Abstract
Biological control of chestnut blight caused by the filamentous ascomycete Cryphonectria parasitica can be achieved with a virus that infects this fungus. This hypovirus causes a perturbation of fungal development that results in low virulence (hypovirulence), poor asexual sporulation, and female infertility without affecting fungal growth in culture. At the molecular level, the virus is known to affect the transcription of a number of fungal genes. Two of these genes, Vir1 and Vir2, produce abundant transcripts in noninfected strains of the fungus, but the transcripts are not detectable in virus-infected strains. We report here that these two genes encode the pheromone precursors of the Mat-2 mating type of the fungus; consequently, these genes have been renamed Mf2/1 and Mf2/2. To determine if the virus affects the mating systems of both mating types of this fungus, the pheromone precursor gene, Mf1/1, of a Mat-1 strain was cloned and likewise was found to be repressed in virus-infected strains. The suppression of transcription of the pheromone precursor genes of this fungus could be the cause of the mating defect of infected strains of the fungus. Although published reports suggest that a Gαi subunit may be involved in this regulation, our results do not support this hypothesis. The prepropheromone encoded by Mf1/1 is structurally similar to that of the prepro-p-factor of Schizosaccharomyces pombe. This is the first description of the complete set of pheromone precursor genes encoded by a filamentous ascomycete.
Chestnut blight, caused by the ascomycete Cryphonectria parasitica, is one of the most devastating plant diseases in recorded history. Because of this disease, only occasional root sprouts grow where the American chestnut (Castanea dentata) was once dominant in eastern North America. A similar fate for the European chestnut (C. sativa) in European forests and orchards was prevented by a naturally occurring biological control of the fungus. The basis of this biological control is infection of C. parasitica by double-stranded RNA viruses which reduce the virulence of the fungus, causing a condition termed hypovirulence. The effect of the virus on reducing the virulence of this plant pathogen is the basis of an effective biological control of chestnut blight (1, 7, 13, 16, 24, 25).
One of these hypoviruses, CHV1-713, causes no detectable effects on the growth rate of its host in culture but perturbs normal developmental processes such as sporulation and virulence. The visible symptoms of CHV1-infected colonies growing on agar are a white rather than orange colony color and poor asexual sporulation, providing a useful phenotype for the study of this virus. The sexual cycle of the fungus is also perturbed by the virus, with the female parent being sterile. At the molecular level, the virus causes transcriptional down-regulation of a number of host genes (10). The cloning and identification of the function of some of these genes have provided us with a group of molecular markers of virus infection. We have previously reported the cloning of three of these genes: Crp, coding for an abundant hydrophobic protein located on the cell surface (28); Lac1, coding for an extracellular laccase (18); and Vir2, a gene identified as being necessary for sexual reproduction (27).
Vir2 and a related gene, Vir1, were identified as mRNAs found by subtractive hybridization between cDNAs of an uninfected strain of the fungus and the total RNA of an isogenic virus-infected strain (17). Vir2 was cloned and deleted; the Vir2 null mutant (dm18) exhibited a phenotype that partially mimicked virus symptoms of the host, i.e., reduced asexual sporulation and impaired sexual fertility. Attempts at sexual crosses of the null mutant produced barren perithecia. The Vir2 gene encodes a small protein of 23 amino acids with a C-terminal CAAX motif, a prenylation signal common to all known fungal lipopeptide pheromones (27). We report here that Vir2 encodes a mating-type specific pheromone precursor, and we describe the cloning of the other two pheromone genes of this fungus. Our results demonstrate that a filamentous ascomycete contains pheromone precursor genes encoding precursors with structural characteristics and maturation processing signals similar to those reported from the ascomyceteous yeasts. We also report that the virus represses the transcription of each of the fungal pheromone precursor genes of both mating types of C. parasitica, which could cause the mating defects exhibited by the virus-infected strains of the fungus.
MATERIALS AND METHODS
Fungal and bacterial strains.
The strains of C. parasitica that were used in this study are listed in Table 1. The fungal strains were maintained as previously described (27). Vectors were constructed with pBluescript (Stratagene, La Jolla, Calif.) and maintained in Escherichia coli DH10B (Gibco BRL, Gaithersburg, Md.).
TABLE 1.
Strains of C. parasitica used in this study
| Strain | Source | Mating type | Virus |
|---|---|---|---|
| EP 42 | ATCC 38751 | 2 | —a |
| EP 44 (EP 3.6) | ATCC 22511 | 1 | — |
| EP 67 | ATCC 38753 | 1 | — |
| EP 155 | ATCC 38755 | 2 | — |
| EP 802 | ATCC 52574 | 1 | CHV1-713 |
| EP 368 | S. Anagnostakisb | 1 | — |
| EP 370 | S. Anagnostakis | 2 | — |
| EP 372 | S. Anagnostakis | 2 | — |
| UEP 1 | Powell and Van Alfen (17) | 2 | CHV1-713 |
| dm 18 | Zhang et al. (27) | 2 | — |
—, not infected with CHV1-713.
Connecticut Agricultural Experiment Station, New Haven, Conn.
Nucleic acid isolation and analysis.
Isolation of fungal genomic DNA for Southern analysis RNA isolation, Southern and Northern analyses, DNA sequencing, and primer extension reactions were conducted as previously described (27).
cDNA library construction.
Total RNA of EP155 and EP67 was isolated from 5-day-old liquid cultures; from this RNA, polyATtract system 1000 of Promega (Madison, Wis.) was used to isolate mRNA. The cDNA libraries were constructed with the SuperScript lambda system for cDNA synthesis and cloning (Gibco BRL) as specified by the manufacturer.
Genomic DNA library construction.
Total genomic DNA of EP155 and EP67 was isolated from lysed spheroplasts by the method of Sweigard et al. (22). The lambda FIX II/XhoI partial fill-in vector system of Stratagene was used to clone the DNA libraries.
Cloning of pheromone precursor genes. (i) Cloning of Mf2/1.
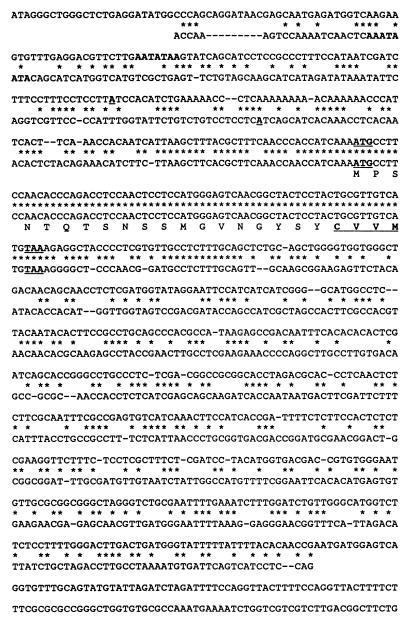
Total genomic DNA from strain dm18 was isolated and restricted with BamHI. The DNA was separated on a 0.9% Tris-borate-EDTA (TBE)–agarose gel, and the DNA corresponding in size to approximately 1.3 kb was eluted. A BamHI-EcoRI adapter (New England Biolabs) was then ligated to the eluted fragments. These fragments were ligated to lambda gt11 EcoRI arms and packaged (Stratagene packaging kit). This subgenomic library was screened with a 32P-labelled fragment of Mf2/2. After selected clones were sequenced, a fragment that had no sequence homology to Mf2/2 was chosen to screen genomic and cDNA libraries of strain EP155. From the genomic library, a 3.4-kb SmaI fragment was cloned into pBluescript at the EcoRV site. Four cDNA clones were selected and sequenced. The primer extension method was used to determine the transcription initiation site of Mf2/1. The primer used was the 21-mer 5′-GAGGTCTGGGTGTTGGAAGGC, which is complementary to the sequence starting 75 bp downstream from the transcription initiation site (Fig. 1).
FIG. 1.
Sequence alignment of Mf2/1 (upper sequence) and Mf2/2 (lower sequence). The sequences shown are of the immediate 5′ end of the untranscribed regions and the transcribed regions, ending with the poly(A) addition sites, as determined from cDNA sequences; the sequences contain, respectively, 731 and 591 bp. In both sequences, the putative TATA box is in boldface type and the transcription initiation site, translation initiation codon (ATG), and termination codon (TAA) are in boldface type and underlined. The derived amino acid sequence of the first ORF of each gene is shown; the prenylation signal sequence (CAAX) is in boldface type and underlined.
(ii) Cloning of Mf1/1.
Mf1/1 was cloned by PCR-mediated subtractive hybridization between cDNA libraries of strains EP155 (Mat-2) and EP67 (Mat-1), using the method of Wang and Brown (26). Total RNA was isolated from 5-day-old mycelia. The mRNA of each strain was isolated with PolyATtract system 1000, and cDNAs were synthesized with the SuperScript lambda system for cDNA synthesis and cloning. The cDNAs were restricted with AluI, and then PCR primers were ligated to the blunt-end restriction fragments. The following primer sequences were used: EP67 (..TAGAACTAGTGGATCCCC-3′ and AGATCTTGATCACCTAGGGG-5′) and EP155 (5′-CGATACCGTCGACCTCGAGG and 3′-GCTATGGCAGCTGGAGCT..).
After PCR amplification, 70 μg of EP155 cDNA was biotinylated and then 18 μg of the biotinylated EP155 cDNA and 1 μg of EP67 cDNA were hybridized together at 68°C for 2 h. After extraction with streptavidin, the remaining EP67 cDNA was again hybridized with 18 μg of biotinylated cDNA of EP155 for 24 h at 68°C. The two-hybridization processes described above were repeated once. The resulting EP67 cDNA was amplified by PCR, digested with BamHI, and ligated into pBluescript. Individual clones from this EP67 subtractive library were used to screen Northern blots of total RNA isolated from strains EP155 and EP67. Clones confirmed to be Mat-1 specific were used to isolate the genomic and cDNA clones from genomic and cDNA libraries.
To determine if multiple copies of Mf1/1 exist in the genome, a degenerate oligonucleotide based on the repeated decapeptide portion of the sequence was designed. The following oligonucleotide was used: CCAGCAVCCYTCDCCGTGGAAGAGRCACCA, where V = G + A + C, Y = C + T, D = G + A + T, and R = A + G. The oligonucleotide was 32P labelled with T4 nucleotide kinase and used to probe a Southern blot of genomic DNA.
Nucleotide sequence accession numbers.
The sequence data reported for Mf1/1 and Mf2/1 have been assigned GenBank accession no. U92042 and U92043, respectively.
RESULTS
Cloning of a second Mat-2 pheromone precursor gene.
Based on the work of Zhang et al. (27) and the results reported here, the names Vir1 and Vir2 have been changed to Mf2/1 and Mf2/2 (mating factors 2/1 and 2/2), respectively. Previous studies had shown that the two mRNAs Mf2/1 (Vir1) and Mf2/2 (Vir2) hybridize to the same DNA probe (17). Deletion of the Mf2/2 gene did not, however, result in loss of the Mf2/1 transcript, showing that these mRNAs were encoded by separate genes. When used as a probe, a DNA fragment of the Mf2/2 gene coding region consistently detected a 1.3-kb BamHI fragment on Southern blots of total genomic DNA from C. parasitica EP155 and the Mf2/2 deletion strain dm18 (27). To clone this 1.3-kb BamHI fragment, a subgenomic library of 1.3-kb BamHI fragments of genomic DNA from strain dm18 was constructed and the resultant clones were screened with a fragment of the Mf2/2 coding region.
The clone identified in this manner was sequenced; an 83-bp region of this 1.3-kb fragment was identical to the first open reading frame (ORF) of Mf2/2 (Fig. 1). Beyond this region, no significant similarity was found. By using a portion of the clone without similarity to Mf2/2 as a probe, a genomic clone and cDNA clones of Mf2/1 were identified from genomic and cDNA libraries of strain EP155. The transcription initiation site was determined by primer extension. Sequence comparisons of genomic DNA and cDNA of Mf2/1 show that there are no introns in this gene. The sequence comparison in Fig. 1 shows that the only region of similarity between the two genes is the identical 83-bp first ORF of each gene. This ORF, common to both Mf2/1 and Mf2/2, encodes a product with the structural features of known fungal lipopeptide sex pheromones, including a common C-terminal CAAX box and an asparagine within 8 to 11 amino acids of the CAAX box. Some of the known fungal sex pheromone precursors are cleaved at this asparagine during the maturation process (15, 19).
Cloning of the Mat-1 pheromone precursor gene.
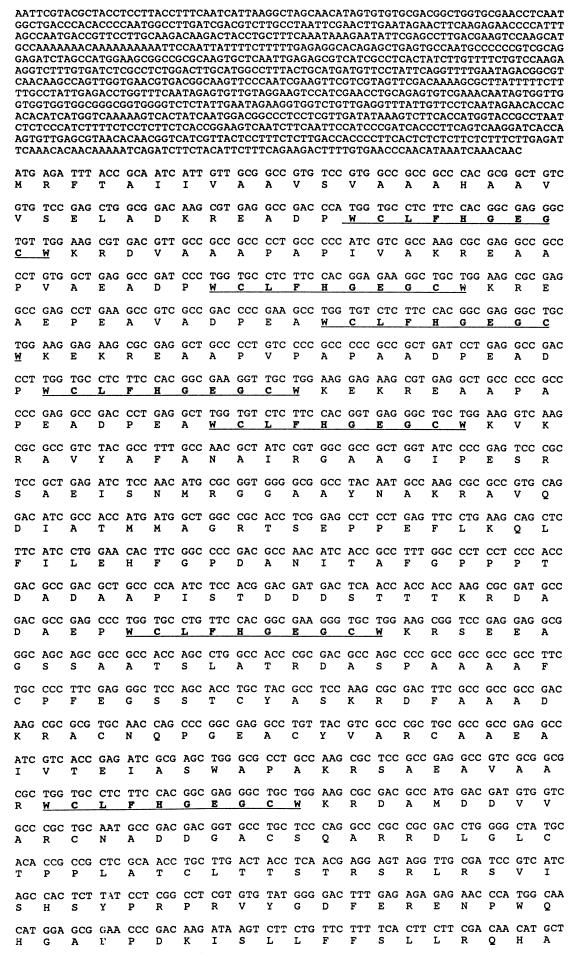
A subtractive cDNA procedure was used to identify Mat-1-specific clones. By this procedure, a genomic clone containing a 4.6-kb EcoRI fragment was selected and partially sequenced (Fig. 2). Comparison of sequences of the genomic and cDNA clones revealed no evidence for introns in this gene. A single ORF coding for a 530-amino-acid peptide was identified within this gene. Within the peptide were seven repeats of the decapeptide sequence WCLFHGEGCW. All seven copies of the decapeptide were preceded by two to eight repeats of a dipeptide XA (or XP), except for the seventh copy, which has an XR directly before the decapeptide. All of these stretches of dipeptide repeats were preceded by the basic dipeptide KR. At the C terminus of the decapeptides was the dipeptide KR; in some cases, the sequence KEKR or KVKR was present at the C terminus (Fig. 2).
FIG. 2.
Sequence of Mf1/1. The putative TATA box, transcription initiation site (position 817), and poly(A) addition site (123 bp from the translation termination codon) are in boldface type and underlined. The derived amino sequence is indicated below the nucleotide sequence. The seven repeats of the decapeptide are underlined. Two putative N-glycosylation sites are starred.
The structure of the polypeptide encoded by this gene and the maturation signals of the polypeptide are similar to those of the polypeptides encoded by the precursor genes of the α-pheromone of Saccharomyces cerevisiae (21) and p-pheromone of Schizosaccharomyces pombe (8). Because of these similarities to the yeast pheromone precursor genes, this gene was named mating factor 1/1 (Mf1/1).
To determine if there are other copies of Mf1/1 in the genome of EP67, a probe that spans five copies of the decapeptide sequence encoded by Mf1/1 was used to probe Southern blots of genomic DNA from EP67 that had been digested with a number of different restriction endonucleases. A degenerate probe based on the nucleotide sequences of the decapeptide repeat was also synthesized. Both strategies failed to detect anything other than the copy of Mf1/1 described above.
Mating-type-specific expression of pheromone precursor genes.
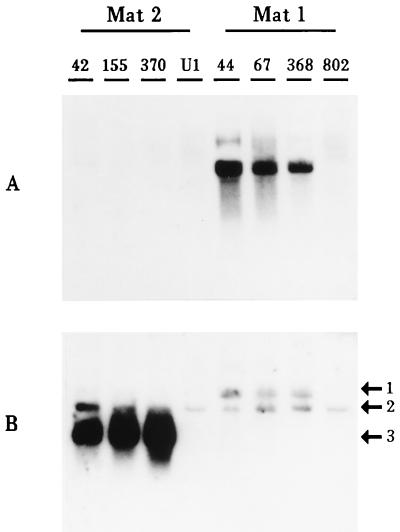
Bipolar, heterothallic fungi, such as C. parasitica, should display mating-type-specific expression of pheromone precursor genes. To determine if Mf2/1, Mf2/2, and Mf1/1 are expressed in a mating-type-specific manner, clones of these genes were used to probe Northern blots of total RNA from a number of different strains of both mating types of C. parasitica. The results showed that each of these genes is expressed in only one or the other mating type (Fig. 3): the clones of Mf2/1 and Mf2/2 detected mRNAs only in Mat-2 strains, and Mf1/1 mRNA was detected only in Mat-1 strains. In addition, we have tested more than 60 field strains of C. parasitica collected from Italy and Switzerland. In each case, the expression of the respective mRNAs correlated with the mating type determined by laboratory crosses (3a).
FIG. 3.
Mating-type-specific expression of pheromone precursor genes. A Northern blot of total RNA extracted from liquid cultures 5 days after inoculation of C. parasitica EP42, EP155, and EP370 (Mat-2) and EP44, EP67, and CP368 (Mat-1) is shown. Also included is RNA from two CHV1-infected strains: UEP1 (isogenic with EP155) and EP802 (isogenic with EP67). The lanes containing RNA from these strains are labeled with only the number designation for the strain or, in the case of UEP1, the label U1. (A) This blot was probed with a clone of Mf1/1. (B) The blot used in panel A was stripped and then probed with a clone of C. parasitica glyceraldehyde phosphate dehydrogenase and also with Mf2/1 and Mf2/2 clones. Arrow 1 shows the Mf1/1 transcript that remains after stripping, arrow 2 shows the Gpd transcript, and arrow 3 shows the Mf2/1 and Mf2/2 transcripts.
Nonexpressed pheromone precursor genes.
In S. cerevisiae and S. pombe, the structural genes for the pheromones and for their receptors are present in both mating types, although only one set of these genes is expressed in each mating type (20). To determine if copies of nonexpressed pheromone precursor genes are present in C. parasitica, Southern blots of genomic DNA from strains of both Mat-1 and Mat-2 mating types were probed with clones of each of the pheromone precursor genes. In each case, the genomic DNA was digested with a variety of restriction endonucleases. In all cases, the resulting hybridization patterns of the DNA from the two mating types were always the same. These results demonstrate that nonexpressed copies of Mf2/1 and Mf2/2 exist in Mat-1 strains and that a nonexpressed copy of Mf1/1 exists in Mat-2 strains. In addition to comparing restriction patterns in Southern blots, we cloned and sequenced the Mf2/2 gene from EP67, a Mat-1 strain. The 1.6 kb that was sequenced included 0.8 kb upstream of the transcription initiation site, the 0.6-kb coding region, and 0.2 kb downstream of the poly(A) addition site. The sequence from EP67 was identical to that of the Mf2/2 gene of EP155 (Mat-2).
Down-regulation of pheromone precursor gene expression in virus-infected strains.
Two virus-infected isogenic strains were used to determine the effects of CHV1 on pheromone gene expression. Northern blots showed that the pheromone precursor genes are significantly down-regulated in both virus-containing strains, EP802 and UEP1. Under identical conditions, these genes are highly expressed in the respective isogenic virus-free strains, EP67 and EP155 (Fig. 3). No transcripts of these genes could be detected on Northern blots of the virus-infected strains at any time during the normal growth cycle of the fungus in liquid culture (data not shown).
DISCUSSION
Mating systems of fungi provide excellent models for understanding developmental processes in simple, genetically tractable eukaryotic organisms. The yeasts S. cerevisiae and S. pombe, in particular, have served as the primary models for understanding mating-regulated development. Unfortunately, knowledge of the mating systems in the filamentous fungi has lagged behind that for the yeasts. The dimorphism of gametes and the multicellular structures involved in filamentous ascomycete fungal mating systems increase the complexity of their mating process compared with yeasts. Although mating-type loci have been cloned from a number of filamentous ascomycetes, pheromone precursor genes have not yet been found in this group of fungi (3). Additionally, pheromone precursor genes analogous to those encoding the α-factor of yeast have not yet been found in any filamentous fungus.
Sex pheromone precursor genes from C. parasitica.
Based on our studies, the three genes Mf1/1, Mf2/1, and Mf2/2 encode the sex pheromone precursors of C. parasitica. Our evidence in support of this conclusion is that (i) these genes are expressed in a mating-type-specific manner (i.e., Mf1/1 is expressed only by Mat-1 strains and Mf2/1 and Mf2/2 are expressed only by Mat-2 strains, even though both mating types of the fungus contain all three genes), (ii) the structure and posttranslational processing signals of the genes are conserved compared with the pheromone precursor genes of the yeasts, and (iii) a null mutation of Mf2/2 results in sexual sterility (27). Additionally, a synthetic peptide based on the Mf1/1 predicted pheromone sequence inhibits the germination of conidia (spermatia) in a mating-type-specific manner reminiscent of G1 arrest in yeast (10a). The names of the pheromone precursor genes were assigned according to the new Mat gene designations developed from sequence comparisons with other filamentous fungal mating-type loci (2). In this new mating-type designation, the old MatA and its synonym Mat1-1 become Mat-2 based on a conserved HMG domain found in the sequence of this mating locus; this brings the nomenclature of C. parasitica into agreement with that of other fungi containing HMG domains in their mating-type loci. The other mating-type locus is then Mat-1.
Structure of pheromone precursor genes.
A TATA box is present in all three pheromone precursor genes 40 to 53 bp upstream of the transcription initiation sites. All sequenced genes of C. parasitica also have transcription initiation sites immediately following a CT-rich region. The Mf2/1 and Mf2/2 transcription initiation sites have been determined by S1 analysis or primer extension analysis. The transcription initiation site of Mf1/1 has not been determined experimentally, but the gene sequence predicts that the A immediately following the CT-rich region is the initiation site. The 5′ end of the longest cDNA clone is 14 bp downstream from this A site.
The Mat-2-specific pheromone precursor genes each contain an identical 83-bp sequence containing the translation initiation signal and an ORF encoding a 23-amino-acid peptide. Otherwise, these two genes differ from each other in sequence. The transcripts of both genes contain ORFs other than this common one, but in each case the 83-bp ORF is the first one; it is not known if the other ORFs present on the transcripts of these genes are translated. The C-terminal sequence of the deduced 23-amino-acid peptide is the conserved CAAX prenylation signal that is found in a variety of different peptides, including some fungal pheromone precursors. In the few cases where fungal pheromones with such posttranslational signals have been isolated, farnesyl groups are present on the active peptide (3).
A comparison of the sequences of those fungal pheromone precursors with prenylation signals indicates that there is also a conserved asparagine within 9 to 12 amino acids of the terminal prenylation signal. This amino acid is predicted to be 11 amino acids from the prenylation signal in the Mat-2-specific pheromone of C. parasitica. In the yeasts S. cerevisiae and S. pombe, this asparagine may predict the N-terminal processing site of the pheromone. The pheromone of Ustilago maydis, however, either is processed at a different site or undergoes additional cleavage at the N terminus during the maturation process (19).
Although pheromone precursor genes have been found in a number of different fungi, particularly the basidiomycetes, there have been no previous reports that filamentous fungi have pheromone precursor genes equivalent to the α-factor pheromone precursor genes of S. cerevisiae. The Mf1/1 gene of C. parasitica, which is structurally similar to the yeast Mfα1 gene, contains an ORF that encodes 530 amino acids and has all of the features of the yeast prepro-α-factor gene of S. cerevisiae and prepro-p-factor gene of S. pombe.
The peptide encoded by Mf1/1 contains a hydrophobic N-terminal sequence of 28 amino acids. Starting from the sixth amino acid, an 8-amino-acid stretch (IIVAAVSV) has an average hydropathy (Kyte-and-Doolittle) index of 2.62. This number is characteristic of leader peptides involved in protein secretion. This putative 28-amino-acid leader sequence is terminated with a C terminus of the dibasic sequence KR, a signal for cleavage by the endopeptidase Kex2p. Previous studies of the peptide cryparin, secreted by C. parasitica, showed that its leader peptide was cleaved at a Kex2p site (28). In this case, the secreted peptide was sequenced, providing direct evidence for Kex2p processing by this fungus.
Within the product of the Mf1/1 ORF, there are seven copies of identical decapeptides with the sequence WCLFHGEGCW. These decapeptide sequences are surrounded by the signal motifs for the three specific proteases used in the S. cerevisiae α-factor maturation process. Six of the seven decapeptides are preceded by two to eight copies of the dipeptide XA or XP, a signal for cleavage by the dipeptidyl aminopeptidase Ste13p. These dipeptide repeats are preceded by the dibasic sequence KR, which is the Kex2p cleavage signal. Each of the decapeptides is followed by signals for processing by Kex2p and Kex1p. One decapeptide is preceded by a single basic amino acid; whether this indicates a difference in endoprotease specificity or whether it indicates that another protease with specificity for single basic amino acid sites may be involved in processing this propheromone is not known. The p-factor propheromone of S. pombe also has a structure that predicts a single basic amino acid cleavage signal site for propheromone N-terminal processing (8).
The conservation in gene structure of the α-factor precursor genes of S. cerevisiae and Mf1/1 of C. parasitica is clear, but there are also some differences. The leader sequence encoded by Mf1/1 has Kex2p processing signals, whereas Kex2p is not involved in cleavage of the leader sequence of the α-factor of S. cerevisiae. There are two genes encoding precursors of the α-factor in S. cerevisiae, but only one copy of the pheromone precursor gene is present in C. parasitica and S. pombe. The N-glycosylation sites of S. cerevisiae closely follow the leader sequence; however, in S. pombe and C. parasitica, these sites are distant from the leader.
Expression of pheromone precursor genes.
The three pheromone precursor genes are each detectable by Southern blot analysis in genomic DNA from both mating types. The presence of identical copies of the Mf2/2 gene in the two mating types was confirmed by sequencing. Northern blot analyses, however, showed that transcripts of the pheromone precursor genes are present only in a mating-type-specific manner; i.e., Mf1/1 is expressed only by Mat-1 strains. As in the ascomyceteous yeast, it is expected that pheromone expression in C. parasitica will be regulated by the mating-type locus.
Viral suppression of pheromone precursor gene expression.
When C. parasitica is infected by CHV1, the pheromone precursor gene transcripts cannot be detected by Northern blot analysis. Nuclear run-on studies of Mf2/1 and Mf2/2 transcription have shown that this lack of transcript detection in virus-infected strains can be attributed primarily to down-regulation of transcription rather than to turnover of transcripts (10). This effect of the virus on its host pheromone precursor genes is not mating-type specific; the pheromone precursor genes of both mating types are suppressed when the virus is present. These results suggest that a symptom of virus infection would be sterility of both mating-type parents, however, the symptom of virus infection is female sterility but male fertility. The most obvious reason for the male fertility is that a significant percentage of the conidia (spermatia) of infected strains are free of the virus (9). These conidia would thus act as normal, fertile male spermatia, even though they were derived from an infected strain. Fertility of female parents is a more complex process than male fertility, and so it is also conceivable that environmental factors could overcome the effects of the virus on spermatia or that virus infection has a greater effect on female parents than male ones under the natural conditions required for successful mating by this fungus.
The implication of these studies is that the virus is perturbing a normal mating-regulatory system of this fungus. The finding that there are significant similarities in structure between the pheromone precursor genes of C. parasitica and the yeasts suggests that features of the mating systems of S. cerevisiae and S. pombe have been conserved in the filamentous ascomycetes. Since so much is known about mating in these model systems, our search for understanding of the mechanism of virus perturbation of sporulation and mating of C. parasitica should be facilitated.
It has been hypothesized that CHV1 causes its range of symptoms by perturbing normal host signal transduction pathways (4, 11, 12). A G-protein α-subunit (the Cpg1 product) from C. parasitica, detectable by Western blot analysis in an uninfected strain of the fungus, was not detected by this method in an infected strain (5). Deletion mutants of the fungus were made by transformation, with the resultant phenotype of poor growth in culture, avirulence, and no sporulation (6). The conclusion of these authors was that the Cpg1 product plays a role in virus-induced symptoms. By this hypothesis, viral perturbation of mating would be a consequence of the proposed effect of the virus on Cpg1 expression.
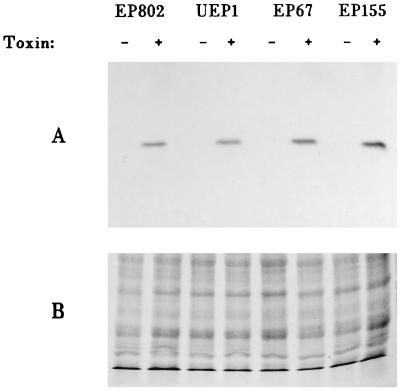
To determine if CHV1 repression of the pheromone precursor genes coincides with a reduction of Cpg1 expression, we chose to measure the relative levels of the Cpg1 product by using the susceptibility of Gαi subunits to ADP-ribosylation by pertussis toxin. Similar to Neurospora crassa gna1 (23), Cpg1 contains the consensus sequences for ADP-ribosylation by pertussis toxin. This is a specific and sensitive method for detecting some members of the Gi family of Gα subunits. Our results (Fig. 4) showed that only a single membrane protein was labelled by pertussis toxin, as was reported for N. crassa (23). This N. crassa protein has a predicted 98% amino acid identity to the Cpg1 gene product of C. parasitica; membranes from N. crassa were used to demonstrate that the protein detected in C. parasitica was of the same size as that detected in N. crassa (data not shown). These results suggest that we were detecting the Cpg1 product by this method. Unlike the results reported by Choi et al. (5), we found no significant difference between pertussis toxin labeling of the Cpg1 product between infected and uninfected strains of the virus after 2 and 5 days of growth; under the same conditions, the pheromone precursor gene transcripts could not be detected in virus-infected strains. Viral repression of pheromone precursor gene expression thus is not likely to be the result of a reduction of Cpg1 gene product concentration. Our results do not negate the possibility that the virus affects G-protein function. The debilitated phenotypes of the Cpg1 mutants (5, 6), however, are not typical of those caused by the virus; unlike the mutant, the virus has no effect on growth of the fungus in culture. It is thus likely that if the virus affects the G protein, the effect would be more subtle than that reported (5, 6).
FIG. 4.
Detection of the Gαi subunit of C. parasitica by pertussis toxin ADP-ribosylation. Crude plasma membranes were extracted from virulent strains EP155 and EP67 and their respective isogenic CHV1-infected strains UEP1 and EP802 and labeled with pertussis toxin by the method of Turner and Borkovich (23). (A) Protein labeled with [32P]NAD in the presence (+) or absence (−) of pertussis toxin. (B) Polyacrylamide gel of the same membrane preparation as in panel but stained with Coomassie blue. A total of threefold more protein was loaded in each lane of panel B than in each lane of panel A.
Implications of viral effects on fungal mating.
Viruses of filamentous fungi are not infectious; they can be transferred from strain to strain only by hyphal anastomoses. Vegetative compatibility group (vcg) genes function to prevent cytoplasmic transfer of fungal viruses. Only individuals with identical or compatible vcg alleles are able to establish cytoplasmic bridges of anastomosing hyphae, which serve as the conduits for movement of fungal viruses. Sexual recombination of the multilocus vcg alleles ensures that the progeny are phenotypically diverse with respect to compatibility type. Diversity of compatibility phenotypes in populations of the fungus are thought to be the primary defense of fungi against viruses. By perturbing the ability of the host to reassort these alleles through the sexual cycle, the virus impairs the long-term ability of the host to limit viral movement. Since its introduction into Europe, most of the populations of the fungus on that continent have become naturally infected with the virus, resulting in a biological control of the disease. In North America, however, virus-containing strains are rare. A number of different studies have reported that vcg diversity in populations of C. parasitica from North America is much greater than in those from Europe (14). The repression of the fungal sexual cycle by viral infection may be responsible for this difference in phenotypic diversity of vcg types between European and North American populations of the fungus.
ACKNOWLEDGMENTS
This work was supported by grants from the USDA National Research Initiative (90-37290-5671) and the National Science Foundation (MCB-9205818).
The technical assistance of Y.-B. Sun, D.-G. Tang, and G. Meyer is greatly appreciated, as is the advice and gift of strains from Kathy Borkovich.
REFERENCES
- 1.Anagnostakis S L. Biological control of chestnut blight. Science. 1982;215:466–471. doi: 10.1126/science.215.4532.466. [DOI] [PubMed] [Google Scholar]
- 2.Arie T, Christiansen S K, Yoder O C, Turgeon B G. Efficient cloning of ascomycete mating type genes by PCR amplification of the conserved MAT HMG box. Fungal Genet Biol. 1997;21:118–130. [PubMed] [Google Scholar]
- 3.Bolker M, Kahmann R. Sexual pheromones and mating responses in fungi. Plant Cell. 1993;5:1461–1469. doi: 10.1105/tpc.5.10.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Carparelli, A., L. Zhang, D. Rigling, E. Gobbi, and N. K. Van Alfen. Unpublished data.
- 4.Chen B, Gao S, Choi G H, Nuss D L. Extensive alteration of fungal gene transcript accumulation and elevation of G-protein-regulated cAMP levels by a virulence-attenuating hypovirus. Proc Natl Acad Sci USA. 1996;93:7996–8000. doi: 10.1073/pnas.93.15.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi G H, Chen B, Nuss D L. Virus-mediated or transgenic suppression of a G-protein α subunit and attenuation of fungal virulence. Proc Natl Acad Sci USA. 1995;92:305–309. doi: 10.1073/pnas.92.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao S, Nuss D L. Distinct roles for two G protein α subunits in fungal virulence, morphology, and reproduction revealed by targeted gene disruption. Proc Natl Acad Sci USA. 1996;93:14122–14127. doi: 10.1073/pnas.93.24.14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heiniger U, Rigling D. Biological control of chestnut blight in Europe. Annu Rev Phytopathol. 1994;32:581–99. [Google Scholar]
- 8.Imai Y, Yamamoto M. The fission yeast mating pheromone P-factor: its molecular structure, gene structure, and ability to induce gene expression and G1 arrest in the mating partner. Genes Dev. 1994;8:328–338. doi: 10.1101/gad.8.3.328. [DOI] [PubMed] [Google Scholar]
- 9.Jaynes R A, Elliston J E. Hypovirulent isolates of Endothia parasitica associated with large American chestnut trees. Plant Dis. 1982;66:769–772. [Google Scholar]
- 10.Kazmierczak P, Pfeiffer P, Zhang L, Van Alfen N K. Transcriptional repression of specific host genes by the mycovirus Cryphonectria hypovirus 1. J Virol. 1996;70:1137–1142. doi: 10.1128/jvi.70.2.1137-1142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a.Kazmierczak, P., P. McCabe, and N. K. Van Alfen. Unpublished data.
- 11.Larson T G, Choi G H, Nuss D L. Regulatory pathways governing modulation of fungal gene expression by a virulence-attenuating mycovirus. EMBO J. 1992;11:4539–4548. doi: 10.1002/j.1460-2075.1992.tb05555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larson T G, Nuss D L. Altered transcriptional response to nutrient availability in hypovirus-infected chestnut blight fungus. EMBO J. 1994;13:5616–5623. doi: 10.1002/j.1460-2075.1994.tb06899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacDonald W L, Fulbright D W. Biological control of chestnut blight: use and limitations of transmissible hypovirulence. Plant Dis. 1991;75:656–661. [Google Scholar]
- 14.Milgroom M G. Population biology of the chestnut blight fungus, Cryphonectria parasitica. Can J Bot. 1995;73:S311–S319. [Google Scholar]
- 15.Moore T D E, Edman J C. The α-mating type locus of Cryptococcus neoformans contains a peptide pheromone gene. Mol Cell Biol. 1993;13:1962–1970. doi: 10.1128/mcb.13.3.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nuss D L. Biological control of chestnut blight: an example of virus-mediated attenuation of fungal pathogenesis. Microbiol Rev. 1992;56:561–576. doi: 10.1128/mr.56.4.561-576.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Powell W A, Van Alfen N K. Differential accumulation of poly(A)+ RNA between virulent and double-stranded RNA-induced hypovirulent strains of Cryphonectria (Endothia) parasitica. Mol Cell Biol. 1987;7:3688–3693. doi: 10.1128/mcb.7.10.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rigling D, Van Alfen N K. Regulation of laccase biosynthesis in the plant-pathogenic fungus Cryphonectria parasitica by double-stranded RNA. J Bacteriol. 1991;173:8000–8003. doi: 10.1128/jb.173.24.8000-8003.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spellig T, Bolker M, Lottspeich F, Frank R W, Kahmann R. Pheromones trigger filamentous growth in Ustilago maydis. EMBO J. 1994;13:1620–1627. doi: 10.1002/j.1460-2075.1994.tb06425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sprague G F., Jr Combinatorial associations of regulatory proteins and the control of cell type in yeast. Adv Genet. 1990;27:33–62. doi: 10.1016/s0065-2660(08)60023-1. [DOI] [PubMed] [Google Scholar]
- 21.Sprague G F, Jr, Thorner J W. Pheromone response and signal transduction during the mating process of Saccharomyces cerevisiae. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 657–744. [Google Scholar]
- 22.Sweigard J A, Orbach M J, Valent B, Chumley F G. A miniprep procedure for isolating genomic DNA from Magnaporthe grisea. Fungal Genet Newsl. 1990;31:44. [Google Scholar]
- 23.Turner G E, Borkovich K A. Identification of a G protein α subunit from Neurospora crassa that is a member of the Gi family. J Biol Chem. 1993;268:14805–14811. [PubMed] [Google Scholar]
- 24.Van Alfen N K. Hypovirulence of Endothia (Cryphnectria) parasitica and Rhizoctonia solani. In: Buck K W, editor. Fungal virology. Boca Raton, Fla: CRC Press, Inc.; 1986. pp. 143–162. [Google Scholar]
- 25.Van Alfen N K, Jaynes R A, Anagnostakis S L, Day P R. Chestnut blight: biological control by transmissible hypovirulence in Endothia parasitica. Science. 1975;189:890–891. doi: 10.1126/science.189.4206.890. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, Brown D D. A gene expression screen. Proc Natl Acad Sci USA. 1991;88:11505–11509. doi: 10.1073/pnas.88.24.11505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L, Churchill A C L, Kazmierczak P, Kim D, Van Alfen N K. Hypovirulence-associated traits, induced by a mycovirus of Cryphonectria parasitica, mimicked by target inactivation of a host gene. Mol Cell Biol. 1993;13:7782–7793. doi: 10.1128/mcb.13.12.7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, Villalon D, Sun Y, Kazmierczak P, Van Alfen N K. Viral-associated down-regulation of the gene encoding cryparin, an abundant hydrophobic protein from the filamentous fungus, Cryphonectria parasitica. Gene. 1994;139:59–64. doi: 10.1016/0378-1119(94)90523-1. [DOI] [PubMed] [Google Scholar]