Abstract
The X-ray repair cross complementing 1 (XRCC1) protein is required for viability and efficient repair of DNA single-strand breaks (SSBs) in rodents. XRCC1-deficient mouse or hamster cells are hypersensitive to DNA damaging agents generating SSBs and display genetic instability after such DNA damage. The presence of certain polymorphisms in the human XRCC1 gene has been associated with altered cancer risk, but the role of XRCC1 in SSB repair (SSBR) in human cells is poorly defined. To elucidate this role, we used RNA interference to modulate XRCC1 protein levels in human cell lines. A reduction in XRCC1 protein levels resulted in decreased SSBR capacity as measured by the comet assay and intracellular NAD(P)H levels, hypersensitivity to the cell killing effects of the DNA damaging agents methyl methanesulfonate (MMS), hydrogen peroxide and ionizing radiation and enhanced formation of micronuclei following exposure to MMS. Lowered XRCC1 protein levels were also associated with a significant delay in S-phase progression after exposure to MMS. These data clearly demonstrate that XRCC1 is required for efficient SSBR and genomic stability in human cells.
INTRODUCTION
DNA single-strand breaks (SSBs) are among the most frequent DNA lesions, arising directly from damage to the deoxyribose moieties or indirectly as intermediates of DNA base excision repair (BER) (1–3). Left unrepaired, SSBs are a major threat to genetic stability and cell survival, accelerating mutation rates and increasing levels of chromosomal aberrations (4–7). The pathways for SSB repair (SSBR) in mammalian cells involve a number of co-ordinated, sequential reactions responsible for damage detection, end processing, gap filling and ligation. In the case of the short-patch pathway of BER for example, a damaged base is recognized and removed by a damage-specific glycosylase, thus creating an abasic site whose 5′ phosphodiester bond is cut by apurinic/apyridinic endonuclease 1 (APE1). Subsequently, DNA polymerase β adds 1 nt to the 3′-OH end of the cleaved abasic site and excises the base-free sugar phosphate residue. Finally, this ligatable nick is sealed by DNA ligase IIIα.
Substantial evidence indicates an important role for X-ray repair cross complementing 1 (XRCC1) in SSBR and BER. Apparently devoid of any enzymatic activity, this protein is thought to act as a scaffolding protein for other repair factors. XRCC1 has been shown to physically interact with several enzymes known to be involved in the repair of SSBs, including DNA ligase IIIα, DNA polymerase β, APE1, polynucleotide kinase/phosphatase, poly(ADP-ribose) polymerases 1 and 2 (PARP-1 and 2) and 8-oxoguanine DNA glycosylase (OGG1) (8–14). Recently, XRCC1 has also been reported to be associated with aprataxin, the protein mutated in Ataxia-oculomotor apraxia 1 (15,16).
XRCC1-deficiency in mice results in embryonic lethality (17). Mutant mouse or CHO cells with no functional XRCC1 protein are hypersensitive to a broad range of DNA damage induced by alkylating agents, reactive oxygen species or ionizing radiation (17–19). SSB rejoining in XRCC1-deficient rodent cells is severely impaired, indicating a defect in DNA repair. Additionally, these cells display increased rates of spontaneous sister-chromatid exchange and chromosomal aberrations (4,5,17,19–21). Interestingly, XRCC1 transgene complemented XRCC1−/− mice that express XRCC1 at highly reduced levels, develop apparently normally and fibroblasts from these animals exhibit almost normal sensitivity to alkylating agents (22). Based on these experiments, it was concluded that XRCC1 is not the rate-limiting factor in SSBR in mouse cells.
Considerable evidence from CHO cells suggests that the hypersensitivity of XRCC1 mutant cells to genotoxins reflect perturbations of DNA replication. This is probably due to a greater number or the longer persistence of unrepaired SSBs encountered by the replication fork or to a deficiency in replication origin firing (4,23,24). The observations that XRCC1 foci increases during S-phase and co-localize with PCNA at replication sites underlines the importance of XRCC1 at this stage of the cell cycle (25–27).
As no human cell line lacking XRCC1 has been identified, few functional studies have investigated the role of the human XRCC1 protein in a cellular context. In HeLa cells, XRCC1 is recruited to laser irradiation-induced sites of SSB (28). In vivo evidence that XRCC1 is indeed necessary for SSBR in human cells was reported by Luo et al. (16), who downregulated XRCC1 expression by RNA interference (RNAi) and found that partial loss of XRCC1 renders HeLa cells sensitive to methyl methanesulfonate (MMS).
In the last few years, many molecular epidemiological studies have investigated the possible associations between XRCC1 polymorphisms and altered cancer risk. The presence of certain polymorphisms seem to be associated with either increased or decreased cancer susceptibility, depending on the type of cancer and the levels of environmental exposure to DNA damaging agents (29–36). These results suggest that the variant alleles may modify XRCC1's function. As a first step to investigate this phenomenon, we have used an RNAi approach to modulate XRCC1 levels in human breast cancer cell lines and report here that XRRC1 is necessary for survival, efficient DNA repair and genomic stability in human cells after DNA damage.
MATERIALS AND METHODS
Cells and cell culture
Human breast cancer cell lines BT20 and MDA-MB-453 were obtained from the American Tissue Culture collection (ATCC HTB-19 and HTB-131, respectively). MDA-MB-549 was a gift from Alain Puisieux (Centre Leon Berard, Lyon, France). They were cultured in DMEM (Gibco, Invitrogen Corporation) supplemented with 10% heat-inactivated fetal calf serum, 100 U/ml penicillin and 100 μg/ml streptomycin.
RNA interference
The short interfering RNA (siRNA) duplexes were designed and synthesized by Eurogentec. The sequences were as follows: 5′-AGGGAAGAGGAAGUUGGAU-3′ to target the XRCC1 transcript and 5′-AAACCCGAUAAUAACGUUGCG-3′ (scrambled control, containing the nucleotides of a region of the XRCC1 transcript in a random order). Cells were transfected using Oligofectamine (Invitrogen) according to the manufacturer's protocol, with a final oligonucleotide concentration of 100 nM. If not otherwise stated, cells were split after 48 h and treated with indicated concentrations of DNA damaging agents 72 h after the start of transfection.
RT–PCR and immunoblotting
In order to monitor the modulation of XRCC1 mRNA levels, a quantitative RT–PCR approach was used. Total RNA was extracted using Trizol (Invitrogen) according to the manufacturer's protocol and cDNA was prepared with 1 μg total RNA using the iScript cDNA Synthesis kit (Bio-Rad). Real-time PCR was run in a Stratagene Mx 3000P system and consisted of a 10 min initial denaturation at 95°C, followed by 40 cycles of 30 s at 95°C and 60 s at 60°C. The reactions were prepared with TaqMan Master Mix (Applied Biosystems). Assay-on-demand GAPDH control reagent (Applied Biosystems) was used to quantify the expression of a reference gene. The XRCC1-specific primers and probe were designed using the Primer Express software (Applied Biosystems) and were as follows: 5′-GGGACCGGGTCAAAATTGTT-3′, 5′-ACCGTACAAAACTCAAGCCAAAG-3′ and 5′-6FAM-AGCCCTACAGCAAGGA-MGB-3′.
Protein extraction and immunoblotting were carried out as described previously (37), with 30 μg of protein being loaded per lane. The membranes were incubated with antibodies to XRCC1 (R&D Systems; 1:1250 dilution), PARP-1 (Alexis Biochemicals; 1:5000 dilution), ligase III (abcam; 1:1000 dilution), APE1 (BD Biosciences; 1:250 dilution) and actin (ICN Biomedicals; 1:5000 dilution). The antigen–antibody complexes were detected by ECL western blotting detection reagent (Amersham). Protein levels were quantified using the Fluor-S MultiImager (Bio-Rad) and the Quantity One 4.2.1 software (Bio-Rad).
Survival curves and determination of intracellular NAD(P)H
The siRNA-transfected and control cells were split and plated into 96-well dishes (3000–5000 cells/well). The next day, they were exposed to H2O2 for 40 min or MMS for 1 h in complete medium, at 37°C. After washing with phosphate-buffered saline (PBS), cells were incubated for 3 days in drug-free medium. Survival was assayed using the CellTiter Aqueous One Solution Cell Proliferation Assay (Promega) according to the manufacturer's protocol. To measure survival after exposure to ionizing radiation, 0.5–1 × 105 cells were plated into T25 flasks and irradiated the following day. They were further incubated for 3–5 days and survival was assessed by Trypan Blue exclusion. All measurements were done in triplicate and experiments repeated at least twice.
Depletion of intracellular NAD(P)H was monitored as described (38) using CCK-8 solution (Dojindo Molecular Technology). Briefly, transfected or mock-transfected cells were split and seeded into 96-well plates. The next day, CCK-8 was added and cells were treated with MMS in the presence or absence of 10 mM 3-aminobenzamide (3-AB) (Sigma). Absorbance at 450 and 600 nm was measured 4 h after the start of the treatment. Measurements were done in triplicate.
Micronucleus assay
Following MMS treatment for 1 h, cells were washed with PBS and reincubated in normal medium containing 3 μg/ml cytochalasin B (Sigma) for 48 h before harvesting. Fixing, staining and scoring was carried out as described previously (39). A total of 400 binucleated cells per slide were scored for each transfection condition. Micronuclei were assessed in two independent experiments.
Comet assay
DNA damage was evaluated using the alkaline (pH > 13) single-cell gel electrophoresis or comet assay (40,41). Transfected cells were MMS-treated, washed and reincubated in normal medium for 3 h before processing. Harvested cells (1 × 105) were mixed with 0.5% low melting agarose (Bio Whittaker Molecular Applications) and layered onto agarose-coated slides. Slides were then submerged into lysis buffer [2.5 M NaCl, 100 mM EDTA, 10 mM Tris (pH 10.0) and 1% Triton X-100] overnight at 4°C. After lysis, slides were incubated for 1 h in electrophoresis buffer (300 mM NaOH and 1 mM EDTA, pH 13). After electrophoresis (1 h, 25 V, 300 mA), slides were neutralized with 0.4 M Tris, pH 7.5, for 30 min, placed into 100% ethanol and then air-dried. Slides were stained with 2 μg/ml ethidium bromide (Sigma). Average Comet Tail Moment (40) was scored (50 cells/slide) under an Axioplan2 Microscope (Zeiss) by using the Comet Imager 1.2.10 software (MetaSystems). All measurements were done on duplicate slides. Repair was quantified by calculating the percentage of MMS-induced strand breakage remaining after 3 h incubation in drug-free medium with the equation [(mean tail moment after repair − mean tail moment untreated cells)/(mean tail moment initial damage − mean tail moment untreated cells)] × 100 (27).
Flow cytometric analysis
Cells were MMS-treated for 1 h and reincubated in normal medium. At the indicated time points, they were processed using the CycleTest PLUS DNA reagent Kit (Becton Dickinson). Analysis was carried out in a Facscalibur (Becton Dickinson), cell-cycle distribution was assessed using the ModFit LT 2.0 software (Verity Software House).
RESULTS
XRCC1-deficient human cell lines are hypersensitive to DNA damaging agents
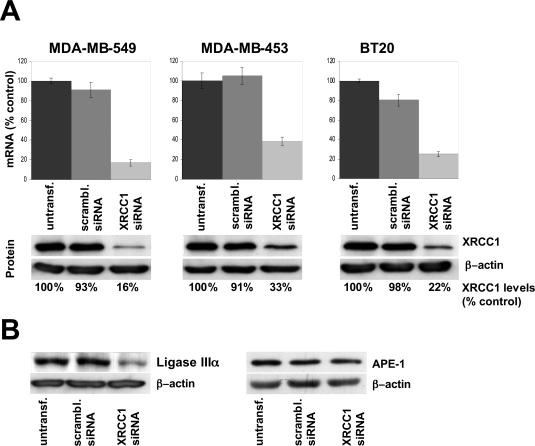
We adopted an RNAi approach to modulate XRCC1 expression in three human breast cancer cell lines, BT20, MDA-MB-453 and MDA-MB-549. Seventy-two hours after the start of transfection, quantitative RT–PCR revealed a reduction of XRCC1 mRNA levels of ∼80% (in MDA-MB-549 and BT20) and 65% (in MDA-MB-453) in XRCC1 siRNA-transfected cells compared with levels in control cells (Figure 1A, upper panels). Western blotting confirmed these results and showed that mRNA and protein levels were closely correlated (Figure 1A, lower panels).
Figure 1.
siRNA targeting the XRCC1 transcript efficiently reduces XRCC1 mRNA and protein levels in the three human breast cancer cell lines MDA-MB-549, MDA-MB-453 and BT20. (A) Cells were transfected with siRNA against the XRCC1 transcript, a scrambled siRNA or untransfected. mRNA (upper panels) and the corresponding protein levels (lower panels) were analyzed 72 h after the start of transfection by quantitative RT–PCR or western blotting, respectively. mRNA and protein levels were calculated by dividing the levels of transfected cells by the levels of untransfected cells. (B) Decreased XRCC1 expression leads to a significant reduction of ligase IIIα protein levels in cells transfected with the siRNA targeting XRCC1 (left panel). APE1 protein levels are unaffected by modulated XRCC1 expression (right panel). Proteins were extracted from cell line BT20 72 h after transfection.
In CHO cells, XRCC1 has been shown to form a complex with DNA ligase IIIα (Lig IIIα) and to act as an important stabilizing factor of this protein (42,43). In agreement with these findings, the level of DNA Lig IIIα protein detected in XRCC1-deficient human cells was significantly reduced compared with control cells (Figure 1B). In contrast, no difference in APE1 protein levels in cells with modulated XRCC1 expression was found 72 h after the start of transfection. This result is in contrast with observations in XRCC1 mutant CHO cells, where upregulated APE1 levels were noted (11).
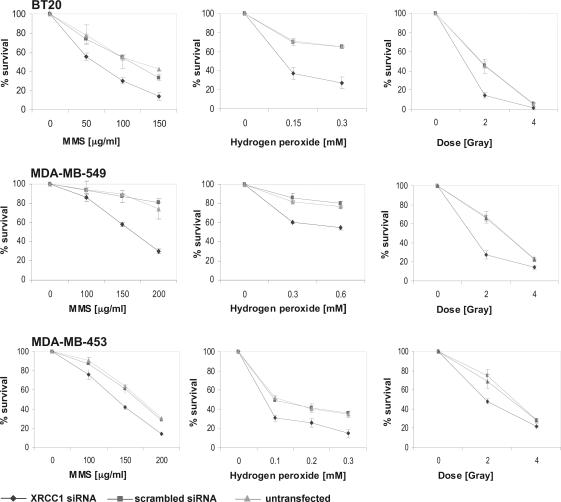
In order to examine the biological consequences of reduced XRCC1 levels, we measured cell survival after induction of DNA damage by MMS, hydrogen peroxide and ionizing radiation (Figure 2). In all three lines, decreased XRCC1 levels were associated with increased sensitivity to the three genotoxins tested. The relative increase in sensitivity was lower in XRCC1 siRNA-transfected MDA-MB-453 cells as compared with the other two cell lines tested. For example, MMS-sensitivity of siRNA-transfected MDA-MB-549 and BT20 cells increases 1.8-fold (LD20), while the sensitivity of MDA-MB-453 cells with reduced XRCC1 increases 1.4-fold compared with their control counterparts. This probably reflects the fact that in MDA-MB-453 cells, RNAi is the least effective and ∼35% of XRCC1 protein is still present after transfection.
Figure 2.
Reduced cell survival of XRCC1-deficient cells after treatment with the DNA damaging agents MMS, hydrogen peroxide or after exposure to ionizing radiation. Cells were treated 72 h after the start of transfection. Survival was assessed 3 days after treatment. All measurements were done in triplicate.
Increased sensitivity to MMS was also observed in the untransformed human fibroblast lines MRC5 and FD104 after siRNA-mediated reduction of XRCC1 levels (data not shown).
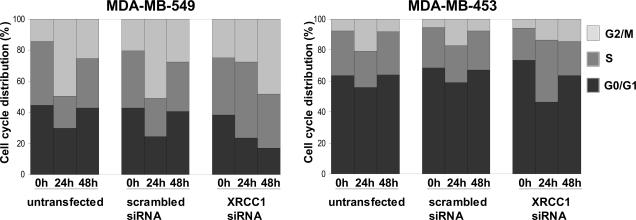
XRCC1-deficient cells display delayed cell-cycle progression following exposure to MMS
As XRCC1-deficiency has been linked to the perturbation of DNA replication, we next assessed whether reduced XRCC1 levels impacted on cell-cycle progression after DNA damage. Untransfected and siRNA-transfected MDA-MB-453 and MDA-MB-549 cells were exposed to MMS and their cell-cycle distribution analyzed before, 24 and 48 h after treatment (Figure 3). Twenty-four hours post-treatment, MMS causes a G2/M arrest in MDA-MB-549 control cells with unmodulated XRCC1 levels. Fifty percent of these cells accumulate at this stage of the cell cycle, while the fraction of cells in G0/G1 or S phases decreases at this time point. Forty-eight hours after MMS treatment, the cells resume progression through the cell cycle and display an almost identical cell-cycle distribution to that seen before the treatment. In contrast, MDA-MB-549 cells with reduced XRCC1 levels progress through S phase at a significantly reduced rate after MMS-induced DNA damage. While the fraction in G0/G1 decreases, 50% of these cells are found in S phase 24 h after the MMS treatment, suggesting that the limited quantity of XRCC1 leads to a reduced capacity to replicate the genome under DNA damage conditions, which is reminiscent of the observations in XRCC1-deficient CHO cells (23). Forty-eight hours after exposure to MMS, 50% of MDA-MB-549 cells with reduced XRCC1 levels arrest at G2/M, while only 17% are found in G0/G1, indicating that these cells do not resume progression through the cell cycle at this time point. No changes in XRCC1 mRNA levels in response to MMS were observed and XRCC1 transcript levels in XRCC1 siRNA-transfected cells remained constant over the analyzed time (data not shown).
Figure 3.
Cell-cycle progression of cells with differential expression of XRCC1. Cells were treated with 100 μg/ml (MDA-MB-453) or 150 μg/ml (MDA-MB-549) MMS for 1 h and reincubated in drug-free medium for 24 and 48 h before cell-cycle distribution was assessed by flow cytometry. Cell-cycle distribution at the time of exposure to MMS was determined in mock-treated cells (0 h).
The MDA-MB-453 cells showed similar, although less marked changes in the cell-cycle profile 24 h after MMS treatment. The number of cells in G2/M increases ∼3-fold while the number of cells in S phase and in G0/G1 decreases when no modulation of XRCC1 protein levels has occurred. Similar to MDA-MB-549 cells, the number of XRCC1-deficient MDA-MB-453 cells in S phase increases 24 h after MMS treatment: 40% of these cells are found in S phase at this time point, which is a 1.8-fold increase compared with untreated cells. Expression analysis by RT–PCR showed that XRCC1 mRNA levels at this time point were not significantly higher than at the time of MMS treatment (data not shown). After 48 h, however, XRCC1 mRNA levels in transfected MDA-MB-453 cells were almost back to normal levels and we could not detect major differences in cell-cycle distribution among the siRNA-transfected and untransfected cells, which resumed an almost normal cell-cycle progression.
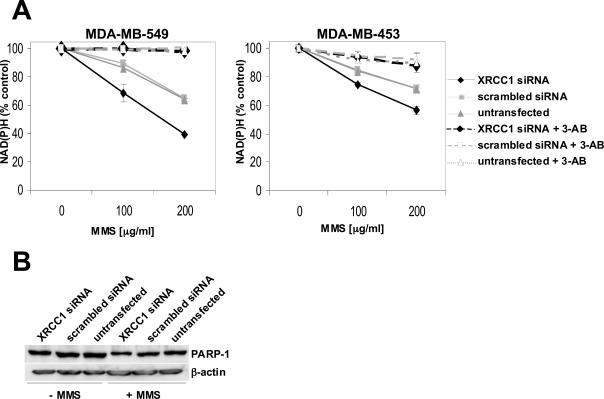
Quantification of intracellular NAD(P)H
To confirm the requirement for XRCC1 in damage repair in human cell lines, we investigated additional SSBR endpoints. First, we used a recently described method that indirectly measures the activity of PARP-1 by quantifying intracellular NAD(P)H (38). At the sites of SSB, activated PARP-1 catalyzes the transfer of the ADP-ribose moiety from NAD+ to a number of protein acceptors, resulting in intracellular NAD+ and NAD(P)H depletion (14,44). Translocation of XRCC1 to sites of SSB in HeLa cells has been reported to be dependent on poly(ADP-ribosyl)ation, while this recruitment in turn seems to negatively regulate PARP-1 activity (14,45). We monitored the reduction of intracellular NAD(P)H after a 4 h incubation in the presence or absence of MMS (Figure 4A). In agreement with the findings in CHO cells, human cells with reduced XRCC1 expression responded to MMS exposure with an enhanced reduction of intracellular NAD(P)H. The presence of 3-AB, a specific PARP-inhibitor, almost completely blocked the MMS-induced decrease of NAD(P)H in all tested cells, strongly suggesting that the observed depletion is due to increased PARP-1 activation. No significant differences in cell killing at the MMS concentrations used were noted, implicating that the depletion of NAD(P)H is not due to a reduction in the number of viable cells (data not shown). Nor were these differences in NAD(P)H depletion due to lower levels of PARP-1 protein itself, as PARP-1 levels were similar in cells with normal or ablated XRCC1 expression (Figure 4B).
Figure 4.
Enhanced depletion of intracellular NAD(P)H levels in cells with reduced XRCC1 expression (A) NAD(P)H depletion was measured after 4 h of MMS treatment either in the presence or in the absence of the PARP-1 inhibitor 3-AB (10 mM). The decrease was calculated by dividing the measured values of MMS-treated cells by the values of mock-treated cells. Measurements were done in triplicate. Shown are the values of one typical experiment. (B) Expression levels of PARP-1 in cells used for the depletion experiments were verified by western blotting of proteins extracted from MDA-MB-549 cells, which were either mock-treated or exposed to 200 μg/ml MMS for 1 h, followed by an incubation in drug-free medium for 3 h.
Cells lacking XRCC1 display SSBR deficiency
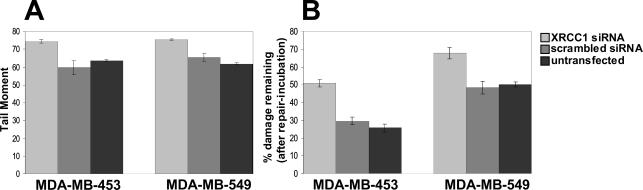
To confirm the role of XRCC1 in DNA repair in human cells, we next measured SSBR proficiency in individual cells by alkaline single-cell agarose gel electrophoresis. DNA damage was assessed before and immediately after a 1 h exposure of the cells to MMS and after a subsequent 3 h repair incubation in drug-free medium. For both the cell lines MDA-MB-453 and 549, average tail moments immediately after treatment were increased ∼20% in cells with reduced XRCC1 levels (Figure 5A), suggesting that these cells are SSBR defective. This was confirmed during the subsequent repair incubation in drug-free medium. In both cell lines analyzed (Figure 5B), DNA damage persists at higher levels in cells with reduced as compared with normal XRCC1 expression, indicating that DNA repair in these cells is impaired. Thus, we concluded that the presence of XRCC1 after the induction of DNA damage is essential for efficient single-strand resealing in human cells. However, cells with diminished XRCC1 expression were still able to repair DNA, albeit at a reduced rate. This very likely reflects the fact that XRCC1 gene silencing is incomplete but could also suggest the existence of an XRCC1-independent repair mechanism.
Figure 5.
Reduced SSBR capacity of cells with decreased XRCC1 levels as assayed by alkaline single-cell gel electrophoresis. siRNA-transfected and untransfected MDA-MB-453 and MDA-MB-549 cells were mock-treated or treated with 100 or 150 μg/ml MMS, respectively, and SSBs (expressed as the tail moment) were quantified before and immediately after the treatment. Cells were also mock-treated or treated with the same concentrations of MMS and subsequently incubated in drug-free medium for 3 h before measuring tail moments. (A) SSBs present immediately after MMS treatment (B) Percentages of MMS-induced SSBs which remain after the 3 h repair incubation (calculated from the tail moment present after the repair incubation).
RNAi of XRCC1 expression leads to increased genetic instability
A hallmark of XRCC1-deficient CHO cells is their increased levels of spontaneous sister-chromatid exchange rates (19,21). Recently, such cells have also been found to exhibit elevated levels of spontaneous formation of micronuclei (MN) (46). To further assess the effect of altered XRCC1 levels on genetic stability, we counted numbers of binucleated cells with induced MN and total numbers of MN 48 h after MMS treatment. In both MDA-MB-549 and MDA-MB-453 cell lines, siRNA-mediated downregulation of XRCC1 was associated with elevated numbers of binucleated cells with induced MN and increased total numbers of micronuclei (Table 1). These results demonstrate unambiguously that the absence of XRCC1 reduces DNA repair capacity and enhances genetic instability of human cells exposed to DNA damaging agents.
Table 1.
Enhanced induction of micronuclei in XRCC1-deficient cells
| Cell line | siRNA | Binucleated cells | Total micronuclei | |
|---|---|---|---|---|
| Scored | With micronuclei | |||
| MDA-MB-549 | XRCC1 | 800 | 243 | 444 |
| MDA-MB-549 | Scrambled | 800 | 190 | 262 |
| MDA-MB-549 | — | 800 | 174 | 245 |
| MDA-MB-453 | XRCC1 | 800 | 251 | 331 |
| MDA-MB-453 | Scrambled | 800 | 180 | 234 |
| MDA-MB-453 | — | 800 | 175 | 230 |
siRNA-transfected or untransfected MDA-MB-453 and MDA-MB-549 cells were treated with 100 or 150 μg/ml MMS, respectively, for 1 h, followed by an incubation in normal medium containing 3 μg/ml cytochalasin B for 48 h before analysis.
DISCUSSION
Human XRCC1 has been successfully used for reconstitution experiments in XRCC1−/− CHO cells and for identification of binding partners, e.g. by co-immunoprecipitation or yeast-two hybrid approaches (18). Owing to the lack of mutant cell lines, the role of XRCC1 in SSBR in human cells has not extensively been studied in vivo. We report here that XRCC1-deficiency in human cells recapitulates most, but not all of the phenotypes observed in XRCC1 mutant rodent cells. RNAi-mediated downregulation of XRCC1 resulted in increased sensitivity of three breast cancer cell lines to DNA damaging agents. Hypersensitivity is significant, but least pronounced in the cell line MDA-MB-453, most probably reflecting the fact that XRCC1 levels are typically only reduced to 35% of the normal levels. Nevertheless, these results indicate that XRCC1 might be a rate-limiting factor for SSBR in human cells and are in contrast with findings in XRCC1-deficient mouse cells. Upon transgene-complementation, <10% of wild-type XRCC1 expression levels almost completely rescue the MMS-hypersensitive phenotype of XRCC1−/− cells, suggesting that XRCC1 is not rate limiting in mouse SSBR (22).
The hypersensitivity of XRCC1−/− CHO cells to DNA damaging agents has been linked to perturbation of DNA replication which leads to a delay of S phase progression. XRCC1-deficient CHO cells accumulate large numbers of SCEs after incorporation of BrdUrd for two consecutive cell cycles and they stop to proliferate (4,23). After the first round of replication, SSBs were found in the halogenated template while nascent DNA, which was copied from this strand, was delayed in maturation as compared with DNA synthesized on an undamaged template. These studies strongly suggested that this delay was due to replication forks encountering SSBs. A different type of perturbation of replication was reported by Kubota and Horiuchi, who observed a decrease in newly fired DNA replication in XRCC1-deficient CHO cells upon MMS treatment (24). This either reflects an unidentified role for XRCC1 in origin firing or an indirect effect due to an increased number or longer persistence of SSBs at replication forks and, as a consequence, a prolonged intra S phase checkpoint. Our experimental set-up for measuring cell-cycle progression is not capable of distinguishing between these different types of perturbations. However, the fact that both MDA-MB-549 and MDA-MB-453 cells with decreased XRCC1 levels progress through S phase considerably more slowly than control cells after MMS treatment strongly suggests that like in rodent cells, XRCC1 is required in human cells to allow unperturbed replication and progression through S phase after DNA damage. We did not detect an accumulation of cells in G0/G1 at any time point, which is in line with observations in CHO cells that XRCC1 function is dispensable during G1 but not S phase (27). After a perturbed S phase, cells with low levels of XRCC1 also arrest at G2/M, indicating that the DNA is still damaged after S phase exit and suggesting that cells are tolerant to a certain degree of damage during S phase progression. This could also explain the rapid progression of control cells to a transient G2/M arrest after MMS treatment, where DNA repair presumably takes place.
We further demonstrated the imbalance of DNA SSBR in human cells with lowered XRCC1 levels by showing that during exposure to MMS, reduced XRCC1 levels are associated with an increased intracellular NAD(P)H depletion, due to enhanced PARP-1 activity. These results are in agreement with findings in XRCC1-deficient CHO cells and provide further evidence for an involvement of XRCC1 and PARP-1 in in vivo BER following MMS-induced DNA damage. Moreover, they support a role of XRCC1 as a negative regulator of PARP-1 activity. Such a regulatory role has been suggested after overexpression of XRCC1 in HeLa cells, which leads to impaired poly(ADP-ribose) synthesis in response to DNA damage (14). The biological consequences of this inhibition remain to be elucidated.
We also provide direct proof that XRCC1 is required for DNA strand break resealing by showing that reduced XRCC1 levels partially inhibit DNA rejoining (as measured by the comet assay) and increase MMS-induced micronuclei formation. Again, this is coherent with findings in XRCC1-deficient rodent cells, which display delayed repair kinetics and elevated rates of markers of genetic instability (7,19,22,27,47). Genetic instability as a consequence of DNA repair deficiency is generally regarded as an enabling trait for cancerogenesis (48), and biomarkers of genetic damage such as micronuclei formation are considered to be causally related to early stages of chronic diseases, especially cancer. The involvement of XRCC1 in tumorigenesis is difficult to assess since in mice XRCC1-deficiency results in embryonic lethality. A large number of molecular epidemiological studies have analyzed the impact of polymorphisms in the human XRCC1 gene on cancer risk. Depending on the type of the cancer, these polymorphisms have been reported to be associated with altered cancer predisposition (29,31–34,49–51). One study reported a positive correlation between the presence of a XRCC1 polymorphism and the number of micronuclei in peripheral lymphocytes of individuals that are occupationally exposed to chronic low doses of ionizing radiation, suggesting that XRCC1 might indeed play a role in the maintenance of genetic stability and hence the prevention of tumorigenesis (52). No direct correlation can be drawn between the observations we report here and a putative involvement of XRCC1 in cancer progression. It is nevertheless tempting to speculate that reduced SSBR and genomic instabilities caused by XRCC1-deficiency might facilitate cancer development.
Acknowledgments
We thank Norman Moullan and Caroline Fayolle for technical assistance and Dr Alain Puisieux for generously providing certain cell lines. We also thank Dr Marie Fernet for helpful suggestions and Dr Magali Olivier for critical reading of the manuscript. The financial support of ‘Electricité de France’ and the ‘Comité Départemental du Rhône, La Ligue Nationale Contre le Cancer’ are gratefully acknowledged. The work of R. Brem was carried out during the tenure of a Special Training Award from the International Agency for Research on Cancer. Funding to pay the Open Access publication charges for this article was provided by the IARC regular budget.
Conflict of interest statement. None declared.
REFERENCES
- 1.Xu Y.J., Kim E.Y., Demple B. Excision of C-4′-oxidized deoxyribose lesions from double-stranded DNA by human apurinic/apyrimidinic endonuclease (Ape1 protein) and DNA polymerase beta. J. Biol. Chem. 1998;273:28837–28844. doi: 10.1074/jbc.273.44.28837. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 3.Beckman K.B., Ames B.N. Oxidative decay of DNA. J. Biol. Chem. 1997;272:19633–19636. doi: 10.1074/jbc.272.32.19633. [DOI] [PubMed] [Google Scholar]
- 4.Carrano A.V., Minkler J.L., Dillehay L.E., Thompson L.H. Incorporated bromodeoxyuridine enhances the sister-chromatid exchange and chromosomal aberration frequencies in an EMS-sensitive Chinese hamster cell line. Mutat. Res. 1986;162:233–239. doi: 10.1016/0027-5107(86)90090-4. [DOI] [PubMed] [Google Scholar]
- 5.Dominguez I., Daza P., Natarajan A.T., Cortes F. A high yield of translocations parallels the high yield of sister chromatid exchanges in the CHO mutant EM9. Mutat. Res. 1998;398:67–73. doi: 10.1016/s0027-5107(97)00241-8. [DOI] [PubMed] [Google Scholar]
- 6.Thompson L.H., Brookman K.W., Dillehay L.E., Mooney C.L., Carrano A.V. Hypersensitivity to mutation and sister-chromatid-exchange induction in CHO cell mutants defective in incising DNA containing UV lesions. Somatic Cell Genet. 1982;8:759–773. doi: 10.1007/BF01543017. [DOI] [PubMed] [Google Scholar]
- 7.Trucco C., Oliver F.J., de Murcia G., Menissier-de Murcia J. DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res. 1998;26:2644–2649. doi: 10.1093/nar/26.11.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caldecott K.W., Aoufouchi S., Johnson P., Shall S. XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res. 1996;24:4387–4394. doi: 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kubota Y., Nash R.A., Klungland A., Schar P., Barnes D.E., Lindahl T. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J. 1996;15:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 10.Marsin S., Vidal A.E., Sossou M., Menissier-de Murcia J., Le Page F., Boiteux S., de Murcia G., Radicella J.P. Role of XRCC1 in the coordination and stimulation of oxidative DNA damage repair initiated by the DNA glycosylase hOGG1. J. Biol. Chem. 2003;278:44068–44074. doi: 10.1074/jbc.M306160200. [DOI] [PubMed] [Google Scholar]
- 11.Vidal A.E., Boiteux S., Hickson I.D., Radicella J.P. XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein–protein interactions. EMBO J. 2001;20:6530–6539. doi: 10.1093/emboj/20.22.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitehouse C.J., Taylor R.M., Thistlethwaite A., Zhang H., Karimi-Busheri F., Lasko D.D., Weinfeld M., Caldecott K.W. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell. 2001;104:107–117. doi: 10.1016/s0092-8674(01)00195-7. [DOI] [PubMed] [Google Scholar]
- 13.Schreiber V., Ame J.C., Dolle P., Schultz I., Rinaldi B., Fraulob V., Menissier-de Murcia J., de Murcia G. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem. 2002;277:23028–23036. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- 14.Masson M., Niedergang C., Schreiber V., Muller S., Menissier-de Murcia J., de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clements P.M., Breslin C., Deeks E.D., Byrd P.J., Ju L., Bieganowski P., Brenner C., Moreira M.C., Taylor A.M., Caldecott K.W. The ataxia-oculomotor apraxia 1 gene product has a role distinct from ATM and interacts with the DNA strand break repair proteins XRCC1 and XRCC4. DNA Repair (Amst.) 2004;3:1493–1502. doi: 10.1016/j.dnarep.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 16.Luo H., Chan D.W., Yang T., Rodriguez M., Chen B.P., Leng M., Mu J.J., Chen D., Songyang Z., Wang Y., et al. A new XRCC1-containing complex and its role in cellular survival of methyl methanesulfonate treatment. Mol. Cell. Biol. 2004;24:8356–8365. doi: 10.1128/MCB.24.19.8356-8365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tebbs R.S., Flannery M.L., Meneses J.J., Hartmann A., Tucker J.D., Thompson L.H., Cleaver J.E., Pedersen R.A. Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Dev. Biol. 1999;208:513–529. doi: 10.1006/dbio.1999.9232. [DOI] [PubMed] [Google Scholar]
- 18.Thompson L.H., Brookman K.W., Jones N.J., Allen S.A., Carrano A.V. Molecular cloning of the human XRCC1 gene, which corrects defective DNA strand break repair and sister chromatid exchange. Mol. Cell. Biol. 1990;10:6160–6171. doi: 10.1128/mcb.10.12.6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson L.H., Brookman K.W., Dillehay L.E., Carrano A.V., Mazrimas J.A., Mooney C.L., Minkler J.L. A CHO-cell strain having hypersensitivity to mutagens, a defect in DNA strand-break repair, and an extraordinary baseline frequency of sister-chromatid exchange. Mutat. Res. 1982;95:427–440. doi: 10.1016/0027-5107(82)90276-7. [DOI] [PubMed] [Google Scholar]
- 20.Shen M.R., Zdzienicka M.Z., Mohrenweiser H., Thompson L.H., Thelen M.P. Mutations in hamster single-strand break repair gene XRCC1 causing defective DNA repair. Nucleic Acids Res. 1998;26:1032–1037. doi: 10.1093/nar/26.4.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zdzienicka M.Z., van der Schans G.P., Natarajan A.T., Thompson L.H., Neuteboom I., Simons J.W. A Chinese hamster ovary cell mutant (EM-C11) with sensitivity to simple alkylating agents and a very high level of sister chromatid exchanges. Mutagenesis. 1992;7:265–269. doi: 10.1093/mutage/7.4.265. [DOI] [PubMed] [Google Scholar]
- 22.Tebbs R.S., Thompson L.H., Cleaver J.E. Rescue of Xrcc1 knockout mouse embryo lethality by transgene-complementation. DNA Repair (Amst.) 2003;2:1405–1417. doi: 10.1016/j.dnarep.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Dillehay L.E., Thompson L.H., Minkler J.L., Carrano A.V. The relationship between sister-chromatid exchange and perturbations in DNA replication in mutant EM9 and normal CHO cells. Mutat. Res. 1983;109:283–296. doi: 10.1016/0027-5107(83)90053-2. [DOI] [PubMed] [Google Scholar]
- 24.Kubota Y., Horiuchi S. Independent roles of XRCC1's two BRCT motifs in recovery from methylation damage. DNA Repair (Amst.) 2003;2:407–415. doi: 10.1016/s1568-7864(02)00242-2. [DOI] [PubMed] [Google Scholar]
- 25.Taylor R.M., Thistlethwaite A., Caldecott K.W. Central role for the XRCC1 BRCT I domain in mammalian DNA single-strand break repair. Mol. Cell. Biol. 2002;22:2556–2563. doi: 10.1128/MCB.22.8.2556-2563.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan J., Otterlei M., Wong H.K., Tomkinson A.E., Wilson D.M., III XRCC1 co-localizes and physically interacts with PCNA. Nucleic Acids Res. 2004;32:2193–2201. doi: 10.1093/nar/gkh556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor R.M., Moore D.J., Whitehouse J., Johnson P., Caldecott K.W. A cell cycle-specific requirement for the XRCC1 BRCT II domain during mammalian DNA strand break repair. Mol. Cell. Biol. 2000;20:735–740. doi: 10.1128/mcb.20.2.735-740.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lan L., Nakajima S., Oohata Y., Takao M., Okano S., Masutani M., Wilson S.H., Yasui A. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc. Natl Acad. Sci. USA. 2004;101:13738–13743. doi: 10.1073/pnas.0406048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harms C., Salama S.A., Sierra-Torres C.H., Cajas-Salazar N., Au W.W. Polymorphisms in DNA repair genes, chromosome aberrations, and lung cancer. Environ. Mol. Mutagen. 2004;44:74–82. doi: 10.1002/em.20031. [DOI] [PubMed] [Google Scholar]
- 30.Nagasubramanian R., Innocenti F., Ratain M.J. Pharmacogenetics in cancer treatment. Annu. Rev. Med. 2003;54:437–452. doi: 10.1146/annurev.med.54.101601.152352. [DOI] [PubMed] [Google Scholar]
- 31.Goode E.L., Ulrich C.M., Potter J.D. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol. Biomarkers Prev. 2002;11:1513–1530. [PubMed] [Google Scholar]
- 32.Han J., Hankinson S.E., Colditz G.A., Hunter D.J. Genetic variation in XRCC1, sun exposure, and risk of skin cancer. Br. J. Cancer. 2004;91:1604–1609. doi: 10.1038/sj.bjc.6602174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L.E., Bondy M.L., Shen H., El Zein R., Aldape K., Cao Y., Pudavalli V., Levin V.A., Yung W.K., Wei Q. Polymorphisms of DNA repair genes and risk of glioma. Cancer Res. 2004;64:5560–5563. doi: 10.1158/0008-5472.CAN-03-2181. [DOI] [PubMed] [Google Scholar]
- 34.Moullan N., Cox D.G., Angele S., Romestaing P., Gerard J.P., Hall J. Polymorphisms in the DNA repair gene XRCC1, breast cancer risk, and response to radiotherapy. Cancer Epidemiol. Biomarkers Prev. 2003;12:1168–1174. [PubMed] [Google Scholar]
- 35.Ito H., Matsuo K., Hamajima N., Mitsudomi T., Sugiura T., Saito T., Yasue T., Lee K.M., Kang D., Yoo K.Y., et al. Gene-environment interactions between the smoking habit and polymorphisms in the DNA repair genes, APE1 Asp148Glu and XRCC1 Arg399Gln, in Japanese lung cancer risk. Carcinogenesis. 2004;25:1395–1401. doi: 10.1093/carcin/bgh153. [DOI] [PubMed] [Google Scholar]
- 36.Tae K., Lee H.S., Park B.J., Park C.W., Kim K.R., Cho H.Y., Kim L.H., Park B.L., Shin H.D. Association of DNA repair gene XRCC1 polymorphisms with head and neck cancer in Korean population. Int. J. Cancer. 2004;111:805–808. doi: 10.1002/ijc.20338. [DOI] [PubMed] [Google Scholar]
- 37.Fernet M., Gribaa M., Salih M.A., Seidahmed M.Z., Hall J., Koenig M. Identification and functional consequences of a novel MRE11 mutation affecting 10 Saudi Arabian patients with the ataxia telangiectasia-like disorder. Hum. Mol. Genet. 2005;14:307–318. doi: 10.1093/hmg/ddi027. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura J., Asakura S., Hester S.D., de Murcia G., Caldecott K.W., Swenberg J.A. Quantitation of intracellular NAD(P)H can monitor an imbalance of DNA single strand break repair in base excision repair deficient cells in real time. Nucleic Acids Res. 2003;31:e104. doi: 10.1093/nar/gng105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gutierrez-Enriquez S., Hall J. Use of the cytokinesis-block micronucleus assay to measure radiation-induced chromosome damage in lymphoblastoid cell lines. Mutat. Res. 2003;535:1–13. [PubMed] [Google Scholar]
- 40.Olive P.L., Johnston P.J., Banath J.P., Durand R.E. The comet assay: a new method to examine heterogeneity associated with solid tumors. Nature Med. 1998;4:103–105. doi: 10.1038/nm0198-103. [DOI] [PubMed] [Google Scholar]
- 41.Singh N.P., McCoy M.T., Tice R.R., Schneider E.L. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 42.Caldecott K.W., McKeown C.K., Tucker J.D., Ljungquist S., Thompson L.H. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol. Cell. Biol. 1994;14:68–76. doi: 10.1128/mcb.14.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caldecott K.W., Tucker J.D., Stanker L.H., Thompson L.H. Characterization of the XRCC1-DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res. 1995;23:4836–4843. doi: 10.1093/nar/23.23.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oei S.L., Griesenbeck J., Schweiger M. The role of poly(ADP-ribosyl)ation. Rev. Physiol. Biochem. Pharmacol. 1997;131:127–173. doi: 10.1007/3-540-61992-5_7. [DOI] [PubMed] [Google Scholar]
- 45.Okano S., Lan L., Caldecott K.W., Mori T., Yasui A. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol. Cell. Biol. 2003;23:3974–3981. doi: 10.1128/MCB.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sossou M., Flohr-Beckhaus C., Schulz I., Daboussi F., Epe B., Radicella J.P. APE1 overexpression in XRCC1-deficient cells complements the defective repair of oxidative single strand breaks but increases genomic instability. Nucleic Acids Res. 2005;33:298–306. doi: 10.1093/nar/gki173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Op het Veld C.W., Zdzienicka M.Z., Vrieling H., Lohman P.H., van Zeeland A.A. Molecular analysis of ethyl methanesulfonate-induced mutations at the hprt gene in the ethyl methanesulfonate-sensitive Chinese hamster cell line EM-C11 and its parental line CHO9. Cancer Res. 1994;54:3001–3006. [PubMed] [Google Scholar]
- 48.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 49.Hao B., Wang H., Zhou K., Li Y., Chen X., Zhou G., Zhu Y., Miao X., Tan W., Wei Q., et al. Identification of genetic variants in base excision repair pathway and their associations with risk of esophageal squamous cell carcinoma. Cancer Res. 2004;64:4378–4384. doi: 10.1158/0008-5472.CAN-04-0372. [DOI] [PubMed] [Google Scholar]
- 50.Kelsey K.T., Park S., Nelson H.H., Karagas M.R. A population-based case-control study of the XRCC1 Arg399Gln polymorphism and susceptibility to bladder cancer. Cancer Epidemiol. Biomarkers Prev. 2004;13:1337–1341. [PubMed] [Google Scholar]
- 51.Ratnasinghe L.D., Abnet C., Qiao Y.L., Modali R., Stolzenberg-Solomon R., Dong Z.W., Dawsey S.M., Mark S.D., Taylor P.R. Polymorphisms of XRCC1 and risk of esophageal and gastric cardia cancer. Cancer Lett. 2004;216:157–164. doi: 10.1016/j.canlet.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 52.Angelini S., Kumar R., Carbone F., Maffei F., Forti G.C., Violante F.S., Lodi V., Curti S., Hemminki K., Hrelia P. Micronuclei in humans induced by exposure to low level of ionizing radiation: influence of polymorphisms in DNA repair genes. Mutat. Res. 2005;570:105–117. doi: 10.1016/j.mrfmmm.2004.10.007. [DOI] [PubMed] [Google Scholar]