ABSTRACT
Acetate is a major intermediate in the anaerobic digestion of organic waste to produce CH4. In methanogenic systems, acetate degradation is carried out by either acetoclastic methanogenesis or syntrophic degradation by acetate oxidizers and hydrogenotrophic methanogens. Due to challenges in the isolation of syntrophic acetate-oxidizing bacteria (SAOB), the diversity and metabolism of SAOB and the mechanisms of their interactions with methanogenic partners are not fully characterized. In this study, the in situ activity and metabolic characteristics of potential SAOB and their interactions with methanogens were elucidated through metagenomics and metatranscriptomics. In addition to the reported SAOB classified in the genera Tepidanaerobacter, Desulfotomaculum, and Thermodesulfovibrio, we identified a number of potential SAOB that are affiliated with Clostridia, Thermoanaerobacteraceae, Anaerolineae, and Gemmatimonadetes. The potential SAOB possessing the glycine-mediated acetate oxidation pathway dominates SAOB communities. Moreover, formate appeared to be the main product of the acetate degradation by the most active potential SAOB. We identified the methanogen partner of these potential SAOB in the acetate-fed chemostat as Methanosarcina thermophila. The dominated potential SAOB in each chemostat had similar metabolic characteristics, even though they were in different fatty-acid-fed chemostats. These novel syntrophic lineages are prevalent and may play critical roles in thermophilic methanogenic reactors. This study expands our understanding of the phylogenetic diversity and in situ biological functions of uncultured syntrophic acetate degraders and presents novel insights into how they interact with methanogens.
IMPORTANCE
Combining reactor operation with omics provides insights into novel uncultured syntrophic acetate degraders and how they perform in thermophilic anaerobic digesters. This improves our understanding of syntrophic acetate degradation and contributes to the background knowledge necessary to better control and optimize anaerobic digestion processes.
KEYWORDS: thermophilic anaerobic digestion, syntrophic acetate oxidation, Wood-Ljungdahl pathway, glycine-mediated acetate oxidation pathway, energy conservation
INTRODUCTION
Anaerobic digestion (AD) of organic waste to produce methane offers opportunities to deliver multiple environmental benefits, as it encompasses biological waste treatment and renewable energy production. The degradation of organic matter to methane consists of four basic steps: hydrolysis, acidogenesis, acetogenesis, and methanogenesis. Volatile fatty acids (VFAs, e.g., acetate, propionate, butyrate, and isovalerate) are the main intermediates in the AD system (1). The degradation of VFAs in anaerobic digesters becomes thermodynamically unfavorable even with a slight accumulation of metabolic products (i.e., acetate, H2, and formate) (2, 3). The syntrophic interaction of VFA oxidizers with partner methanogenic archaea that consume the products is essential in AD (4–6). Thus, syntrophic fatty acid oxidation is thought to be a critical step in AD. Acetate is the product of syntrophic propionate, butyrate, and isovalerate degradation (equations 1-3) (2, 7). Theoretically, one, two, and three molecules of acetate can be produced from the degradation of one molecule of propionate, butyrate, and isovalerate, respectively. Therefore, acetate serves as an important intermediate metabolite and the major precursor of methane in the AD of organic matter (8, 9). Metabolic disorders within anaerobic digesters would lead to the accumulation of acetate and/or H2, which may cause acidification and reduce methane production, destabilizing the AD system. Therefore, uncovering the underlying mechanism of anaerobic acetate metabolism is fundamental to managing the microbial AD system for better performance.
| (1) |
| (2) |
| (3) |
Under methanogenic conditions, methane production from acetate follows two routes (i) acetoclastic methanogenesis (equation 4) and (ii) acetate oxidation (equation 5) coupled with hydrogenotrophic methanogenesis (equation 6) (9).
| (4) |
| (5) |
| (6) |
In the first instance, acetate is cleaved into carbonyl and methyl groups, then, respectively, oxidized to CO2 and reduced to CH4 by multitrophic methanogen Methanosarcina or acetoclastic methanogen Methanothrix (10). In the second instance, both methyl and carbonyl groups of acetate are oxidized to CO2, associated with the generation of H2. This reaction is thermodynamically unfavorable under standard conditions (equation 5). Thus, “syntrophic” cooperation with hydrogen-scavenging methanogenic partners (equation 6) is necessary to maintain thermodynamic favorability (9). Previous studies have observed syntrophic acetate oxidation under specific conditions [e.g., high concentration of ammonia (11–14), high temperature (14, 15), or low loading rate and long retention time (16)], suggesting that this metabolism may play a critical role in diverse methanogenic systems and that there may be challenges in supporting acetoclastic methanogens.
Although six strains of SAOB have been cultured, the diversity and metabolism of SAOB are not fully understood. Among the described SAOB, three species [Thermacetogenium phaeum (17), Syntrophaceticus schinkii (18), and Tepidanaerobacter acetatoxydans (19)] possess the well-known reverse Wood-Ljungdahl (WL) pathway for syntrophic acetate oxidation (20, 21), but two others, Pseudothermotoga lettingae (22) and Schnuerera ultunensis (23), lack genes for this classical WL pathway and are suspected to possess an alternative metabolism, potentially mediated by a glycine cleavage pathway (24). Moreover, these cultured SAOB are generally detected at low abundances in anaerobic bioreactors (less than 1%) (25, 26). Previous studies based on DNA or protein stable isotope probing point toward the presence of other phylogenetically distinct uncultured acetate oxidizers in anaerobic digestors (27, 28). Therefore, uncovering the diversity, ecology, metabolism, and symbiotic interactions of these yet-to-be-cultured SAOB is essential for improving our understanding of methanogenic bioreactors under various conditions.
In our previous studies, we operated chemostats fed with acetate, propionate, or isovalerate as the sole carbon source to enrich syntrophic fatty acid oxidizers (28–30). Some unclassified bacterial populations were detected at high abundances in all acetate-, propionate-, and isovalerate-fed thermophilic chemostats based on 16S ribosomal RNA (rRNA) analysis (28–30). Since acetate is the product of propionate and isovalerate oxidation, those unclassified bacterial populations may be involved in acetate degradation. Using the DNA stable-isotope probing (DNA-SIP) method, some of those unclassified bacterial populations in acetate- and propionate-fed thermophilic chemostats were found to be potential acetate oxidizers (28). However, the metabolic characteristics and prevalence of these potential acetate oxidizers remain unclear. To address this, four additional thermophilic fatty-acid-fed chemostats were constructed and analyzed in this study. To systematically investigate the diversity and metabolism of potential acetate oxidizers, we employed metagenomics to recover genomes (metagenome-assembled genomes, MAGs) of potential acetate oxidizers from acetate-, propionate-, butyrate-, and isovalerate-fed thermophilic chemostats. Metagenomics- and metatranscriptomics-based metabolic reconstruction of these populations uncovered the catabolic pathways of acetate oxidation, the mechanisms for electron transduction from acetate oxidation to H2 and formate generation, and the interactions of these populations with their methanogenic partners. This study provides insights into uncultured novel thermophilic acetate oxidizers and their in situ performance.
MATERIALS AND METHODS
Operation of thermophilic anaerobic chemostats
In our previous work, four thermophilic (55°C) chemostats (ATL chemostat, acetate-fed, dilution rate of 0.025 day−1; ATH chemostat, acetate-fed, dilution rate of 0.05 day−1; PTL chemostat, propionate-fed, dilution rate of 0.025 day−1; and VTL chemostat, isovalerate-fed, dilution rate of 0.025 day−1) were built (28–30). In this study, we constructed four additional thermophilic chemostats (PTH chemostat, propionate-fed, dilution rate of 0.05 day−1; BTL chemostat, butyrate-fed, dilution rate of 0.025 day−1; BTH chemostat, butyrate-fed, dilution rate of 0.05 day−1; and VTH chemostat, isovalerate-fed, dilution rate of 0.05 day−1). Overall, eight thermophilic anaerobic chemostats operated with different carbon sources (acetate, propionate, butyrate, or isovalerate), two dilution rates [0.025 day−1, hydraulic retention time (HRT) = 40 days; 0.05 day−1, HRT = 20 days] (Table 1). The HRT of 20 and 40 days were selected based on the typical HRT of 15–40 days needed for the anaerobic digestion of agricultural and municipal wastes.
TABLE 1.
Operational performance of thermophilic chemostats during the steady operation perioda
| Chemostat name | ATL | ATH | PTL | PTH | BTL | BTH | VTL | VTH |
|---|---|---|---|---|---|---|---|---|
| Carbon source | Acetate | Acetate | Propionate | Propionate | Butyrate | Butyrate | Isovalerate | Isovalerate |
| Dilution rate (day−1) | 0.025 | 0.05 | 0.025 | 0.05 | 0.025 | 0.05 | 0.025 | 0.05 |
| HRT (day) | 40 | 20 | 40 | 20 | 40 | 20 | 40 | 20 |
| Inflow concentration (mg·L−1) | 20,000 | 20,000 | 16,444 | 16,444 | 14,667 | 14,667 | 13,600 | 13,600 |
| Gas production rate (mL·L−1·day−1) | 115 ± 22 | 294 ± 67 | 103 ± 17 | 383 ± 43 | 155 ± 28 | 396 ± 35 | 143 ± 37 | 365 ± 18 |
| CH4 content (%) | 58 ± 4 | 60 ± 5 | 62 ± 2 | 63 ± 3 | 66 ± 2 | 73 ± 4 | 67 ± 4 | 76 ± 6 |
| H2 partial pressure (Pa) | 0.8 ± 0.4 | 1.4 ± 0.6 | 2.9 ± 1.2 | 3.1 ± 1.3 | 2.3 ± 1.4 | 2.9 ± 1.3 | 2.0 ± 1.2 | 2.5 ± 0.9 |
| pH | 8.04 ± 0.14 | 7.85 ± 0.14 | 7.95 ± 0.14 | 7.78 ± 0.14 | 7.78 ± 0.08 | 7.63 ± 0.11 | 7.84 ± 0.14 | 7.66 ± 0.10 |
| Outflow TOC (mg·L−1) | 139 ± 83 | 132 ± 78 | 84 ± 49 | 118 ± 46 | 87 ± 74 | 123 ± 91 | 84 ± 49 | 114 ± 44 |
| Formate (mg·L−1)b | ND | ND | ND | ND | ND | ND | ND | ND |
| Acetate (mg·L−1) | 11 ± 22 | 12 ± 35 | 6 ± 8 | 2 ± 4 | 31 ± 47 | 27 ± 69 | 23 ± 53 | 20 ± 20 |
| Propionate (mg·L−1) | 0 | 0 | 11 ± 13 | 15 ± 48 | ~1.0 | ~1.0 | ~1.0 | ~1.0 |
| Butyrate (mg·L−1) | 0 | 0 | 0 | 0 | ~1.0 | ~1.0 | ~1.0 | ~1.0 |
| Valerate (mg·L−1) | 0 | 0 | 0 | 0 | 0 | 0 | ~1.0 | ~1.0 |
| Iso-valerate (mg·L−1) | 0 | 0 | 0 | 0 | 0 | 0 | ~1.0 | ~1.0 |
| SS (g·L−1) | 0.421 ± 0.08 | 0.543 ± 0.14 | 0.468 ± 0.11 | 0.68 ± 0.07 | 1.43 ± 0.49 | 1.44 ± 0.38 | 1.35 ± 0.44 | 1.38 ± 0.25 |
| VSS (g·L−1) | 0.308 ± 0.07 | 0.392 ± 0.11 | 0.352 ± 0.10 | 0.513 ± 0.07 | 0.62 ± 0.21 | 0.65 ± 0.18 | 0.57 ± 0.21 | 0.67 ± 0.19 |
HRT, hydraulic retention time; TOC, total organic carbon; SS, suspended solid; VSS, volatile suspended solid; ND, not detected. The operational parameters (e.g., pH, gas production rate, CH4 content, etc.) were the averages (mean ± SD, n > 30) during the steady-state period in ATL (day 100–550), ATH (day 80–550), PTL (day 100–650), PTH (day 300–450), BTL (day 100–650), BTH (day 300–650), VTL (day 150–650), and ATL (day 300–650) chemostats (Fig. S2). The steady state was defined as the operational parameters (i.e., biogas production, pH, TOC, VSS, and concentrations of VFAs) being relatively stable.
Formate concentration was below the detective limit (10 mg L−1) of the high-performance liquid chromatography.
The eight chemostats were constructed using continuous stirred tank reactors, each with a working volume of 1.8 L (Fig. S1). The seed sludge for inoculating acetate- and propionate-degrading chemostats was obtained from a thermophilic anaerobic digester treating kitchen waste (Sichuan Province, China), and the seed sludge for inoculating butyrate- and isovalerate-degrading chemostats was from a swine manure treatment plant (Sichuan Province, China) (Table S1). The thermophilic chemostats were fed with artificial wastewater containing acetate (20.0 g·L−1), propionate (16.4 g·L−1), butyrate (14.7 g·L−1), or isovalerate (13.6 g·L−1) as the sole carbon source, respectively [total organic carbon (TOC) = 8,000 mg·L−1]. The artificial wastewater contained 0.3 g·L−1 KH2PO4, 4.0 g·L−1 KHCO3, 1.0 g·L−1 NH4Cl, 0.6 g·L−1 NaCl, 0.82 g·L−1 MgCl2·6H2O, 0.08 g·L−1 CaCl2·2H2O, 0.1 g·L−1 cysteine-HCl·H2O, 10 mL trace element solution; and 10 mL vitamin solution (28, 29).
Cultures from the chemostats were used for fluorescence microscopic observation and the analyses of pH, suspended solids, volatile suspended solids, TOC, and VFAs using the same protocols described previously (28). CH4, CO2, and H2 contents of the biogas were determined using gas chromatography (GC-2014C, Shimadzu, Kyoto, Japan). During the steady operation period, biomass collected from the chemostats was used for DNA and RNA extraction.
DNA and RNA extraction, 16S rRNA gene, metagenomics, and metatranscriptomics sequencing
Samples for microbial analysis were taken at random time intervals when the chemostats operated stably. The steady-state operation periods of chemostats were defined based on the stability of the reactor performance (i.e., biogas production was stable and VFA accumulation was negligible; Fig. S2) and the microbial community structure (16S rRNA gene sequencing). Sampling time points for 16S rRNA gene amplicon, metagenomics, and metatranscriptomics sequencing in this study are shown in Table S1. Total DNA and RNA were extracted via the cetyl-trimethyl ammonium bromide method (31). Total RNA was reverse transcribed using PrimeScript RT reagent Kit with gDNA Eraser (Perfect Real Time) according to the manufacturer’s protocol (Takara, Kusatsu, Japan). DNA and cDNA samples were subjected to 16S rRNA gene amplicon sequencing. The V4−V5 hypervariable regions of the bacterial and archaeal 16S rRNA genes were amplified with universal primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 909R (5′-CCCCGYCAATTCMTTTRAGT-3′). PCR product purification and Illumina sequencing were conducted using the protocol previously reported (28). Metagenomic DNA was sequenced on an Illumina HiSeq 2000 platform (Illumina). For metatranscriptomics sequencing, total RNA was purified and then sequenced on an Illumina HiSeq 2000 platform (Illumina). Before sequencing, rRNA was removed from the DNase-treated RNA via the Ribo-Zero rRNA Removal Kits (Illumina, San Diego, CA, USA).
Bioinformatics analyses
The data processing of 16S rRNA gene sequences was conducted using the protocol previously reported (28). The paired-end metagenomics reads (2 × 150 bp) were trimmed via Trimmomatic v0.36 (32), co-assembled via SPAdes v.3.5.0 (33), binned through MetaBAT v0.26.3 (34) and MaxBin 2.0 (35), and checked for completeness and contamination using CheckM v1.0.5 (36). Taxonomy affiliation of MAGs was processed through GTDB-tk (v1.3.1) (37). Phylogenetic trees were built with PhyloPhlAn v0.99 (38), and the tree was edited using iTOL (39). All genomes were annotated through a combination of Prokka v1.13 (kingdom = Bacteria or Archaea) (40), KEGG KAAS (https://www.genome.jp/tools/kaas/), and manual curation (see Supplementary Methods for details of bioinformatics analyses).
The paired-end metatranscriptomic reads (2 × 150 bp) were trimmed as the DNA-trimming step described above and mapped to MAGs using the BBMap with the parameters as: minid = 1 (v35.85; http://sourceforge.net/projects/bbmap/). The gene expression levels of functional genes from each MAG were calculated as reads per kilobase transcript per million reads mapped to the MAG (RPKM) averaged from duplicate samples. The RPKM was further normalized to the median gene expression level in the heat map illustration for each MAG (RPKM-NM) averaged from duplicate samples (29). Genes exhibiting high expression levels indicate that their RPKM-NM values ranked within the top quartile among all genes' RPKM-NM values.
RESULTS AND DISCUSSION
Chemostat operation and performance
Eight thermophilic anaerobic chemostats were successfully operated with artificial wastewater, containing acetate, propionate, butyrate, or isovalerate as the sole carbon/energy source at two hydraulic retention times. These chemostats were continuously operated for more than 450 days, and the performance was kept stable during the steady-state period in each chemostat [i.e., biogas production was stable and effluent concentrations of VFAs in the eight chemostats were low (10–30 mg L−1)] (Table 1; Fig. S2). The methane content reached approximately 54%–77%, and the H2 partial pressure was below 5 Pa. These indicated that the influent VFAs were almost completely degraded by these microbial communities. Additionally, various fatty acids were used as the sole carbon sources in this study, and the reactors were operated in chemostat mode over 450 days, which seems to be favorable for enriching fatty acid oxidizers.
Microbial diversity and community composition of thermophilic anaerobic chemostats
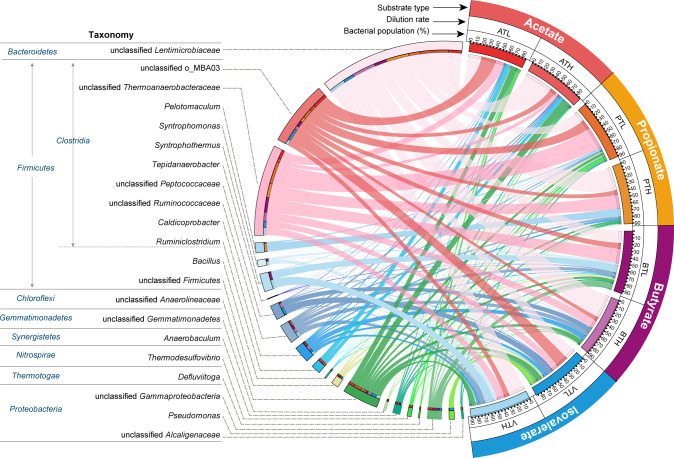
DNA- and RNA-based 16S ribosomal RNA gene analysis revealed that the bacterial community of the thermophilic chemostats contained a diverse population belonging to uncultured lineages (Fig. 1; Fig. S3). The dominant bacterial populations included Firmicutes (e.g., order MBA03 and family Thermoanaerobacteraceae), Bacteroidetes (Lentimicrobiaceae), and Chloroflexi (Anaerolineaceae), which were at high relative abundance (up to 72%) and RNA-based activity (up to 41%) in all of the thermophilic chemostats. Thermodesulfovibrio displayed a low DNA-based relative abundance but high activity (up to 18% of transcriptome). Notably, the genus associated with previously isolated SAOB (Tepidanaerobacter) (19) was detected but only comprised less than 2% of the total bacterial community (DNA-based abundance, 0.04%–0.32%; RNA-based activity, 0.10%–1.89%). In our previous study, members of MBA03, Thermoanaerobacteraceae, Anaerolineae, Lentimicrobiaceae, and Thermodesulfovibrio were determined to be potential SAOB in ATL and PTL by using DNA-stable isotope probing (28). These populations were also detected in other chemostats (ATH, PTH, BTL, BTH, VTL, and VTH; Fig. 1; Fig. S3), suggesting the potential contribution of the syntrophic acetate oxidization pathway for methane generation. However, the metabolic characteristics of these potential SAOB require further investigation. In regard to the archaeal community, according to 16S rRNA gene analysis, Methanosarcina whose relative abundance accounted for 26%–94% and 48%–99% of the archaeal community at the DNA and RNA level, respectively, was the active methanogen across all thermophilic chemostats (Fig. S4). Operational taxonomic units (OTUs) of Methanosarcina held a 99.5% sequence similarity to the multitrophic methanogen M. thermophila TM-1, which is able to convert H2, acetate, and methanol to methane (41, 42). The activities of Methanoculleus and Methanothermobacter were negligible in ATL. Methanothermobacter was dominant in PTL (67% DNA and 51% RNA), PTH (47% DNA and 42% RNA), and BTL (25% DNA and 24% RNA), while Methanoculleus predominated in BTH (33% DNA and 29% RNA) and VTH (59% DNA and 9% RNA).
Fig 1.
Relative abundance of bacterial genera in the thermophilic chemostats. The relative abundance was calculated based on 16S rRNA gene amplicon sequencing (DNA level). ATL chemostat, acetate-fed, dilution rate of 0.025 day−1; ATH chemostat, acetate-fed, dilution rate of 0.05 day−1; PTL chemostat, propionate-fed, dilution rate of 0.025 day−1; PTH chemostat, propionate-fed, dilution rate of 0.05 day−1; BTL chemostat, butyrate-fed, dilution rate of 0.025 day−1; BTH chemostat, butyrate-fed, dilution rate of 0.05 day−1; VTL chemostat, isovalerate-fed, dilution rate of 0.025 day−1; VTH chemostat, isovalerate-fed, dilution rate of 0.05 day−1.
The structures of the microbial community in low- and high-dilution rate chemostats fed with the same carbon source were similar (Fig. 1; Fig. S3 and S4). Therefore, we only conducted metagenomics and metatranscriptomics analyses on the chemostats with low dilution rates (ATL, PTL, BTL, and VTL) to profile the metabolic characteristics of such bacteria and methanogens (potential partners and competitors). A total of 173 Gbp metagenomic clean sequences (ATL, 35 Gbp; PTL, 69 Gbp; BTL, 34 Gbp; VTL, 35 Gbp) were obtained. Binning of the assembled contigs from the metagenomes yielded 45, 41, 52, and 56 high-quality (≥70% genome completeness and <10% contamination) prokaryotic MAGs from ATL, PTL, BTL, and VTL, respectively. To obtain gene expression profiles of the bacteria and archaea in the chemostats, a total of 287 million metatranscriptomic reads (33.6 Gbp, approximately 4.2 Gbp for each RNA sample) were sequenced and mapped to the MAGs for each chemostat (78%–95% of reads mapped using a 100% nucleotide similarity cutoff).
Syntrophic metabolism and energy conservation of acetate-degrading community in ATL
Based on 16S ribosomal RNA gene analysis (DNA- and RNA-based), the relative abundance and RNA-based activity of bacteria were higher than that of archaea in all chemostats, with the exception of PTH (Fig. S5A and B). Based on mapping metagenomic reads to the obtained MAGs, the bacterial populations retrieved accounted for 79%, 52%, 67%, and 67% of the metagenomic reads obtained from ATL, PTL, BTL, and VTL, respectively (Fig. S5C). In addition, these bacterial populations accounted for 80%, 74%, 65%, and 73% of the metatranscriptomic reads from ATL, PTL, BTL, and VTL, respectively (Fig. S5D). In PTL, BTL, and VTL, bacteria displayed higher abundance and activity than archaea. The explanation for this phenomenon was that propionate, butyrate, and isovalerate were degraded by syntrophic fatty acid-oxidizing bacteria, and then the products (i.e., acetate and H2/formate) were handed off to partnering acetate- and H2-consuming methanogenic archaea. In ATL, acetate could be converted by Methanosarcina to produce methane. However, the microbial consortia were still dominated by bacterial populations. This result together with the low concentration of VFAs (10–30 mg L−1) suggested that some bacterial populations (e.g., unclassified MBA03, unclassified Thermoanaerobacteraceae, unclassified Lentimicrobiaceae, and unclassified Anaerolineaceae) may be potential acetate oxidizers and play a significant role in acetate degradation at least in our acetate-fed chemostats, which was consistent with our previous DNA-SIP experiments (28).
Activity of CO2-reducing methanogenesis in Methanosarcina in ATL
MAGs of methanogens, including multitrophic Methanosarcina (MAG.ATL014), hydrogenotrophic Methanoculleus (MAG.ATL103) and Methanothermobacter (MAG.ATL045), and methylotrophic Methanomassiliicoccus (MAG.ATL089) were recovered from ATL (Fig. S6; Table S2). Based on metatranscriptomics, the activity of Methanoculleus (MAG.ATL103) was not detected. Methanothermobacter (MAG.ATL045) displayed low activity (0.22%) and did not express genes involved in hydrogenotrophic methanogenesis (Fig. 2 and 3B; Tables S3 and S4). Methanomassiliicoccus MAG.ATL089 (activity of 1.64%) highly expressed methylotrophic methanogenesis (top octile). On the other hand, Methanosarcina (99.5% rRNA sequence similarity to M. thermophila TM-1; MAG.ATL014) was highly abundant (17%) and active (18%) in the ATL. It highly expressed genes for CO2 reduction (in addition to acetate catabolism; top quartile of expressed genes in the corresponding MAG; Fig. 2 and 3B; Tables S3 and S4). Thus, only Methanosarcina MAG.ATL014 may be capable of consuming H2 in ATL and possibly act as a syntrophic partner of SAOB. Even though some Methanosarcina species are known to significantly downregulate the expression of genes for CO2 reduction during acetate degradation as they are not necessary (43), this was not the case for Methanosarcina (MAG.ATL014) in ATL. A decrease in the activity of the CO2-reducing pathway also results in decreased cellular concentrations (up to 10-fold) of coenzyme F420, an electron carrier for the CO2 branch during growth on acetate (44). Though only qualitatively, Methanosarcina-like cells showed higher autofluorescence (at 420 nm) in chemostats where Methanosarcina highly expressed the CO2-reducing pathway [i.e., ATL and ATH compared to PTL in Fig. S7; (30)]. Thus, in situ, the Methanosarcina likely were involved in hydrogenotrophic methanogenesis.
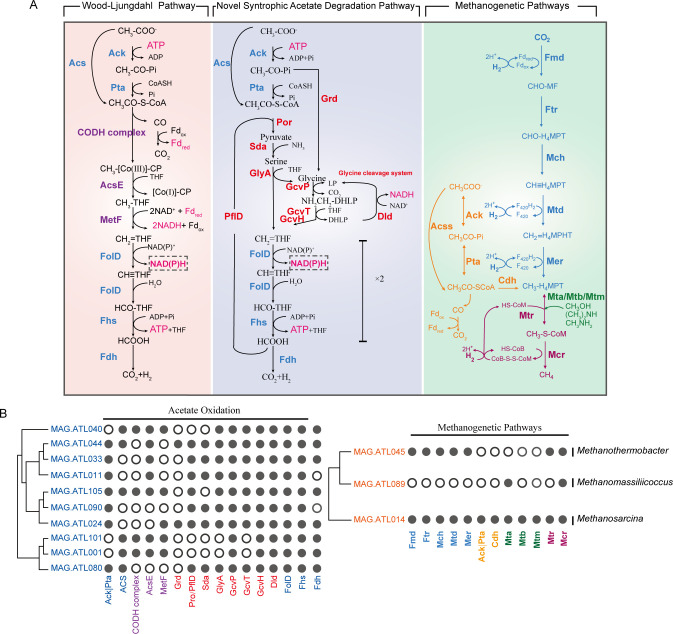
Fig 2.
(A) Metabolic pathways of acetate oxidation and methanogenic pathways in the thermophilic acetate-fed chemostat, and (B) distribution of catabolic pathways among the studied contributors. For each syntroph and methanogen, we show the presence (indicated by filled circles) of genes encoding pathways for acetate catabolism and methanogenesis. Enzyme abbreviations are as follows: Ack, acetate kinase; Pta, phosphate acetyltransferase; Acs, acetyl-CoA synthetase; CODH complex, acetyl CoA synthetase complex; AcsE, methyltetrahydrofolate:corrinoid/iron-sulfur protein methyltransferase; MetF, methylenetetrahydrofolate reductase; FolD, methylenetetrahydrofolate dehydrogenase/cyclohydrolase; Fhs, formate-tetrahydrofolate ligase; Fdh, formate dehydrogenase; Grd, glycine reductase; Por, pyruvate dehydrogenase; PflD, pyruvate-formate lyase; Sda, serine dehydratase; GlyA, glycine hydroxymethyltransferase; GcvPA, glycine dehydrogenase subunit A; GcvPB, glycine dehydrogenase subunit B; GcvT, glycine cleavage system T protein; GcvH, glycine cleavage system H protein; Dld, dihydrolipoyl dehydrogenase. Fmd, formylmethanofuran dehydrogenase; Ftr, formylmethanofuran-tetrahydromethanopterin N-formyltransferase; Mch, methenyltetrahydromethanopterin cyclohydrolase; Mtd, methylenetetrahydromethanopterin dehydrogenase; Mer, F420-dependent 5,10-methenyltetrahydromethanopterin reductase; Cdh, acetyl-CoA decarbonylase/synthase complex; Mta, [methyl-Co(III) methanol-specific corrinoid protein]:CoM methyltransferase; Mtb, [methyl-Co(III) dimethylamine-specific corrinoid protein]:CoM methyltransferase; Mtm, [methyl-Co(III) monomethylamine-specific corrinoid protein]:CoM methyltransferase; Mtr, tetrahydromethanopterin S-methyltransferase; Mcr, methyl-CoM reductase. CHO-MF, formyl-methanofuran; CHO-H4MPT, formyl-tetrahydromethanopterin; CH≡H4MPT, methenyl-tetrahydromethanopterin; CH2=H4MPT, methylene-tetrahydromethanopterin; CH3-H4MPT, methyl-tetrahydromethanopterin; CH3-S-CoM, methyl-coenzyme M; HS-CoM, coenzyme M; HS-CoB, coenzyme B; CoB-S-S-CoM, mixed disulfide of CoM and CoB. Enzyme abbreviations and their corresponding genes are elaborated in Tables S3 and S5.
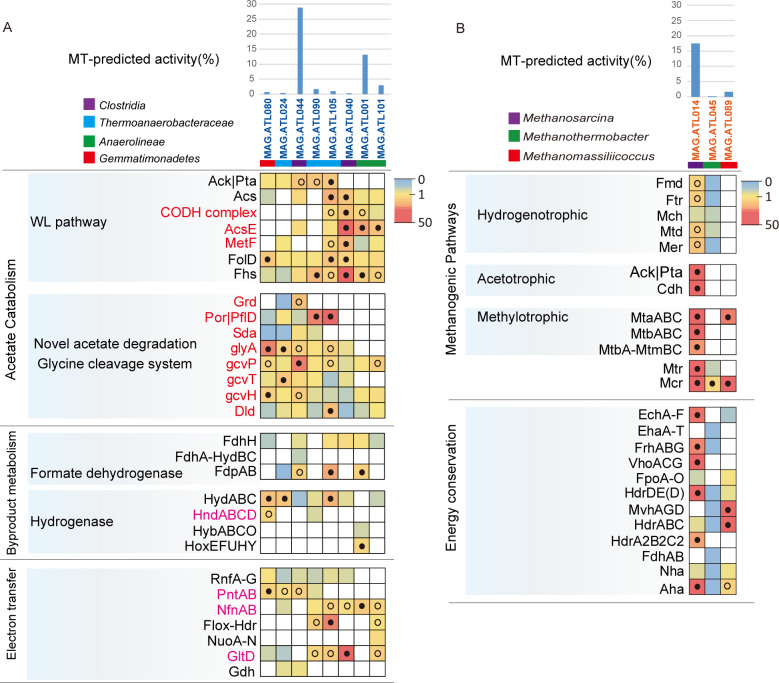
Fig 3.
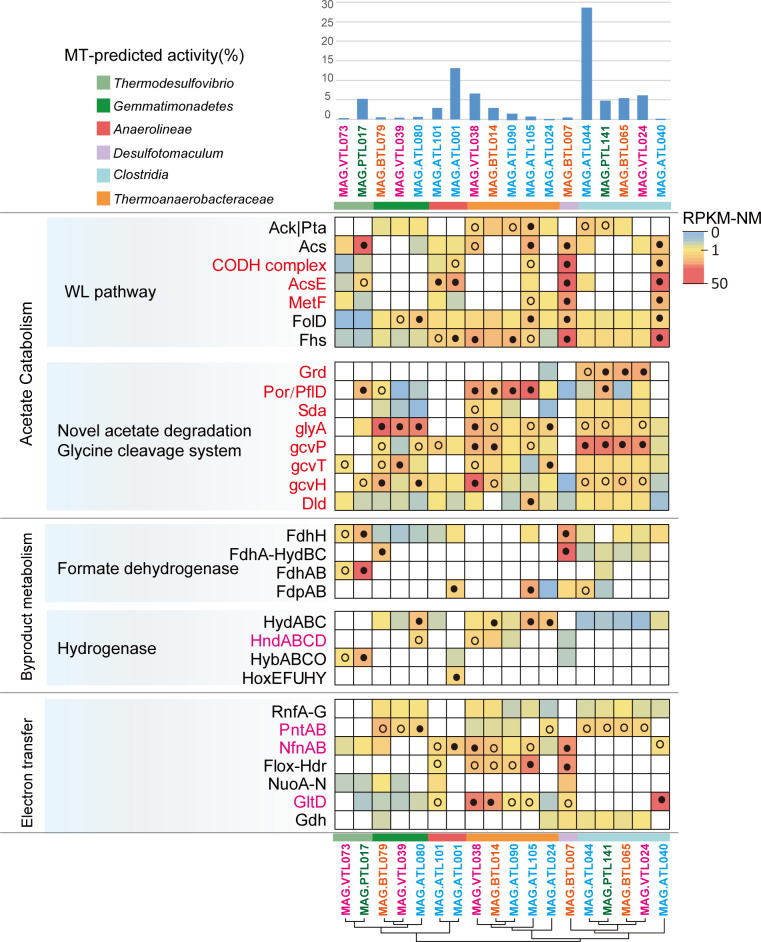
Gene expression level for acetate oxidation, H2/formate metabolism, and electron transfer genes in (A) potential SAOB, which may syntrophically degrade acetate and (B) methanogens in ATL. For each MAG, the percentages of the metatranscriptomic (MT) reads mapped to the MAG out of the metatranscriptomics mapped to all MAGs (both Bacteria and Archaea) are shown. Pathways containing genes with RPKM-NM greater than the octile and quartile are marked (filled and open dots, respectively). Enzyme abbreviations and their corresponding genes are elaborated in Tables S3 to S6.
Given that H2 was not added to the system, this may indicate the presence of some bacterial populations in the chemostat catabolizing acetate and syntrophically transferring H2 to Methanosarcina. In support of this, (i) acetate-degrading M. thermophila cells are capable of consuming H2 (41, 42), and (ii) Methanosarcina MAG.ATL014 highly expressed hydrogenases (i.e., VhoGAC, EchA-F, and FrhABG; Fig. 3B and 4; Table S4). This is consistent with one previous study where Methanosarcina had been observed together with potential SAOB in acetate-fed thermophilic anaerobic digesters (27).
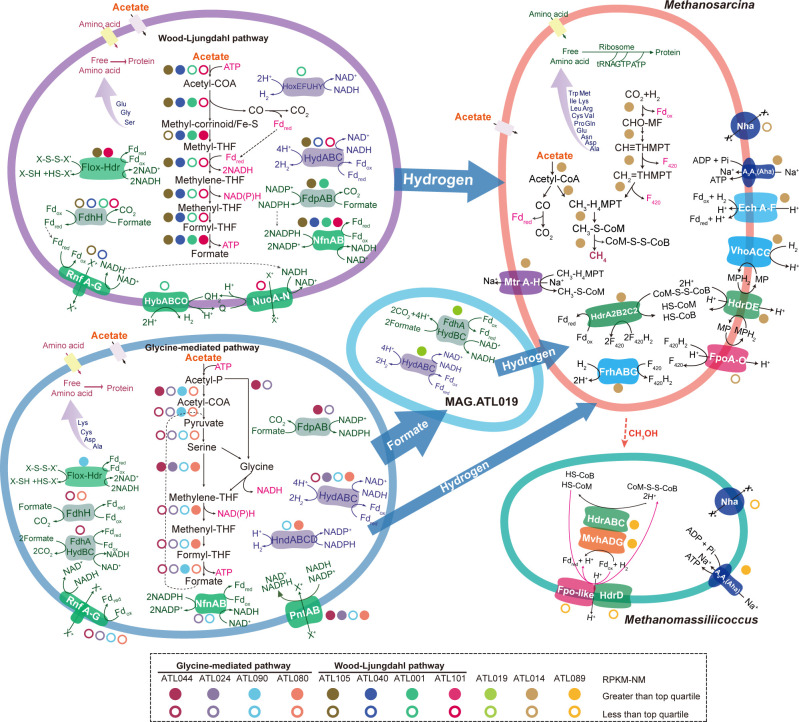
Fig 4.
Overview of the metabolism of potential SAOB and methanogens in ATL. Enzyme abbreviations and their corresponding genes are elaborated in Tables S3 to S6.
Putative syntrophic acetate metabolizers in ATL
To identify potential uncultured SAOB that may interact with the above-mentioned methanogens, we performed metabolic reconstruction of the MAGs recovered from abundant and active bacterial populations. We found that 10 bacterial MAGs that are associated with uncultured Clostridia, Thermoanaerobacteraceae, Anaerolineae, and Gemmatimonadetes encode the complete WL or glycine-mediated acetate-oxidizing pathway (Fig. 2; Table S5). Phylogenetic analysis revealed that these bacterial populations were distantly related to isolated SAOB and pure-cultured organisms (Fig. S8). Genome- and transcriptome-based prediction of SAOB is challenging given that (i) the conventional acetate oxidation pathway (WL pathway) can be used for carbon fixation, and (ii) the previously proposed glycine-mediated pathway can be used for serine/glycine biosynthesis. To identify genotypic features associated with SAOB, we performed comparative genomics of isolated SAOB and homoacetogens. All isolated SAOB (T. phaeum, S. schinkii, and T. acetatoxydans) that possess the WL pathway harbor NAD(P) transhydrogenase (PntAB) (20, 21, 24), while species only capable of homoacetogenesis do not encode genes for this enzyme. Both SAOB (S. ultunensis and P. lettingae) that possess glycine-mediated pathway (24) harbor NADPH re-oxidizing complexes, albeit different enzymes: NADPH-dependent FeFe hydrogenase (S. ultunensis) and NADH-dependent NADP:ferredoxin oxidoreductase (NfnAB, P. lettingae). Interestingly, S. ultunensis encodes the glycine dehydrogenase (CUESP1_0060-63) directly upstream of NADPH-dependent FeFe hydrogenase (CUESP1_0067-68), suggesting the potential association of glycine metabolism, NADPH re-oxidation, and H2 generation. To further increase the stringency of these potential SAOB possessing the glycine-mediated pathway, we excluded any populations that highly expressed amino acid catabolism (which is often NADP-dependent) using the expression of glutamate dehydrogenase (gdhA/gdhB) as a marker (i.e., gdhA/gdhB in the top quartile of expression profile). Thus, we restricted our analysis to populations that encoded and highly expressed the WL pathway or glycine-mediated pathway along with NADPH re-oxidation and H2/formate generation (expression in the top quartile of each population’s expression profile) and had low expression of amino acid catabolism.
Based on the criteria above, 8 of the 10 populations were identified as potential SAOB (Fig. 3A; Tables S5 and S6). Thermoanaerobacteraceae (MAG.ATL105), Anaerolineae (MAG.ATL001 and MAG.ATL101), and Clostridia (MAG.ATL040) may syntrophically degrade acetate via the WL pathway, while Thermoanaerobacteraceae (MAG.ATL024 and MAG.ATL090), Clostridia population (MAG.ATL044), and Gemmatimonadetes (MAG.ATL080) may syntrophically degrade acetate via the glycine-mediated acetate-oxidizing pathway. These results suggested that these novel potential SAOB play a critical role in syntrophic acetate oxidation.
Energy conservation and electron flow in potential acetate oxidizers in ATL
Syntrophic acetate oxidation necessitates the complementation of substrate oxidation with electron balance (24, 45). Thus, we explored energy conservation systems (e.g., intracellular electron transfer and electron confurcation/bifurcation) of the putative acetate oxidizers. For formate metabolism, six of the eight potential SAOB MAGs possessed a ferredoxin-dependent formate dehydrogenase (FdhH) (Fig. 3A and 4; Table S6). The Clostridia (MAG.ATL044), Thermoanaerobacteraceae (MAG.ATL105 and MAG.ATL024), and Anaerolineae (MAG.ATL001) related members possessed a putative NADPH-dependent formate dehydrogenase (FdpAB). MAG.ATL044 also harbored a putative NAD+-dependent electron-bifurcating complex (FdhA-hydBC: formate dehydrogenase). As for H2 generation, most of the putative SAOB possessed cytoplasmic [FeFe]-type electron-confurcating hydrogenases (HydABC) (Fig. 3A and 4; Table S6) that use exergonic oxidation of reduced ferredoxin (Fdred) (E0′ = −430 mV) to drive unfavorable H2 generation from NADH oxidation (E0′ = −230 mV) (46), an energy conservation strategy associated with syntrophic fatty acid oxidizers (21, 47). In addition, some populations (MAG.ATL080 and MAG.ATL090) performed H2 generation via NADPH-dependent [FeFe] hydrogenase (HndABCD). The Anaerolineae member (MAG.ATL001) also harbored a cytochrome b-linked NiFe hydrogenase (HybABCO) and a cytosolic NiFe hydrogenase (HoxEFUHY).
As discussed above, reducing equivalents (i.e., NADH, NADPH, and reduced ferredoxin [Fdred]) are involved in the actions of hydrogenases and formate dehydrogenases. To complete acetate degradation, SAOB in acetate-degrading communities encode redox complexes that support electron transfer between (i) NAD(H) and Fd (Rhodobacter nitrogen fixation complex Rnf; NADH:Fd oxidoreductase) (48), (ii) NADP(H) and NAD(H) and Fd (NADH-dependent NADP:Fd oxidoreductase NfnAB) (24, 46), (iii) NAD(H) and NADP(H) [NAD(P) transhydrogenase PntAB] (49), and (iv) Fd and unknown electron carriers (uncharacterized oxidoreductase Flox:Hdr) (24) (Fig. 3A and 4; Table S6). Using these complexes, the potential acetate oxidizers may employ reverse electron transport and electron bifurcation to generate H2.
In addition, we found that Clostridia member MAG.ATL044 contained a more energy-efficient pathway compared to the WL pathway, a potential explanation for its 25% high abundance and 29% activity in ATL based on total community (Fig. 3A and 4; Table S6). Analyses of the metatranscriptome indicated that MAG.ATL044 did not express hydrogenase (HydABC), suggesting it may convert acetate to formate but not further to H2 or it may possess unknown hydrogenase. A previous study has observed that some potential acetate-degrading species expressing the glycine-mediated pathway just oxidized acetate to formate (no H2 generation) in full-scale anaerobic digesters (50). Formate was not detected in ATL, indicating that acetate was completely oxidized without accumulation of this metabolic intermediate. In addition, we did not detect F420-reducing formate oxidation (lacking F420-dependent formate dehydrogenases) in Methanosarcina in ATL. Thus, we suspected other syntrophic species may convert the acetate-derived formate to H2. In agreement with this, metagenomics analyses showed that Clostridia (MAG.ATL106) and Anaerolineaceae (MAG.ATL019) highly expressed formate dehydrogenases and hydrogenases (Fig. 4; Table S6), suggesting potential involvement in syntrophic formate oxidation (51). The glycine cleavage pathway avoids endergonic 5-methyl-THF oxidation, generating a yield of one ATP per acetate (24). In comparison, the WL pathway holds a theoretical yield of zero ATP per acetate (Fig. 2A) (46, 52, 53). This optimal energy generation strategy might partially explain the high abundance and activity of MAG.ATL044.
Syntrophic metabolism and energy conservation of potential SAOB in propionate-, butyrate-, and isovalerate-fed chemostats
Acetate is an important by-product of syntrophic fatty acid degradation. To investigate potential SAOB in PTL, BTL, and VTL, we analyzed the acetate metabolic pathway of the MAGs in the other three chemostats. We found 10 (PTL), 9 (BTL), and 10 (VTL) bacterial MAGs encoding a complete WL or glycine-mediated acetate-oxidizing pathways (Fig. S9; Table S5). These populations associated with uncultured Clostridia (16 MAGs), Thermoanaerobacteraceae (5), Anaerolineae (1), Gemmatimonadetes (2), Desulfotomaculum (1), Tepidanaerobacter (2), and Thermodesulfovibrio (2) (Fig. S8). Notably, populations related to the isolated acetate-oxidizing genus Tepidanaerobacter (MAG.BTL055 and MAG.VTL084) displayed low activity and did not express acetate-oxidizing activity in BTL and VTL. Based on the criteria in the “Putative syntrophic acetate metabolizers in ATL” section above, 10 of the 29 populations were identified as potential SAOB (Fig. 5; Fig. S9; Tables S5 and S6). Thermodesulfovibrio (MAG.PTL017 and MAG.VTL073), Desulfotomaculum (MAG.BTL007), and Thermoanaerobacteraceae (MAG.VTL038 and MAG.BTL014) may syntrophically degrade acetate via WL pathway, while Clostridia (MAG.PTL141, MAG.BTL065, and MAG.VTL024) and Gemmatimonadetes (MAG.BTL079 and MAG.VTL039) may syntrophically degrade acetate via the glycine-mediated acetate-oxidizing pathway. Our results were consistent with our previous study in which members of Clostridia, Thermoanaerobacteraceae, Anaerolineae, and Thermodesulfovibrio were labeled by 13C2-acetate and determined to be potential SAOB in ATL and PTL (28). Therefore, these microorganisms were potential acetate degraders.
Fig 5.
Gene expression level for acetate oxidation, H2/formate metabolism, and electron transfer genes in potential SAOB, which may syntrophically degrade acetate in ATL, PTL, BTL, and VTL. For each MAG, the percentages of the metatranscriptomic (MT) reads mapped to the MAG out of the metatranscriptomics mapped to all MAGs (both Bacteria and Archaea) are shown. Pathways containing genes with RPKM-NM greater than the octile and quartile are marked (filled and open dots, respectively). Enzyme abbreviations and their corresponding genes are elaborated in Tables S3 to S6.
Potential SAOB showed higher diversity, abundance, and activity in ATL compared to the other three chemostats (PTL, BTL, and VTL). Eight MAGs were identified as potential SAOB in ATL, four MAGs in BTL and VTL, and only two MAGs in PTL. The relative abundance of potential SAOB in ATL, PTL, BTL, and VTL accounted for 38.2%, 9.4%, 8.2%, and 27.2%, respectively. The activity of potential SAOB in ATL, PTL, BTL, and VTL accounted for 48.8%, 10.2%, 9.8%, and 13.8%, respectively. These potential SAOB displayed the highest relative abundance and activity in ATL, but their abundance and activity were reduced in other chemostats, especially in PTL and BTL. Since acetate is the carbon/energy source for SAOB, this phenomenon might be attributed to the differences in the acetate availability in four chemostats. The TOC fed to each chemostat was the same; as acetate was supplied as the substrate in ATL, the acetate availability of SAOB in ATL was higher than that in the other three chemostats, which enabled SAOB to dominate in ATL. The carbon converting ratios of propionate, butyrate, and isovalerate to acetate are 2/3, 4/4, and 6/5, respectively (equations 1–3). As a result, the accessibility of acetate in VTL was higher than that in PTL and BTL, which enabled SAOB in VTL to obtain more energy and had a higher abundance and activity than those in PTL and BTL.
In the PTL, BTL, and VTL, most of the populations identified as potential SAOB were closely related to those identified in the ATL, suggesting the contribution of the acetate oxidization pathway for methane generation in the chemostats (Fig. S8). For instance, Clostridia members (MAG.ATL044, MAG.PTL141, MAG.BTL065, and MAG.VTL024) reconstructed from chemostats were phylogenetically closely related to each other. Among these potential acetate oxidizers, SAOB possessing glycine-mediated pathway dominated in SAOB communities in all four chemostats (Fig. 5; Tables S5 and S6). Clostridia (MAG.ATL044, MAG.PTL141, MAG.BTL065, and MAG.VTL024) were the most active SAOB across four chemostats and displayed high metabolic similarity (e.g., acetate oxidation pathway, H2 generation, formate metabolism, and electron transduction mechanism). Gemmatimonadetes (MAG.ATL080, MAG.BTL079, and MAG.VTL039) exhibited similar activity and metabolic characteristics in ATL, BTL, and VTL. Thermoanaerobacteraceae (MAG.ATL090, MAG.VTL038, and MAG.BTL014) were at higher activity in BTL and VTL than in ATL. Anaerolineae (MAG.ATL001 and MAG.ATL101) displayed high activity and highly expressed genes for the WL pathway in ATL. Desulfotomaculum only had highly expressed genes for the WL pathway in BTL. ATL and PTL were inoculated with the same seed sludge; however, a big difference was found in the species of potential SAOB detected in the two chemostats. In addition, SAOB in Desulfotomaculum was found in BTL but not VTL, and SAOB in Thermoanaerobacteraceae was found in VTL but not BTL, though BTL and VTL were inoculated with the same seed sludge. These results indicated that compared to the inocula, the type of fatty acid used for enrichment had a greater effect on the structure and metabolic characteristics of SAOB community. However, the most dominant potential SAOB in each chemostat were those in Clostridia, which had similar metabolic characteristics even though they were in different fatty-acid-fed chemostats.
Biosynthetic metabolism of potential acetate oxidizers
In our chemostats, the potential SAOB used acetate as the carbon and energy source for producing H2/formate, reducing equivalents (i.e., NADH, NADPH, and reduced ferredoxin [Fdred]), and ATP, which provided the cell with energy for biomass biosynthesis. These potential acetate degraders encode pathways for converting acetyl-CoA to pyruvate and other important precursors for the biosynthesis of sugars, amino acids (AAs), and nucleotides, as well as the pathways for AA degradation (Tables S7 to S9). The potential SAOB that were phylogenetically closely related to each other possessed similar capabilities for AA biosynthesis (Table S7). Most of the Clostridia, Thermoanaerobacteraceae, Anaerolineae, and Gemmatimonadetes members encode and highly express the majority of the genes involved in glycolysis/gluconeogenesis, as well as pentose phosphate pathway for nucleotide biosynthesis (Table S9).
The prevalence of the novel potential SAOB across diverse AD communities
The potential acetate oxidizers (Clostridia, Thermoanaerobacteraceae, Anaerolineae, and Gemmatimonadetes) from ATL, PTL, BTL, and VTL were distantly related (16S rRNA gene sequence similarity < 88% and amino acid identity < 65%) to any currently isolated species (Fig. S8). These potential SAOB displayed high phylogenetic diversity and metabolic diversity (e.g., acetate oxidation pathway and formate metabolism; Fig. 5; Tables S5 and S6).
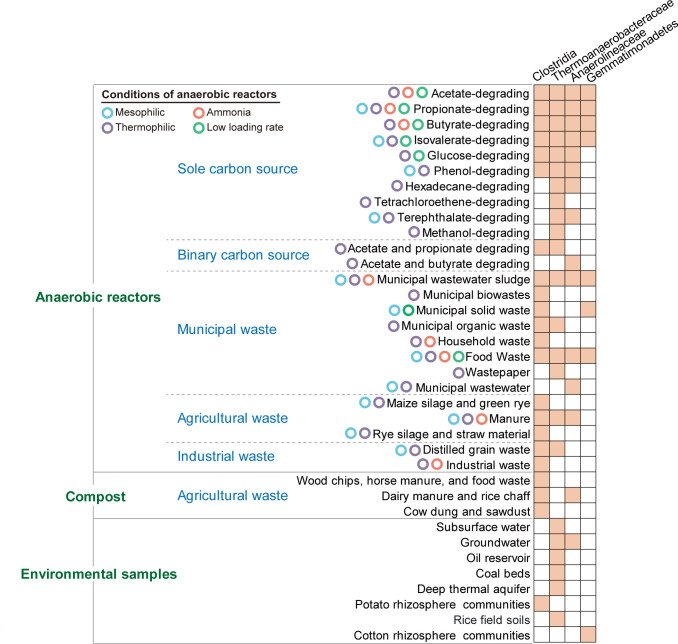
To investigate whether the potential SAOB identified in this study also occur in other habitats, we compared 16S rRNA genes of these potential SAOB with the NCBI nr/nt database using BLASTn. The results showed that the potential SAOB (Clostridia, Thermoanaerobacteraceae, Anaerolineae, and Gemmatimonadetes) in our chemostats were also detected in multiple environments. OTUs of these potential acetate oxidizers were closely related (≥97% sequence similarity) to uncultured populations detected in anaerobic digesters under various conditions (e.g., thermophilic, mesophilic, ammonia stress, or low loading rate) (Fig. 6; Table S10). These anaerobic digesters were fed with a wide diversity of substrates [e.g., sole carbon source (54), binary carbon source (55), municipal waste (56), agricultural waste (57), and industrial waste (58)]. The in situ bioactivation or bioaugmentation of these potential bacterial clades may be beneficial to the start-up process and resilience of anaerobic digestion (59). Moreover, the potential acetate oxidizers presented high sequence similarity (≥97%) with uncultured populations detected in different environmental communities [e.g., compost (60), groundwater (61), oil reservoir (62), and rhizosphere (63)] (Fig. 6; Table S10). These results suggest that these potential SAOB may be extensively present in diverse environments.
Fig 6.
Ecological characterization of the novel potential acetate oxidizers. The pairwise connections represent the existence of the relatives (≥97% similarity of 16S rRNA gene sequences) of the potential acetate oxidizers in various environments. The accession numbers of all reference sequences used in this study are provided in Table S10.
Conclusion
In summary, we combined metagenomics and metatranscriptomics to characterize novel potential syntrophic acetate oxidizers, including Clostridia, Thermoanaerobacteraceae, Anaerolineae, Gemmatimonadetes, Desulfotomaculum, and Thermodesulfovibrio members, in thermophilic fatty-acid-fed anaerobic chemostats. The high expression of genes involved in acetate oxidation and energy conservation based on metatranscriptomics revealed that these potential acetate-oxidizing species played an essential role in acetate degradation. These potential acetate oxidizers displayed high phylogenetic diversity and differences in the acetate oxidation pathway and formate metabolism. Our results expand the phylogenetic diversity and in situ metabolic characteristics of potential syntrophic acetate degraders in thermophilic anaerobic digesters, thus providing helpful insights to better control and optimize AD processes.
ACKNOWLEDGMENTS
This study was funded by the National Natural Science Foundation of China (No. 51678378) and the Ministry of Science and Technology of China (No. 2016YFE0127700).
Contributor Information
Ya-Ting Chen, Email: cytscu1101@scu.edu.cn.
Masaru Konishi Nobu, Email: mnobu@jamstec.go.jp.
Yue-Qin Tang, Email: tangyq@scu.edu.cn.
Arpita Bose, Washington University in St. Louis, St. Louis, Missouri, USA.
DATA AVAILABILITY
Raw sequence data reported in this paper are accessible at https://ngdc.cncb.ac.cn/gsa/browse/CRA004311.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aem.01090-23.
Supplemental text, Fig. S1 to S10, and legends of Tables S1 to S11.
Tables S1 to S11.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Pind PF, Angelidaki I, Ahring BK. 2003. Dynamics of the anaerobic process: effects of volatile fatty acids. Biotechnol Bioeng 82:791–801. doi: 10.1002/bit.10628 [DOI] [PubMed] [Google Scholar]
- 2. Stieb M, Schink B. 1986. Anaerobic degradation of isovalerate by a defined methanogenic coculture. Arch Microbiol 144:291–295. doi: 10.1007/BF00410965 [DOI] [Google Scholar]
- 3. de Bok FA, Stams AJ, Dijkema C, Boone DR. 2001. Pathway of propionate oxidation by a syntrophic culture of Smithella propionica and Methanospirillum hungatei. Appl Environ Microbiol 67:1800–1804. doi: 10.1128/AEM.67.4.1800-1804.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schink B. 1997. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev 61:262–280. doi: 10.1128/mmbr.61.2.262-280.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McInerney MJ, Struchtemeyer CG, Sieber J, Mouttaki H, Stams AJM, Schink B, Rohlin L, Gunsalus RP. 2008. Physiology, ecology, phylogeny, and genomics of microorganisms capable of syntrophic metabolism. Ann N Y Acad Sci 1125:58–72. doi: 10.1196/annals.1419.005 [DOI] [PubMed] [Google Scholar]
- 6. Kato S, Watanabe K. 2010. Ecological and evolutionary interactions in syntrophic methanogenic consortia. Microbes Environ 25:145–151. doi: 10.1264/jsme2.me10122 [DOI] [PubMed] [Google Scholar]
- 7. Stams AJM, Plugge CM. 2009. Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat Rev Microbiol 7:568–577. doi: 10.1038/nrmicro2166 [DOI] [PubMed] [Google Scholar]
- 8. Smith PH, Mah RA. 1966. Kinetics of acetate metabolism during sludge digestion. Appl Microbiol 14:368–371. doi: 10.1128/am.14.3.368-371.1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hattori S. 2008. Syntrophic acetate-oxidizing microbes in methanogenic environments. Microbes Environ 23:118–127. doi: 10.1264/jsme2.23.118 [DOI] [PubMed] [Google Scholar]
- 10. Ferry JG. 1992. Methane from acetate. J Bacteriol 174:5489–5495. doi: 10.1128/jb.174.17.5489-5495.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sun L, Müller B, Westerholm M, Schnürer A. 2014. Syntrophic acetate oxidation in industrial CSTR biogas digesters. J Biotechnol 171:39–44. doi: 10.1016/j.jbiotec.2013.11.016 [DOI] [PubMed] [Google Scholar]
- 12. Werner JJ, Garcia ML, Perkins SD, Yarasheski KE, Smith SR, Muegge BD, Stadermann FJ, DeRito CM, Floss C, Madsen EL, Gordon JI, Angenent LT. 2014. Microbial community Dynamics and stability during an ammonia-induced shift to Syntrophic acetate oxidation. Appl Environ Microbiol 80:3375–3383. doi: 10.1128/AEM.00166-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Westerholm M, Levén L, Schnürer A. 2012. Bioaugmentation of syntrophic acetate-oxidizing culture in biogas reactors exposed to increasing levels of ammonia. Appl Environ Microbiol 78:7619–7625. doi: 10.1128/AEM.01637-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Müller B, Sun L, Westerholm M, Schnürer A. 2016. Bacterial community composition and fhs profiles of low-and high-ammonia biogas digesters reveal novel syntrophic acetate-oxidising bacteria. Biotechnol Biofuels 9:48. doi: 10.1186/s13068-016-0454-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhu X, Kougias PG, Treu L, Campanaro S, Angelidaki I. 2017. Microbial community changes in methanogenic granules during the transition from mesophilic to thermophilic conditions. Appl Microbiol Biotechnol 101:1313–1322. doi: 10.1007/s00253-016-8028-0 [DOI] [PubMed] [Google Scholar]
- 16. Shigematsu T, Tang YQ, Kawaguchi H, Ninomiya K, Kijima J, Kobayashi T, Morimura S, Kida K. 2003. Effect of dilution rate on structure of a mesophilic acetate-degrading methanogenic community during continuous cultivation. J Biosci Bioeng 96:547–558. doi: 10.1016/S1389-1723(04)70148-6 [DOI] [PubMed] [Google Scholar]
- 17. Hattori S, Kamagata Y, Hanada S, Shoun H. 2000. Thermacetogenium phaeum gen. nov., sp. nov., a strictly anaerobic, thermophilic, syntrophic acetate-oxidizing bacterium. Int J Syst Evol Microbiol 50 Pt 4:1601–1609. doi: 10.1099/00207713-50-4-1601 [DOI] [PubMed] [Google Scholar]
- 18. Westerholm M, Roos S, Schnürer A. 2010. Syntrophaceticus schinkii gen. nov., sp. nov., an anaerobic, syntrophic acetate-oxidizing bacterium isolated from a mesophilic anaerobic filter. FEMS Microbiol Lett 309:100–104. doi: 10.1111/j.1574-6968.2010.02023.x [DOI] [PubMed] [Google Scholar]
- 19. Westerholm M, Roos S, Schnürer A. 2011. Tepidanaerobacter acetatoxydans sp. nov., an anaerobic, syntrophic acetate-oxidizing bacterium isolated from two ammonium-enriched mesophilic methanogenic processes. Syst Appl Microbiol 34:260–266. doi: 10.1016/j.syapm.2010.11.018 [DOI] [PubMed] [Google Scholar]
- 20. Manzoor S, Bongcam-Rudloff E, Schnürer A, Müller B. 2016. Genome-guided analysis and whole transcriptome profiling of the mesophilic syntrophic acetate oxidising bacterium Syntrophaceticus schinkii. PLoS One 11:e0166520. doi: 10.1371/journal.pone.0166520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Oehler D, Poehlein A, Leimbach A, Müller N, Daniel R, Gottschalk G, Schink B. 2012. Genome-guided analysis of physiological and morphological traits of the fermentative acetate oxidizer Thermacetogenium phaeum. BMC Genomics 13:723. doi: 10.1186/1471-2164-13-723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Balk M, Weijma J, Stams AJM. 2002. Thermotoga lettingae sp. nov., a novel thermophilic, methanol-degrading bacterium isolated from a thermophilic anaerobic reactor. Int J Syst Evol Microbiol 52:1361–1368. doi: 10.1099/00207713-52-4-1361 [DOI] [PubMed] [Google Scholar]
- 23. Schnurer A, Schink B, Svensson BH. 1996. Clostridium ultunense sp. nov., a mesophilic bacterium oxidizing acetate in syntrophic association with a hydrogenotrophic methanogenic bacterium. Int J Syst Bacteriol 46:1145–1152. doi: 10.1099/00207713-46-4-1145 [DOI] [PubMed] [Google Scholar]
- 24. Nobu MK, Narihiro T, Rinke C, Kamagata Y, Tringe SG, Woyke T, Liu WT. 2015. Microbial dark matter ecogenomics reveals complex synergistic networks in a methanogenic bioreactor. ISME J 9:1710–1722. doi: 10.1038/ismej.2014.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Narihiro T, Terada T, Ohashi A, Kamagata Y, Nakamura K, Sekiguchi Y. 2012. Quantitative detection of previously characterized syntrophic bacteria in anaerobic wastewater treatment systems by sequence-specific rRNA cleavage method. Water Res 46:2167–2175. doi: 10.1016/j.watres.2012.01.034 [DOI] [PubMed] [Google Scholar]
- 26. Vanwonterghem I, Jensen PD, Rabaey K, Tyson GW. 2016. Genome-centric resolution of microbial diversity, metabolism and interactions in anaerobic digestion. Environ Microbiol 18:3144–3158. doi: 10.1111/1462-2920.13382 [DOI] [PubMed] [Google Scholar]
- 27. Mosbæk F, Kjeldal H, Mulat DG, Albertsen M, Ward AJ, Feilberg A, Nielsen JL. 2016. Identification of syntrophic acetate-oxidizing bacteria in anaerobic digesters by combined protein-based stable isotope probing and metagenomics. ISME J 10:2405–2418. doi: 10.1038/ismej.2016.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zheng D, Wang HZ, Gou M, Nobu MK, Narihiro T, Hu B, Nie Y, Tang YQ. 2019. Identification of novel potential acetate-oxidizing bacteria in thermophilic methanogenic chemostats by DNA stable Isotope probing. Appl Microbiol Biotechnol 103:8631–8645. doi: 10.1007/s00253-019-10078-9 [DOI] [PubMed] [Google Scholar]
- 29. Chen YT, Zeng Y, Li J, Zhao XY, Yi Y, Gou M, Kamagata Y, Narihiro T, Nobu MK, Tang YQ. 2020. Novel syntrophic Isovalerate-degrading bacteria and their energetic cooperation with methanogens in methanogenic chemostats. Environ Sci Technol 54:9618–9628. doi: 10.1021/acs.est.0c01840 [DOI] [PubMed] [Google Scholar]
- 30. Chen YT, Zeng Y, Wang HZ, Zheng D, Kamagata Y, Narihiro T, Nobu MK, Tang YQ. 2020. Different interspecies electron transfer patterns during mesophilic and thermophilic syntrophic propionate degradation in chemostats. Microb Ecol 80:120–132. doi: 10.1007/s00248-020-01485-x [DOI] [PubMed] [Google Scholar]
- 31. Griffiths RI, Whiteley AS, O’Donnell AG, Bailey MJ. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl Environ Microbiol 66:5488–5491. doi: 10.1128/AEM.66.12.5488-5491.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kang DD, Froula J, Egan R, Wang Z. 2015. Metabat, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 3:e1165. doi: 10.7717/peerj.1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu YW, Simmons BA, Singer SW. 2016. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32:605–607. doi: 10.1093/bioinformatics/btv638 [DOI] [PubMed] [Google Scholar]
- 36. Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and Metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Parks DH, Chuvochina M, Chaumeil PA, Rinke C, Mussig AJ, Hugenholtz P. 2020. A complete domain-to-species taxonomy for bacteria and archaea. Nat Biotechnol 38:1079–1086. doi: 10.1038/s41587-020-0501-8 [DOI] [PubMed] [Google Scholar]
- 38. Segata N, Börnigen D, Morgan XC, Huttenhower C. 2013. Phylophlan is a new method for improved phylogenetic and taxonomic placement of microbes. Nat Commun 4:1–11. doi: 10.1038/ncomms3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 41. Lackner N, Hintersonnleitner A, Wagner AO, Illmer P. 2018. Hydrogenotrophic methanogenesis and autotrophic growth of Methanosarcina thermophila. Archaea 2018:4712608. doi: 10.1155/2018/4712608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ahring BK, Westermann P, Mah RA. 1991. Hydrogen inhibition of acetate metabolism and kinetics of hydrogen consumption by Methanosarcina thermophila TM-1. Arch Microbiol 157:38–42. doi: 10.1007/BF00245332 [DOI] [Google Scholar]
- 43. Hovey R, Lentes S, Ehrenreich A, Salmon K, Saba K, Gottschalk G, Gunsalus RP, Deppenmeier U. 2005. DNA microarray analysis of Methanosarcina mazei Gö1 reveals adaptation to different methanogenic substrates. Mol Genet Genomics 273:225–239. doi: 10.1007/s00438-005-1126-9 [DOI] [PubMed] [Google Scholar]
- 44. Heine-Dobbernack E, Schoberth SM, Sahm H. 1988. Relationship of intracellular coenzyme F(420) content to growth and metabolic activity of Methanobacterium bryantii and Methanosarcina barkeri. Appl Environ Microbiol 54:454–459. doi: 10.1128/aem.54.2.454-459.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sieber JR, McInerney MJ, Gunsalus RP. 2012. Genomic insights into syntrophy: the paradigm for anaerobic metabolic cooperation. Annu Rev Microbiol 66:429–452. doi: 10.1146/annurev-micro-090110-102844 [DOI] [PubMed] [Google Scholar]
- 46. Buckel W, Thauer RK. 2013. Energy conservation via electron bifurcating ferredoxin reduction and proton/Na(+) translocating ferredoxin oxidation. Biochim Biophys Acta 1827:94–113. doi: 10.1016/j.bbabio.2012.07.002 [DOI] [PubMed] [Google Scholar]
- 47. Hidalgo-Ahumada CAP, Nobu MK, Narihiro T, Tamaki H, Liu WT, Kamagata Y, Stams AJM, Imachi H, Sousa DZ. 2018. Novel energy conservation strategies and behaviour of pelotomaculum schinkii driving syntrophic propionate catabolism. Environ Microbiol 20:4503–4511. doi: 10.1111/1462-2920.14388 [DOI] [PubMed] [Google Scholar]
- 48. Biegel E, Schmidt S, González JM, Müller V. 2011. Biochemistry, evolution and physiological function of the Rnf complex, a novel ion-motive electron transport complex in prokaryotes. Cell Mol Life Sci 68:613–634. doi: 10.1007/s00018-010-0555-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sauer U, Canonaco F, Heri S, Perrenoud A, Fischer E. 2004. The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J Biol Chem 279:6613–6619. doi: 10.1074/jbc.M311657200 [DOI] [PubMed] [Google Scholar]
- 50. Nobu MK, Narihiro T, Mei R, Kamagata Y, Lee PKH, Lee PH, McInerney MJ, Liu WT. 2020. Catabolism and interactions of uncultured organisms shaped by eco-thermodynamics in methanogenic bioprocesses. Microbiome 8:111. doi: 10.1186/s40168-020-00885-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dolfing J, Jiang B, Henstra AM, Stams AJM, Plugge CM. 2008. Syntrophic growth on formate: a new microbial niche in anoxic environments. Appl Environ Microbiol 74:6126–6131. doi: 10.1128/AEM.01428-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Huang H, Wang S, Moll J, Thauer RK. 2012. Electron bifurcation involved in the energy metabolism of the acetogenic bacterium Moorella thermoacetica growing on glucose or H2 plus CO2. J Bacteriol 194:3689–3699. doi: 10.1128/JB.00385-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Poehlein A, Schmidt S, Kaster AK, Goenrich M, Vollmers J, Thürmer A, Bertsch J, Schuchmann K, Voigt B, Hecker M, Daniel R, Thauer RK, Gottschalk G, Müller V. 2012. An ancient pathway combining carbon dioxide fixation with the generation and utilization of a sodium ion gradient for ATP synthesis. PLoS One 7:e33439. doi: 10.1371/journal.pone.0033439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tang YQ, Matsui T, Morimura S, Wu XL, Kida K. 2008. Effect of temperature on microbial community of a glucose-degrading methanogenic consortium under hyperthermophilic chemostat cultivation. J Biosci Bioeng 106:180–187. doi: 10.1263/jbb.106.180 [DOI] [PubMed] [Google Scholar]
- 55. Ueno Y, Tatara M. 2008. Microbial population in a thermophilic packed-bed reactor for methanogenesis from volatile fatty acids. Enzyme Microb Tech 43:302–308. doi: 10.1016/j.enzmictec.2008.04.007 [DOI] [Google Scholar]
- 56. Tang Y, Shigematsu T, Ikbal, Morimura S, Kida K. 2004. The effects of micro-aeration on the phylogenetic diversity of microorganisms in a thermophilic anaerobic municipal solid-waste digester. Water Res 38:2537–2550. doi: 10.1016/j.watres.2004.03.012 [DOI] [PubMed] [Google Scholar]
- 57. Podmirseg SM, Gadermaier M, Franke-Whittle IH, Wett B, Insam H, Goberna M. 2016. Prokaryotic community dynamics during the start-up of a full-scale BIO4GAS digester. J Environ Eng 142:04015055. doi: 10.1061/(ASCE)EE.1943-7870.0001011 [DOI] [Google Scholar]
- 58. Wang TT, Sun ZY, Huang YL, Tan L, Tang YQ, Kida K. 2018. Biogas production from distilled grain waste by thermophilic dry anaerobic digestion: pretreatment of feedstock and dynamics of microbial community. Appl Biochem Biotechnol 184:685–702. doi: 10.1007/s12010-017-2557-6 [DOI] [PubMed] [Google Scholar]
- 59. Li MT, Rao L, Wang L, Gou M, Sun ZY, Xia ZY, Song WF, Tang YQ. 2022. Bioaugmentation with syntrophic volatile fatty acids-oxidizing consortia to alleviate the ammonia inhibition in continuously anaerobic digestion of municipal sludge. Chemosphere 288:132389. doi: 10.1016/j.chemosphere.2021.132389 [DOI] [PubMed] [Google Scholar]
- 60. Sizova MV, Izquierdo JA, Panikov NS, Lynd LR. 2011. Cellulose- and xylan-degrading thermophilic anaerobic bacteria from biocompost. Appl Environ Microbiol 77:2282–2291. doi: 10.1128/AEM.01219-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Moser DP, Gihring TM, Brockman FJ, Fredrickson JK, Balkwill DL, Dollhopf ME, Lollar BS, Pratt LM, Boice E, Southam G, Wanger G, Baker BJ, Pfiffner SM, Lin LH, Onstott TC. 2005. Desulfotomaculum and Methanobacterium spp. Dominate a 4- to 5-kilometer-deep fault. Appl Environ Microbiol 71:8773–8783. doi: 10.1128/AEM.71.12.8773-8783.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mayumi D, Mochimaru H, Yoshioka H, Sakata S, Maeda H, Miyagawa Y, Ikarashi M, Takeuchi M, Kamagata Y. 2011. Evidence for syntrophic acetate oxidation coupled to hydrogenotrophic methanogenesis in the high-temperature petroleum reservoir of Yabase oil field (Japan). Environ Microbiol 13:1995–2006. doi: 10.1111/j.1462-2920.2010.02338.x [DOI] [PubMed] [Google Scholar]
- 63. Liu F, Conrad R. 2010. Thermoanaerobacteriaceae oxidize acetate in methanogenic rice field soil at 50°C. Environ Microbiol 12:2341–2354. doi: 10.1111/j.1462-2920.2010.02289.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental text, Fig. S1 to S10, and legends of Tables S1 to S11.
Tables S1 to S11.
Data Availability Statement
Raw sequence data reported in this paper are accessible at https://ngdc.cncb.ac.cn/gsa/browse/CRA004311.