peroxisome biogenesis events.
Peroxisomes are ubiquitous, eukaryotic subcellular organelles whose biogenesis can be induced in response to nutritional (37) and developmental (6) cues. Notably, their size, number, biochemical composition, and cellular role are greatly influenced by the environmental milieu of the cells that house them. These properties are particularly striking in yeast species for which peroxisome-requiring carbon and/or nitrogen sources have been identified. For example, Pichia pastoris cells grown in glucose-containing medium harbor only a few (1 to 3) small (<0.1-μm) peroxisomes; however, a switch to medium containing methanol induces cells to produce several (5 to 10) large (0.5-μm) peroxisomes, and peroxisomal proteins can constitute up to 40% of total cellular protein (34).
All of the proteins required for peroxisome biogenesis are encoded by nuclear genes. The lipids essential for the growth of their membranes must be derived from extraperoxisomal sources. Peroxisomes are believed to be maintained by the growth and division of preexisting peroxisomes (18). Recently, however, the possibility of de novo peroxisome biogenesis was raised by studies which demonstrated that some yeast (pex mutant) strains defective in peroxisome biogenesis or function have no detectable peroxisomal remnants and that the organelle can be restored in these strains by introduction of the complementing gene (4, 41) or, in the case of temperature-sensitive strains, by a shift from a nonpermissive to a permissive temperature (39). So far, no data describing the steps required for de novo peroxisome biogenesis have been presented.
Several yeast model systems have been exploited to unravel the mechanism of peroxisome biogenesis and import, leading to the description of a number of genes and proteins, collectively termed peroxins (7), that play a role in these processes. These and other studies have led, for example, to the characterization of peroxisomal targeting signals for peroxisomal matrix and membrane proteins, the receptors that recognize some of these peroxisomal targeting signals, docking proteins for the PTS1 and PTS2 receptors, and cytosolic and peroxisomal membrane proteins involved in the import process (2, 13, 32).
Despite the impressive progress in the description of peroxins involved in the biogenesis process, our knowledge of the biochemical functions of these proteins is only rudimentary. The search for this information is driven both by the desire to understand the biochemistry of peroxisome biogenesis and because defects in peroxisome biogenesis are the underlying molecular cause of a number of devastating human peroxisomal disorders (40).
Among the many peroxins (over 17) implicated in peroxisome biogenesis are Pex1p and Pex6p, which belong to the AAA family (ATPases associated with diverse cellular activities) (15) of proteins (8, 11, 23, 29, 35, 38, 42). Our work on these proteins in P. pastoris has shown that they are involved in peroxisome biogenesis (11, 29). PpΔpex1 and PpΔpex6 strains contain only vesicular remnants of peroxisomes (“ghosts”) that import some proteins but exclude most peroxisomal matrix proteins in the cytosol. Pex1p and Pex6p comprise part of the peroxisome biogenesis machinery of all eukaryotes because they are conserved in yeasts and, in the case of Pex6p, the mammalian homolog has also been described (8, 11, 23, 29, 35, 38, 42). Their importance is exemplified by the fact that mutations in HsPex6p are one of the causes of Zellweger syndrome in humans (35, 42).
In this paper, we show that there is an Mg2+- and ATP-dependent interaction between PpPex1p and PpPex6p. We also demonstrate that these proteins associate with vesicles distinct from peroxisomes, a result which suggests a heretofore-unrecognized role for such vesicles in peroxisome biogenesis.
MATERIALS AND METHODS
Yeast strains and culture conditions.
Liquid cultures of P. pastoris cells were grown in YPD (1% yeast extract, 2% Bacto Peptone, 2% dextrose), YPM (1% yeast extract, 2% Bacto Peptone, 0.5% methanol), or mineral medium (36) containing either 0.5% (wt/vol) glucose (MMG), 0.5% (vol/vol) methanol (MMM), or 0.2% (vol/vol) oleate and 0.02% (vol/vol) Tween 40 (MMOT). For growth on plates, defined synthetic medium consisting of yeast nitrogen base at 0.67% (wt/vol) and supplemented with a carbon source to a final concentration of either 2% dextrose (SD) or 0.5% methanol (SM) was used. For auxotrophic strains requiring arginine, histidine, or both, the required amino acids were included at 40 mg/ml.
Culturing, mating, sporulation, and random spore analysis for P. pastoris were done as previously described (9). Strains PPY301 (arg4 his4 Pppex1::PpARG4) and SEW1 (arg4 his4 Pppex3::PpARG4) were described previously (11, 41). PPY20 (arg4 Pppex1-2) (9) was crossed with PPY4 (his4; Northern Regional Research Laboratories, Peoria, Ill.) to create PPY303 (arg4 his4 Pppex1-2). SMD1163 (his4 pep4 prb1) is referred to in this paper as “WT” and was obtained from Phillips Petroleum, Bartlesville, Okla. A Pppex1::HIS4 strain (his4 pep4 prb1 Pppex1::PpHIS4) was obtained through targeted disruption of the genomic PpPEX1 locus of SMD1163. The DNA fragment used for the gene disruption was created through the following steps. A 2.4-kb BglII fragment of the PpPEX1 open reading frame (ORF) was cloned into the BamHI site of pUC19; this plasmid was digested with XhoI and made blunt with Klenow polymerase. A blunted 2.7-kb DNA fragment (EcoRI-SphI) which contained the P. pastoris HIS4 gene was cloned into this site, creating a plasmid containing the HIS4 gene flanked on each side by 1.2 kb of the PpPEX1 ORF. This gene disruption fragment was excised by SmaI-PstI digestion and used to transform SMD1163, and transformants which contained the correct chromosomal configuration of the integrated DNA were selected. The resultant strain did not synthesize PpPex1p.
Strains with pep4 and prb1 mutations in vacuolar proteases were used when necessary because uncomplexed PpPex1p and PpPex6p are unstable in strains with wild-type vacuolar enzymes.
Molecular biological techniques.
Escherichia coli DH5αF′ was used for the propagation of plasmids. Recombinant DNA procedures were performed essentially as described previously (26). Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) was done according to Laemmli and Favre (16), and Western blotting was done according to Towbin et al. (33). For Western blotting, horseradish peroxidase-conjugated secondary antibody was detected with an ECL kit (Amersham Corp., Arlington Heights, Ill.). Protein concentrations were determined by the Bradford assay (Bio-Rad Laboratories, Hercules, Calif.).
Nonmutagenic PCR conditions were 5 min at 94°C followed by 27 cycles of 1 min at 94°C, 2 min at 50°C, 3 min at 72°C, and a final cycle of 10 min at 72°C. Vent polymerase and buffer (New England Biolabs, Beverly, Mass.) were used for the amplification. The product was treated with GIBCO Taq polymerase according to the procedure described in the New England Biolabs catalog if it was to be ligated into the pCRII cloning vector (Invitrogen, San Diego, Calif.).
Plasmid rescue from yeast strains.
Plasmids were rescued from yeast strains bearing episomal plasmids following growth to the stationary phase in selective medium. A 1.5-ml sample from the culture was pelleted in an Eppendorf Microfuge and resuspended in 200 μl of breaking buffer (2% Triton X-100, 1% SDS, 100 mM NaCl, 10 mM Tris-HCl, 1 mM EDTA [pH 8.0]). Glass beads (200 μl) were added, followed by the addition of 200 μl of 25:24:1 phenol-chloroform-isoamyl alcohol, and the mixture was vortexed at high speed for 2 min. Lysates were centrifuged at high speed for 5 min, and competent E. coli cells were transformed with 5 μl of the aqueous supernatant.
Construction of temperature-sensitive strains.
Temperature-sensitive alleles of PpPEX1 were generated by a procedure described earlier (22). pJAH17 (11) is an autonomously replicating plasmid containing HIS4 as a selectable marker, the 3,471-bp PpPEX1 ORF, and PpPEX1 promoter and terminator sequences. pJAH17 was digested with XhoI and AflII to remove bp 1796 to 2854 of PpPEX1 (coding for amino acids 599 to 952, a region which contains the entire 3′ ATP-binding domain). Random point mutations were introduced in the DNA fragment spanning bp 607 to 3009 (amino acids 203 to 1003) of the PpPEX1 coding sequence by low-fidelity PCR (27 cycles of 1 min at 94°C, 2 min at 50°C, and 3 min at 72°C with Taq polymerase in standard buffer adjusted to 0.1 mM MnCl2). The gapped plasmid and PCR products were coelectroporated into PPY301 cells (25), and transformants were screened to obtain strains that grew on SM plates at 22°C but not at 30°C. Plasmids encoding temperature-sensitive PpPex1p were rescued and integrated into the his4 locus of strain PPY303 to create stable Pppex1-ts strains, including SJH242. SJH217 is a strain in which BamHI-linearized pJAH17 is integrated into the his4 locus of PPY303. SJH200 is a strain in which BamHI-linearized pJAH16 is integrated into the his4 locus of PPY303. pJAH16 and pJAH17 are the same except that pJAH16 does not contain the PpPEX1 gene (11).
Construction of strains that synthesize GFP-SKL and overproduce PpPex6p.
A two-step cloning procedure was used to introduce a 3.5-kb ScaI-NotI (ends made blunt with Klenow polymerase) DNA fragment encoding the full-length PpPex6p downstream of the alcohol oxidase promoter into pPIC3K (Invitrogen) which had been digested with BamHI and HindIII (both sites made blunt with Klenow polymerase). The resulting plasmid was called pKNSD67. pJAH125 was constructed by modification of pKNSD67. First, pKNSD67 was digested with MscI and XhoI to remove the HIS4 and kanamycin resistance genes and treated with Klenow polymerase and phosphatase, and ligated to a 2,075-bp blunt fragment (obtained by HindIII digestion, filling in with Klenow polymerase, and SmaI digestion) encoding the P. pastoris ARG4 gene to create pJAH61. Next, a 1.5-kb fragment containing the P. pastoris GAPDH promoter driving expression of a gene encoding GFP-SKL (green fluorescent protein with C-terminal amino acids SKL) was amplified from pTW74 (39a) with primers JH10 (5′-GCT TAT CGA TAA GCA TGC AC-3′) and JH11 (5′-ACA TGC ATG CTT TTT TGT AGA AAT GTC-3′), digested with SphI (the SphI site was encoded by the PCR primers), and cloned into the SphI site of pJAH61 to form pJAH125. pJAH125 and pJAH127 are the same except that pJAH127 does not contain any PpPEX6 coding sequence. pJAH125 and pJAH127 were digested with PmeI to direct integration into the methanol oxidase locus of strains SJH242, SJH200, and SJH217. SJH242 with integrated pJAH127 is called SJH242-GFP, and SJH242 with integrated pJAH125 is called SJH242-GFP6.
Two-hybrid system.
Full-length clones coding for PpPex1p, PpPex3p, and PpPex6p were inserted into derivatives of plasmids pVP16 (containing the transactivation domain) and pBTM116 (containing the DNA-binding domain) (12). Through insertion of double-stranded oligonucleotides, three derivatives each of pVP16 and pBTM116 were constructed, introducing a multiple cloning site (mcs) harboring NotI, BamHI, SpeI and/or NheI (or neither), PstI, EcoRI, and SalI sites in three different reading frames downstream of the functional (transactivation or DNA-binding) domain. The three pVP16 derivatives were named pKNSD50, pKNSD51, and pKNSD52; the three pBTM116 derivatives were called pKNSD53, pKNSD54, and pKNSD55. Starting from the BamHI site, the reading frame in pKNSD50 and pKNSD53 is GG.ATC.C (not containing an NheI or an SpeI site in the mcs), that in pKNSD51 and pKNSD54 is G.GAT.CC (containing an SpeI site in the mcs), and that in pKNSD52 and pKNSD55 is GGA.TCC (containing an NheI site in the mcs).
These plasmids were subsequently used for insertion of the PpPEX genes through the following cloning steps. For PpPEX1, through subcloning, an Asp718 site was introduced preceding the PpPEX1 initiation codon. A 4.0-kb Asp718 (ends made blunt with Klenow polymerase)-XbaI fragment containing the full-length PpPEX1 coding sequence was inserted into pKNSD51 and pKNSD54 that had been digested with BamHI (ends made blunt with Klenow polymerase) and NheI, generating plasmids pKNSD85 and pKNSD86, respectively. For PpPEX3, a 1.8-kb BglII-SpeI DNA fragment from pKNSD44 (41) was inserted into pKNSD52 and pKNSD55 that had been digested with BamHI and SpeI, generating plasmids pKNSD97 and pKNSD98, respectively. For PpPEX6, a 3.5-kb ScaI (genomic site preceding the PpPEX6 coding sequence)-SalI DNA fragment containing the full-length PpPEX6 coding sequence was inserted into pKNSD51 and pKNSD54 that had been digested with BamHI (ends made blunt with Klenow polymerase) and SalI, generating plasmids pKNSD91 and pKNSD92, respectively.
All combinations of pVP16 and pBTM116 plasmids containing the PpPEX genes were used to cotransform Saccharomyces cerevisiae L40 (MATa trp1 leu2 his3 LYS2::lexA-HIS3 URA3::lexA-lacZ). Individual transformants were screened for expression of the chromosomal HIS4 and lacZ marker genes.
Microscopy.
To observe GFP in vivo, cells were grown overnight in YPM medium, immobilized on a microscope slide in a 3-μl drop of 100% glycerol, and observed by fluorescence or phase-contrast microscopy. Both phase-contrast and fluorescence images were viewed with a Leitz Fluotar 100× PL 1.3 objective under oil immersion on a Leitz Laborlux 12 microscope. The GFP fluorochrome was viewed with the Leitz selective filter set L/3.
Organelle-free sucrose gradients for detection of protein complexes.
P. pastoris cells were cultured overnight in YPM medium, collected by centrifugation, treated for 20 min at room temperature with 140 mM β-mercaptoethanol in 100 mM Tris–50 mM EDTA (pH 7.5), converted to spheroplasts, lysed by resuspension in lysis buffer (20 mM HEPES [pH 7.4], 2 mM EDTA, 2 mM dithiothreitol [DTT], 0.5% Triton X-100, 100 mM KCL, 1 mM phenylmethylsulfonyl fluoride, (PMSF); supplemented with 4 mM Mg2+, 0.5 mM ATP, or 0.5 mM ATPγS [a nonhydrolyzable analog of ATP]), and centrifuged for 20 min at 20,000 × g to produce the cell-free lysate. Approximately 4 mg of protein was loaded on continuous 10 to 20% (wt/vol) sucrose gradients made in lysis buffer and centrifuged for 28 h at 32,000 rpm in a Beckman SW41 rotor (125,000 × g), and the tubes were drained from the top into 28 fractions of 430 μl. Equal volumes of fractions were analyzed by Western blot analysis with affinity-purified α-PpPex1p or α-PpPex6p antibodies prepared as described earlier (19).
Immunoprecipitations.
Lysates were prepared as described above for organelle-free sucrose gradients in lysis buffer, and 4 mM MgCl2, 0.5 mM ATP, or 5 U of apyrase (Sigma, St. Louis, Mo.) per ml was added. Lysate protein (1 mg) was incubated for 1 h at 4°C with 2 μl of serum and 30 μl of swelled protein A beads (Pharmacia Biotechnology, Piscataway, N.J.) and was washed two times with 1 ml of lysis buffer. These conditions were chosen so that over 90% of the target protein would be obtained in the pellet fraction. Twenty percent of the pellet fraction was subjected to SDS-PAGE and subsequently transferred to nitrocellulose for Western blot analysis.
Differential centrifugation and cell fractionation.
Strains were precultured in MMG, used to inoculate 1.5-liter MMOT cultures, and grown overnight to an optical density at 600 nm (OD600) of approximately 2. Cells were harvested by centrifugation, pretreated in 100 mM Tris-HCl–50 mM EDTA–140 mM β-mercaptoethanol (pH 7.5) for 20 min, and pelleted by centrifugation. Cells were washed once in 20 mM potassium phosphate–1.2 M sorbitol (pH 7.5), resuspended in the same buffer, and converted to spheroplasts by a 30-min incubation at 30°C following the addition of 6 mg of Zymolyase 20T (ICN Biochemicals, Inc., Aurora, Ohio) per 1,000 OD600 units of cells. Spheroplasts were pelleted by centrifugation, resuspended in ice-cold Dounce buffer (0.8 M sorbitol, 5 mM morpholineethanesulfonic acid [MES], 0.5 mM EDTA [pH 6.0], 0.1% ethanol; supplemented with 1 mM PMSF, 5 mM NaF, and 12.5 mg of leupeptin per ml), and broken with 10 strokes in a Dounce homogenizer. The resulting homogenate was subjected to centrifugation for 10 min at 2,000 × g (two times) to clear unbroken cells and cell debris, and the supernatant (postnuclear supernatant [PNS]) was centrifuged for 30 min at 27,000 × g. The resulting supernatant was centrifuged for 1 h at 100,000 × g, and equal volumes of fractions of the pellet and supernatant were analyzed by Western blot analysis.
Flotation gradient centrifugation.
The 100,000 × g pellet fraction (obtained from the 27,000 × g supernatant fraction) was resuspended in 0.5 ml of 65% (wt/wt) sucrose in Dounce buffer without sorbitol. A total of 2.25 ml of 50% (wt/wt) sucrose and 2.25 ml of 30% (wt/wt) sucrose (in Dounce buffer) were layered on top of the sample and spun for 24 h at 100,000 × g in an SW50.1 (Beckman) rotor. The gradients were drained from the top into 13 fractions of approximately 400 μl, and equal volumes of fractions were analyzed by SDS-PAGE and Western blot analysis.
Subcellular fractionation with Nycodenz gradients.
Nycodenz gradients were loaded with PNS fractions prepared as described above from cells grown overnight in 3 liters of MMOT (16 h) or MMG (8 h) to an OD600 of 2. Then, 5 ml of the supernatant (PNS) was loaded on a 35-ml continuous Nycodenz gradient (15 to 35%, with a cushion of 5 ml of 50% [wt/vol]), centrifuged as described before (20), and drained from the bottom into 24 fractions of approximately 1.6 ml. Equal volumes of every second fraction were analyzed by SDS-PAGE and Western blot analysis. Antibody dilutions were as follows: α-PpAcyl-coenzyme A oxidase, 1:10,000; α-Scthiolase, 1:10,000; α-F1β subunit of mitochondrial ATPase, 1:10,000; α-PpPex1p, 1:4,000; α-PpPex3p, 1:10,000; and α-PpPex6, 1:4,000.
RESULTS
Generation of temperature-sensitive alleles of PpPex1p.
The first step in identifying the cellular partners of PpPex1p was the generation of temperature-sensitive PpPEX1 alleles for use in a suppressor screen. The PpPEX1 ORF was subjected to a PCR-based mutagenesis procedure, and the mutagenized DNA was used to cotransform P. pastoris PPY301 (PpΔpex1 his4) together with linearized plasmid pJAH17. Transformants able to utilize methanol at 22°C but not at 30°C and/or 34°C were selected and considered to encode temperature-sensitive PpPex1p. Following rescue from the PPY301 cells, the plasmids were integrated into the his4 locus of strain PPY303 (arg4 his4 Pppex1-2), which contains a nonfunctional chromosomal Pppex1 gene, to create stable strains (SJH241 to SJH246 and SJH248 to SJH250) with a temperature-sensitive PpPEX1 allele. SJH242 was used in the studies that follow.
Overexpression of PpPex6p suppresses a temperature-sensitive mutation in PpPex1p.
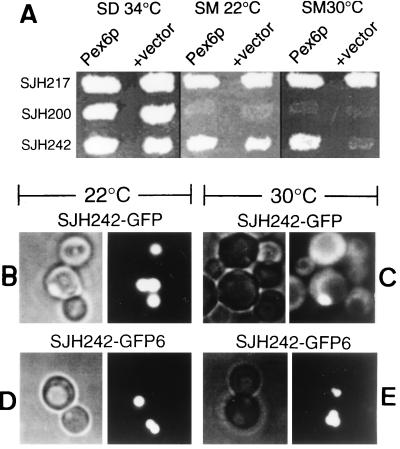
To identify proteins that interact with PpPex1p, we transformed strain SJH242 (Pppex1-ts) with plasmids that overproduce PpPex3p (a peroxisomal membrane protein) (41), PpPex5p (the PTS1 receptor) (19, 31), or PpPex6p (29). Overproduction of PpPex6p but not of PpPex5p or PpPex3p rescued the methanol utilization defect of SJH242 at 30°C. PpPex6p overproduction did not enable a Pppex1 inactive point mutant strain, SJH200 (Pppex1), or a PpΔpex1 strain (not shown) to utilize methanol, nor did it affect the growth of wild-type strain SJH217 (WT PpPEX1) cells on methanol (Fig. 1A).
FIG. 1.
Overproduction of PpPex6p rescues the methanol utilization and peroxisome morphology defects of a Pppex1-ts strain. (A) Overexpression of PpPEX6 from plasmid pAS001 (29) in strains SJH217 (WT PpPEX1), SJH200 (Pppex1), and SJH242 (Pppex1-ts). Strains were replica plated onto SM or SD plates and grown for 3 days at the indicated temperature. (B to E) Fluorescence microscopy (right panels) of SJH242 synthesizing GFP-SKL (SJH242-GFP) from integrated plasmid pJAH127 grown at 22°C (B) and 30°C (C) and SJH242 overproducing PpPex6p and synthesizing GFP-SKL (SJH242-GFP6) from integrated plasmid pJAH125 grown at 22°C (D) and 30°C (E). Nomarski images are shown in the left panels.
We analyzed the import of a reporter protein into peroxisomes in the Pppex1-ts strains in the presence and absence of overproduced PpPex6p. A hybrid protein (GFP-SKL) consisting of GFP with a C-terminal peroxisomal targeting signal, Ser-Lys-Leu (20), was expressed in strain SJH242 either in the absence (SJH242-GFP) or in the presence (SJH242-GFP6) of overexpressed PpPex6p, and the localization of GFP-SKL was visualized in vivo by fluorescence microscopy. At the permissive temperature (22°C), virtually all cells of SJH242-GFP and SJH242-GFP6 (Fig. 1B and D, respectively) contained GFP-SKL-labeled peroxisomes equivalent to those observed in the wild-type strain (STW3) (20). At the restrictive temperature (30°C), SJH242-GFP cells showed cytosolic staining and only occasionally exhibited punctate structures containing GFP-SKL (Fig. 1C). In contrast, SJH242-GFP6 cells grown at 30°C contained bright, GFP-SKL-labeled peroxisomes (Fig. 1E) indistinguishable from those seen in wild-type cells and showed no cytosolic labeling. Thus, the overproduction of PpPex6p restored growth as well as normal peroxisomal morphology and import to SJH242 cells cultured on methanol under restrictive conditions.
Interaction between PpPex1p and PpPex6p in the yeast two-hybrid system.
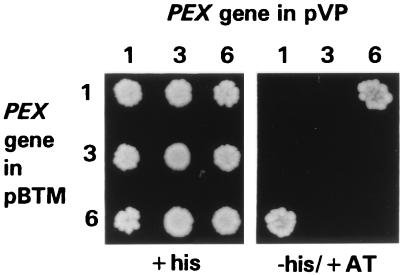
Further evidence for an interaction between PpPex1p and PpPex6p was obtained by a yeast two-hybrid analysis (12), in which an interaction between two fusion proteins expressed from specific plasmids allows the growth of the reporter yeast strain on medium lacking histidine. One of the plasmids expresses a protein fused to the DNA-binding domain of the LexA repressor, while the other expresses a fusion with the transcriptional-transactivation domain of a viral protein, VP16. S. cerevisiae L40 cells containing either combination of the PpPEX1 and PpPEX6 expression plasmids were able to grow on medium lacking histidine (Fig. 2) and expressed a second marker, β-galactosidase, in qualitative assays for this enzyme (data not shown). Other combinations of the PpPEX1, PpPEX3, PpPEX5, and PpPEX6 plasmids and the parental plasmids alone were unable to produce either histidine prototrophy or β-galactosidase above background levels. These data show that PpPex1p and PpPex6p interact with each other but not with themselves or with PpPex3p (Fig. 2) or PpPex5p (data not shown).
FIG. 2.
PpPex1p and PpPex6p interact in the yeast two-hybrid system. Full-length clones coding for PpPex1p, PpPex3p, and PpPex6p were inserted into derivatives of plasmids pVP16 (transactivation domain) and pBTM116 (DNA-binding domain) (12). All combinations of pVP16 and pBTM116 plasmids containing the PpPEX genes were used to cotransform S. cerevisiae L40. Individual transformants were screened for expression of the chromosomal HIS4 marker gene (+his: media containing histidine [left panel]; −his/+AT: media lacking histidine and containing 25 mM 3-aminotriazole [right panel]).
The interaction between PpPex1p and PpPex6p is ATP dependent.
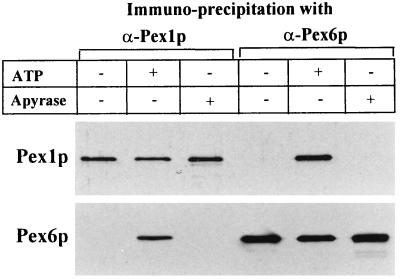
Antibodies against PpPex1p and PpPex6p were used to immunoprecipitate the proteins from total extracts of methanol-grown P. pastoris WT cells. Under standard conditions, where no ATP was present in the immunoprecipitation buffer (which contains 4 mM MgCl2 to allow ATP binding if ATP is added), only PpPex1p was precipitated when anti-PpPex1p antibody was used, and only PpPex6p was precipitated when anti-PpPex6p antibody was used (Fig. 3, lanes 1 and 4). These results show that each antibody is specific for the appropriate native target Pex protein. Strikingly, when 0.5 mM ATP was included in the immunoprecipitation (Fig. 3, lanes 2 and 5), either antibody coimmunoprecipitated both proteins, indicating that ATP binding and/or hydrolysis is essential for the formation of the complex containing PpPex1p and PpPex6p. Depletion of ATP in the extracts, through the addition of apyrase, resulted in no coimmunoprecipitation of PpPex1p and PpPex6p (Fig. 3, lanes 3 and 6), in keeping with the role of ATP in complex formation. It should be noted that the ATP-dependent complexes that were immunoprecipitated with either anti-PpPex1p or anti-PpPex6p antibody contained roughly equal amounts of PpPex1p and PpPex6p, suggesting that these proteins are present in a 1:1 ratio in the complexes.
FIG. 3.
Coimmunoprecipitation of PpPex1p and PpPex6p is ATP dependent. Shown are immunoblots (antibodies are indicated) of immunoprecipitates in which α-PpPex1p or α-PpPex6p was incubated with lysates from P. pastoris WT. The addition of ATP (0.5 mM) or apyrase (5 U/ml) to the immunoprecipitation buffer and wash buffer was as indicated.
PpPex1p and PpPex6p are present in a heterodimeric protein complex.
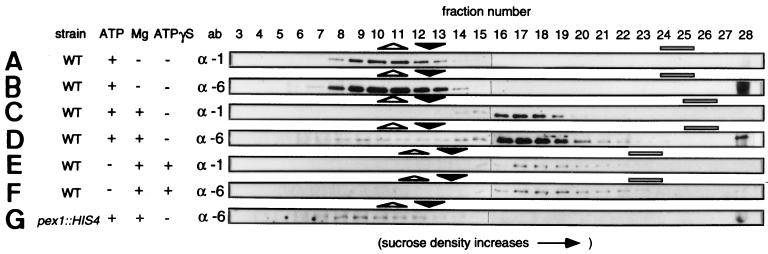
To further determine the molecular requirements for the PpPex1p-PpPex6p interaction and the size of the protein complexes, we generated total organelle-free extracts of methanol-grown WT cells in the absence or presence of either Mg2+, ATP, or both and determined the migration of PpPex1p and PpPex6p after continuous sucrose velocity gradient centrifugation. Triton X-100 and 100 mM KCl were included in the lysates to ensure that Pex1p and Pex6p were not associated with subcellular structures. In the presence of ATP (Fig. 4A and B) or Mg2+ alone (data not shown), both PpPex1p and PpPex6p migrated as proteins (or complexes) of approximately 150 kDa, similar to the migration of the endogenous 150-kDa standard dihydroxyacetone synthase (Fig. 4A and B). Bacterially expressed His6-tagged PpPex6p (deduced mass, 129 kDa) migrated similarly (data not shown), suggesting that PpPex1p and PpPex6p do not interact when either ATP or Mg2+ is absent. In contrast, PpPex1p and PpPex6p from lysates of wild-type yeast cells prepared in the presence of ATP and Mg2+ migrated as protein complexes with an estimated mass of between 320 and 400 kDa (Fig. 4C and D). ATP hydrolysis was not required to stabilize these complexes, because in the presence of Mg2+ and ATPγS, a nonhydrolyzable analog of ATP, the high-molecular-weight complex was observed (Fig. 4E and F). The amount of the complex was diminished in this particular experiment relative to that seen in the presence of ATP and Mg2+, but this was not the case in other experiments. Complex formation required both PpPex1p and PpPex6p because in the presence of Mg2+ and ATP, PpPex1p from a PpΔpex6 lysate (data not shown) and PpPex6p from a PpΔpex1 lysate migrated as monomers (Fig. 4G). These data demonstrate that PpPex1p and PpPex6p interact physically and that this interaction requires both proteins, ATP and Mg2+, but not ATP hydrolysis.
FIG. 4.
PpPex1p and PpPex6p are present in a protein complex of approximately 320 to 400 kDa. Shown is a Western blot analysis of equal volumes of fractions from sucrose gradients loaded with lysates prepared from P. pastoris WT (A to F) or a Pppex1::HIS4 strain (G). ATP (0.5 mM) (A to D and G), 4 mM MgCl2 (C to G), or the nonhydrolyzable analog of ATP, ATPγS (E and F), was added to the lysis buffer, and the effect on the migration of PpPex1p and PpPex6p was determined. To assess protein separation within each gradient, the locations of dihydroxyacetone synthase (DHAS, 150-kDa dimer), catalase (242-kDa tetramer), and methanol oxidase (640-kDa octamer, but not correctly assembled in Pppex1 and Pppex6 cells) were determined by Western blot analysis. The two fractions containing peak levels of DHAS (open triangle), catalase (closed triangle), and methanol oxidase (bar) are indicated above each panel. Ab, antibody. α-1 and α-6, α-PpPex1p and α-PpPex6p antibodies, respectively.
PpPex1p and PpPex6p are associated with membranous subcellular structures distinct from mature peroxisomes.
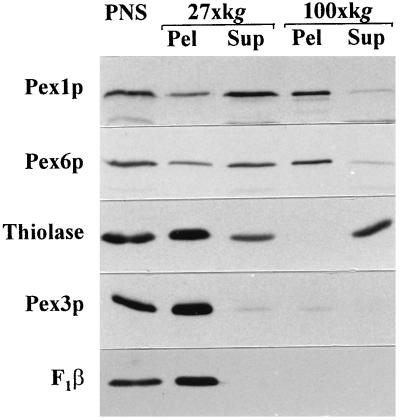
In order to gain insights into the roles of PpPex1p and PpPex6p in peroxisome biogenesis, their subcellular locations were examined in detail. Differential centrifugation of a PNS prepared from oleate-grown WT cells showed PpPex1p and PpPex6p to be predominantly in the 27,000 × g supernatant fraction; smaller amounts of PpPex1p and PpPex6p appeared in the 27,000 × g organellar pellet fraction, which consists primarily of peroxisomes and mitochondria (Fig. 5), as shown for the peroxisomal matrix protein thiolase, the peroxisomal membrane protein PpPex3p, and the F1β subunit of mitochondrial ATPase. The bulk of both PpPex1p and PpPex6p in the 27,000 × g supernatant fraction was subsequently pelleted at 100,000 × g, suggesting that these proteins might be associated with subcellular structures distinct from peroxisomes or might be part of a large protein complex (Fig. 5).
FIG. 5.
PpPex1p and PpPex6p are enriched in the 100,000 × g pellet fraction. A PNS prepared from oleate-grown WT cells was subjected to differential centrifugation. Equal volumes of the PNS, 27,000 × g pellet (27×kg Pel) or supernatant (27×kg Sup), and 100,000 × g pellet (100×kg Pel) or supernatant (100×kg Sup) were loaded in each lane and analyzed by Western blot analysis with antibodies to the indicated proteins.
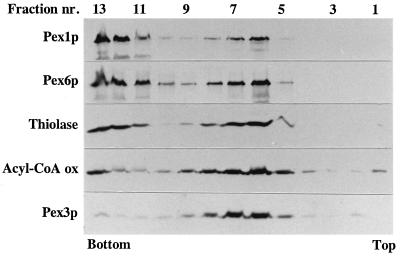
PpPex1p and PpPex6p present in the 100,000 × g pellet fraction are membrane-associated rather than aggregated proteins, since these proteins migrated up in a flotation gradient (Fig. 6). Peroxisomal matrix proteins (acyl coenzyme A oxidase and thiolase), which were found only in relatively low amounts in the 100,000 × g pellet fraction compared to the 27,000 × g pellet fraction, showed similar behavior in these experiments; PpPex3p, a peroxisomal integral membrane protein, floated almost completely into the gradient (Fig. 6). Some of PpPex1p and PpPex6p remained at the bottom of the gradient, perhaps because they were partially dissociated from the membranous structures with which they were associated. In control experiments, a soluble protein, bovine serum albumin, placed at the bottom of such flotation gradients, did not float into the gradient (data not shown). These experiments demonstrate that PpPex1p and PpPex6p are associated with membranes.
FIG. 6.
PpPex1p and PpPex6p are membrane associated. The 100,000 × g pellet fraction (obtained from the 27,000 × g supernatant fraction) prepared from oleate-grown WT cells was subjected to flotation gradient centrifugation. The gradient was drained from the top into 13 fractions (1, top; 13, bottom), and equal volumes of fractions were analyzed by Western blot analysis with antibodies to the indicated proteins. Very low amounts of thiolase, PpPex3p, and acyl coenzyme A oxidase (Acyl-CoA ox) were present in the 100,000 × g pellet fraction (relative to the 27,000 × g pellet fraction) used for flotation (Fig. 5). The Western blots analyzed for these proteins had to be overexposed, but clear signals were obtained due to the high quality of the antibodies. nr., number.
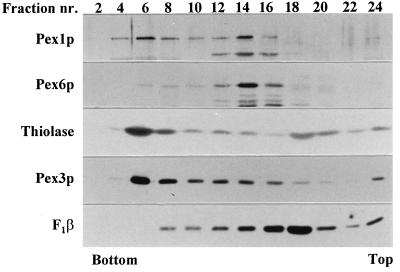
The association of these proteins with peroxisomes and/or other subcellular structures was analyzed by subjecting a PNS fraction from oleate-grown WT cells to isopycnic gradient centrifugation (Fig. 7). A clear separation was seen between intact peroxisomes (thiolase and PpPex3p: fraction 6) and mitochondria (F1β subunit of mitochondrial ATPase: fraction 18). Peak amounts of PpPex6p were observed in fraction 14, with trailing to fraction 6. PpPex1p showed a bimodal distribution in these gradients, with peak amounts in fractions 14 and 6. These data indicate that PpPex6p and, at least in part, PpPex1p are associated with membrane structures distinct from mature peroxisomes. Although the distribution of PpPex6p was very reproducible in all experiments, the relative amounts of PpPex1p in fractions 6 through 16 varied from experiment to experiment. Also, the peak fraction in the dense part of the gradient (fraction 6) did not always exactly colocalize with the peroxisomal peak fraction. Therefore, we analyzed the sedimentation behavior of these proteins under conditions in which the densities of peroxisomes and the structures containing PpPex1p and PpPex6p were more distinct.
FIG. 7.
PpPex1p and PpPex6p are associated with subcellular structures distinct from peroxisomes. Thiolase, the F1β subunit of mitochondrial ATPase, PpPex3p, PpPex1p, and PpPex6p in fractions of a linear Nycodenz gradient loaded with a PNS fraction of oleate-grown WT cells were subjected to immunodetection. The gradient was drained from the bottom into 24 fractions (2, bottom; 24, top), and equal volumes of every second fraction were analyzed by Western blot analysis. nr., number.
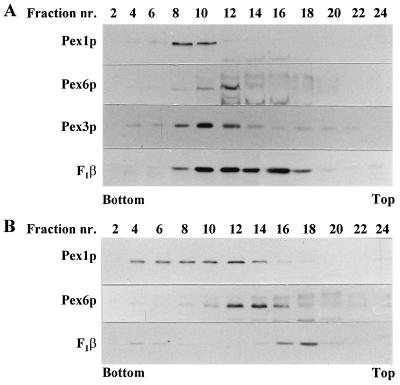
We made use of the fact that PpPex1p, PpPex3p, and PpPex6p are biochemically detectable under peroxisome-repressed conditions (i.e., in glucose-grown cells). Analysis of a Nycodenz gradient loaded with a PNS of glucose-grown WT cells showed the peak of PpPex3p in fraction 10. PpPex1p and PpPex6p were both concentrated in one part of the gradient only: PpPex1p in fractions 8 to 10 and PpPex6p in fraction 12 (Fig. 8A). Although partly overlapping, all three peroxins showed distinct sedimentation behaviors in these experiments and therefore are at least partly present on distinct subcellular membranous structures.
FIG. 8.
PpPex1p and PpPex6p are present on different subcellular structures independent of the presence or absence of peroxisomes. A PNS fraction was prepared from glucose-grown P. pastoris WT cells (A) or PpΔpex3 cells (B) and loaded onto a linear Nycodenz gradient. Equal volumes of every second fraction of the gradient drained from the bottom (fraction 1) were analyzed for the presence of the indicated proteins by Western blot analysis. nr., number.
The bimodal distribution observed for PpPex1p in the Nycodenz gradient prepared from oleate-grown cells is consistent with two alternative possibilities: (i) part of PpPex1p is associated with peroxisomes, and the rest is associated with another, unidentified subcellular membranous structure, or (ii) all of PpPex1p is associated with heterogeneous membrane structures distinct from peroxisomes. In order to distinguish between these possibilities and to obtain stronger evidence that the localizations of PpPex1p and PpPex6p were not peroxisomal, we analyzed the locations of these proteins in PpΔpex3 cells, which completely lack peroxisomes and peroxisomal remnants, as analyzed morphologically and biochemically (41).
Analysis of a Nycodenz gradient loaded with a PNS of glucose-grown PpΔpex3 cells revealed that both PpPex1p and PpPex6p migrated to densities similar to those in the gradient for WT cells (compare Fig. 8A and B). This result indicates that the sedimentation of PpPex1p and PpPex6p is independent of the presence of peroxisomes or remnants thereof and therefore that the structures containing these proteins could not correspond to peroxisomes. The result also suggests that PpPex1p is not necessarily present on peroxisomes in the gradient for WT cells but rather may be present on membranous structures with a density similar to that of peroxisomes.
DISCUSSION
Our results provide genetic and physical evidence for an interaction between PpPex1p and PpPex6p, two related peroxins of the AAA family. Genetic evidence for an interaction between these proteins is supported by the fact that the overexpression of PpPex6p suppressed a temperature-sensitive defect in PpPex1p (Fig. 1) and by the yeast two-hybrid experiments (Fig. 2). Physical evidence for an interaction between these proteins comes from the observation that they coimmunoprecipitated with each other in a 1:1 complex only in the presence of ATP (Fig. 3). In addition, in the presence of both proteins, ATP, and Mg2+, a heterodimeric complex of 320 to 400 kDa was detected in sucrose velocity gradients (Fig. 4). This interaction does not require ATP hydrolysis because it was seen even in the presence of ATPγS, a nonhydrolyzable analog. Like the bacterially expressed monomeric proteins, uncomplexed PpPex1p and PpPex6p migrated as approximately 150-kDa proteins in sucrose velocity gradients. Since the observed size of the complex was somewhat larger than the sum of the sizes of the interacting peroxins, we cannot rule out the possibility that some other protein(s) is also part of this complex.
PpPex1p and PpPex6p are associated with subcellular membranous structures distinct from peroxisomes, and only small amounts of these proteins may be peroxisome associated. During differential centrifugation, very little PpPex1p or PpPex6p was found in the 100,000 × g supernatant corresponding to the cytosol (Fig. 5). Instead, most of these proteins were associated with the 27,000 × g and 100,000 × g pellets. The presence of these proteins in the 100,000 × g pellet distinguishes their localization from that of other peroxisomal markers, most of which are associated with the 27,000 × g pellet. The structures with which these proteins associate in the 100,000 × g pellet contain membranes because they float in a sucrose flotation gradient (Fig. 6). Additional evidence for the localization of PpPex1p and PpPex6p to structures distinct from peroxisomes comes from their migration in isopycnic gradients. Their distribution not only is distinct from that of peroxisomes (Fig. 7) but also is independent of peroxisomes (Fig. 8), because in the PpΔpex3 strain, determined to lack peroxisomal remnants by morphological and biochemical criteria (41), the structures containing PpPex1p and PpPex6p persist and behave essentially as they do in wild-type cells.
The structures containing PpPex1p and PpPex6p are likely to be vesicles, based on their enrichment in the 100,000 × g pellet fraction, their behavior in flotation gradients, and their migration in Nycodenz gradients. Consistent with the lack of transmembrane segments in PpPex1p and PpPex6p, the association of these proteins with membranes appears to be peripheral, because we observed that these proteins are easily removed by salt washes (data not shown). This is the first report of the localization of peroxins to subcellular structures other than peroxisomes themselves and clearly implicates nonperoxisomal structures, which we believe to be vesicles, in peroxisome biogenesis.
Previous studies have shown that mutations in the most conserved ATP-binding domain of ScPex1p (homolog of PpPex1p) (8, 14) and mammalian Pex6p (homolog of PpPex6p) (35, 42) abolish the ability of these proteins to function in biogenesis. This result suggests that the ATP-dependent interaction between PpPex1p and PpPex6p may be impaired in these mutants. The PpPex1p-PpPex6p interaction and the location of these peroxins on membranous structures of different densities also suggest that each protein has a distinct function and explain why the expression of either protein cannot substitute for the absence of the other (data not shown), despite the fact that the proteins are structurally similar (each is approximately 130 kDa, and they have 29% sequence identity and 49% sequence similarity) (11).
Different subcellular locations have been reported for Pex6p in different organisms. Rat Pex6p was reported to be solely peroxisome associated (35), human Pex6p was found to be mainly cytosolic (42), and P. pastoris Pex6p is, as described here, primarily associated with vesicular structures distinct from peroxisomes. The reason for this discrepancy may relate to variations in the methods used for cell fractionation and protein detection. Tsukamoto et al. (35) noted that the levels of Pex6p were so low in rat liver that they were unable to detect the protein in various subcellular fractions, except for those highly enriched in peroxisomes. They did not describe the distribution of the protein in the 100,000 × g pellet fraction. Our data showing the enrichment of PpPex6p in the 100,000 × g pellet fraction suggest that the corresponding rat liver fraction might have contained rat Pex6p. Yahraus et al. (42) expressed myc epitope-tagged Pex6p from the strong cytomegalovirus promoter in human cells and found it to be predominantly cytoplasmic by indirect immunofluorescence. This result might have been observed if the level of this protein was substantially higher than that seen endogenously or if the location of the protein was altered by the epitope tag.
Our studies highlight two important questions that remain the goal of our future work. What is the subcellular origin of the membrane vesicles containing Pex1p and Pex6p? What role do these vesicles play in the biogenesis of peroxisomes? Purification and characterization of the vesicles will reveal whether they are derived from the endoplasmic reticulum, as proposed recently based on a reexamination of available data (30), or from some other source. With respect to the second question, our earlier data show that P. pastoris mutants lacking PpPex1p and PpPex6p accumulate peroxisomal remnants that import some matrix and membrane proteins, but these remnants are smaller and fewer in number than peroxisomes in wild-type yeast cells under similar conditions (11, 29). Similarly, human patients lacking Pex6p contain peroxisome ghosts capable of importing peroxisomal membrane proteins, as well as reduced levels of PTS1 and PTS2 proteins (27, 42). These results suggest that the defect in cells lacking Pex1p or Pex6p may be in the growth of the peroxisomal compartment and not directly in import per se. Because Pex5p, the PTS1 receptor, is unstable in some human cell lines lacking Pex6p, it has been suggested that Pex6p may be involved in the assembly of the peroxisomal matrix protein import machinery (42). Although this idea cannot be completely ruled out at present, it is difficult to reconcile a direct role of Pex1p and Pex6p in the stability of Pex5p and therefore in import, in view of the following facts. (i) Some amount of matrix protein import is still observed in yeast mutants with deletions of the PEX1 and PEX6 genes. (ii) The majority of Pex1p and Pex6p does not colocalize (in the absence of ATP) to identical subcellular structures or to the cytosolic and peroxisomal locations reported for Pex5p. (iii) An impairment in biogenesis may indirectly affect the assembly of the import machinery. (iv) Our yeast two-hybrid analysis revealed no interactions between either PpPex1p and PpPex5p or PpPex6p and PpPex5p. (v) Our published work has shown that in a PpΔpex1 strain, the induction and steady-state levels of PpPex5p are comparable to those seen for wild-type cells, whether the cells are grown on methanol or oleate (11). Our data, which localize most of PpPex1p and PpPex6p not to the cytosol or peroxisomes but to distinct membranous structures, coupled with the Mg2+- and ATP-dependent association of these proteins and the requirement of the ATP-binding domains on these proteins for peroxisome biogenesis, suggest that the interaction between Pex1p and Pex6p could serve to juxtapose the vesicles, perhaps facilitating the assembly of components required for membrane fusion or protein import into peroxisomes. Successive rounds of fusion would generate larger vesicles, which could then assemble the import machinery on the membrane, import matrix and membrane proteins, and mature into larger peroxisomes. Alternatively, the larger vesicles could fuse with preexisting peroxisomes to allow their growth. This model, involving a role for vesicles in peroxisome biogenesis, provides a mechanism for lipid and/or membrane (protein and lipid) addition to peroxisome membranes during biogenesis and explains the observation that mutant cells which apparently lack all peroxisomal remnants (4, 41) can generate peroxisomes after reintroduction of the complementing gene. The recovery of peroxisomes in such complemented mutants would be difficult to explain with the currently favored model for peroxisome biogenesis, in which the organelles are believed to arise exclusively by budding and fission of preexisting organelles.
Although this proposed role of vesicle fusion in peroxisome biogenesis remains to be tested, it appears reasonable because PpPex1p and PpPex6p have high homology to proteins thought to facilitate membrane fusion events (NSF and its yeast homolog, Sec18p [10, 28], as well as p97 and its yeast homolog, Cdc48p [17, 24]). These NSF-like ATPases of the AAA family act as chaperones by activating SNARE proteins, which then participate in membrane fusion events, including Golgi reassembly (1, 24), endoplasmic reticulum fusion (17), intracellular protein trafficking (5), and neuronal secretion (21).
Recently, an ATP-dependent interaction was described for another pair of proteins belonging to the AAA family (3). These mitochondrial proteins, Yta10p and Yta12p of S. cerevisiae, are proteases whose activity is regulated by an ATP-dependent interaction and by ATP hydrolysis, with the protease being active in the complexed but not in the uncomplexed state. Independent of the proteolytic function, they also have a chaperonelike activity required for the assembly of membrane-associated mitochondrial ATP synthase. PpPex1p and PpPex6p are required for peroxisome biogenesis, have no known protease domains, and are therefore unlikely to be involved in protein degradation. However, their activity as molecular chaperones that facilitate peroxisome assembly or in promoting vesicle fusion directly or indirectly is likely to be regulated by their ATP-dependent association and by ATP hydrolysis, analogous to the way in which Yta10p and Yta12p functioning is modulated.
ACKNOWLEDGMENTS
K.N.F. and J.A.H. contributed equally to this work.
We thank Michael P. Yaffe for the F1β antiserum and the use of his microscope and Stan Hollenberg, Oregon Health Sciences University, Portland, for the two-hybrid vectors. We thank Georg Lüers for the development of the gradient protocols using the PNS, help with experiments, and valuable advice during the course of this work. We also thank Alan Spong, Thibaut J. Wenzel, Chris Burd, and Oliver Tuescher for helpful discussions and experimental observations.
This work was supported by a fellowship from HFSPO (to K.N.F.) and by a grant from the NIH (DK41737).
REFERENCES
- 1.Acharya U, Jacobs R, Peters J M, Watson N, Farquhar M G, Malhotra V. The formation of Golgi stacks from vesiculated Golgi membranes requires two distinct fusion events. Cell. 1995;82:895–904. doi: 10.1016/0092-8674(95)90269-4. [DOI] [PubMed] [Google Scholar]
- 2.Albertini M, Rehling P, Erdmann R, Girzalsky W, Kiel J A K W, Veenhuis M, Kunau W-H. Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell. 1997;89:83–92. doi: 10.1016/s0092-8674(00)80185-3. [DOI] [PubMed] [Google Scholar]
- 3.Arlt H, Tauer R, Feldmann H, Neupert W, Langer T. The YTA10-12 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell. 1996;85:875–885. doi: 10.1016/s0092-8674(00)81271-4. [DOI] [PubMed] [Google Scholar]
- 4.Baerends R J S, Rasmussen S W, Hilbrands R E, van der Heide M, Faber K N, Reuvekamp P T W, Kiel J A K W, Cregg J M, van der Klei I J, Veenhuis M. The Hansenula polymorpha PER9 gene encodes a peroxisomal membrane protein essential for peroxisome assembly and integrity. J Biol Chem. 1996;271:8887–8894. doi: 10.1074/jbc.271.15.8887. [DOI] [PubMed] [Google Scholar]
- 5.Beckers C J, Block M R, Glick B S, Rothman J E, Balch W E. Vesicular transport between the endoplasmic reticulum and the Golgi stack requires the NEM-sensitive fusion protein. Nature. 1989;339:397–398. doi: 10.1038/339397a0. [DOI] [PubMed] [Google Scholar]
- 6.Berteaux L V, Picard M, Thompson C C, Zickler D, Panvier A A, Simonet J M. A nonmammalian homolog of the PAF1 gene (Zellweger syndrome) discovered as a gene involved in caryogamy in the fungus Podospora anserina. Cell. 1995;81:1043–1051. doi: 10.1016/s0092-8674(05)80009-1. [DOI] [PubMed] [Google Scholar]
- 7.Distel B, Erdmann R, Gould S J, Blobel G, Crane D I, Cregg J M, Dodt G, Fujiki Y, Goodman J M, Just W W, Kiel J A K W, Kunau W-H, Lazarow P B, Mannaerts G P, Moser H, Osumi T, Rachubinski R A, Roscher A, Subramani S, Tabak H F, Valle D, van der Klei I J, van Veldhoven P, Veenhuis M. A unified nomenclature for peroxisome biogenesis. J Cell Biol. 1996;135:1–3. doi: 10.1083/jcb.135.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erdmann R, Wiebel F F, Flessau A, Rytka J, Beyer A, Frohlich K U, Kunau W-H. PAS1, a yeast gene required for peroxisome biogenesis, encodes a member of a novel family of putative ATPases. Cell. 1991;64:499–510. doi: 10.1016/0092-8674(91)90234-p. [DOI] [PubMed] [Google Scholar]
- 9.Gould S J, McCollum D, Spong A P, Heyman J A, Subramani S. Development of the yeast Pichia pastoris as a model organism for a genetic and molecular analysis of peroxisome assembly. Yeast. 1992;8:613–628. doi: 10.1002/yea.320080805. [DOI] [PubMed] [Google Scholar]
- 10.Graham T R, Emr S D. Compartmental organization of Golgi-specific protein modification and vacuolar protein sorting events defined in a yeast sec18 (NSF) mutant. J Cell Biol. 1991;114:207–218. doi: 10.1083/jcb.114.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heyman J A, Monosov E, Subramani S. Role of the PAS1 gene of Pichia pastoris in peroxisome biogenesis. J Cell Biol. 1994;127:1259–1273. doi: 10.1083/jcb.127.5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hollenberg S M, Sternglanz R, Cheng P F, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komori M, Rasmussen S W, Kiel J A K W, Baerends R S J, Cregg J M, van der Klei I J, Veenhuis M. The Hansenula polymorpha PEX14 gene encodes a novel peroxisomal membrane protein essential for peroxisome biogenesis. EMBO J. 1996;16:44–53. doi: 10.1093/emboj/16.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krause T, Kunau W H, Erdmann R. Effect of site-directed mutagenesis of conserved lysine residues upon Pas1 protein function in peroxisome biogenesis. Yeast. 1994;10:1613–1620. doi: 10.1002/yea.320101210. [DOI] [PubMed] [Google Scholar]
- 15.Kunau W H, Beyer A, Franken T, Gotte K, Marzioch M, Saidowsky J, Skaletz R A, Wiebel F F. Two complementary approaches to study peroxisome biogenesis in Saccharomyces cerevisiae: forward and reversed genetics. Biochimie. 1993;75:209–224. doi: 10.1016/0300-9084(93)90079-8. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli U K, Favre M. Maturation of the head of bacteriophage T4. I. DNA packaging events. J Mol Biol. 1973;80:575–599. doi: 10.1016/0022-2836(73)90198-8. [DOI] [PubMed] [Google Scholar]
- 17.Latterich M, Frohlich K U, Schekman R. Membrane fusion and the cell cycle: Cdc48p participates in the fusion of ER membranes. Cell. 1995;82:885–893. doi: 10.1016/0092-8674(95)90268-6. [DOI] [PubMed] [Google Scholar]
- 18.Lazarow P B, Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- 19.McCollum D, Monosov E, Subramani S. The pas8 mutant of Pichia pastoris exhibits the peroxisomal protein import deficiencies of Zellweger syndrome cells—the PAS8 protein binds to the COOH-terminal tripeptide peroxisomal targeting signal, and is a member of the TPR protein family. J Cell Biol. 1993;121:761–774. doi: 10.1083/jcb.121.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monosov E Z, Wenzel T J, Luers G H, Heyman J A, Subramani S. Labeling of peroxisomes with green fluorescent protein in living P. pastoris cells. J Histochem Cytochem. 1996;44:581–589. doi: 10.1177/44.6.8666743. [DOI] [PubMed] [Google Scholar]
- 21.Morgan A, Burgoyne R D. A role for soluble NSF attachment proteins (SNAPs) in regulated exocytosis in adrenal chromaffin cells. EMBO J. 1995;14:232–239. doi: 10.1002/j.1460-2075.1995.tb06996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- 23.Nuttley W M, Brade A M, Eitzen G A, Veenhuis M, Aitchison J D, Szilard R K, Glover J R, Rachubinski R A. PAY4, a gene required for peroxisome assembly in the yeast Yarrowia lipolytica, encodes a novel member of a family of putative ATPases. J Biol Chem. 1994;269:556–566. [PubMed] [Google Scholar]
- 24.Rabouille C, Levine T P, Peters J M, Warren G. An NSF-like ATPase, p97, and NSF mediate cisternal regrowth from mitotic Golgi fragments. Cell. 1995;82:905–914. doi: 10.1016/0092-8674(95)90270-8. [DOI] [PubMed] [Google Scholar]
- 25.Rickey C. Efficient electroporation of yeast. Mol Biol Rep. 1990;9:2–3. [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Slawecki M L, Dodt G, Steinberg S, Moser A B, Moser H W, Gould S J. Identification of three distinct peroxisomal protein import defects in patients with peroxisome biogenesis disorders. J Cell Sci. 1995;108:1817–1829. doi: 10.1242/jcs.108.5.1817. [DOI] [PubMed] [Google Scholar]
- 28.Söllner T, Bennett M K, Whiteheart S W, Scheller R H, Rothman J E. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- 29.Spong A P, Subramani S. Cloning and characterization of PAS5: a gene required for peroxisome biogenesis in the methylotrophic yeast Pichia pastoris. J Cell Biol. 1993;123:535–548. doi: 10.1083/jcb.123.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramani S. Protein translocation into peroxisomes. J Biol Chem. 1996;271:32483–32486. doi: 10.1074/jbc.271.51.32483. [DOI] [PubMed] [Google Scholar]
- 31.Terlecky S R, Nuttley W M, McCollum D, Sock E, Subramani S. The Pichia pastoris peroxisomal protein PAS8p is the receptor for the C-terminal tripeptide peroxisomal targeting signal. EMBO J. 1995;14:3627–3634. doi: 10.1002/j.1460-2075.1995.tb00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terlecky S R, Nuttley W M, Subramani S. The requisite cytosolic and membrane components of peroxisomal protein import. Cell Mol Life Sci. 1996;52:1050–1054. doi: 10.1007/BF01952101. [DOI] [PubMed] [Google Scholar]
- 33.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tschopp J F, Cregg J M. Heterologous gene expression in methylotrophic yeast. Bio/Technology. 1991;18:305–322. doi: 10.1016/b978-0-7506-9188-8.50020-6. [DOI] [PubMed] [Google Scholar]
- 35.Tsukamoto T, Miura S, Nakai T, Yokota S, Shimozawa N, Suzuki Y, Orii T, Fujiki Y, Sakai F, Bogaki A, Yasumo H, Osumi T. Peroxisome assembly factor-2, a putative ATPase cloned by functional complementation on a peroxisome-deficient mammalian cell mutant. Nature Genet. 1995;11:395–401. doi: 10.1038/ng1295-395. [DOI] [PubMed] [Google Scholar]
- 36.Veenhuis M, Keizer I, Harder W. Characterization of peroxisomes in glucose-grown Hansenula polymorpha and their development after the transfer of cells into methanol-containing media. Arch Microbiol. 1979;120:167–175. [Google Scholar]
- 37.Veenhuis M, van Dijken J, Harder W. The significance of peroxisomes in the metabolism of one-carbon compounds in yeasts. Adv Microb Physiol. 1983;24:1–82. doi: 10.1016/s0065-2911(08)60384-7. [DOI] [PubMed] [Google Scholar]
- 38.Voorn-Brouwer T, van der Leij I, Hemrika W, Distel B, Tabak H F. Sequence of the PAS8 gene, the product of which is essential for biogenesis of peroxisomes in Saccharomyces cerevisiae. Biochim Biophys Acta. 1993;1216:325–328. doi: 10.1016/0167-4781(93)90166-b. [DOI] [PubMed] [Google Scholar]
- 39.Waterham H R, Titorenko V I, Swaving G J, Harder W, Veenhuis M. Peroxisomes in the methylotrophic yeast Hansenula polymorpha do not necessarily derive from pre-existing organelles. EMBO J. 1993;12:4785–4794. doi: 10.1002/j.1460-2075.1993.tb06167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39a.Wenzel, T., and S. Subramani. Unpublished data.
- 40.Wiemer E A, Subramani S. Protein import deficiencies in human peroxisomal disorders. Mol Genet Med. 1994;4:119–152. doi: 10.1016/b978-0-12-462004-9.50008-6. [DOI] [PubMed] [Google Scholar]
- 41.Wiemer E A C, Lüers G, Faber K N, Wenzel T J, Veenhuis M, Subramani S. Isolation and characterization of Pas2p, a peroxisomal membrane protein essential for peroxisome biogenesis in the methylotrophic yeast Pichia pastoris. J Biol Chem. 1996;271:18973–18980. doi: 10.1074/jbc.271.31.18973. [DOI] [PubMed] [Google Scholar]
- 42.Yahraus T, Braverman N, Dodt G, Kalish J, Morell J C, Moser H W, Valle D, Gould S J. The peroxisome biogenesis disorder group 4 gene, PXAAA1, encodes a cytoplasmic ATPase required for stability of the PTS1 receptor. EMBO J. 1996;15:2914–2923. [PMC free article] [PubMed] [Google Scholar]