ABSTRACT
Despite the significant presence of plant-derived tricarboxylic acids in some environments, few studies detail the bacterial metabolism of trans-aconitic acid (Taa) and tricarballylic acid (Tcb). In a soil bacterium, Acinetobacter baylyi ADP1, we discovered interrelated pathways for the consumption of Taa and Tcb. An intricate regulatory scheme tightly controls the transport and catabolism of both compounds and may reflect that they can be toxic inhibitors of the tricarboxylic acid cycle. The genes encoding two similar LysR-type transcriptional regulators, TcuR and TclR, were clustered on the chromosome with tcuA and tcuB, genes required for Tcb consumption. The genetic organization differed from that in Salmonella enterica serovar Typhimurium, in which tcuA and tcuB form an operon with a transporter gene, tcuC. In A. baylyi, tcuC was not cotranscribed with tcuAB. Rather, tcuC was cotranscribed with a gene, designated pacI, encoding an isomerase needed for Taa consumption. TcuC appears to transport Tcb and cis-aconitic acid (Caa), the presumed product of PacI-mediated periplasmic isomerization of Taa. Two operons, tcuC-pacI and tcuAB, were transcriptionally controlled by both TcuR and TclR, which have overlapping functions. We investigated the roles of the two regulators in activating transcription of both operons in response to multiple effector compounds, including Taa, Tcb, and Caa.
IMPORTANCE
Ingestion of Taa and Tcb by grazing livestock can cause a serious metabolic disorder called grass tetany. The disorder, which results from Tcb absorption by ruminants, focuses attention on the metabolism of tricarboxylic acids. Additional interest stems from efforts to produce tricarboxylic acids as commodity chemicals. Improved understanding of bacterial enzymes and pathways for tricarboxylic acid metabolism may contribute to new biomanufacturing strategies.
KEYWORDS: ADP1, Acinetobacter baylyi, TcuR, aconitate isomerase, LTTR, LysR, transcriptional regulator, tricarboxylic acids, tricarballylate, aconitate
INTRODUCTION
Tricarboxylic acids, produced by grasses and legumes, can be relatively abundant in plant-based forage. These compounds, such as tricarballylic acid (Tcb) and trans-aconitic acid (Taa), have been of interest because of problems that occur when grazing ruminants ingest them. Tcb in the rumen gets absorbed by the animal wherein the chelation properties of Tcb can cause a serious magnesium deficiency that manifests as a malady called grass tetany (1). This problem, which can be fatal to livestock, was at one time attributed to Taa but later shown to result from the conversion of Taa to Tcb by microbial fermentation in the rumen (2, 3).
The biochemical basis for the bacterial uptake and use of Tcb as a sole carbon source has, to date, been reported for only Salmonella enterica subspecies enterica serovar Typhimurium LT2 (hereafter LT2; 4–7). Fewer studies have been published on the microbial consumption of Taa as a growth substrate. The only biochemical studies center on a constitutively expressed periplasmic aconitate isomerase involved in Taa assimilation by Pseudomonas sp. WU-0701 (8, 9). Further information about Taa assimilation or transport has not been reported. Interest in this aconitate isomerase stems from biomanufacturing because tricarboxylic acids are important commodity chemicals that have the potential to be synthesized by bacteria (10, 11). Heterologous expression of the aconitate isomerase was used in a two-step bioconversion of citric acid (Cit) to trans-aconitic acid (Taa) by whole bacterial cells (12).
As current understanding of these pathways remains incomplete, we characterized the consumption of cis-aconitic acid (Caa), Taa, Tcb, and Cit by a soil bacterium, Acinetobacter baylyi ADP1. This metabolically versatile bacterium was chosen because its environmental habitat is replete with plant-derived growth substrates. Moreover, ADP1 has a uniquely powerful genetic system that facilitates investigation using methods to exploit its exceptionally high efficiency of natural transformation and homologous recombination. Hypotheses concerning metabolism can be readily tested with mutational approaches. This soil bacterium is gaining traction as a model organism for metabolic engineering (13, 14).
Genome sequence and a global metabolic model of ADP1 (15) identified genes for Tcb catabolism based on homology to genes characterized in LT2, tcuABC, and tcuR (4–7). Studies of the Tcu proteins in LT2 demonstrate that TcuA is an FAD-dependent Tcb dehydrogenase and TcuB is a transmembrane protein that re-oxidizes the flavin cofactor of the dehydrogenase (Fig. 1; 6, 7). In LT2, tcuC, which encodes a Tcb transporter, is cotranscribed with tcuA and tcuB. An adjacent regulatory gene, tcuR, encodes a LysR-type transcriptional regulator (LTTR) that activates the tcuABC operon in response to Tcb (5). LT2 does not use Taa as a sole carbon source, and the consumption of this compound by ADP1 was not previously determined.
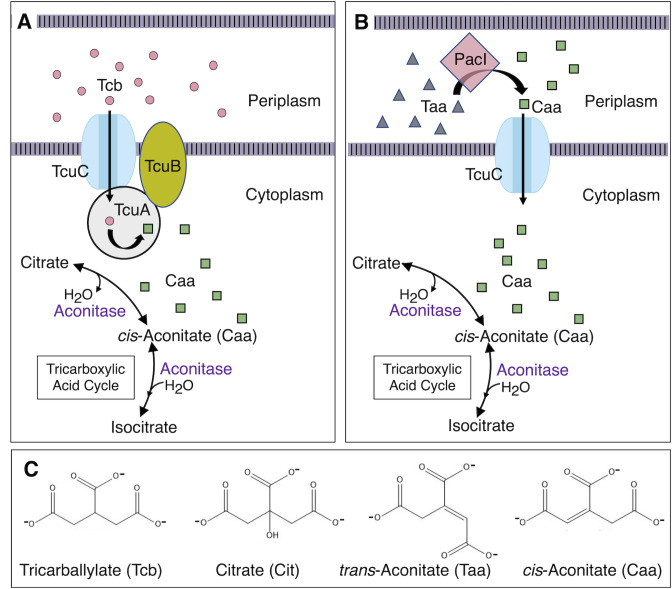
Fig 1.
Tcb, Taa, and Caa utilization. (A) The functions of TcuA and TcuB in converting Tcb (circles) to Caa (squares) and the role of TcuC in Tcb transport, which were demonstrated in strain LT2 (4–7), are predicted to be the same in ADP1. (B) In ADP1, our results support a model in which PacI, which does not have a counterpart in LT2, converts Taa (triangles) to Caa (squares) in the periplasm, prior to transport into the cytoplasm by TcuC. (C) Chemical structures are shown for tricarboxylic acids investigated in this report.
The tcu gene order in ADP1 is similar but not identical to that of LT2 (Fig. 2). Notably, tcuC is not adjacent to tcuA and tcuB. As described in this report, we investigated the roles of two tcuR paralogs near tcuA in the regulation of tricarboxylate metabolism in ADP1 and renamed one tclR (TcuR-like regulator). Studies of genetic regulation and function revealed that the uncharacterized gene immediately downstream of tcuC (locus tag ACIAD_RS07095) in ADP1 enables the use of Taa as a sole carbon source. Based on the homology of its gene product to the aconitase of Pseudomonas sp. WU-0701 (8, 9), we designated this gene pacI (for periplasmic aconitate isomerase). Here, we present a novel metabolic and regulatory scheme whereby the growth of ADP1 on Taa involves the periplasmic PacI-mediated conversion of Taa to Caa followed by the TcuC-mediated transport of Caa into the cytoplasm (Fig. 1B). The TcuC transport protein appears to have dual specificity for Caa and Tcb.
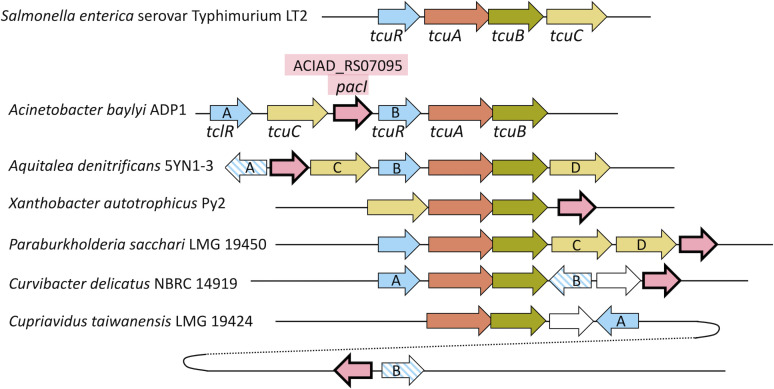
Fig 2.
Chromosomal regions from diverse bacteria (drawn to scale), aligned relative to tcuB. Homologs, shown in the same color and pattern, were aligned using SyntTax and BioCyc programs (16, 17). Similarity searches based on pacI (ACIAD_RS07095) revealed four bacteria in which homologs (in pink) were clustered in the vicinity of tcuB homologs. In Cupriavidus taiwanensis LMG 19424, a pacI homolog is adjacent and divergent to an LTTR gene, but it is distal to the tcu-like region. In this case, tcuA is at map position 1,262,684–1,264,114, and the pacI homolog (RALTA_RS24780) is at map position 2,072,478–2,073,302 (NCBI Reference Sequence: NC_010530.1). Sequence identifiers for genomes are listed in Materials and Methods. Labels A and B (arbitrarily assigned) indicate two LTTR paralogs. Striped patterns mark three LTTR genes that are more like each other in sequence than they are to the solid-colored LTTR genes (Fig. S2 and S3). Labels C and D (arbitrarily assigned) denote two tcuC-like paralogs in the same region predicted to encode transporters in the major facilitator superfamily (MFS), entry IPR011701 of InterPro (18); their sequences are aligned in Fig. S5. In each bacterium, the tcuA and tcuB genes have overlapping stop and start codons, a configuration that may couple translation to produce equal levels of both interrelated proteins (19).
RESULTS
Organization and bioinformatic analysis of clustered tcu genes in A. baylyi ADP1
Database searches revealed many bacterial homologs of TcuA, TcuB, TcuC, and TcuR that shared sequence similarity comparable to that between the gene products in ADP1 and LT2 (Table S1). However, the gene downstream of tcuC in ADP1 has no counterpart in LT2, and we questioned its role. Genome annotation and homology searches suggested only that this gene, ACIAD_RS07095, encodes a substrate-binding periplasmic transport protein of unknown function. We posited that if ACIAD_RS07095 homologs clustered with tcu genes in diverse bacteria, it might signify that this gene has a function related to Tcb metabolism. To identify genomic synteny among bacteria with tcu genes near homologs of ACIAD_RS07095, chromosomal neighborhoods near tcuB were searched with the online STRING program (20). Regions in diverse bacteria that might be of interest were examined individually, and several different genetic arrangements were found with tcuAB homologs in the vicinity of homologs of the gene of unknown function from ADP1 (Fig. 2).
The protein products of the homologs of unknown function were aligned, and conserved sequences guided additional homology searches. An iterative approach revealed homology to the aconitate isomerase of Pseudomonas sp. WU-0701 (9), as shown in Fig. S1. The genomic context in strain WU-0701 was unclear because surrounding DNA sequences are unavailable. However, a protein encoded by Pseudomonas oryzihabitans was found to differ from that of WU-0701 by a single amino acid (P or S at position 202, Fig. S1). There were no tcu genes in the genome of P. oryzihabitans and no LTTR genes close to this putative aconitate isomerase gene. Nevertheless, we explored the possibility that Tcb and aconitate (Taa and Caa) metabolism were linked in ADP1. We hypothesized that ACIAD_RS07095 encodes an aconitate isomerase, designated PacI, that converts Taa to Caa. Periplasmic localization was predicted by SignalP analysis (21). This program identified the first 24 amino acids of PacI as a cleavable signal used to export the enzyme. Therefore, our model depicts the production of Caa from Taa in the periplasm followed by transport into the cytoplasm (Fig. 1B).
Effects of gene-specific mutation: growth on tricarboxylic acids as sole carbon sources
We tested whether genes in the tcu region of ADP1 affect the regulation and metabolism of tricarboxylic acids after constructing mutants with specific deletions (described in Table 1; Table S2 through S4). These strains were compared with ADP1 for growth using Cit, Tcb, Taa, Caa, or pyruvate as the carbon source. ADP1 grows well on pyruvate, unlike sugars, and pyruvate is typically used as a growth substrate. The growth rates of ADP1 and all mutants on pyruvate were comparable (Table 1). Isocitrate was also tested, but no growth was observed.
TABLE 1.
Growth of ADP1 and mutants on different sole carbon sources
| Strain | ADP1 | ACN1388 | ACN3225 | ACN1456 | ACN3168 | ACN3123 | ACN3178 | ACN1376 | ACN3171 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Genotypea | WT | ΔpacI | ΔpacI | ΔtcuC | ΔtcuC | ΔtcuA::gfpsf | ΔtclR | ΔtcuR | ΔtclR | |
| tcuC-gfp sf | ΔcltA | ΔtcuR | ||||||||
| Carbon source | Parameter | |||||||||
| Pyre | Growth rateb | 1 ± 0.1 | 1.1 ± 0.1 | 1.2 ± 0.1 | 1 ± 0.1 | 1.2 ± 0.1 | 1.2 ± 0.2 | 1.1 ± 0.1 | 1.1 ± 0.1 | 1.2 ± 0.1 |
| Max ODc | 1 ± 0.1 | 1.1 ± 0.2 | 1 ± 0.2 | 1 ± 0.1 | 1.1 ± 0.2 | 1.1 ± 0.2 | .9 ±0.2 | 1 ± 0.2 | .9 ±0.2 | |
| Lag (h)d | 2.25 | 2.75 | 3.25 | 2.75 | 2.75 | 3.25 | 2.25 | 2.25 | 2.75 | |
| Cit | Growth rateb | 1.1 ± 0.2 | 1.1 ± 0.1 | 1.2 ± 0.2 | 1.4 ± 0.1 | 1.2 ± 0.2 | 1.2 ± 0.2 | 1.2 ± 0.1 | 1 ± 0.03 | 1.3 ± 0.2 |
| Max ODc | 1 ± 0.2 | 1.1 ± 0.2 | 1.2 ± 0.2 | 1 ± 0.1 | 1 ± 0.2 | 1.1 ± 0.2 | 1 ± 0.1 | 1 ± 0.1 | 1 ± 0.1 | |
| Lag (h)d | 3.25 | 2.75 | 2.75 | 3.25 | 4.75 | 3.25 | 3.75 | 3.75 | 4.25 | |
| Tcb | Growth rateb | 1 ± 0.1 | 1 ± 0.1 | .2 ±0.01 | NGf | NG | NG | .2 ±0.03 | NG | NG |
| Max ODc | .9 ±0.1 | .6 ±0.1 | .7 ±0.1 | NG | NG | NG | .5 ±0.05 | NG | NG | |
| Lag (h)d | 7.75 | 10.25 | 18.75 | NG | NG | NG | 43.25 | NG | NG | |
| Taa | Growth rateb | .9 ±0.1 | NG | NG | NG | NG | 1.1 ± 0.03 | NG | 1.1 ± 0.1 | NG |
| Max ODc | 1 ± 0.2 | NG | NG | NG | NG | 1.1 ± 0.2 | NG | 1 ± 0.2 | NG | |
| Lag (h)d | 2.75 | NG | NG | NG | NG | 3.25 | NG | 2.75 | NG | |
| Caa | Growth rateb | 1.3 ± 0.2 | 1.5 ± 0.1 | 1.26 ± 0.36 | NG | NG | 1.2 ± 0.1 | NG | .9 ±0.1 | NG |
| Max ODc | 1.1 ± 0.2 | 1 ± 0.1 | 1.02 ± 0.14 | NG | NG | 1.1 ± 0.2 | NG | 1.1 ± 0.2 | NG | |
| Lag (h)d | 2.25 | 2.25 | 2.75 | NG | NG | 2.75 | NG | 2.25 | NG |
Additional information on strain genotypes is provided in Table 2; Tables S2 to S4.
Ratio of the growth rate of each strain on the indicated carbon source to that of ADP1 grown on pyruvate. A 95% CI calculated from three replicates is indicated by ±values. The generation time of pyruvate-grown ADP1 is 1.3 h when grown in shaking tubes/flasks and 3.0 h when grown in the plate reader.
Max growth assessed by turbidity (measured as OD595) in comparison to ADP1 grown on Pyr. A 95% CI calculated from three replicates is represented by ±values.
A graphing program (Prism, GraphPad Software) was used to calculate average growth rates across replicates at 30-min intervals. Lag time is here defined as the time taken for the growth rate to increase by ≥0.01 (absorbance unit/h), which corresponds to the beginning of the log phase. Standard deviations of lag times are <15% of the average values.
Carbon sources were added to the minimal medium at the following final concentrations: 15-mM pyruvate (Pyr), 5-mM Cit, 5-mM Tcb, 5-mM Taa, and 5-mM Caa.
No growth (NG) is defined as an increase in OD595 ≤0.05 within 72 h following inoculation.
Deletion of pacI (in ACN1388) prevented growth on Taa as the carbon source but not growth on Tcb or Caa (Taa-, Tcb+, and Caa+). In contrast, a tcuA mutant (ACN3123) was Taa+ and Caa+ but did not grow on Tcb (Table 1). These results support a model where TcuA is required to grow on Tcb (Fig. 1A), and the loss of PacI only prevents growth on Taa (Fig. 1B). A ΔtcuC mutant (ACN1456) was Tcb-, Taa-, and Caa- but grew on pyruvate and Cit (Table 1). These results are consistent with TcuC transporting Tcb, as in LT2 (4). Failure to grow on both compounds could be explained by a model wherein Taa is converted to Caa in the periplasm and TcuC transports Caa in addition to Tcb (Fig. 1B). Growth on Taa with Caa as an intermediate is supported by the observation that the four mutants that are Caa- are all also Taa- (Table 1).
Growth of the ΔtcuR mutant (ACN1376) on Caa suggests that the TcuR regulator in ADP1 is not required for TcuC expression, unlike in LT2 where transcription of the tcuABC operon requires TcuR (5). Different gene organization also raises regulatory questions about the roles of two tcuR paralogs in ADP1, one designated tclR (Fig. 2). The ΔtclR mutant (ACN3178) and the ΔtclR plus ΔtcuR mutant (ACN3171) were both Caa- and Taa- (Table 1). Therefore, TclR rather than TcuR may control TcuC expression in ADP1 during growth on these substrates.
Since growth on Tcb required tcuR (in ACN1376), TcuR may control the transcription of tcu genes, as does its counterpart in LT2. However, deletion of tclR also affected growth on Tcb, leading to very slow growth and a long lag (ACN3178). When both tcuR and tclR were deleted (ACN3171), no growth was observed with Tcb, Caa, or Taa as the carbon source. Furthermore, while the ΔtcuR mutant (ACN1376) was Tcb-, it gave rise to spontaneous Tcb+ mutants. Two of these, which were isolated independently, ACN1445 and ACN1556, each encoded a TclR variant, TclR(R200Q, L216P) or TclR(R200Q).
Evaluation of tcuA regulation with a fluorescent transcriptional reporter
To clarify regulation, the tcuA coding sequence of ADP1 was replaced on the chromosome by the gene for a green fluorescent protein “super folder” (GFPsf; 24). Because this reporter strain (ACN3123) does not have TcuA, Tcb cannot be catabolized further and can be studied as a non-metabolizable inducer of GFPsf expressed from the tcuA promoter (PtcuA). Other reporter strains were similarly constructed with additional deletions of tclR, tcuR, or both (ACN3184, ACN3122, and ACN3173, respectively). The same chromosomal reporter was also studied in strains with tcuR deleted and the wild-type tclR coding sequence exactly replaced by a mutated allele encoding either TclR(R200Q, L216P) or TclR(R200Q), ACN3190, and ACN3191, respectively. Strains were grown on pyruvate with or without Tcb, Caa, or Taa as inducers. Compared to cultures grown without inducers, the fluorescence of the strain with both regulators (tcuRWT, tclRWT) increased when Tcb, but not Caa or Taa, was added (Fig. 3, solid dark bars).
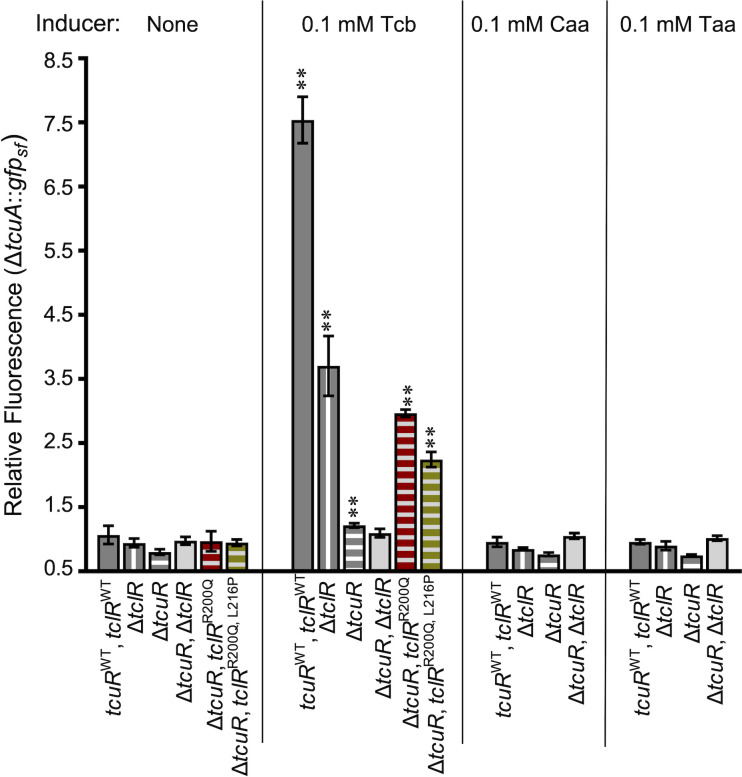
Fig 3.
Transcription of gfpsf controlled by PtcuA. Each strain has a chromosomal gfpsf (allele ΔtcuA::gfpsf -ΩK53122). Differences in genetic backgrounds are indicated. All strains were grown on pyruvate supplemented with inducer compounds as indicated. Relative fluorescence is the ratio of fluorescence/OD for each condition compared to that of ADP1, which does not contain gfpsf, grown on pyruvate. The error bars show the standard deviation (σ) from at least three biological replicates. The relative fluorescence of each strain with an inducer was compared to that of the same strain with no inducer. Unless indicated, the relative fluorescence was not significantly different from uninduced conditions, as calculated by unpaired samples t test analyses. Significant increases in expression compared to uninduced conditions are indicated (**, P < 0.01). Strain names are indicated parenthetically: tcuRWT, tclRWT (ACN3123); ΔtclR (ACN3184); ΔtcuR (ACN3122); ΔtcuR, ΔtclR (ACN3173); ΔtcuR, tclRR200Q (ACN3191); and ΔtcuR, tclRR200Q, L216P (ACN3190).
With both regulators missing (ΔtcuR, ΔtclR), Tcb did not increase fluorescence. In contrast, TcuR without TclR (ΔtclR) was sufficient for Tcb-increased fluorescence, albeit to a lesser extent than with both regulators. The loss of TclR could reduce PtcuA-controlled fluorescence if TclR directly controls tcuA transcription. Alternatively, or in addition, loss of TclR could affect fluorescence through effects on the transcription of tcuC, which could alter the transport of the inducer and affect transcription from PtcuA indirectly. Unlike in a strain without both regulators (ΔtcuR, ΔtclR), there was a small increase in Tcb-dependent induction in a strain with TclR alone (ΔtcuR; Fig. 3), suggesting that TclR directly regulates Tcb-dependent tcuA transcription, but poorly. In strains encoding the TclR variants (ΔtcuR, tclRR200Q and ΔtcuR, tclRR200Q, L216P), Tcb-increased fluorescence relative to no inducer and relative to the induction observed in the strain with only the wild-type TclR (Fig. 3). This increase in Tcb-stimulated expression may account for the Tcb+ phenotypes of the corresponding spontaneous mutants. These results indicate that TcuR is the primary transcriptional activator of tcu genes in response to Tcb, whereas TclR plays a lesser role.
Evaluation of tcuC regulation with a fluorescent transcriptional reporter
To assess the regulation of the tcuC promoter, PtcuC, the GFPsf gene was inserted in the chromosome immediately downstream of tcuC, close enough to ensure cotranscription. This location was designed to preserve the TcuC transport function. Additionally, pacI was deleted to prevent interconversion of Taa and Caa so their individual effects could be distinguished. The Tcb+ phenotype of the resulting strain, ACN3225 (Table 1), indicated that TcuC was functional. However, this strain grew more slowly on Tcb and had a longer lag time than the wild type (ADP1). Additional strains were constructed with this tcuC transcriptional reporter and deletion of ΔtclR or ΔtcuR (ACN3224 and ACN3222) or deletions of both (ACN3223). Strains were also made with ΔtcuR plus tclRR200Q (ACN3228) or ΔtcuR plus tclRR200Q, L216P (ACN3227). With wild-type tcuR and tclR, but not without them (ΔtcuR, ΔtclR), Tcb increased fluorescence (Fig. 4). Without TcuR, the TclR variants (with R200Q or with R200Q plus L216P) increased Tcb-dependent induction relative to wild-type TclR at both promoters. Yet, Tcb-dependent induction differed at PtcuA (Fig. 3) and PtcuC (Fig. 4). Response to Tcb at PtcuC required higher concentrations than at PtcuA (5 mM vs 0.1 mM) and caused a smaller increase in fluorescence (Fig. 3 and 4). The relative importance of TcuR and TclR for Tcb-regulated transcription appears to be reversed at PtcuC and PtcuA, with TclR being more important than TcuR for regulating PtcuC (Fig. 4).
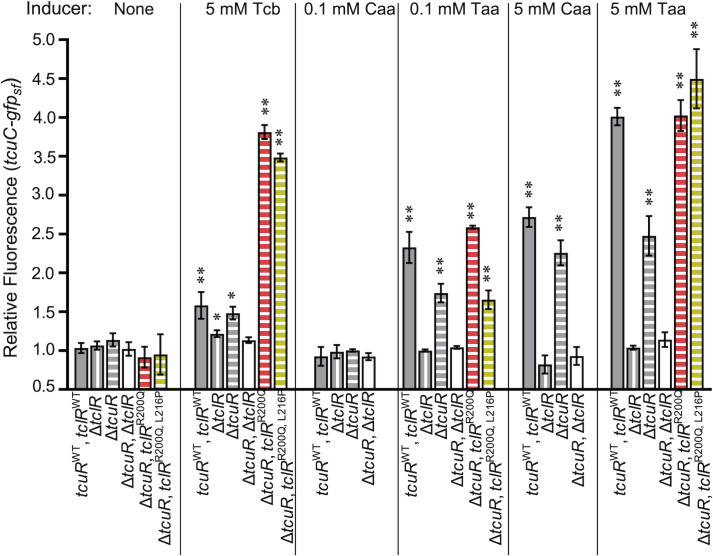
Fig 4.
Transcription of gfpsf controlled by PtcuC. Each strain has a chromosomal gfpsf (allele tcuC-gfpsf-ΩK53222; ∆pacI53222). Differences in genetic backgrounds are indicated. All strains were grown on pyruvate supplemented with inducer compounds as indicated. Relative fluorescence is the ratio of fluorescence/OD for each condition compared to that of ADP1, which does not contain gfpsf, grown on pyruvate. The error bars show the standard deviation (σ) from at least three biological replicates. Unless indicated, fluorescence/OD is not significantly different from uninduced conditions, as calculated by unpaired samples t tests. Significant increases in expression compared to uninduced conditions are indicated (*P < 0.05; **P < 0.01). Strain names are indicated parenthetically: tcuRWT, tclRWT (ACN3225); ΔtclR (ACN3224); ΔtcuR (ACN3222); ΔtcuR, ΔtclR (ACN3223); ΔtcuR, tclRR200Q (ACN3228); and ΔtcuR, tclRR200Q, L216P (ACN3227).
The addition of Caa and Taa increased fluorescence at PtcuC (Fig. 4) but not PtcuA (Fig. 3). With TclR and TcuR, but not without them, Taa (0.1 mM or 5 mM) or Caa (5 mM) increased fluorescence (Fig. 4). This induction did not occur without TclR, suggesting that TclR may be the only regulator that responds to Taa or Caa at PtcuC. In strains with either TclR or the TclR variants, fluorescence increased in response to either Caa or Taa (Fig. 4).
Transcriptional units and start sites
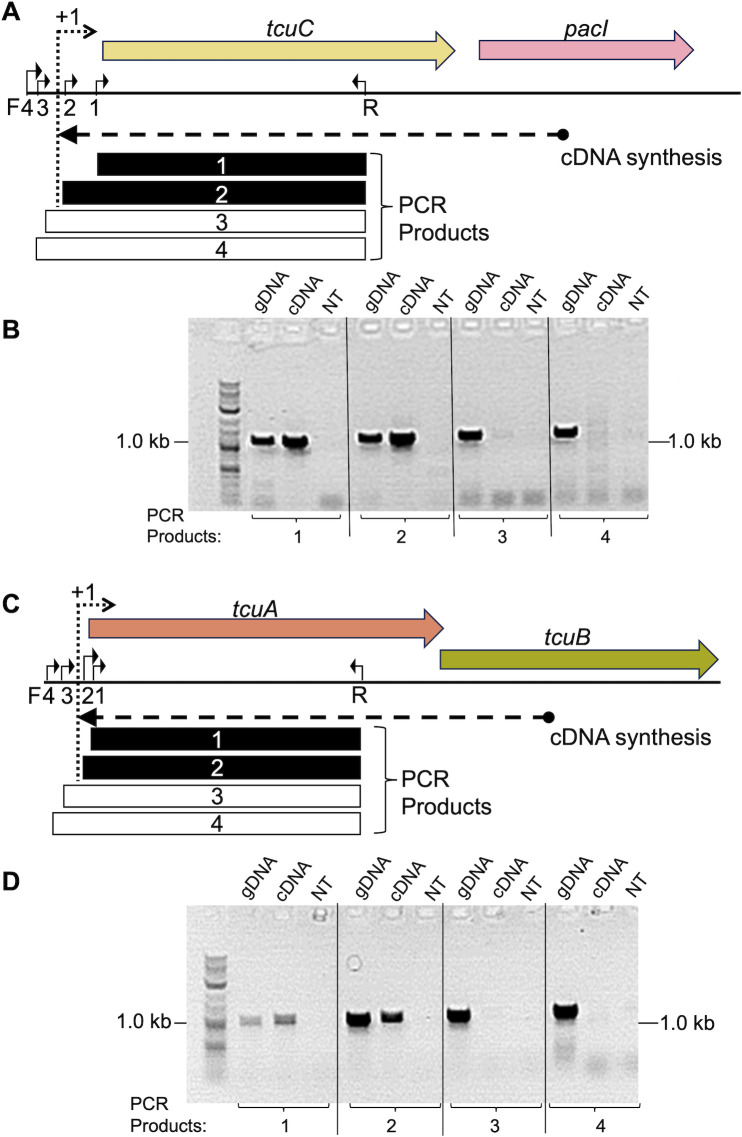
To assess which genes are transcribed in operons, RNA was isolated from Taa-grown ADP1. The cDNA synthesized from a pacI-specific primer was used as a PCR template with tcuC primers (Fig. 5A and B). Products of the expected sizes were observed that could only be generated if tcuC and pacI were cotranscribed. Similar experiments were conducted with Tcb-grown ADP1 to evaluate the cotranscription of tcuA and tcuB. The results showed that tcuA and tcuB form an operon (Fig. 5C and D).
Fig 5.
RT-PCR analysis of operons. (A) With RNA from Taa-grown ADP1, pacI-specific cDNA was made and used in four individual PCR reactions. The same reverse primer, R (oTCB113), was used with different forward primers, F1 (oTCB44), F2 (oTCB45), F3 (oTCB144), or F4 (oTCB145). With cDNA as a template, products were detected from reactions 1 and 2 (black rectangles), but not reactions 3 and 4 (white rectangles). (B). PCR products of the expected size, ~1 kbp, were observed on an agarose gel for the cDNA template using F1 or F2. With F3 or F4, no PCR products were observed. The reactions with genomic DNA (gDNA) as a template (positive controls) gave correctly sized products for all primer pairs, and the reactions with no template (NT) DNA (negative controls) gave no products. (C) Comparable studies of tcuA and tcuB were done with cDNA generated with a tcuB-specific primer and RNA from Tcb-grown ADP1. The reverse primer, R (oTCB154), was used with different forward primers, F1 (oTCB156), F2 (oTCB56), F3 (oTCB55), or F4(oTCB155). (D) PCR products were assessed as indicated for panel B. The transcriptional start site (+1) for the tcuC-pacI operon and that for the tcuAB operon lies between forward primers 2 and 3, since the former but not the latter yielded a PCR product with the cDNA template.
The transcription initiation site (+1) for each operon was localized between two forward primers, one that gave a product (PCR 2) and the other that did not (PCR 3; Fig. 5). In both cases, PCR3 and PCR4 failed to yield a product with the cDNA template. The absence of these PCR products (3 and 4) indicates that tclR is not cotranscribed with its downstream neighbor, tcuC, and that tcuR is not cotranscribed with its downstream neighbor, tcuA.
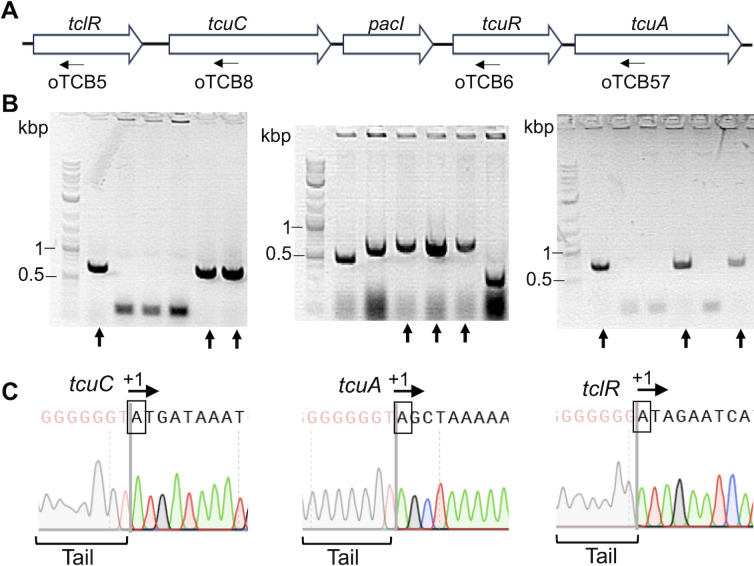
Transcription initiation sites were pinpointed by “Rapid Amplification of cDNA Ends” (5´ RACE; 25). For Taa-grown ADP1, mRNA was isolated, and cDNA was generated using primers for tclR and tcuC in separate reactions. For Tcb-grown ADP1, mRNA was isolated, and cDNA was generated with primers for tcuR and tcuA in separate reactions. The 5´ RACE protocol modifies the cDNA by adding a known DNA sequence (a tail) to its 5′ end. This approach enables subsequent PCR amplification of the region. After cloning the PCR products, plasmids were checked for appropriately sized inserts (Fig. 6B). Individual clones were sequenced to find the junction between the tail and sequences identical to ADP1 DNA. These junctions, Fig. 6C, mark the transcription initiation sites (+1). The experimentally determined +1 site for tcuC was separated from its coding sequence by 187 nt. The transcription start site for tcuA was separated from its coding sequence by 22 nt. Both sites were within the regions localized by RT-PCR (Fig. 5). For tclR, the +1 site was separated from its coding sequence by 40 nt, whereas multiple attempts to identify the start site for tcuR were unsuccessful.
Fig 6.
Transcriptional start sites (+1) for tcuC, tcuA, and tclR. (A) Genetic organization and locations of primers (arrows) used to generate cDNA for tclR, tcuC, tcuR, and tcuA: oTCB5, oTCB8, oTCB6, and oTCB57, respectively. (B) Following the 5’ RACE protocol (Materials and Methods), PCR products corresponding to the regions of transcriptional start sites were cloned into plasmids for tcuC, tcuA, and tclR. Reactions for tcuR were unsuccessful. Individual plasmids were isolated, and PCR was used to detect cloned inserts shown on gels for tcuC, tcuA, and tclR (left to right). Multiple DNA fragments (indicated by arrows) were sequenced. (C) DNA sequencing chromatograms used to identify the +1 sites (boxed and labeled with an arrow) that correspond to the junction between DNA added during the 5’ RACE procedure (the tail) and ADP1 DNA sequence. Identical results were obtained for each gene using two or more independently isolated RNA samples and three or more sequenced clones.
Regulation of the tcuC-pacI and the tcuAB operons by TcuR and TclR
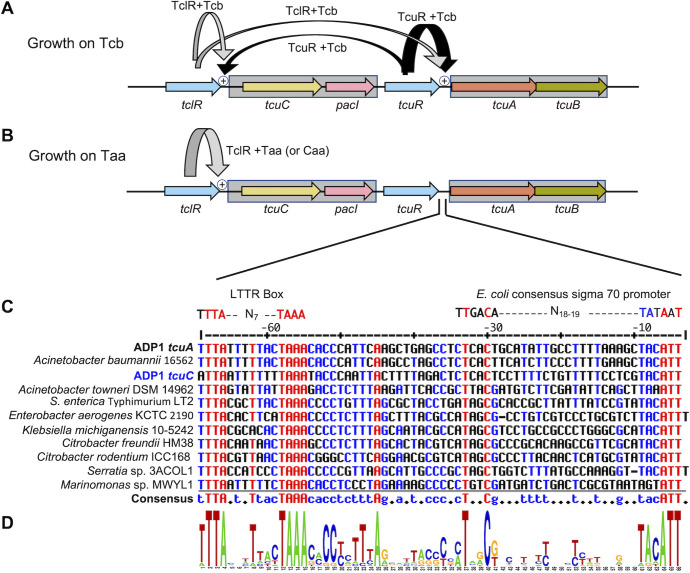
A regulatory model based on the data is presented in Figure 7. The functional overlap of TcuR and TclR at both operator-promoter regions is not surprising considering their sequence similarity. Sequences of TcuR, TclR, and the other LTTRs encoded near tcu genes (Fig. 2) were aligned (Fig. S2). The three LTTRs encoded by genes (striped in Fig. 2) that are divergent from pacI-like genes are more like each other than they are to the TcuR-like homologs. The two LTTRs from ADP1 more closely resemble each other than the regulators from other bacteria (Fig. S2 and S3). The helix-turn-helix region (HTH), involved in DNA sequence recognition and binding by LTTRs (26, 27), is nearly identical in TcuR and TclR (Fig. S2B). Thus, both proteins may recognize similar operator-promoter sequences, as has been shown for other paralogous LTTRs in ADP1 (28, 29).
Fig 7.
Regulatory model. (A) During growth on Tcb, TcuR (black arrow) responds to Tcb to activate transcription of the tcuAB operon and to a lesser extent (thinner arrow) the tcuC-pacI operon. TclR (gray arrow) responds to Tcb for low-level transcriptional activation from PtcuC and PtcuA. (B) During growth on Taa, TclR (gray arrow) activates transcription from PtcuC in response to Taa or Caa. (C) Alignment of the ADP1 tcuA promoter region with bacterial sequences upstream of tcuA from LT2 and other predicted tcuA genes. The rightmost T in the top line corresponds to position −5 relative to the transcriptional start site. The promoter region of ADP1 tcuC was aligned with the tcuA sequences. The rightmost T in the alignment (third line) corresponds to position −9 from the transcriptional start site. The alignment was generated using MultiAlin and default settings (30). Identical residues in all aligned positions are shown in red. Those that are identical in 50% or more of the sequences are shown in blue. (D) Sequence conservation represented with the WebLogo generator; sequence conservation at each position is indicated by the height of the letter (31). Conserved nucleotides in the −10 and −35 regions resemble the patterns of E. coli sigma 70 promoters.
Upstream of tcuA, a site matching a consensus for binding LTTRs (26), an LTTR box, was discovered using sequence alignments and pattern searches near tcuA homologs (Fig. 7C; 32). A putative binding site for TcuR was found to be TTTA-N7-TAAA. The sequence upstream of tcuC also aligns well with the tcuA regions (Fig. 7C). No LTTR box was found upstream of tclR, but one potential binding site lies within the tclR coding sequence (Fig. S4).
Transport of tricarboxylic acids
TcuC, a member of the major facilitator superfamily (MFS), entry IPR011701 of InterPro (18, 33), affects Tcb consumption (Table 1). Its counterpart in LT2, originally identified as a Cit transporter, was initially designated CitA (34). In most databases, tcuC remains annotated as citA, as it is for ADP1. To clarify whether loss of tcuC affects Cit consumption, the tcuC mutant (ACN1456) was tested and found to be Cit+ (Table 1). However, the impact of tcuC deletion could be masked by a paralog, also annotated as citA (ACIAD_RS0183), which encodes a protein 57% identical to TcuC. We designated this gene cltA (for the citA-like gene) and compared strains with deletions in tcuC (ACN1456) or both cltA and tcuC (ACN3168). Since Cit+ growth was comparable for both (Table 1), no role for TcuC in Cit transport was evident. Furthermore, the primary role of CltA remains unclear. Some bacteria have two nearby genes encoding TcuC-like transporters, labeled C and D (Fig. 2), which have sequences that align well with the other MFS transporters discussed here (Fig. S5).
DISCUSSION
We discovered interrelated metabolic pathways in A. baylyi ADP1 for the consumption of Tcb, Taa, and Caa (Fig. 1). Few studies have addressed the consumption of these compounds as growth substrates for soil bacteria. Nevertheless, the clustered genetic arrangement of tcu genes with an aconitate isomerase-like gene in diverse bacteria (Fig. 2) suggests that ADP1 is not unique in co-regulating Tcb and Taa consumption. Bacteria with these genes were isolated from different environments, ranging from wetland peat (Aquitalea denitrificans 5YN1-3) to root nodules (Cupriavidus taiwanensis LMG 19424). The wide distribution of bacteria capable of consuming plant-derived tricarboxylic acids presumably corresponds to the varied fates of plants themselves. In addition to the presence of Taa and Tcb in grasses and plant forage, these compounds have been detected in barley, maize, beets, and cane molasses, and they can be derived in animals from consuming carbohydrates (35). Therefore, Tcb utilization by gut microbes, such as LT2, most likely reflects exposure from the diet of the host.
In LT2, the regulation of Tcb utilization is tightly controlled at the transcriptional level by TcuR. The organization of the tcuABC genes in an operon may help to couple Tcb transport to its metabolism. Tethering TcuC-mediated transport to TcuAB-mediated conversion to Caa, as depicted in Figure 1A, appears to prevent the cellular accumulation of Tcb, a compound that can inhibit aconitase and isocitrate dehydrogenase (36). Similarly, the regulated cotranscription of tcuC and pacI could coordinate the PacI-mediated conversion of Taa to Caa in the periplasm with the TcuC-mediated uptake of Caa, as depicted in Fig. 1B. This regulation might limit the cellular accumulation of Taa, a compound long known to inhibit aconitase (37).
The inference from growth patterns of ADP1-derived mutants (Table 1) that TcuC can transport both Caa and Taa is consistent with transport studies in Salmonella published 50 years ago. At that time it was shown that a Tcb-inducible transporter could also transport Caa (38). Although the associated genes/protein sequences were not identified at that time, the Tcb-inducible transporter presumably corresponds to TcuC in LT2.
TcuC-like transporters
It seems unlikely that TcuC would import Taa, yet some of this compound must enter the cell for it to be an inducer in strains that do not express PacI (Fig. 4). In its protonated form, Taa likely diffuses into the cell in amounts limited by pH and concentration. MFS family members are often stereospecific (33), so it is likely that TcuC is able to distinguish the cis- and trans-forms of aconitate. TcuC of LT2 also transports Cit and does so in preference to isocitrate (34). However, seven single amino acid replacements increase the transport of isocitrate (Fig. S5). ADP1 cannot use isocitrate as a sole carbon source, but it grows well on Cit, even in deletion mutants without tcuC and cltA. In Pseudomonas aeruginosa, deletion of a citA homolog did not affect the growth on Cit, even in conjunction with the inactivation of another putative transporter gene (39). Redundant paralogs with overlapping transport functions, often with different regulatory patterns, are frequently observed in bacteria, including ADP1 (29, 40). A few bacteria had two nearby tcuC paralogs (Fig. 2, labeled C and D), yet alignments failed to suggest any functional, phylogenetic, or regulatory significance to sequence differences, including at positions shown to affect the substrate specificity of TcuC from LT2 (Fig. S5).
Regulation by TcuR and TclR: effector compound recognition and response
The ten aligned LTTRs formed two branches in a phylogram (Fig. S2 and S3). TcuR and TclR from ADP1 are more like each other and TcuR in LT2 (6) than to the three LTTRs encoded near the pacI-like genes of other bacteria. These differences raise questions about regulation mediated by the three distinct LTTRs. Nevertheless, the sequence similarity between TcuR and TclR correlates with their regulatory overlap (Fig. 7A and B). Overall, these ADP1 paralogs appear to function similarly to members of the large LTTR family (26).
Typically, LTTRs respond to one or more small chemicals that serve as inducers, coinducers, and/or coactivators, collectively referred to as effectors to acknowledge that LTTRs can be transcriptional repressors, activators, or both (26). Binding an effector in the C-terminal region, the effector-binding domain (EBD), of an LTTR subunit causes a conformational change that propagates throughout the oligomer, usually a homotetramer (26). While both TcuR and TclR responded to Tcb as an effector, two TclR variants were independently selected that increased Tcb-dependent transcriptional activation (Fig. 3 and 4). In the absence of characterized atomic-level structures of any Tcb-binding LTTRs, we used models of TclR-EBD and TcuR-EBD to compare with crystal structures of two Cit-responsive LTTRs, CcpE and CcpC (Fig. S6; 41, 42). In these structures, Cit binds between two EBD subdomains (Fig. S6A). Superimposition of the TclR model indicates that the R200 side chain corresponds to the position of Cit. Therefore, the R200Q replacement in the TclR variant may have a direct impact on the Tcb-binding site (Fig. S6C and D). The possible effect of the L216P replacement is less clear, based on its position in the protein and because it only appeared together with R200Q.
Positions of amino acids that render TcuR of LT2 able to activate transcription without Tcb (5) were also compared to the CcpC structure (Fig. S7). The R264 residue in the LT2 TcuR, which is conserved in all sequences in Fig. S2, is predicted to be centrally positioned in one of two beta-strands forming a hinge between the EBD subdomains (Fig. S7D). In LTTRs, an effector-binding pocket forms between these subdomains (26). The R264L replacement appears to lock the protein in an active conformation without Tcb. Other changes in constitutive TcuR variants (5) also cluster near the predicted effector-binding site (Fig. S7C).
While TcuR and TclR responded to Tcb, TclR additionally responded to Taa, under conditions where it was not metabolized. Caa may also activate transcription by TclR, but this compound was only tested in strains that could metabolize it. Thus, Caa-dependent changes could be mediated by a downstream metabolite. This broader range of effectors for TclR apparently helps induce TcuC expression for growth on Taa, Tcb, and Caa.
Regulation by TcuR and TclR: operator-promoter recognition
The transcriptional initiation sites (+1) for the tcuAB operon and the tcuC-pacI operon were aligned with each other and comparable regions near other predicted tcuA genes (Fig. 7C). A conserved sequence, TTTA-N7-TAAA, matches the consensus sequence for LTTR binding (26, 27, 43). Based on its sequence, conservation, and position (−63 relative to the +1 site of tcuA of ADP1), we inferred that this site, called an LTTR box, anchors the binding of two subunits of a TcuR (or TclR) tetramer. For transcriptional activation, interactions of the LTTR are also expected in the vicinity of the −35 region of the promoter. However, sequences that typically indicate such interactions (43) were not observed for tcuA or tcuC. There are many examples of atypical operator-promoter sequences regulated by LTTRs (26).
The tclR operator-promoter region was aligned with those of tcuA and tcuC, and no evident LTTR box was found upstream of the coding sequence (Fig. S4). Although a common method of negative autoregulation occurs when LTTR genes are divergent from the corresponding target, autoregulation sometimes occurs differently (26). The significance of an LTTR box sequence in the tclR coding region remains to be tested (Fig. S4). No potential LTTR box was identified for tcuR of ADP1, and studies in LT2 ruled out autoregulation in that microbe (5). The tcuR transcriptional start site was not identified. This failure of the 5′ RACE attempts may reflect that the levels of LTTR transcripts tend to be low and difficult to detect (44).
Overlapping regulation by similar LTTR pairs
The nuanced control by TcuR and TclR parallels that of other pairs of LTTR paralogs in ADP1. For example, DarR and AalR have interrelated but distinct functions in a shared regulon for aspartate metabolism (29). Each LTTR responds to a distinct enantiomer of aspartate, but both recognize the same LTTR box, which controls several different genes and operons. Two other LTTR paralogs, BenM and CatM, jointly regulate multiple loci involved in aromatic compound metabolism (45, 46). Both recognize similar DNA sequences and the same effector molecule. An added level of fine-tuned control in this example is the integration of multiple cellular signals via the synergistic response of BenM to a second effector compound (28).
These elaborate regulatory schemes underscore the importance of balanced transport and consumption of growth substrates. For aromatic compound catabolism, there are very complicated multistep pathways in which each step may generate a toxic metabolite, and the accumulation of such intermediates is carefully regulated. For the consumption of Tcb, Taa, and Caa, there are few catabolic steps prior to entry to the tricarboxylic acid cycle. Nevertheless, the potential of Taa and Tcb to inhibit the tricarboxylic acid cycle, and other characteristics of these compounds, such as metal chelation, may account for multiple layers of transcriptional control governed by TcuR and TclR.
MATERIALS AND METHODS
Media and growth conditions
Cultures were grown in minimal medium (MM; 47) or lysogeny broth (LB), also known as Luria-Bertani medium (10 g of Bacto Tryptone, 5 g of yeast extract, and 10 g of NaCl per L; 48), as previously described (29). Unless otherwise noted, carbon sources were added to MM at final concentrations of 15 mM for pyruvate or 5 mM for Tcb, Taa, Caa, or Cit. Antibiotics were added as needed at final concentrations of 25-mg/mL for kanamycin (Km), 12.5-mg/mL each for streptomycin (Sm) and spectinomycin (Sp), and 150-mg/mL for ampicillin (Ap).
Bacteria were incubated at 37°C or 30°C. Liquid cultures, except in microtiter plates, were aerated by shaking at 250 rpm, at 37°C. Culture volumes of 5 mL were grown in 15-mL tubes, and larger cultures of 25 mL were grown in 250-mL flasks. To inoculate, a dilution ratio of 1:100 was typical. For monitoring growth in a Synergy H1 plate reader (BioTek), 2 µL of an inoculum (from a liquid culture that was grown for at least 12 h) was added to 198 µL of medium in the well of a Costar 3603 96-well plate. The total volume of each well was 360 µL. Readings were taken every 30 min (OD595) while the plate was incubated at 30°C, with orbital shaking at 282 rpm and a 3-mm orbit. All comparative data that we report used strains grown under identical conditions (e.g., Table 1). Strains grew approximately threefold slower in the plate reader, but the growth rates of different strains relative to each other were the same regardless of condition.
Strains, plasmids, and PCR primers
A. baylyi strains were derived from the wild type, ADP1 (22, 23), as listed in Table 2 with additional details in Tables S2 to S4. Escherichia coli strains XL1-Blue (Agilent Technologies) and DH5α (49) were used as plasmid hosts. Plasmids are listed in Table S3 and PCR primers in Table S4. Drug resistance cassettes were obtained from pUI1637 (ΩK, confers KmR) and pUI1638 (ΩS, confers SmR and SpR; 50). Standard vectors were used for cloning in E. coli, such as pUC18/pUC19 (51). The gfpsf transcriptional reporter gene encodes a green fluorescent protein (24). A selectable/counter-selectable cassette (sacB-KmR) was obtained from pRMJ1 (52).
TABLE 2.
| Strain | Relevant characteristics | Source |
|---|---|---|
| ADP1 | Wild-type strain (BD413) | (22, 23) |
| ACN1376 | ΔtcuR51376 | This study |
| ACN1388 | ΔpacI51388 | This study |
| ACN1445 | ΔtcuR51376, tclR51445 | This study |
| ACN1456 | ΔtcuC51456 | This study |
| ACN1556 | ΔtcuR51376, tclR51556 | This study |
| ACN3122 | ΔtcuR51376, ΔtcuA::gfpsf -ΩK53122 | This study |
| ACN3123 | ΔtcuA::gfpsf -ΩK53122 | This study |
| ACN3125 | ΔcltA::sacB-ΩS53125 | This study |
| ACN3168 | ΔtcuC51456, ΔcltA::sacB-ΩS53125 | This study |
| ACN3171 | ΔtcuR51376, ΔtclR53171 | This study |
| ACN3173 | ΔtcuR51376, ΔtclR53171, ΔtcuA::gfpsf -ΩK53122 | This study |
| ACN3178 | ΔtclR53171 | This study |
| ACN3179 | ΔtcuR51376, tclR51445 [TclR(R200Q, L216P)] | This study |
| ACN3184 | ΔtclR53171, ΔtcuA::gfpsf -ΩK53122 | This study |
| ACN3187 | ΔtcuR51376, tclR51556 [TclR(R200Q)] | This study |
| ACN3190 | ΔtcuR51376, tclR51445, ΔtcuA::gfpsf -ΩK53122 [TclR(R200Q, L216P)] | This study |
| ACN3191 | ΔtcuR51376, tclR51556, ΔtcuA::gfpsf -ΩK53122 [TclR(R200Q)] | This study |
| ACN3222 | ΔtcuR51376, tcuC-gfpsf-ΩK53222, ∆pacI53222 | This study |
| ACN3223 | ΔtcuR51376, ΔtclR53171, tcuC-gfpsf-ΩK53222, ∆pacI53222 | This study |
| ACN3224 | ΔtclR53171, tcuC-gfpsf-ΩK53222, ∆pacI53222 | This study |
| ACN3225 | tcuC-gfpsf-ΩK53222, ∆pacI53222 | This study |
| ACN3226 | ΔcltA::sacB-ΩS53125, tcuC-gfpsf-ΩK53222, ∆pacI53222 | This study |
| ACN3227 | ΔtcuR51376, tclR51445, tcuC-gfpsf-ΩK53222, ∆pacI53222 [TclR(R200Q, L216P)] | This study |
| ACN3228 | ΔtcuR51376, tclR51556, tcuC-gfpsf-ΩK53222, ∆pacI53222 [TclR(R200Q)] | This study |
Strains and plasmids were constructed by standard molecular biology methods (48) and techniques developed for ADP1 (13, 14). High-fidelity polymerases were used for PCR, PrimeSTAR Max (Takara Biosciences) and Phusion (New England Biolabs), with primers shown in Table S4. Plasmids were constructed by overlapping sequence assembly (53), by restriction digest and ligation (Quick Ligation Kit; New England Biolabs), with the NEBuilder kit (New England Biolabs) and/or by overlap extension PCR (54). Plasmids were confirmed by restriction mapping and/or regional DNA sequencing (Eurofins Genomics). A. baylyi genomic changes were made by allelic replacement using linear DNA to transform naturally competent recipients (13, 14, 55, 56). When PCR products were used as donor DNA, digestion by DpnI (New England Biolabs) was used to degrade template DNA. Genotypes of the recipient strains were confirmed by PCR with LongAmp polymerase (New England Biolabs).
Fluorescence assays of transcriptional fusions
Growth and fluorescence were assessed for strains with a chromosomal copy of gfpsf under the transcriptional control of PtcuC or PtcuA using a Synergy H1 plate reader (BioTek). Cultures were grown with pyruvate as the carbon source and diluted (1:100) in the same medium with and without an exogenous inducer (Tcb, Taa, or Caa at concentrations of 0.1 mM or 5 mM). Green fluorescence measurements (to assess GFPsf expression) were taken, at excitation 479 nm and emission 520 nm, concurrently to growth measurements at OD595. Fluorescence values, corresponding to cultures that reached an OD595 of 0.35–0.50, were recorded relative to cell density (OD595). To account for a high background fluorescence in ADP1, the data in Fig. 3 and 4 are reported in comparison to that of the wild-type strain, which has no gfpsf gene.
Transcript analysis
For RNA isolation, cultures were grown and harvested at the mid-log phase with an OD595 of approximately 0.4–0.6. The cells were diluted to a concentration of approximately 108 cells. The cells were lysed using 200 µL of lysozyme into 100 µL of liquid culture. RNA was preserved using RNAprotect (Qiagen) and isolated using the RNEasy Mini Kit (Qiagen). RNA was further treated with dsDNase (Thermo Fisher) and purified (Monarch) or RQ1 DNase (Promega). RNA was confirmed to be free of DNA using PCR. cDNA was generated using the Maxima H Minus First Strand cDNA Synthesis Kit (Thermo Scientific). A 5′ RACE DNA purification system kit (Invitrogen) was used for cDNA purification and to add a homopolymeric tail of nucleotides (dCTP) with terminal deoxynucleotidyltransferase. Nested PCR was performed on the tailed DNA as described in the kit protocol using a Taq polymerase and primers from the kit along with gene-specific primers (Table S4). Fragments were visualized on agarose gels, and the largest observed fragments were cloned in the pCR2.1-TOPO vector using a TOPO TA cloning kit (Invitrogen). Plasmids were isolated, after the transformation of E. coli, and DNA sequencing was used to identify sequences that correspond to transcriptional start sites. Multiple plasmids were sequenced and shown to indicate the same start site. All transcript analyses were confirmed with repetitions using two or more independently isolated RNA samples.
Selection of spontaneous mutants that grow on Tcb without TcuR
A ΔtcuR mutant, ACN1376, did not grow on Tcb as the sole carbon source. Pyruvate-grown cultures of ACN1376 were concentrated approximately 10-fold and spread on plates with Tcb as the carbon source. Tcb+ colonies arose (at a frequency of 10−9). Two independently isolated Tcb+ derivatives of ACN1376, ACN1445 and ACN1556, were streak-purified and further characterized. Localized DNA sequencing revealed mutations in tclR.
DNA sequence analysis
The reference sequences, from the National Center for Biotechnology Information (NCBI; 57), for bacteria discussed in this report or referenced in figures are indicated parenthetically: A. baylyi ADP1 (NC_005966), Acinetobacter baumannii 1656–2 (NC_017162), Acinetobacter towneri DSM 14962 (NZ_KB849689), Aquitalea denitrificans 5YN1-3 (NZ_CP047241), Citrobacter freundii HM38 (NZ_CP024672), Citrobacter rodentium ICC168 (NC_013716), Cupriavidus taiwanensis LMG 19424 (NC_010530), Curvibacter delicatus NBRC 14919 (NZ_BCWP01000009), Enterobacter aerogenes KCTC 2190 (NC_015663), Klebsiella michiganensis strain 10–5242 (NZ_JH603153), Marinomonas sp. MWYL1 (NC_009654), Paraburkholderia sacchari LMG 19450 (NZ_JTDB02000009), S. enterica LT2 (NC_003197), Serratia sp. 3ACOL1 (CP033055.1), and Xanthobacter autotrophicus Py2 (NC_009720). For homology searches, we used the Basic Local Alignment Search Tool (BLAST; 58) and the sequence similarity database (SSDB) with associated search tools of the Kyoto Encyclopedia of Genes and Genomes (59). Protein sequences were aligned, and data were exported using programs on the UniProt website (30).
In the A. baylyi ADP1 genome, our new gene designations correspond to the following locus tags: cltA (ACIAD_RS01830), tclR (ACIAD_RS07105), tcuC (ACIAD_RS07100), pacI (ACIAD_RS07095), tcuR (ACIAD_RS07090), tcuA (ACIAD_RS07085), and tcuB (ACIAD_RS07080). Protein sequence alignments (in Fig. S1, S2 and S5) were generated using a Sequence Manipulation Suite program, Multiple Align Show (60). Promoter sequences shown in Fig. 7 and Fig. S4 were aligned and displayed using the MultAlin program (61). The Signal P program (21) was used online: https://biolib.com/DTU/SignalP-6/. The STRING database program (20) was used online: https://string-db.org.
ACKNOWLEDGMENTS
Some of these studies were conducted during several semesters of a University of Georgia Microbiology laboratory course. The following students contributed to generating and characterizing plasmids and strains: Diana Ambrocio, John Hunter Bowen, Lindsey Brock, Gary Kevin Davis, Nicholas Evans, Destiny Finnell, John Patrick Goldun, Mark Goodwin, Grigsby Gordy, Amanda Helton, Maliha Ishaq, Brandy Le, Connor Lombardi, Mengzhuo (Jen) Ma, Kieli Mills, Alex Moosariparambil, Maria Najera, Nneka Okoro, Destiny Okpomo, Rashidatu Olorunsola, Emma Osborne, Cathryn Paul, Quynh Phan, Christopher Perez, Mason Pyles, Delaney Ragsdale, Liam Richardson, Tonya Brook Ringle, Emma Grace Turner, Keiana Watkins, Kaitlin Whitely, Winsen Wijaya, Domenic Won, and Christofer Zepeda. Emily McIntyre and Andrew Wiggins contributed as teaching assistants for the course. Additional students in the Neidle laboratory helped to design and conduct experiments: Stephanie Thurmond, Silke Andresen, Stacy Bedore, and Hannah Toutkoushian.
We thank Drs. Cory Momany and Jorge Escalante-Semerena for their helpful discussions and advice.
This work was supported by grants from the National Science Foundation (MCB2225858 to E.L.N. and DBI-1757720 and DBI-1062589 for Research Experience for Undergraduate Site programs for two participants, Kayla Figatner and Hannah Toutkoushian, respectively). Funding was also provided from the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, Genomic Science Program (DE-SC0022220 to E.L.N.). Additional funding was provided by the UGA Microbiology Department.
Contributor Information
Ellen L. Neidle, Email: eneidle@uga.edu.
Isaac Cann, University of Illinois Urbana-Champaign, Champaign, Illinois, USA.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aem.02111-23.
Tables S1 to S4 and Fig. S1 to S7.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Cook GM, Wells JE, Russell JB. 1994. Ability of Acidaminococcus fermentans to oxidize trans-aconitate and decrease the accumulation of tricarballylate, a toxic end product of ruminal fermentation. Appl Environ Microbiol 60:2533–2537. doi: 10.1128/aem.60.7.2533-2537.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Russell JB, Forsberg N. 1986. Production of tricarballylic acid by rumen microorganisms and its potential toxicity in ruminant tissue metabolism. Br J Nutr 56:153–162. doi: 10.1079/bjn19860095 [DOI] [PubMed] [Google Scholar]
- 3. Russell JB, Mayland HF. 1987. Absorption of tricarballylic acid from the rumen of sheep and cattle fed forages containing trans‐aconitic acid. J Sci Food Agric 40:205–212. doi: 10.1002/jsfa.2740400303 [DOI] [Google Scholar]
- 4. Lewis JA, Horswill AR, Schwem BE, Escalante-Semerena JC. 2004. The tricarballylate utilization (tcuRABC) genes of Salmonella enterica serovar Typhimurium LT2. J Bacteriol 186:1629–1637. doi: 10.1128/JB.186.6.1629-1637.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lewis JA, Stamper LW, Escalante-Semerena JC. 2009. Regulation of expression of the tricarballylate utilization operon (tcuABC) of Salmonella enterica. Res Microbiol 160:179–186. doi: 10.1016/j.resmic.2009.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lewis JA, Escalante-Semerena JC. 2006. The FAD-dependent tricarballylate dehydrogenase (TcuA) enzyme of Salmonella enterica converts tricarballylate into cis-aconitate. J Bacteriol 188:5479–5486. doi: 10.1128/JB.00514-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lewis JA, Escalante-Semerena JC. 2007. Tricarballylate catabolism in Salmonella enterica. The TcuB protein uses 4Fe-4S clusters and heme to transfer electrons from FADH2 in the tricarballylate dehydrogenase (TcuA) enzyme to electron acceptors in the cell membrane. Biochemistry 46:9107–9115. doi: 10.1021/bi7006564 [DOI] [PubMed] [Google Scholar]
- 8. Takiguchi A, Yoshioka I, Oda Y, Ishii Y, Kirimura K. 2021. Constitutive production of aconitate isomerase by Pseudomonas sp. WU-0701 in relation to trans-aconitic acid assimilation. J Biosci Bioeng 131:47–52. doi: 10.1016/j.jbiosc.2020.09.003 [DOI] [PubMed] [Google Scholar]
- 9. Yuhara K, Yonehara H, Hattori T, Kobayashi K, Kirimura K. 2015. Enzymatic characterization and gene identification of aconitate isomerase, an enzyme involved in assimilation of trans-aconitic acid, from Pseudomonas sp. WU-0701. FEBS J 282:4257–4267. doi: 10.1111/febs.13494 [DOI] [PubMed] [Google Scholar]
- 10. Bruni GO, Klasson KT. 2022. Aconitic acid recovery from renewable feedstock and review of chemical and biological applications. Foods 11:573. doi: 10.3390/foods11040573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Di Lorenzo RD, Serra I, Porro D, Branduardi P. 2022. State of the art on the microbial production of industrially relevant organic acids. Catalysts 12:234. doi: 10.3390/catal12020234 [DOI] [Google Scholar]
- 12. Kobayashi K, Maruebi J, Kirimura K. 2016. Bioproduction of trans-aconitic acid from citric acid by whole-cell reaction of Escherichia coli heterologously expressing the aconitate isomerase gene from Pseudomonas sp WU-0701. ChemistrySelect 1:1467–1471. doi: 10.1002/slct.201600234 [DOI] [Google Scholar]
- 13. Bedore SR, Neidle EL, Pardo I, Luo J, Baugh AC, Duscent-Maitland CV, Tumen-Velasquez MP, Santala V, Santala S. 2023. Chapter 8 - natural transformation as a tool in Acinetobacter baylyi: streamlined engineering and mutational analysis, p 207–234. In Gurtler V, Calcutt M (ed), Methods in microbiology. Vol. 52. Academic Press. 10.1016/bs.mim.2023.01.001. [DOI] [Google Scholar]
- 14. Biggs BW, Bedore SR, Arvay E, Huang S, Subramanian H, McIntyre EA, Duscent-Maitland CV, Neidle EL, Tyo KEJ. 2020. Development of a genetic toolset for the highly engineerable and metabolically versatile Acinetobacter baylyi ADP1. Nucleic Acids Res. 48:5169–5182. doi: 10.1093/nar/gkaa167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Durot M, Le Fèvre F, de Berardinis V, Kreimeyer A, Vallenet D, Combe C, Smidtas S, Salanoubat M, Weissenbach J, Schachter V. 2008. Iterative reconstruction of a global metabolic model of Acinetobacter baylyi ADP1 using high-throughput growth phenotype and gene essentiality data. BMC Syst Biol 2:85. doi: 10.1186/1752-0509-2-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oberto J. 2013. Synttax: a web server linking synteny to prokaryotic taxonomy. BMC Bioinformatics 14:4. doi: 10.1186/1471-2105-14-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Karp PD, Billington R, Caspi R, Fulcher CA, Latendresse M, Kothari A, Keseler IM, Krummenacker M, Midford PE, Ong Q, Ong WK, Paley SM, Subhraveti P. 2019. The biocyc collection of microbial genomes and metabolic pathways. Brief Bioinform 20:1085–1093. doi: 10.1093/bib/bbx085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Paysan-Lafosse T, Blum M, Chuguransky S, Grego T, Pinto BL, Salazar GA, Bileschi ML, Bork P, Bridge A, Colwell L, et al. 2023. Interpro in 2022. Nucleic Acids Res 51:D418–D427. doi: 10.1093/nar/gkac993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huber M, Faure G, Laass S, Kolbe E, Seitz K, Wehrheim C, Wolf YI, Koonin EV, Soppa J. 2019. Translational coupling via termination-reinitiation in archaea and bacteria. Nat Commun 10:4006. doi: 10.1038/s41467-019-11999-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, Gable AL, Fang T, Doncheva NT, Pyysalo S, Bork P, Jensen LJ, von Mering C. 2023. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res 51:D638–D646. doi: 10.1093/nar/gkac1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Teufel F, Almagro Armenteros JJ, Johansen AR, Gíslason MH, Pihl SI, Tsirigos KD, Winther O, Brunak S, von Heijne G, Nielsen H. 2022. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat Biotechnol 40:1023–1025. doi: 10.1038/s41587-021-01156-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vaneechoutte M, Young DM, Ornston LN, De Baere T, Nemec A, Van Der Reijden T, Carr E, Tjernberg I, Dijkshoorn L. 2006. Naturally transformable Acinetobacter sp strain ADP1 belongs to the newly described species Acinetobacter baylyi. Appl Environ Microbiol 72:932–936. doi: 10.1128/AEM.72.1.932-936.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Juni E, Janik A. 1969. Transformation of Acinetobacter calco-aceticus (Bacterium anitratum). J Bacteriol 98:281–288. doi: 10.1128/jb.98.1.281-288.1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pédelacq J-D, Cabantous S, Tran T, Terwilliger TC, Waldo GS. 2006. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol 24:79–88. doi: 10.1038/nbt1172 [DOI] [PubMed] [Google Scholar]
- 25. Frohman MA, Dush MK, Martin GR. 1988. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci U S A 85:8998–9002. doi: 10.1073/pnas.85.23.8998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Baugh AC, Momany C, Neidle EL. 2023. Versatility and complexity: common and uncommon facets of LysR-type transcriptional regulators. Annu Rev Microbiol 77:317–339. doi: 10.1146/annurev-micro-050323-040543 [DOI] [PubMed] [Google Scholar]
- 27. Alanazi AM, Neidle EL, Momany C. 2013. The DNA-binding domain of BenM reveals the structural basis for the recognition of a T-N11-A sequence motif by LysR-type transcriptional regulators. Acta Crystallogr D Biol Crystallogr 69:1995–2007. doi: 10.1107/S0907444913017320 [DOI] [PubMed] [Google Scholar]
- 28. Bundy BM, Collier LS, Hoover TR, Neidle EL. 2002. Synergistic transcriptional activation by one regulatory protein in response to two metabolites. Proc Natl Acad Sci U S A 99:7693–7698. doi: 10.1073/pnas.102605799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bedore SR, Schmidt AL, Slarks LE, Duscent-Maitland CV, Elliott KT, Andresen S, Costa FG, Weerth RS, Tumen-Velasquez MP, Nilsen LN, Dean CE, Karls AC, Hoover TR, Neidle EL, Alexandre G. 2022. Regulation of L - and D -aspartate transport and metabolism in Acinetobacter baylyi ADP1. Appl Environ Microbiol 88:e0088322. doi: 10.1128/aem.00883-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. UniProt Consortium . 2021. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res 49:D480–D489. doi: 10.1093/nar/gkaa1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mrázek J, Xie S. 2006. Pattern locator: a new tool for finding local sequence patterns in genomic DNA sequences. Bioinformatics 22:3099–3100. doi: 10.1093/bioinformatics/btl551 [DOI] [PubMed] [Google Scholar]
- 33. Drew D, North RA, Nagarathinam K, Tanabe M. 2021. Structures and general transport mechanisms by the major facilitator superfamily (MFS). Chem Rev 121:5289–5335. doi: 10.1021/acs.chemrev.0c00983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shimamoto T, Negishi K, Tsuda M, Tsuchiya T. 1996. Mutational analysis of the CitA citrate transporter from Salmonella typhimurium: altered substrate specificity. Biochem Biophys Res Commun 226:481–487. doi: 10.1006/bbrc.1996.1381 [DOI] [PubMed] [Google Scholar]
- 35. Altekar WW, Rao MR. 1963. Microbiological dissimilation of tricarballylate and trans-aconitate. J Bacteriol 85:604–613. doi: 10.1128/jb.85.3.604-613.1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Boyd JM, Teoh WP, Downs DM. 2012. Decreased transport restores growth of a Salmonella enterica apbC mutant on tricarballylate. J Bacteriol 194:576–583. doi: 10.1128/JB.05988-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Saffran M, Prado JL. 1949. Inhibition of aconitase by trans-aconitate. J Biol Chem 180:1301–1309. [PubMed] [Google Scholar]
- 38. Imai K, Iijima T, Hasegawa T. 1973. Transport of tricarboxylic acids in Salmonella typhimurium . J Bacteriol 114:961–965. doi: 10.1128/jb.114.3.961-965.1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Underhill SAM, Cabeen MT. 2022. Redundancy in citrate and cis-aconitate transport in Pseudomonas aeruginosa. J Bacteriol 204:e0028422. doi: 10.1128/jb.00284-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. D’Argenio DA, Vetting MW, Ohlendorf DH, Ornston LN. 1999. Substitution, insertion, deletion, suppression, and altered substrate specificity in functional protocatechuate 3,4-dioxygenases. J Bacteriol 181:6478–6487. doi: 10.1128/JB.181.20.6478-6487.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen J, Shang F, Wang L, Zou L, Bu T, Jin L, Dong Y, Ha NC, Quan C, Nam KH, Xu Y. 2018. Structural and biochemical analysis of the citrate-responsive mechanism of the regulatory domain of catabolite control protein E from Staphylococcus aureus. Biochemistry 57:6054–6060. doi: 10.1021/acs.biochem.8b00671 [DOI] [PubMed] [Google Scholar]
- 42. Liu W, Chen J, Jin L, Liu ZY, Lu M, Jiang G, Yang Q, Quan C, Nam KH, Xu Y. 2021. Functional and structural analysis of catabolite control protein C that responds to citrate. Sci Rep 11:20285. doi: 10.1038/s41598-021-99552-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Oliver P, Peralta-Gil M, Tabche ML, Merino E. 2016. Molecular and structural considerations of TF-DNA binding for the generation of biologically meaningful and accurate phylogenetic footprinting analysis: the LysR-type transcriptional regulator family as a study model. BMC Genomics 17:686. doi: 10.1186/s12864-016-3025-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stoudenmire JL, Schmidt AL, Tumen-Velasquez MP, Elliott KT, Laniohan NS, Walker Whitley S, Galloway NR, Nune M, West M, Momany C, Neidle EL, Karls AC. 2017. Malonate degradation in Acinetobacter baylyi ADP1: operon organization and regulation by MdcR. Microbiology (Reading) 163:789–803. doi: 10.1099/mic.0.000462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tumen-Velasquez MP, Laniohan NS, Momany C, Neidle EL. 2019. Engineering CatM, a LysR-type transcriptional regulator, to respond synergistically to two effectors. Genes (Basel) 10:421. doi: 10.3390/genes10060421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Craven SH, Ezezika OC, Momany C, Neidle EL. 2008. LysR homologs in Acinetobacter: insights into a diverse and prevalent family of transcriptional regulators. Caister Academic Press Norfolk. [Google Scholar]
- 47. Singh A, Bedore SR, Sharma NK, Lee SA, Eiteman MA, Neidle EL. 2019. Removal of aromatic inhibitors produced from lignocellulosic hydrolysates by Acinetobacter baylyi ADP1 with formation of ethanol by Kluyveromyces marxianus. Biotechnol Biofuels 12:91. doi: 10.1186/s13068-019-1434-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. NY. [Google Scholar]
- 49. Grant SG, Jessee J, Bloom FR, Hanahan D. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci U S A 87:4645–4649. doi: 10.1073/pnas.87.12.4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Eraso JM, Kaplan S. 1994. prrA, a putative response regulator involved in oxygen regulation of photosynthesis gene expression in Rhodobacter sphaeroides. J Bacteriol 176:32–43. doi: 10.1128/jb.176.1.32-43.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Norrander J, Kempe T, Messing J. 1983. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene 26:101–106. doi: 10.1016/0378-1119(83)90040-9 [DOI] [PubMed] [Google Scholar]
- 52. Jones RM, Williams PA. 2003. Mutational analysis of the critical bases involved in activation of the AreR-regulated σ54-dependent promoter in Acinetobacter sp. strain ADP1. Appl Environ Microbiol 69:5627–5635. doi: 10.1128/AEM.69.9.5627-5635.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kostylev M, Otwell AE, Richardson RE, Suzuki Y, Isalan M. 2015. Cloning should be simple: Escherichia coli DH5 alpha-mediated assembly of multiple DNA fragments with short end homologies. PLoS ONE 10:e0137466. doi: 10.1371/journal.pone.0137466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Horton RM, Ho SN, Pullen JK, Hunt HD, Cai Z, Pease LR. 1993. Gene splicing by overlap extension. Methods Enzymol 217:270–279. doi: 10.1016/0076-6879(93)17067-f [DOI] [PubMed] [Google Scholar]
- 55. Seaton SC, Elliott KT, Cuff LE, Laniohan NS, Patel PR, Neidle EL. 2012. Genome-wide selection for increased copy number in Acinetobacter baylyi ADP1: locus and context-dependent variation in gene amplification. Mol Microbiol 83:520–535. doi: 10.1111/j.1365-2958.2011.07945.x [DOI] [PubMed] [Google Scholar]
- 56. Neidle EL, Hartnett C, Ornston LN. 1989. Characterization of Acinetobacter calcoaceticus catM, a repressor gene homologous in sequence to transcriptional activator genes. J Bacteriol 171:5410–5421. doi: 10.1128/jb.171.10.5410-5421.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sayers EW, Bolton EE, Brister JR, Canese K, Chan J, Comeau DC, Connor R, Funk K, Kelly C, Kim S, Madej T, Marchler-Bauer A, Lanczycki C, Lathrop S, Lu Z, Thibaud-Nissen F, Murphy T, Phan L, Skripchenko Y, Tse T, Wang J, Williams R, Trawick BW, Pruitt KD, Sherry ST. 2022. Database resources of the national center for biotechnology information. Nucleic Acids Res 50:D20–D26. doi: 10.1093/nar/gkab1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 59. Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. 2004. The KEGG resource for deciphering the genome. Nucleic Acids Res 32:D277–D280. doi: 10.1093/nar/gkh063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stothard P. 2000. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28:1102–1104. doi: 10.2144/00286ir01 [DOI] [PubMed] [Google Scholar]
- 61. Corpet F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16:10881–10890. doi: 10.1093/nar/16.22.10881 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S4 and Fig. S1 to S7.