ABSTRACT
White-rot fungi, such as Phanerochaete chrysosporium, are the most efficient degraders of lignin, a major component of plant biomass. Enzymes produced by these fungi, such as lignin peroxidases and manganese peroxidases, break down lignin polymers into various aromatic compounds based on guaiacyl, syringyl, and hydroxyphenyl units. These intermediates are further degraded, and the aromatic ring is cleaved by 1,2,4-trihydroxybenzene dioxygenases. This study aimed to characterize homogentisate dioxygenase (HGD)-like proteins from P. chrysosporium that are strongly induced by the G-unit fragment of vanillin. We overexpressed two homologous recombinant HGDs, PcHGD1 and PcHGD2, in Escherichia coli. Both PcHGD1 and PcHGD2 catalyzed the ring cleavage in methoxyhydroquinone (MHQ) and dimethoxyhydroquinone (DMHQ). The two enzymes had the highest catalytic efficiency (kcat/Km) for MHQ, and therefore, we named PcHGD1 and PcHGD2 as MHQ dioxygenases 1 and 2 (PcMHQD1 and PcMHQD2), respectively, from P. chrysosporium. This is the first study to identify and characterize MHQ and DMHQ dioxygenase activities in members of the HGD superfamily. These findings highlight the unique and broad substrate spectra of PcHGDs, rendering them attractive candidates for biotechnological applications.
IMPORTANCE
This study aimed to elucidate the properties of enzymes responsible for degrading lignin, a dominant natural polymer in terrestrial lignocellulosic biomass. We focused on two homogentisate dioxygenase (HGD) homologs from the white-rot fungus, P. chrysosporium, and investigated their roles in the degradation of lignin-derived aromatic compounds. In the P. chrysosporium genome database, PcMHQD1 and PcMHQD2 were annotated as HGDs that could cleave the aromatic rings of methoxyhydroquinone (MHQ) and dimethoxyhydroquinone (DMHQ) with a preference for MHQ. These findings suggest that MHQD1 and/or MHQD2 play important roles in the degradation of lignin-derived aromatic compounds by P. chrysosporium. The preference of PcMHQDs for MHQ and DMHQ not only highlights their potential for biotechnological applications but also underscores their critical role in understanding lignin degradation by a representative of white-rot fungus, P. chrysosporium.
KEYWORDS: lignin, white-rot fungus, vanillic acid, syringic acid, homogentisate dioxygenase
INTRODUCTION
Lignin is an abundant natural phenylpropanoid polymer that constitutes 15%–35% of the total lignocellulosic biomass on Earth (1, 2). Hence, lignin degradation by environmental microbes plays a key role in the biospheric carbon cycle (3–6). White-rot basidiomycete fungi are known for their ability to completely degrade lignin (3–6). Phanerochaete chrysosporium, one of the best-studied white-rot basidiomycetes, produces extracellular lignin peroxidases (LiP) and manganese peroxidases (5–8). These nonspecific one-electron-oxidizing enzymes cleave the carbon-carbon and ether bonds in lignin, resulting in the formation of a variety of lignin-derived aromatics such as benzoquinones, hydroquinones (HQ), benzaldehydes, and aromatic acids (5–8). These lignin fragments are categorized into guaiacyl units, i.e., vanillin (VN) and vanillic acid (VA); syringyl units, i.e., syringaldehyde (SN) and syringic acid (SA); and hydroxyphenyl units, i.e., p-hydroxybenzaldehyde (HBN) and p-hydroxybenzoic acid (HBA) (9). These intermediates are then intracellularly oxidized, decarboxylated, hydroxylated, and/or demethoxylated to form 1,2,4-trihydroxybenzene (THB) (10–12), followed by a ring cleavage by THB dioxygenases (11, 13). Although lignin fragment metabolism in P. chrysosporium has been extensively studied (10–13), the enzymes involved in fragment conversion remain largely obscure (14).
We had previously reported changes in protein expression in P. chrysosporium grown with VN, and a putative dioxygenase was annotated as homogentisic acid 1,2-dioxygenase (HGD; EC 1.13.11.5), which was strongly induced by VN (15). Moreover, a recent multi-omics analysis of the degradation of lignin and lignin-derived aromatic compounds was performed in two white-rot fungi, Trametes versicolor and Gelatoporia subvermispora (16). Among the dioxygenases induced by SA, the lignin-derived aromatic compound and poplar-derived aromatic compounds, two homologous dioxygenases annotated as HGD (TV_20432 and TV_124955), were also induced at the transcript and protein levels (16). However, the biochemical characteristics of HGD-like enzymes in lignin and their derived aromatic compounds have not yet been elucidated.
HGD is widely distributed and characterized in aerobic organisms, including bacteria, fungi, plants, and animals (17, 18). The enzyme is involved in the catabolism of phenylalanine and tyrosine, and catalyzes the oxidative cleavage of the aromatic ring in homogentisic acid (HGA) to produce maleylacetoacetate (19). A deficiency of the HGD gene in humans causes HGA accumulation in urine, which darkens upon exposure to air, a key characteristic of alkaptonuria (20). The accumulation of HGA in mammal tissues results in pigmentation (ochronosis) and eventually causes serious arthropathy (20–22). Human and bacterial HGDs, HGDOHs and HGDOPp, respectively, are highly specific and prefer HGA and its derivatives as substrates (20–22). They form a homohexameric complex with a subunit molecular mass of approximately 50 kDa and possess the structural fold of cupin (22, 23), a functionally diverse superfamily (24) that includes gentisate 1,2-dioxygenases (EC: 1.13.11.4).
In the current study, two HGD homologs from P. chrysosporium, PcHGD1 and PcHGD2, with 69.7% amino acid sequence identity were produced as recombinant proteins in Escherichia coli and characterized. The results are discussed with respect to the physiological roles of HGD homologs and the degradation of lignin-derived aromatics by P. chrysosporium.
RESULTS
Searching for HGD homologs in the P. chrysosporium genome
A BLAST search was conducted with the amino acid sequence of HGD-like protein from T. versicolor as a query to screen the published P. chrysosporium genome database (https://mycocosm.jgi.doe.gov/Phchr4_2/Phchr4_2.home.html) for HGD homologs. The T. versicolor HGD-like protein (TV_20432) shared 62.0% and 52.5% amino acid sequence identity with PcHGD1 (6344326) and PcHGD2 (6344291), respectively. Sequence alignments of PcHGD1 and PcHGD2 with HGDs from Homo sapiens (Q93099), Pseudomonas putida (Q88E47), T. versicolor (TV_20432), and G. subvermispora (GS_117547) are shown in Fig. 1. The two histidine residues and one glutamate residue involved in active-site non-heme ferric iron coordination (22) were highly conserved in all HGDs, whereas one tyrosine residue for interaction with HGA in the active-site lid of HGD was conserved in six HGDs (22), except for PcHGD2, where a phenylalanine is present (Fig. 1).
Fig 1.
Amino acid sequence alignment of HGD homologs. Sequences of PcHGD1 (protein ID 6344326), PcHGD2 (6344291), and HGD from Trametes versicolor (TV_20432), Gelatoporia subvermispora (GS_117547), Homo sapiens (Q93099), and Pseudomonas putida (Q88E47) are shown. The protein IDs for H. sapiens and P. putida were obtained from the UniProt Knowledgebase (http://beta.uniprot.org), and those for the other species were obtained from the JGI Genome Portal (https://mycocosm.jgi.doe.gov/Phchr4_2/Phchr4_2.home.html). The residues in the red boxes are responsible for active-site non-heme ferric iron coordination and catalytic activity in HGD. The sequences were aligned using ClustalW program (https://www.genome.jp/tools-bin/clustalw). The active site lid is underlined.
Catalytic properties of the HGD-like proteins from P. chrysosporium
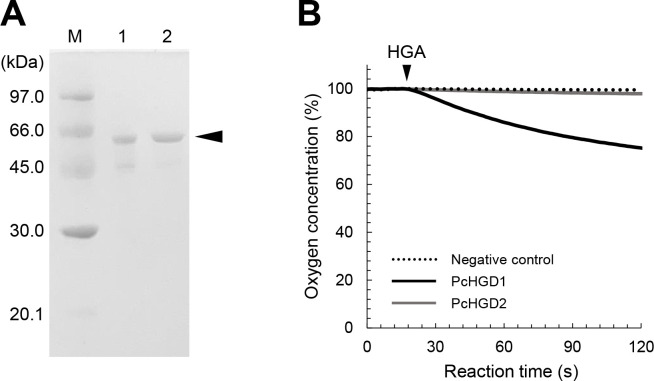
N-terminal 6× His-tagged PcHGD1 and PcHGD2 were produced in E. coli using the pET expression system, and the molecular masses of the protein bands were consistent with those predicted from the deduced amino acid sequences (55.6 and 56.8 kDa, respectively) (Fig. 2A). The activities of recombinant PcHGD1 and PcHGD2 toward HGA were evaluated by polarographic analysis of the catalytic reaction using a Clark O2 electrode (Fig. 2B). O2 consumption was detected in the reaction of HGA with PcHGD1 (Fig. 2B), indicating that PcHGD1 catalyzes the ring cleavage of HGA. In contrast, O2 consumption was not detected in the reaction with PcHGD2, indicating that the enzyme does convert HGA (Fig. 2B). The product obtained from the ring cleavage of HGA by PcHGD1 was determined by gas chromatography-mass spectrometry (GC-MS) analysis (Fig. S1), while no conversion products of HGA were observed with PcHGD2. Mass spectra of the trimethylsilyl (TMS) derivatives of the reaction products by PcHGD1 showed fragmentation patterns identical to those of maleylacetoacetate (Fig. S1).
Fig 2.
Preparation of recombinant PcHGD1 and PcHGD2 and oxygen consumption during HGD reactions with HGA as a substrate. (A) SDS-PAGE analysis of purified PcHGD1 (lane 1) and PcHGD2 (lane 2). (B) Oxygen consumption during HGD reactions with HGA. Consumption of oxygen was monitored using a Clark O2 electrode. The arrow indicates the addition of a substrate to the reaction mixture.
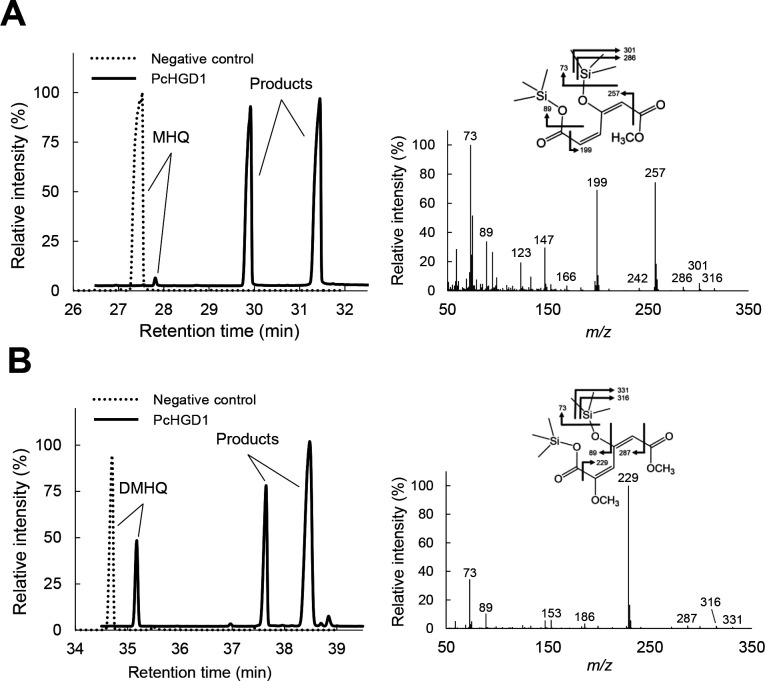
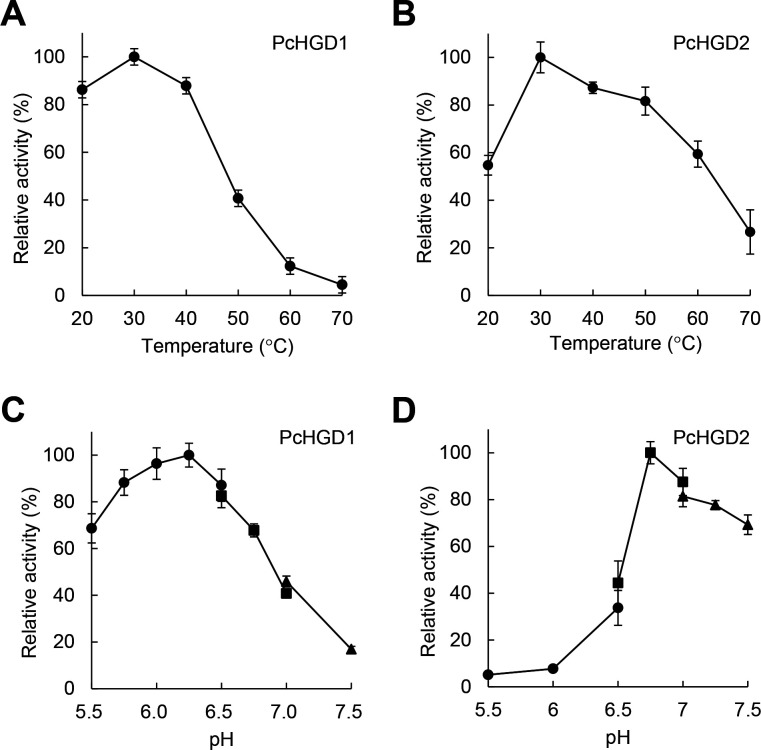
As mentioned earlier, PcHGD1 is induced by VN-, SA-, and poplar-derived aromatic compounds. Several VN derivatives were tested as substrates for the two PcHGDs. Although PcHGD1 did not convert THB, VN, VA, SN, SA, HBN, or HBA, the ring-cleavage products of methoxyhydroquinone (MHQ) and 2,6-dimethoxyhydroquinone (DMHQ) conversion by PcHGD1 were identified using GC-MS (Fig. 3). Similar results were obtained when PcHGD2 was used (data not shown). These findings reveal the uniquely broad substrate spectrum of PcHGD1 and PcHGD2 in comparison with other HGDs (17–19). The effects of pH and temperature on PcHGD1 and PcHGD2 activity were also determined using MHQ as the substrate (Fig. 4). The optimal reaction temperature for PcHGD1 and PcHGD2 was 30°C (Fig. 4A and B), and the optimal pH values were 6.25 and 6.75, respectively (Fig. 4C and D).
Fig 3.
Total ion chromatograms and mass spectra of the reaction products generated by PcHGD1 from MHQ (A) and DMHQ (B) as substrates. The TMS-derivatized reaction products were analyzed using GC-MS. The mass spectra (A, 4-hydroxy-6-methoxy-6-oxohexa-2,4-dienoic acid; B, 4-hydroxy-2,6-dimethoxy-6-oxohexa-2,4-dienoic acid) of the reaction products were obtained from the GC peaks appearing at retention times 29.9 and 31.5 min (A) and 37.6 and 38.5 min (B). The experiment was performed three times, and representative results are shown.
Fig 4.
Optimal temperature and pH of PcHGD1 and PcHGD2. (A and B) Optimal temperatures for PcHGD1 and PcHGD2 determined using MHQ as the substrate. Enzyme reactions proceeded at temperatures ranging from 20°C to 70°C. (C and D) Optimal pH of PcHGD1 and PcHGD2. Enzyme reactions proceeded over a pH range of 5.5–8.0: in 50 mM MES (pH 5.0–6.5; ●), 50 mM MOPS (pH 6.5–7.0; ■), and 50 mM HEPES (pH 7.0–8.0; ▲). Data are presented as mean ± standard deviation of four independent experiments.
Apparent kinetic parameters of PcHGD1 and PcHGD2
The apparent kinetic parameters of PcHGD1 and PcHGD2 were determined using HGA, MHQ, and DMHQ as substrates (Table 1). The rates of O2 consumption and substrate conversion by PcHGD1 and PcHGD2 were measured polarographically by monitoring the changes in O2 concentration and quantifying MHQ using GC-MS. The stoichiometric ratio between O2 consumption and ring cleavage of MHQ was 1.00:1.17. The highest catalytic efficiency (kcat/Km) of PcHGD1 was observed with MHQ (Table 1). The lowest Km value was observed for MHQ among the three substrates, and the highest kcat was observed for MHQ (Table 1). The highest catalytic efficiency (kcat/Km) for PcHGD2 was observed with MHQ (Table 1). The Km value for MHQ was lower than those for HGA and DMHQ, whereas the kcat values for MHQ and DMHQ were indifferent (Table 1). Hence, hereafter, we call PcHGD1 and PcHGD2 as MHQ dioxygenases 1 and 2 (PcMHQD1 and PcMHQD2) from P. chrysosporium, respectively.
TABLE 1.
Apparent kinetic parameters of PcHGD1 and PcHGD2 for MHQ, DMHQ, and HGAa
| Substrate | Km (μM) | kcat (s−1) | kcat/Km (s−1 mM−1) | |
|---|---|---|---|---|
| PcHGD1 | Methoxyhydroquinone | 49 ± 14 | 8.66 ± 0.84 | 177 |
| Dimethoxyhydroquinone | 993 ± 236 | 1.37 ± 0.40 | 1.38 | |
| Homogentisic acid | 628 ± 238 | 0.08 ± 0.02 | 0.12 | |
| PcHGD2 | Methoxyhydroquinone | 34 ± 4 | 1.02 ± 0.01 | 33 |
| Dimethoxyhydroquinone | 2,896 ± 821 | 0.99 ± 0.25 | 0.34 | |
| Homogentisic acid | – | – | – |
The activity levels of PcHGD1 and PcHGD2 were determined in the reaction mixtures (1.0 mL) containing 0.1–5 µM PcHGDs and 5 µL of a substrate solution (0–600 mM in dimethylsulfoxide) in 50 mM MES and MOPS (pH 6.25 and 6.75), 30°C. We used the initial velocity for 30 s after the addition of each substrate for the calculation of apparent kinetic parameters. Data are presented as mean ± standard error of three experiments.
Transcriptional regulation of PcMHQDs
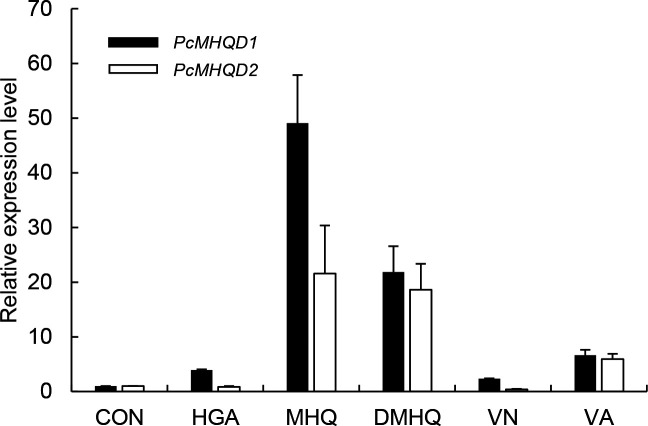
PcMHQD1 gene expression was upregulated when HGA, MHQ, DMHQ, VN, and VA were added to the medium (Fig. 5). In particular, when P. chrysosporium was cultured with MHQ and DMHQ, the transcription levels of PcMHQD1 were 49-fold and 21-fold higher, respectively, than in those grown without the substrates (Control: CON) (Fig. 5). Similar results were obtained for PcHGD2 expression, with the exception of HGA (Fig. 5). Thus, both PcMHQDs are likely involved in the conversion of MHQ and DMHQ in P. chrysosporium.
Fig 5.
Gene expression profiles of PcMHQD1 and PcMHQD2 in P. chrysosporium in response to exogenous HGA, MHQ, DMHQ, VN, and VA. The abundance of amplified cDNA fragments of PcMHQD1 and PcMHQD2 transcripts was normalized using ACT1 as the reference gene. The normalized expression of each gene in the fungus upon 6 h exposure to HGA, MHQ, DMHQ, VN, and VA relative to the normalized expression in the absence of the substrate is shown. Data are presented as mean ± standard deviation (error bars) of three independent experiments.
Fungal metabolism of MHQ and DMHQ
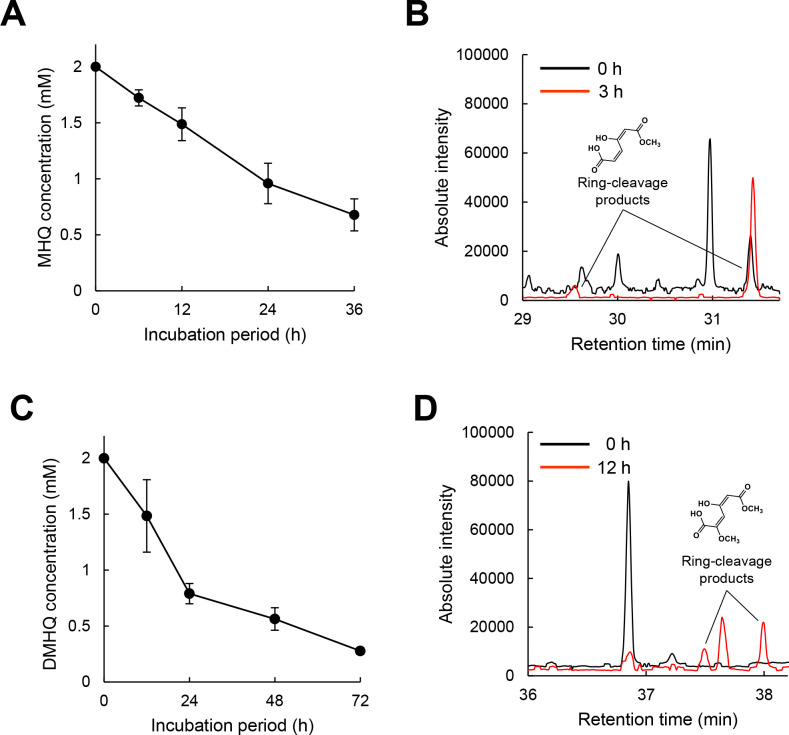
In our evaluation of the metabolism of MHQ and DMHQ in P. chrysosporium, 2 mM MHQ in the initial medium decreased to 0.68 mM after 36 h cultivation, and 2 mM DMHQ in the initial medium decreased to 0.28 mM after 72 h cultivation (Fig. 6A and C). The ring-cleavage products of MHQ and DMHQ were detected in reactions using homogenized fungal cells (Fig. 6B and D), indicating that the substrates were ring cleaved, at least partially, in fungal cells.
Fig 6.
Fungal metabolism of methoxyhydroquinone and dimethoxyhydroquinone. The time course of MHQ and DMHQ (A and C) metabolisms was monitored. After a 2-day pre-incubation, MHQ and DMHQ were added to a final concentration of 2.0 mM. The ring-cleavage products of MHQ and DMHQ (B and D) were detected in the reaction solutions using the fungal cell lysate. MHQ and DMHQ were added to a final concentration of 2.0 mM. After 3 or 12 h of incubation with MHQ or DMHQ, reaction products were identified using GC-MS. Data are presented as mean ± standard deviation of three independent experiments.
DISCUSSION
In the present study, we report the biochemical characterization of two HGD homologs from P. chrysosporium. MHQD1 and MHQD2 were annotated as HGD in P. chrysosporium genome database. MHQD1 was capable of catalyzing ring cleavage in HGA, MHQ, and DMHQ with a preference for MHQ, whereas MHQD2 can catalyze ring cleavage in only MHQ and DMHQ, also with a preference for MHQ as a substrate (Fig. 3; Fig. S1). These results suggest that they play important roles in the degradation of lignin-derived aromatic compounds, such as MHQ and DMHQ, in P. chrysosporium.
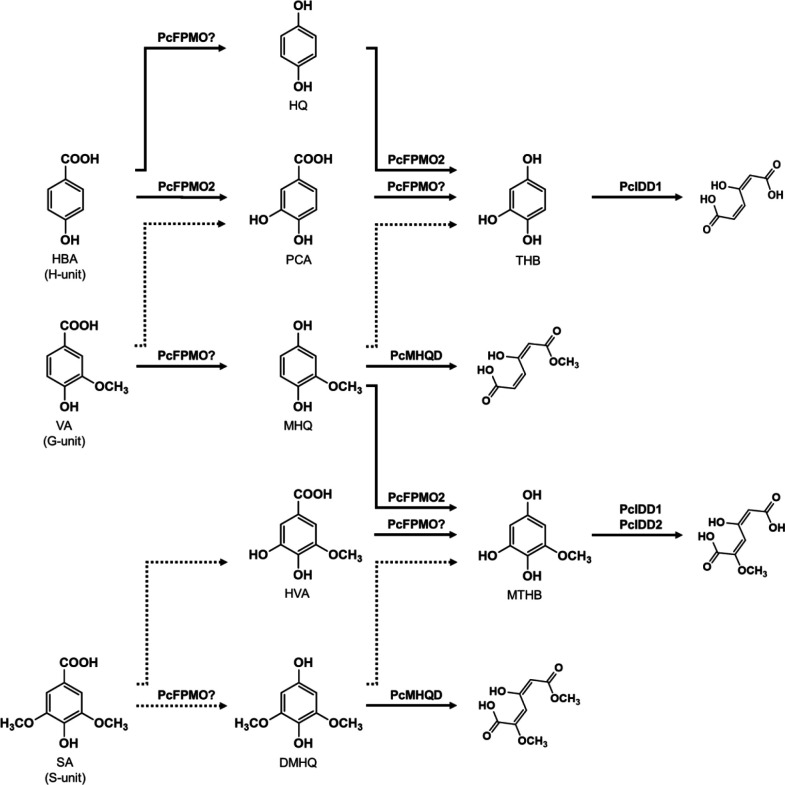
In P. chrysosporium, the G-unit fragment VA is subsequently decarboxylated to MHQ and demethoxylated to THB, which is then degraded via aromatic ring fission catalyzed by THB dioxygenase (10, 11). Although VA decarboxylase has not been identified in P. chrysosporium, 4-hydroxybenzoate 1-hydroxylase (4HB1H: G8B709) with VA decarboxylase activity, which belongs to the flavoprotein monooxygenase (FPMO) group A superfamily, was reported (25). FPMO, which was first isolated from Candida parapsilosis (25), catalyzes an oxidative decarboxylation reaction yielding hydroquinone and MHQ from HBA and VA, respectively. The P. chrysosporium genome harbors 59 putative FPMO-encoding genes (12). The BLAST search results from the published P. chrysosporium genome database indicated that 4HB1H shares 36.0% amino acid sequence identity with 6557855 (protein ID), suggesting that FPMO may have the ability to decarboxylate VA. We recently identified vanillin-induced MHQ hydroxylase (PcFPMO2) and methoxytrihydroxybenzene (MTHB) dioxygenases such as PcIDD1 and PcIDD2 in P. chrysosporium (12, 13). In addition, we identified PcMHQD1 and PcMHQD2 in the current study. Although the main metabolic pathway for MHQ in white-rot fungi has not yet been identified, previous studies have shown that P. chrysosporium possesses diverse metabolic routes for MHQ degradation (Fig. 7). The ring-cleavage product from MHQ was identified during VA metabolism (Fig. 7), indicating that PcMHQD1 and/or PcMHQD2 are involved in the aromatic ring fission of MHQ in P. chrysosporium. Overall, these results suggest that PcMHQD1 and/or PcMHQD2 are involved in the ring cleavage of MHQ, a decarboxylated product of VA, during VN degradation.
Fig 7.
Metabolic pathways of HBA, VA, and SA in the white-rot fungus P. chrysosporium. HBA is estimated to convert to hydroquinone and protocatechuic acid (PCA). HQ is hydroxylated to THB by PcFPMO2 (12), while PCA is decarboxylated to THB, which is then ring cleaved by intradiol dioxygenase 1 (PcIDD1) (11, 13). VA is estimated to convert to PCA and MHQ. MHQ is ring cleaved (this study) or be converted to THB and MTHB, which are then cleaved (10–13). SA is estimated to convert to hydroxyl vanillic acid (HVA) and DMHQ. HVA is converted to MTHB, while DMHQ is ring cleaved (this study) or may be converted to MTHB. Dotted arrows indicate the estimated reactions; solid arrows indicate the reactions by identified enzymes: PcMHQD (this study), PcFPMO2 (12), PcIDD1(13), and PcIDD2 (13).
Basidiomycetes, including P. chrysosporium, can degrade a wide variety of lignin-derived aromatics, including MHQ and DMHQ (10, 26–28). MHQ and DMHQ are intermediates of the major lignin-derived aromatics, VA and SA, respectively, and these might be demethoxylated to MHQ and DMHQ; however, MHQ and DMHQ demethylases have not yet been identified in fungi. In bacteria, it is reported that the GcoAB cytochrome P450 system comprising a coupled monoxygenase (GcoA) and reductase (GcoB) catalyzes the O-demethylation of 3-methoxycatechol to form pyrogallol (29). A total of 154 putative P450-encoding genes have been identified in the P. chrysosporium genome, revealing the genetic diversity of fungal P450s (30–32), and suggesting that some P450s catalyze the O-demethylation of MHQ and DMHQ to form THB and MTHB, respectively. The current study revealed that MHQ-induced PcMHQD1 and PcMHQD2 were capable of catalyzing ring cleavage of MHQ and DMHQ, respectively (Fig. 3A and B). The ring-cleavage products of MHQ and DMHQ were also identified in the reaction (Fig. 6B and D). These results suggest that MHQ and DMHQ directly cleave aromatic rings in fungal cells.
Although the enzymes did not convert THB, the catalytic efficiencies (kcat/Km) of PcMHQD1 and PcMHQD2 for MHQ were significantly higher than those for HGA and DMHQ (Table 1). Additionally, DMHQ was the preferred substrate for the enzymes over HGA (Table 1). The current study indicated that MHQ is the physiological substrate of the HGD homologs PcMHQD1 and PcMHQD2. Whether bacterial and human HGDs also catalyze ring cleavage of MHQ and DMHQ is unclear. PcMHQD1 shares 43.4% and 41.1% amino acid sequence identity with HGDOHs and HGDOPp, respectively (Fig. 1). In addition, PcMHQD2 shared 35.1% and 38.1% amino acid sequence identity with HGDOHs and HGDOPp, respectively (Fig. 1). The two histidine residues and one glutamate residue involved in active-site non-heme ferric iron coordination (22) are highly conserved in all HGDs, whereas the tyrosine residue for interaction with the carboxyl group of HGA in the active-site lid of HGD is conserved in six HGDs (22), except for PcHGD2 (Fig. 1). In fact, PcMHQD2 did not convert HGA, suggesting that the tyrosine in the active-site lid of HGD is important for HGA interaction. Sequence alignment of the active-site lid including the tyrosine residue of PcMHQD1 and PcMHQD2 with HGDs from T. versicolor, G. subvermispora, Coprinopsis cinerea, Aspergillus nidulans, P. putida, Caenorhabditis elegans, Arabidopsis thaliana, Drosophila melanogaster, Danio rerio, Mus musculus, and H. sapiens is shown in Fig. S2. Only PcMHQD2 has a phenylalanine residue instead of the tyrosine among the HGD members. HGDs from G. subvermispora (116559) and A. nidulans (Q5B618) are substituted with the glycine residue, which may compromise their ability to cleave HGA. The amino acid sequences of the active-site lid differ between fungal HGDs and other HGDs (Fig. S2). Except for A. nidulans HGD (Q00667), the active-site lids of all fungal HGDs are mainly composed of DYGGRSD, while the lids of other HGDs are mainly composed of HYEAKEG. Differences in the amino acid residues constituting the active-site lid may be responsible for the difference in activity toward HGA, MHQ, and DMHQ. Putative three-dimensional structure of PcHGD1 was generated using AlphaFold (data not shown). The RMSD value between crystal structure of HGDOPp with HGA (22) and PcMHQD1 was 0.719, indicating high similarity between the two structures. However, the predicted local distance difference test score at the active-site lid of PcMHQD1 was low (data not shown). The active-site lid (G344-E351), when HGA does not bind to HGDOPp, was not defined in the electron density (22). The interaction with HGA and the active-site lid is most likely responsible for the closure of the active-site lid when HGA binds, suggesting that the exact structure of the lid is determined when the substrate binds. Further research using structural analysis is required to confirm this hypothesis. The current study is the first to show the ring-cleavage activity of the G-unit fragment MHQ and S-unit fragment DMHQ in the HGD superfamily. Homologs of PcMHQDs are found in some white-rot fungi, indicating that they may also contribute to the degradation of lignin and lignin-derived aromatics. Although bacteria use an iron (II)-dependent dioxygenase to cleave the aromatic ring of MHQ (33), the small subunit (HqdA) shares 13.5% and 12.9% amino acid sequence identity with PcMHQD1 and PcMHQD2, respectively, while the corresponding values for the large subunit (HqdB) are 10.3% and 13.2%, respectively. These results suggest that in comparison with bacteria, white-rot fungi use different types of enzymes such as PcMHQD1 and PcMHQD2, which belong to the HGD family, to achieve aromatic ring cleavage.
Bacterial and human HGDs that catalyze the ring-opening step in the tyrosine and phenylalanine degradation pathways for HGA have been well studied, and their functions have been elucidated (20, 22). The catalytic efficiency (kcat/Km) of PcHGDs toward HGA was significantly lower than that of HGDOPp (1,494 s−1mM−1) and HGDOHs (1,610 s−1mM−1) (20, 22), suggesting that P. chrysosporium may not degrade tyrosine and phenylalanine like other organisms. White-rot basidiomycetes, including P. chrysosporium, degrade lignin and various aromatic pollutants using lignin peroxidases (34). Veratryl alcohol, a metabolite of P. chrysosporium biosynthesized from phenylalanine, has been implicated as a diffusible oxidant and LiP stabilizer in ligninolysis (35). Thus, the low aromatic amino acid degradation activity may be beneficial for the effective production of veratryl alcohol by P. chrysosporium.
A recent study reported that two HGD homologs from the white-rot fungus T. versicolor (TV_20432 and TV_124955) were also induced at transcript and protein levels by SA and poplar-derived aromatic compounds (16). However, the findings did not clarify whether these HGD homologs catalyze ring cleavage of MHQ and DMHQ. Although HGDs are widely distributed in aerobic organisms (17, 18), identification of the HGDs that catalyze the ring cleavage of MHQ and DMHQ remains a topic of interest. However, further research is required to confirm this hypothesis.
In conclusion, this report presents the biochemical characterization of PcMHQD1 and PcMHQD2, belonging to the HGD superfamily, in a basidiomycetous fungus, providing a better understanding of the degradation of lignin and lignin-derived aromatics in relation to the functional diversity of MHQ dioxygenases in white-rot fungi. The broad substrate specificity and high catalytic efficiency (kcat/Km) of PcMHQDs toward MHQ are attractive for biotechnological applications in the biodegradation of aromatic compounds and provide insights into the biochemistry of this enzyme group.
MATERIALS AND METHODS
Chemicals
HGA, MHQ, VA, SA, and HBA were purchased from Tokyo Chemical Industry Co. Ltd. (Tokyo, Japan). VN, SN, and HBN were purchased from Wako Pure Chemical Industries (Osaka, Japan). THB was purchased from Kanto Chemical Co. Inc. (Tokyo, Japan). DMHQ was purchased from Apollo Scientific, Ltd. (Stockport, UK). All the chemicals were of analytical grade. Deionized water was obtained using a Milli-Q system (Merck Millipore, Billerica, MA, USA).
Strains, cultures, and media
P. chrysosporium (ATCC 34541) was maintained in high-carbon low-nitrogen (HCLN) medium (pH 4.5) containing the following constituents per liter of distilled H2O: KH2PO4, 0.2 g; MgSO4⋅7H2O, 0.05 g; CaCl2, 0.01 g; mineral solution, 1 mL; and vitamin solution, 0.5 mL. The mineral solution contained the following constituents per liter of distilled H2O: nitrilotriacetate, 1.5 g; MgSO4⋅7H2O, 3.0 g; MnSO4⋅H2O, 0.5 g; NaCl, 1.0 g; FeSO4⋅7H2O, 100 mg; CoSO4, 100 mg; CaCl2, 82 mg; ZnSO4, 100 mg; CuSO4⋅5H2O, 10 mg; AlK(SO4)2, 10 mg; H3BO3, 10 mg; and NaMoO4, 10 mg. The vitamin solution contained the following constituents per liter of distilled H2O: biotin, 2 mg; folic acid, 2 mg; thiamine⋅HCl, 5 mg; riboflavin, 5 mg; pyridoxine⋅HCl, 10 mg; cyanocobalamine, 0.1 mg; nicotinic acid, 5 mg; dl-calcium pantothenate, 5 mg; p-aminobenzoic acid, 5 mg; thioctic acid, 5 mg (36). The medium was also supplemented with 28 mM d-glucose and 1.2 mM ammonium tartrate as carbon and nitrogen sources, respectively, and was buffered with 20 mM sodium 2,2-dimethylsuccinate (pH 4.5), as described previously (15, 36). For the experiments, fungal conidia were inoculated in 30 mL of HCLN medium (pH 4.5) in a 300-mL Erlenmeyer flask and incubated at 37°C in stationary culture.
Construction of the gene expression system
Full-length P. chrysosporium PcHGD1 and PcHGD2 genes (6344326 and 6344291) with an amino acid sequence identity of 69.7% were PCR amplified using the primer combinations shown in Table S1 and a DNA thermal cycler 2400 (Takara Bio, Otsu, Japan) as follows: initial denaturation at 95°C for 4 min, followed by 30 cycles of denaturation at 95°C for 2 min, annealing at 60°C for 30 s, and extension at 68°C for 30 s. The primer sets were designed using the genomic sequence of P. chrysosporium (https://mycocosm.jgi.doe.gov/Phchr4_2/Phchr4_2.home.html). PCR products were separated on 1% agarose gels, stained with ethidium bromide, and visualized using Molecular Imager FX (Bio-Rad, Hercules, CA, USA). Amplified PcHGD1 and PcHGD2 were inserted into a pET21a vector (Invitrogen, Carlsbad, CA, USA). Recombinant plasmids were used to transform E. coli BL21 (Invitrogen) using the heat-shock method, and transformants were selected on the basis of ampicillin resistance in Luria-Bertani (LB) medium. The identities of the recombinant plasmids (pET21-PcHGD1 and pET21-PcHGD2) were verified by sequencing.
Heterologous expression and purification of PcHGDs
Escherichia coli BL21 cells harboring each of the PcHGD1 and PcHGD2 gene expression plasmids were grown at 37°C with constant shaking in LB medium supplemented with 100 µg/mL ampicillin until the optical density of the cultures reached 0.6 at 600 nm. PcHGD expression was induced by adding 0.1 mM isopropyl β-D-1-thiogalactopyranoside and 1 mM FeSO4・7H2O to the medium, and the cultures were incubated further up to 24 h at 25°C. Cells were harvested by centrifugation (3,000 × g at 20°C for 5 min), and the cell pellet was resuspended in buffer A [50 mM Tris-HCl buffer pH 8.0, 200 mM NaCl, and 10% (wt/vol) glycerol]. Cells were lysed by sonication (5 × 30 s) using a Q700 sonicator (Qsonica, Melville, NY, USA). After centrifugation (15,000 × g at 4°C for 15 min) to remove the insoluble debris, the supernatant was obtained.
For protein purification, the crude lysate was loaded onto a nickel affinity column (Cytiva, Marlborough, MA, USA) equilibrated with buffer A at 4°C. The column was washed with buffer A, and the proteins were eluted with a 0–0.3 M imidazole gradient in buffer A. Fractions containing PcHGDs were brown collected and directly loaded onto a Superdex 200 HR 10/30 column (Cytiva) equilibrated with buffer A. The resulting eluate contained the purified recombinant proteins. The absorption spectra were analyzed using SpectraMax (Molecular Devices, San Jose, CA, USA).
HGD enzyme assays
HGD activity under air-saturated conditions was determined polarographically by measuring the O2 consumption during the enzyme reaction using Oxytherm + R (Hansatech Instruments, Norfolk, UK), as previously described (20). Briefly, the activity and substrate specificity of the PcHGDs were determined in reaction mixtures (0.5 mL) containing 2 µM PcHGDs and 5 µL of substrate solution (0–600 mM in acetonitrile) in 50 mM MES buffer (pH 6.0). The reaction was initiated by adding the substrate, and O2 consumption in the reaction mixture was monitored using a Clark O2 electrode. To avoid rapid autoxidation of the substrates, superoxide dismutase (6 µg) was added to the reaction mixture (37, 38). The background rate of O2 consumption in the reaction mixture without the substrate was subtracted from the observed rates for various substrates. The apparent kinetic parameters Km and kcat were calculated by fitting the obtained initial rates using the Michaelis-Menten equation in Origin version 6.0 software (OriginLab, Northampton, MA, USA). After enzymatic reaction for 60 min at 30°C, the residual substrate and reaction products were analyzed and identified using GC-MS after extraction with ethyl acetate (0.5 mL ×3), evaporation, and TMS derivation using N,O-bis(TMS)trifluoroacetamide/pyridine (2:1, vol/vol) (13). The optimum temperature for the enzyme reaction was determined by measuring activity over a range of 20°C–70°C. The optimal pH was determined using 50 mM MES buffer (pH 5.5–6.5), 50 mM MOPS (pH 6.5–7.0), and 50 mM HEPES buffer (pH 7.0–7.5).
Analytical methods
GC-MS was performed at 70 eV using a GCMS-QP2010 (Shimadzu, Kyoto, Japan) apparatus equipped with a 30-m fused silica column (DB-5; J & W Scientific, Folsom, CA, USA). The oven temperature was programmed to ramp from 80°C to 320°C at 8°C/min, with an injection temperature of 280°C. Substrate conversion products were identified by comparing their respective GC retention times and mass fragmentation patterns with those of authentic standards (12, 13). The compounds for which authentic standards were unavailable were identified by comparing their mass data with those available from the National Institute of Standard Technology Mass Spectral Library.
Quantitative RT-PCR analysis of PcHGD genes
After a 2-day pre-incubation in 30 mL of HCLN medium (pH 4.5), as described above, HGA, MHQ, DMHQ, VN, and VA in acetonitrile were added to a final concentration of 2 mM, and the mycelia were incubated for an additional 6 h. Total RNA was extracted from the mycelia grown in the absence or presence of substrates using an RNeasy Mini Kit (Qiagen, Venlo, The Netherlands) and then used to synthesize single-stranded cDNA. Quantitative RT-PCR was performed using PcHGD1 and PcHGD2 (PcHGD1; 6344326 and PcHGD2; 6344291, respectively) gene-specific primer sets (Table S1) designed using the genomic sequence of P. chrysosporium, resulting in PCR product of lengths between 130 and 150 bp. HGD gene expression was normalized to Actin expression (13). Real-time PCR was performed in a final volume of 25 µL, using a Smart Cycler II (Sunnyvale, California, USA). TB Green Premix Ex Taq (Takara) was applied according to the manufacturer’s instructions, and then the PCR proceeded as follows: (i) initial denaturation at 95°C for 30 s, (ii) 40 cycles of denaturation at 95°C for 15 s, annealing at 55°C for 30 s, and elongation at 72°C for 30 s. Specific amplification was confirmed by analyzing melting curves from 60°C to 95°C. The expression level of each gene in the presence of a substrate is depicted relative to that in the absence of a substrate.
MHQ and DMHQ metabolism by fungal cells
After a 2-day pre-incubation in 30 mL of HCLN medium, MHQ and DMHQ were added at a final concentration of 2 mM. The fungal cells grown for 24 h were filtered and washed thrice with distilled water and stored at −80°C. For the reaction experiments, fungal cells were crushed with liquid nitrogen, and an equal amount of 50 mM MES buffer (pH 6.25) was added. After centrifugation (15,000 × g at 4°C for 15 min), the supernatant was used for the reaction with 1 mM MHQ and 1 mM DMHQ. The conversion products in the culture were also analyzed by GC-MS after extraction with ethyl acetate at pH 2, drying over MgSO4, evaporation under N2, and TMS derivatization, as described above.
ACKNOWLEDGMENTS
We would like to thank Editage for English language editing.
This study was supported by a Grant-in-Aid for Scientific Research (20K05815 to M.S. and 22K05417 to M.K.) and partially supported by a research grant from the Iwatani Naoji Foundation.
Contributor Information
Motoyuki Shimizu, Email: moshimi@meijo-u.ac.jp.
Marina Lotti, University of Milano-Bicocca, Milan, Italy.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aem.01753-23.
Primers, GC-MS spectrum, and alignment of active site lid.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Sugiarto S, Leow Y, Tan CL, Wang G, Kai D. 2022. How far is lignin from being a biomedical material? Bioact Mater 8:71–94. doi: 10.1016/j.bioactmat.2021.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sarkanen KV, Ludwig CH. 1971. Lignins: occurrence, formation, structure and reactions. J Polym Sci B Polym Phys 10:228–230. [Google Scholar]
- 3. Crawford RL. 1982. Lignin biodegradation and transformation. Enzyme Microb Technol 4:285. doi: 10.1016/0141-0229(82)90047-3 [DOI] [Google Scholar]
- 4. Tien M. 1987. Properties of ligninase from Phanerochaete chrysosporium and their possible applications. Crit Rev Microbiol 15:141–168. doi: 10.3109/10408418709104456 [DOI] [PubMed] [Google Scholar]
- 5. Kirk TK, Farrell RL. 1987. Enzymatic "combustion": the microbial degradation of lignin. Annu Rev Microbiol 41:465–505. doi: 10.1146/annurev.mi.41.100187.002341 [DOI] [PubMed] [Google Scholar]
- 6. Gold MH, Wariishi H, Valli K. 1989. Extracellular peroxidases involved in lignin degradation by the white rot basidiomycete Phanerochaete chrysosporium. ACS Symp Ser 389:127–140. doi: 10.1021/bk-1989-0389 [DOI] [Google Scholar]
- 7. Hammel KE, Moen MA. 1991. Depolymerization of a synthetic lignin in vitro by lignin peroxidase. Enzyme Microb Technol 13:15–18. doi: 10.1016/0141-0229(91)90182-A [DOI] [Google Scholar]
- 8. Wariishi H, Valli K, Gold MH. 1991. In vitro depolymerization of lignin by manganese peroxidase of Phanerochaete chrysosporium. Biochem Biophys Res Commun 176:269–275. doi: 10.1016/0006-291x(91)90919-x [DOI] [PubMed] [Google Scholar]
- 9. Tai D, Terazawa M, Chen CL, Chang H. 1990. Lignin biodegradation products from birch wood by Phanerochaete chrysosporium. Part 1. Fractionation of methanol-extractive and characterization of ether-insoluble low-molecular-weight fraction. Holzforschung 44:257–262. doi: 10.1515/hfsg.1990.44.3.185 [DOI] [Google Scholar]
- 10. Yajima Y, Enoki A, Mayfield MB, Gold MH. 1979. Vanillate hydroxylase from the white-rot basidiomycete Phanerochaete chrysosporium. Arch Microbiol 123:319–321. doi: 10.1007/BF00406669 [DOI] [Google Scholar]
- 11. Rieble S, Joshi DK, Gold MH. 1994. Purification and characterization of a 1,2,4-trihydroxybenzene 1,2-dioxygenase from the basidiomycete Phanerochaete chrysosporium. J Bacteriol 176:4838–4844. doi: 10.1128/jb.176.16.4838-4844.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Suzuki H, Mori R, Kato M, Shimizu M. 2023. Biochemical characterization of hydroquinone hydroxylase from Phanerochaete chrysosporium. J Biosci Bioeng 135:17–24. doi: 10.1016/j.jbiosc.2022.10.001 [DOI] [PubMed] [Google Scholar]
- 13. Kato H, Furusawa TT, Mori R, Suzuki H, Kato M, Shimizu M. 2022. Characterization of two 1,2,4-trihydroxybenzene 1,2-dioxygenases from Phanerochaete chrysosporium. Appl Microbiol Biotechnol 106:4499–4509. doi: 10.1007/s00253-022-12007-9 [DOI] [PubMed] [Google Scholar]
- 14. Nakamura T, Ichinose H, Wariishi H. 2010. Cloning and heterologous expression of two aryl-aldehyde dehydrogenases from the white-rot basidiomycete Phanerochaete chrysosporium. Biochem Biophys Res Commun 394:470–475. doi: 10.1016/j.bbrc.2010.01.131 [DOI] [PubMed] [Google Scholar]
- 15. Shimizu M, Yuda N, Nakamura T, Tanaka H, Wariishi H. 2005. Metabolic regulation at the tricarboxylic acid and glyoxylate cycles of the lignin-degrading basidiomycete Phanerochaete chrysosporium against exogenous addition of vanillin. Proteomics 5:3919–3931. doi: 10.1002/pmic.200401251 [DOI] [PubMed] [Google Scholar]
- 16. Del Cerro C, Erickson E, Dong T, Wong AR, Eder EK, Purvine SO, Mitchell HD, Weitz KK, Markillie LM, Burnet MC, Hoyt DW, Chu RK, Cheng J-F, Ramirez KJ, Katahira R, Xiong W, Himmel ME, Subramanian V, Linger JG, Salvachúa D. 2021. Intracellular pathways for lignin catabolism in white-rot fungi. Proc Natl Acad Sci U S A 118:e2017381118. doi: 10.1073/pnas.2017381118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Arias-Barrau E, Olivera ER, Luengo JM, Fernández C, Galán B, García JL, Díaz E, Miñambres B. 2004. The homogentisate pathway: a central catabolic pathway involved in the degradation of L-phenylalanine, L-tyrosine, and 3-hydroxyphenylacetate in Pseudomonas putida. J Bacteriol 186:5062–5077. doi: 10.1128/JB.186.15.5062-5077.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fernández-Cañón JM, Peñalva MA. 1995. Molecular characterization of a gene encoding a homogentisate dioxygenase from Aspergillus nidulans and identification of its human and plant homologues. J Biol Chem 270:21199–21205. doi: 10.1074/jbc.270.36.21199 [DOI] [PubMed] [Google Scholar]
- 19. Borowski T, Georgiev V, Siegbahn PEM. 2005. Catalytic reaction mechanism of homogentisate dioxygenase: a hybrid DFT study. J Am Chem Soc 127:17303–17314. doi: 10.1021/ja054433j [DOI] [PubMed] [Google Scholar]
- 20. Amaya AA, Brzezinski KT, Farrington N, Moran GR. 2004. Kinetic analysis of human homogentisate 1,2-dioxygenase. Arch Biochem Biophys 421:135–142. doi: 10.1016/j.abb.2003.10.014 [DOI] [PubMed] [Google Scholar]
- 21. Veldhuizen EJA, Vaillancourt FH, Whiting CJ, Hsiao MM-Y, Gingras G, Xiao Y, Tanguay RM, Boukouvalas J, Eltis LD. 2005. Steady-state kinetics and inhibition of anaerobically purified human homogentisate 1,2-dioxygenase. Biochem J 386:305–314. doi: 10.1042/BJ20041370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jeoung JH, Bommer M, Lin TY, Dobbek H. 2013. Visualizing the substrate-, superoxo-, alkylperoxo-, and product-bound states at the nonheme Fe(II) site of homogentisate dioxygenase. Proc Natl Acad Sci U S A 110:12625–12630. doi: 10.1073/pnas.1302144110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Titus GP, Mueller HA, Burgner J, Rodríguez De Córdoba S, Peñalva MA, Timm DE. 2000. Crystal structure of human homogentisate dioxygenase. Nat Struct Biol 7:542–546. doi: 10.1038/76756 [DOI] [PubMed] [Google Scholar]
- 24. Qi Y, Lu J, Lai W. 2016. Insights into the reaction mechanism of aromatic ring cleavage by homogentisate dioxygenase: a quantum mechanical/molecular mechanical study. J Phys Chem B 120:4579–4590. doi: 10.1021/acs.jpcb.6b03006 [DOI] [PubMed] [Google Scholar]
- 25. Eppink MH, Boeren SA, Vervoort J, van Berkel WJ. 1997. Purification and properties of 4-hydroxybenzoate 1-hydroxylase (decarboxylating), a novel flavin adenine dinucleotide-dependent monooxygenase from Candida parapsilosis CBS604. J Bacteriol 179:6680–6687. doi: 10.1128/jb.179.21.6680-6687.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ander P, Eriksson K-E, Yu H. 1983. Vanillic acid metabolism by Sporotrichum pulverulentum: evidence for demethoxylation before ring-cleavage. Arch Microbiol 136:1–6. doi: 10.1007/BF00415600 [DOI] [Google Scholar]
- 27. Kamimura N, Sakamoto S, Mitsuda N, Masai E, Kajita S. 2019. Advances in microbial lignin degradation and its applications. Curr Opin Biotechnol 56:179–186. doi: 10.1016/j.copbio.2018.11.011 [DOI] [PubMed] [Google Scholar]
- 28. Weng C, Peng X, Han Y. 2021. Depolymerization and conversion of lignin to value-added bioproducts by microbial and enzymatic catalysis. Biotechnol Biofuels 14:84. doi: 10.1186/s13068-021-01934-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang S, Wu X, Xiao Y. 2022. Conversion of lignin-derived 3-methoxycatechol to the natural product purpurogallin using bacterial P450 GcoAB and laccase CueO. Appl Microbiol Biotechnol 106:593–603. doi: 10.1007/s00253-021-11738-5 [DOI] [PubMed] [Google Scholar]
- 30. Sakai K, Matsuzaki F, Wise L, Sakai Y, Jindou S, Ichinose H, Takaya N, Kato M, Wariishi H, Shimizu M, Zhou N-Y. 2018. Biochemical characterization of CYP505D6, a self-sufficient cytochrome P450 from the white-rot fungus Phanerochaete chrysosporium. Appl Environ Microbiol 84:e01091-18. doi: 10.1128/AEM.01091-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Doddapaneni H, Chakraborty R, Yadav JS. 2005. Genome-wide structural and evolutionary analysis of the P450 monooxygenase genes (P450ome) in the white rot fungus Phanerochaete chrysosporium: evidence for gene duplications and extensive gene clustering. BMC Genomics 6:92. doi: 10.1186/1471-2164-6-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Syed K, Yadav JS. 2012. P450 monooxygenases (P450ome) of the model white rot fungus Phanerochaete chrysosporium. Crit Rev Microbiol 38:339–363. doi: 10.3109/1040841X.2012.682050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kolvenbach BA, Lenz M, Benndorf D, Rapp E, Fousek J, Vlcek C, Schäffer A, Gabriel FL, Kohler H-PE, Corvini PF. 2011. Purification and characterization of hydroquinone dioxygenase from Sphingomonas sp. strain TTNP3. AMB Express 1:8. doi: 10.1186/2191-0855-1-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kijpornyongpan T, Schwartz A, Yaguchi A, Salvachúa D. 2022. Systems biology-guided understanding of white-rot fungi for biotechnological applications: a review. iScience 25:104640. doi: 10.1016/j.isci.2022.104640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hammel KE, Cullen D. 2008. Role of fungal peroxidases in biological ligninolysis. Curr Opin Plant Biol 11:349–355. doi: 10.1016/j.pbi.2008.02.003 [DOI] [PubMed] [Google Scholar]
- 36. Kirk TK, Schultz E, Connors WJ, Lorenz LF, Zeikus JG. 1978. Influence of culture parameters on lignin metabolism by Phanerochaete chrysosporium. Arch Microbiol 117:277–285. doi: 10.1007/BF00738547 [DOI] [Google Scholar]
- 37. Takenaka S, Koshiya J, Okugawa S, Takata A, Murakami S, Aoki K. 2011. Fe-superoxide dismutase and 2-hydroxy-1,4-benzoquinone reductase preclude the auto-oxidation step in 4-aminophenol metabolism by Burkholderia sp. strain AK-5. Biodegradation 22:1–11. doi: 10.1007/s10532-010-9369-5 [DOI] [PubMed] [Google Scholar]
- 38. Semana P, Powlowski J. 2019. Four aromatic intradiol ring cleavage dioxygenases from Aspergillus niger. Appl Environ Microbiol 85:e01786-19. doi: 10.1128/AEM.01786-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers, GC-MS spectrum, and alignment of active site lid.