Abstract
The transient inactivation of gene regulatory proteins by their sequestration to the cytoplasmic membrane in response to cognate signals is an increasingly recognized mechanism of gene regulation in bacteria. It remained to be shown, however, whether tethering to the membrane per se could be responsible for inactivation, i.e. whether such relocation leads to a spatial separation from the chromosome that results in inactivity or whether other mechanisms are involved. We, therefore, investigated the activity of Lac repressor artificially attached to the Escherichia coli cytoplasmic membrane. We demonstrate that this chimeric protein perfectly represses transcription initiated at the tac operator–promoter present on a plasmid and even in the chromosome. Moreover, this repression is inducible as normal. The data suggest that proteins localized to the inner face of the cytoplasmic membrane in principle have unrestricted access to the chromosome. Thus sequestration to the membrane in terms of physical separation from the chromosome cannot account alone for the inactivation of regulatory proteins. Other mechanisms, like induction of a conformational change or masking of binding domains are required additionally.
INTRODUCTION
In the past decade a novel mode of activity control of regulatory proteins has been revealed in bacteria. In this mode, transcription factors shuttle between their binding sites on the chromosome and the inner face of the cytoplasmic membrane, where they are held inactive. In response to a specific stimulus, these regulators are again released from the membrane and can bind back to their target sites on the chromosome [for a recent review see (1)].
The first example of such a mechanism was the regulation of the proline utilization (put) operon by PutA in Salmonella typhimurium (2). PutA has been shown to be a bi-functional protein. In the absence of proline, PutA remains in the cytoplasm, where it binds to the put operators and represses put gene expression. When an excess of proline is present, proline binds to PutA. Binding of proline, combined with reduction of the PutA protein by the membrane-associated electron transport chain, leads to release of PutA from its operators and its association with the membrane. From this location it catalyzes the degradation of proline to glutamate.
Another elegant system where membrane sequestration of a repressor leads to induction of its assigned genes is represented by the mlc regulon in Escherichia coli. The repressor Mlc controls expression of several genes involved in carbohydrate utilization [for reviews see (1,3)]. One of them is ptsG encoding the EIIGlc transport protein of the phosphotransferase system (PTS). Mlc is in turn regulated by the transport activity of EIIGlc. In the absence of substrate for EIIGlc, Mlc binds to its operator sites and represses transcription. When EIIGlc is engaged in transport, it binds Mlc, thereby sequestering it to the membrane, which leads to concomitant de-repression of the Mlc-controlled genes. It has recently been shown that an artificial attachment of Mlc to the cytoplasmic membrane by fusion to the LacY permease causes de-repression of Mlc-controlled genes even in the absence of EIIGlc, suggesting that tethering of Mlc to the membrane alone, rather than binding to EIIGlc, may lead to its inactivation as a repressor (4). Not only repressors but also activator proteins were shown to be sequestered to the membrane in response to their cognate signals and concomitantly kept inactive. These include MalT of E.coli (1,5), TraR of Agrobacterium tumefaciens (6) and NifL of Klebsiella pneumoniae, in this case an inhibitor of a gene activator protein (7). In most of these cases the molecular reason for inactivity of the membrane-localized regulatory proteins is not clear, but it has been proposed that a spatial gap may exist between the membrane and the chromosome with the consequence that the transcription factors have to ‘shuttle’ back to the nucleoid to gain access to their binding sites in the chromosome (1,7). However, in one case, the DNA binding gene activator protein ToxR of Vibrio cholerae, it has been demonstrated that this protein is naturally membrane-bound (8). In addition, we have previously shown that the transcriptional antiterminator protein BglG from E.coli that is regulated in its activity by a membrane spanning sugar transport protein can be artificially tethered to the membrane without impairing its function. The membrane-bound BglG protein is capable of interacting with its chromosomally encoded nascent RNA target sequence, catalyzing transcriptional read-through with virtually no difference to wild-type BglG, and it is perfectly regulated in its activity (9). These observations suggested that in principle there should be no mechanistic necessity for a regulatory protein like BglG to leave the place of its own regulation, i.e. the membrane, in order to interact with its target sequences.
However, for activator proteins like ToxR or BglG transient interactions with their cognate target sequences could be sufficient to stimulate gene expression. Furthermore, actively transcribed DNA is believed to often reside close to the membrane, especially when co-translational insertion of proteins into the membrane takes place. For efficient transcriptional repression, however, occupation of the operator binding site by the repressor protein for most of the time is required and no or little transcription should occur that could redirect the operator DNA to the membrane. It, therefore, was an open question whether repressor proteins could exist or be engineered that function from the cytoplasmic membrane. Here, we addressed this question and artificially anchored the E.coli Lac repressor to the cytoplasmic membrane. We demonstrate that this membrane-attached LacI protein perfectly represses transcription initiation at a chromosomally located tac operator–promoter and that it can be released from repression by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) as inducer. Our experiments suggest that membrane-attached and soluble Lac repressor proteins find their operator site in the chromosome with similar efficiency. Together, our results suggest that membrane sequestration of a repressor protein per se cannot be the reason for its inactivation. Other mechanisms are required. The concept that membrane sequestration provides a spatial separation from and prevents access to the chromosome has thus to be revised.
MATERIALS AND METHODS
Plasmids, strains and growth conditions
The strains and plasmids used and their relevant characteristics are listed in Table 1. For DNA cloning, standard procedures were followed (10). The correct nucleotide sequences of PCR-derived fragments in the various plasmids were verified by DNA sequencing. Bacteria were routinely grown in 2YT broth supplemented with the appropriate antibiotics (kanamycin at 30 μg/ml, chloramphenicol at 10 μg/ml and ampicillin at 100 μg/ml, if not otherwise indicated).
Table 1.
Strains and plasmids used in this study
| Genotype or relevant structures | Source or reference | |
|---|---|---|
| Strains | ||
| R1279 | CSH50 Δ(pho-bgl)201 Δ(lac-pro) ara thi | (16) |
| R2171 | As R1279 but attB::[tacOP-lacZ, bla]; RBS of phage T7gene10 in front of lacZ | pFDX3552→R1279 |
| Plasmids | ||
| M13mp18 | M13 phage cloning vector | (28) |
| pFDX500 | lacI, neo, ori p15A | (11) |
| pFDX3225 | φ(galK′-bglG) under control of Ptac, tet, ori p15A | (12) |
| pFDX3401 | λ int under control of λPR, λcI857 (cat; ori pSC101-repTS) | (12) |
| pFDX3551 | pLDR10 derivative for the insertion of a tacOP-lacZ cassette into the attB site; RBS of gene bglG in front of lacZ, bla, ori pMB1 | This study |
| pFDX3552 | As pFDX3551, but RBS of phage T7gene10 in front of lacZ | This study |
| pFDX4151 | As pFDX500, but with a NdeI site overlapping with the lacI ATG start codon | This study |
| pFDX4152 | As pFDX4151, but lacI fused to M13gene8 | This study |
| pFDX4153 | As pFDX4151, but lacI fused to λcI′ (codons 96–132) | This study |
| pFDX4154 | As pFDX4151, but lacI fused to galK′ (codons 1–32) | This study |
| pFDX4155 | As pFDX4151, but lacI fused to galK′-λcI′ | This study |
| pFDX4156 | As pFDX4151, but lacI fused to M13gene8-λcI′ | This study |
| pFDX4157 | As pFDX4152, but RBS of phage T7gene10 in front of φ(M13gene8-lacI) | This study |
| pFDX4158 | As pFDX4154, but RBS of phage T7gene10 in front of φ(galK′-lacI) | This study |
| pFDX4159 | As pFDX4155, but RBS of phage T7gene10 in front of φ(galK′-λcI′-lacI) | This study |
| pFDX4160 | As pFDX4156, but RBS of phage T7gene10 in front of φ(M13gene8-λcI′-lacI) | This study |
| pFDX4165 | As pFDX4151, but RBS of phage T7gene10 in front of lacI | This study |
| pLDR10 | Multiple cloning site, λattP, bla, cat, ori pMB1 | (14) |
Construction of chimeric lacI fusion genes
Plasmid pFDX4151 was the ancestor of plasmids carrying the different chimeric lacI derivatives. It was constructed by a three fragment ligation combining the ClaI–PstI- and the Psp1406I–ClaI-fragments of plasmid pFDX500 (11) with a PCR fragment which was amplified with primers #631 (5′-CGTATAACGTTACTGGTTTCATATGCACCACCCTGAATTG) and #632 (5′-GCCGGGAAGCTAGAG) from plasmid pFDX500 as template and digested with Psp1406I and PstI. This construction changed the start codon of lacI from GTG to ATG and simultaneously created an NdeI site overlapping with the start codon that allowed the insertion of foreign DNAs in-frame with lacI. For construction of the M13gene8-lacI fusion gene, the M13gene8 was amplified by using primers #293 (5′-GGGATTTAAACATATGAAAAAGTCTTTAGTCCTCAAAGC) and #294 (5′-TTTAAAGTACTTGCTTTCGAGGTGAATTTCTTAAA) and plasmid M13mp18 as template. The PCR fragment was digested at the NdeI and Csp6I sites within the primers and inserted into the NdeI site of plasmid pFDX4151, resulting in plasmid pFDX4152. For the construction of plasmid pFDX4154 carrying the galK′-lacI fusion gene, the 5′-part of galK encompassing the first 32 codons was amplified by using the primers #636 (5′-GATATACATATGGCTAG) and #637 (5′-CACTGCATTAATAGCAAGGACAGGC) and plasmid pFDX3225 (12) as template. The PCR fragment was digested at the NdeI and AseI sites within the primers and inserted into the NdeI site of plasmid pFDX4151. Plasmids pFDX4155 and pFDX4156 carrying the galK′-λcI′-lacI and the gene8-λcI′-lacI fusion genes, respectively, were constructed in two steps. First, codons 96–132 of the λcI gene encoding a flexible linker were amplified by using primers #634 (5′-AAAGAGTATCATATGGACCCGTCACTT) and #635 (5′-CTCCTCATTAATGCTTACCCATCTCTC) and plasmid λdv1 (13) as template. The PCR fragment was digested at the NdeI and MseI sites within the primers and inserted into the NdeI site of plasmid pFDX4151 resulting in the intermediate construct pFDX4153 carrying a λcI′-lacI fusion gene. Next, the digested PCR fragments from above, encompassing the 5′-part of galK and gene8, respectively, were inserted into the NdeI site of plasmid pFDX4153, resulting in plasmids pFDX4155 and pFDX4156. In order to replace the ribosomal binding site (RBS) in front of lacI and its various chimeric derivatives, the sequence upstream was amplified by using primers #646 (5′-GTTGCCATTGCTGCAGGC) and #647 (5′-GCATATCGCATATGTATATCTCCTTCTTGACTCTCTTCCGGGCGCTAT; contains the RBS of T7gene10) and plasmid pFDX4152 as template. The PCR fragment was digested at the PstI and NdeI sites within the primers and used to replace the corresponding fragment in plasmids pFDX4151, pFDX4152, pFDX4154, pFDX4155 and pFDX4156 resulting in plasmids pFDX4165, pFDX4157, pFDX4158, pFDX4159 and pFDX4160, respectively.
Construction of a tacOP-lacZ reporter cassette and integration into the chromosomal attB site
Plasmid pFDX3551 carries a tacOP-lacZ cassette with the relatively weak RBS sequence of the bglG gene in front of lacZ. It was constructed by replacement of the HindIII–ClaI fragment of plasmid pFDX3157 (12) encompassing gene bglG by the HindIII–ClaI fragment of plasmid pFDX3549 (12) encompassing the lacZ gene. Plasmid pFDX3552 is isogenic with plasmid pFDX3551 but carries the strong RBS sequence of gene10 of phage T7 in front of lacI. Its construction involved several steps, and details are available on request. Integration of the tacOP-lacZ cassette of plasmid pFDX3552 by site-specific recombination into the bacteriophage λ attachment site attB, was achieved with the integration system provided by Diederich et al. (14) as described previously (12).
Determination of β-galactosidase activities
Enzyme assays were performed according to the method of Miller (15) as described previously (16). The bacteria were grown in M9 minimal medium supplemented with 1% (w/v) glycerol, 20 μg/ml proline, 1 μg/ml thiamine, 0.66% (w/v) casamino acids and the appropriate antibiotics. If not otherwise indicated, overnight cultures were inoculated to an OD600 of 0.15 and grown for 1 h at 37°C. Then, 1 mM IPTG was added where necessary and growth was continued for 2 h before the cultures were harvested. Samples were assayed in triplicate and each experiment was repeated three times using independent transformants. Enzyme activities are expressed in Miller units.
In vivo pulse-chase [35S]methionine labeling of proteins
The pulse-chase labeling of proteins with [35S]methionine was carried out as described previously (9).
Subcellular fractionation of [35S]methionine-labeled proteins
Separation of [35S]methionine-labeled soluble and membrane proteins was performed by sucrose gradient centrifugation as described previously (9).
RESULTS
Construction, stability and subcellular localization of chimeric Lac repressor proteins
It has previously been shown that the 50 amino acid residue major coat protein of bacteriophage M13 encoded by M13gene8 is a suitable tool to stably anchor a protein to the cytoplasmic membrane. For this purpose the protein to be anchored is fused to the C-terminus of the coat protein (9,17). We made use of this tool to attach the Lac repressor to the membrane. To this end, a translational fusion of the lacI gene encoding Lac repressor to the exact 3′ end of gene8 was constructed and placed on a low copy plasmid under control of the lacI promoter. In order to determine whether an N-terminal extension would generally impair the function of Lac repressor, a control fusion comprising the first 32 codons of the galK gene fused to lacI was also constructed. The galK gene encodes galactokinase, a soluble protein (9).
The DNA binding domain of Lac repressor encompasses the first 60 N-terminal amino acid residues. It thus appeared conceivable that the tight tethering of this domain to the membrane could negatively interfere with its DNA binding ability. We, therefore, constructed additional fusion genes in which codons 96–132 of the λcI repressor gene encoding a flexible linker (18) were placed between the lacI- and the gene8- or galK-part of the respective fusion genes.
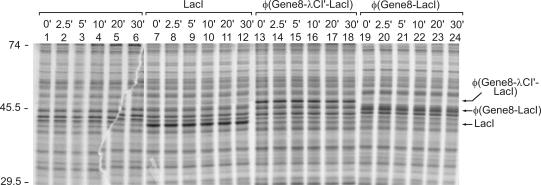
In order to allow investigation by gel electrophoresis of whether the fusion proteins are stable in vivo and indeed attached to the membrane, a second set of isogenic vectors was constructed in which the RBS of lacI was replaced by the stronger RBS of gene10 of phageT7, leading to a higher expression of the different Lac repressor derivatives. Transformants carrying the respective expression plasmids were analyzed by [35S]methionine labeling and subsequent SDS–PAGE (Figure 1). As a control, the untransformed strain was also employed. Labeled protein bands became visible, which were exclusively present in the extracts of the respective transformants expressing LacI (39.6 kDa), the φ(Gene8–λCI–LacI) fusion (49.6 kDa) or the φ(Gene8–LacI) fusion (45.3 kDa). The positions of these proteins on the gel fitted well with the calculated molecular weight of the respective LacI derivatives, indicating that they are properly expressed (Figure 1, compare lanes 1, 7, 13 and 19; data not shown for the GalK–LacI and the GalK–λCI′–LacI fusion proteins).
Figure 1.
Synthesis and stability of LacI derivatives. Strain R1279 was transformed with plasmid pFDX4160 carrying the φ(gene8-λcI′-lacI) fusion gene (lanes 13–18) or with plasmid pFDX4157 carrying the φ(gene8-lacI) fusion gene (lanes 19–24) under control of the wild-type lacI promoter, respectively. For comparison, a transformant with the isogenic plasmid pFDX4165 carrying wild-type lacI (lanes 7–12) as well as the untransformed strain (lanes 1–6) were also employed. Aliquots of the cultures were pulse-labeled with [35S]methionine and subsequently chased with unlabeled methionine for the times indicated in the figure. Proteins were separated by SDS–PAGE and analyzed by autoradiography and phospho-imager analysis. The positions of molecular weight standards are given on the left (in kDa).
In order to test the stability of the different LacI derivatives, the labeled cultures were chased with unlabeled methionine for different times prior to protein extraction and SDS–PAGE. All LacI derivatives were stable over the tested period of 30 min, as was the wild-type LacI protein (Figure 1, compare lanes 14–18 and 20–24 with lanes 8–12; data not shown for the GalK–LacI and the GalK–λCI′–LacI fusion proteins).
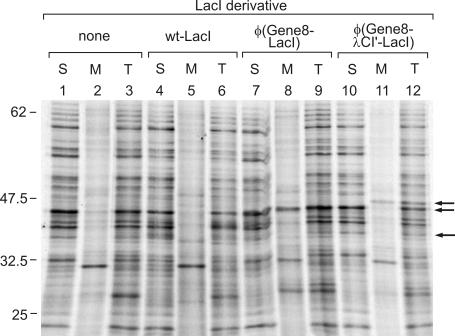
Next, we wanted to verify that the Gene8 fusion proteins were indeed tightly attached to the cytoplasmic membrane. We, therefore, performed a subcellular fractionation of [35S]methionine-labeled transformants harboring the wild-type lacI gene or the φ(gene8-lacI) and the φ(gene8-λcI′-lacI) fusion genes, respectively, by sucrose gradient centrifugation, as described previously (9). The untransformed strain served as a control. The soluble and membrane fractions were analyzed by SDS–PAGE along with the unfractionated samples (Figure 2). It can be seen that both the φ(Gene8-LacI) as well as the φ(Gene8–λCI′–LacI) fusion proteins were present in the membrane fraction (Figure 2, lanes 8 and 11) but not in appreciable amounts in the soluble fraction (Figure 2, lanes 7 and 10). The wild-type LacI protein, on the other hand, was only visible in the soluble fraction, as expected (Figure 2, lane 4). Its amount was, however, lower than that detected with the [35S]methionine-labeled whole cell extracts analyzed in Figure 1. Since the amount of LacI was similarly reduced in case of the unfractionated control sample (Figure 2, lane 6), it is feasible that this reduction was due to the TCA precipitation step which was performed with all samples analyzed in Figure 2 but not with those presented in Figure 1. This difference has, however, no impact on the main conclusion that both, the φ(Gene8–LacI) as well as the φ(Gene8–λCI′–LacI) fusion protein are stable and are anchored to the cytoplasmic membrane.
Figure 2.
Subcellular fractionation demonstrating that both, the φ(Gene8–LacI) as well as the φ(Gene8–λCI′–LacI) fusion proteins, are membrane-anchored. The untransformed strain R1279 as well as its various transformants (Figure 1) were labeled with [35S]methionine. Subsequently the cells were gently lysed, and half of the lysates were subjected to sucrose gradient centrifugation to separate soluble (S) and membrane (M) proteins. The remaining halves were kept on ice during the fractionation procedure and served as samples representing total protein (T). The samples were separated by SDS–PAGE, followed by autoradiography and phospho-imager analysis. Lanes 1–3, strain R1279 (untransformed); lanes 4–6, R1279/pFDX4165 (wild-type lacI); lanes 7–9, R1279/pFDX4157 [φ(gene8-lacI)]; lanes 10–12, R1279/pFDX4160 [φ(gene8-λcI′-lacI)]. The positions of molecular weight standards are given on the left (in kDa), and the positions of the respective LacI derivatives are indicated by arrows on the right.
Membrane-attached Lac repressor is active in transcriptional repression
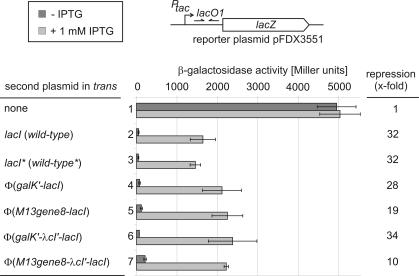
The important question was whether or not the membrane-attached LacI proteins are functional in repression and thus in operator binding, and if so, whether or not repression by these proteins could be relieved by the addition of IPTG, a gratuitous inducer for wild-type Lac repressor. To this end, strain R1279 in which the chromosomal lac operon is deleted, was co-transformed with the various plasmids carrying the different lacI derivatives under control of the wild-type lacI RBS and in addition with reporter plasmid pFDX3551. This plasmid carries a lacZ reporter gene under control of the tac promoter which is controlled by the main lac operator O1 that overlaps with its transcriptional start site. When strain R1279 was transformed with this reporter plasmid alone, high β-galactosidase activities of ∼5000 U were produced which were unaffected by the absence or presence of IPTG, as expected (Figure 3, bars 1). The additional presence of plasmid pFDX500 delivering wild-type Lac repressor led to the synthesis of 51 U in the absence and of 1640 U in the presence of IPTG. Thus, the expression of lacZ was repressed 32-fold by wild-type Lac repressor in this reporter system (Figure 3, bars 2). Nearly identical values were obtained when the LacI delivering plasmid pFDX4151 carrying an NdeI site that overlaps with the lacI start codon was present (Figure 3, bars 3). Thus, the introduction of this site and the exchange of the GTG start codon against ATG had no appreciable effect on repression. The expression of the φ(GalK–LacI) fusion protein yielded activities which were similar to those obtained with the wild-type LacI protein (Figure 3, compare bars 3 and 4), indicating that the fusion of a foreign polypeptide to the N-terminus of LacI may in general not have deleterious effects on repression and thus on binding of the Lac repressor to its operator. Expression of the membrane-anchored φ(Gene8–LacI) fusion protein led to the synthesis of 2250 U of β-galactosidase activities in the presence of IPTG which were reduced to 120 U in its absence. Thus, the membrane-anchored φ(Gene8–LacI) protein was indeed able to efficiently repress expression of the lacZ reporter gene. Moreover, this repression could be relieved 19-fold by the addition of IPTG (Figure 3, bars 5). The presence of the flexible protein linker of the λCI protein had no effect on the repression activity of the φ(GalK–λCI′–LacI) protein when compared with the φ(GalK–LacI) protein without this linker (Figure 3, compare bars 4 and 6). In contrast, the φ(Gene8–λCI′–LacI) protein was less active in transcriptional repression when compared with the φ(Gene8–LacI) fusion protein lacking the λCI-linker. Therefore, induction was only 10-fold (Figure 3, compare bars 7 and 5). Obviously, the λCI-linker negatively interfered with the operator binding ability of this membrane-attached Lac repressor protein.
Figure 3.
The membrane-anchored LacI fusion proteins regulate expression of a plasmid encoded tacOP-lacZ reporter cassette in an inducer-dependent manner. Strain R1279 was transformed with reporter plasmid pFDX3551 which carries the tacOP-lacZ cassette and in addition with one of various plasmids delivering LacI or its fusion derivatives. β-Galactosidase activities of these transformants were determined in the absence or presence for 2 h of IPTG, as indicated. Bars 1: R1279/pFDX3551 (single transformant); bars 2: R1279/pFDX3551/pFDX500 (wild-type lacI); bars 3: R1279/pFDX3551/pFDX4151 (wild-type lacI with ATG-start codon and NdeI site); bars 4: R1279/pFDX3551/pFDX4154 [φ(galK-lacI)]; bars 5: R1279/pFDX3551/pFDX4152 [φ(gene8-lacI)]; bars 6: R1279/pFDX3551/pFDX4155 [φ(galK-λcI′-lacI)]; and bars 7: R1279/pFDX3551/pFDX4156 [φ(gene8-λcI′-lacI)]. The reason for the high enzyme activities seen in the absence of a Lac repressor delivering plasmid (bars 1) as compared with those obtained in its presence when IPTG as inducer was additionally present is unclear. Similar high activities were seen when the vector plasmid without a lacI gene was present (data not shown). One reason could be that induction by IPTG is incomplete.
The membrane-attached LacI proteins regulate expression of a chromosomal gene
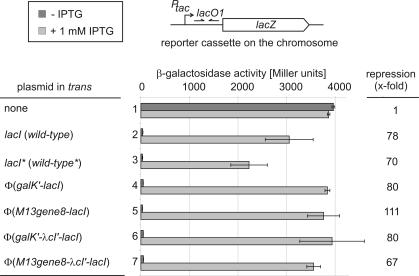
The experiments presented above (Figure 3) demonstrate that the membrane-attached Lac repressor proteins are indeed active in transcriptional repression. However, in these tests the lacO1 target site was encoded on a plasmid. Although plasmids appear to be clustered and partitioned within a cell (19) they may be more readily accessible by membrane-localized DNA binding proteins than sequences within the nucleoid. The question was thus still open whether the membrane-located Lac repressor protein would have free access and tightly bind to a chromosomally located operator site. In order to address this question, the tacOP-lacZ reporter cassette present in plasmid pFDX3551 was integrated into the λ attB site in the chromosome by site-specific recombination. However, this strain yielded very low β-galactosidase activities [<100 Miller units already in the absence of Lac repressor (data not shown)], making it difficult to precisely compare activities of the various Lac repressor derivatives. Therefore, the isogenic strain R2171 was constructed in which the more efficient ribosome binding site of gene10 of phage T7 was placed in front of lacZ. This strain yielded ∼4000 U of β-galactosidase, a convenient activity for the intended experiments. As expected, the presence of IPTG had no effect on this activity (Figure 4, bars 1). Transformants of this strain harboring plasmids pFDX500 and pFDX4151, respectively, that both encode the wild-type Lac repressor yielded 30–40 U of β-galactosidase. These activities increased 70–80-fold in the presence of IPTG (Figure 4, bars 2 and 3), verifying that expression of the chromosomal lacZ reporter cassette is perfectly regulated by the wild-type repressor in an inducer-dependent manner. Very similar results were obtained in the presence of plasmids pFDX4154 and pFDX4155 encoding the φ(GalK–LacI) and the φ(GalK–λCI′–LacI) fusion proteins, respectively, demonstrating once again that this fusion of a foreign peptide to the N-terminus of LacI has no negative effect on Lac repressor function (Figure 4, bars 4 and 6). When strain R2171 was transformed with plasmid pFDX4152 that encodes the membrane-anchored φ(Gene8-LacI) chimeric repressor, the transformants synthesized only 34 U of β-galactosidase. Thus, the membrane-anchored repressor is able to repress the tac promoter present in the chromosome as efficiently as the wild-type repressor. Addition of IPTG resulted in 3760 U or a 111-fold induction (Figure 4, bars 5). Thus, the membrane-attached LacI derivative is capable of regulating the expression of the chromosomal lacZ reporter gene as efficiently as the wild-type repressor. Finally, the membrane-anchored φ(Gene8–λCI–LacI) fusion protein delivered by plasmid pFDX4156 was tested. Presence of this plasmid led to 53 U of β-galactosidase in the absence of IPTG and to 3560 U in its presence, which is a 67-fold induction (Figure 4, bars 7). This result corroborates the above observation (Figure 3, bars 7) that the presence of the λCI-linker may slightly impair operator binding of the membrane-anchored repressor.
Figure 4.
Repression and de-repression of a chromosomal tacOP-lacZ reporter cassette by the membrane-anchored LacI fusion proteins. Strain R2171 carrying a tacOP-lacZ reporter cassette integrated into the λattB site in the chromosome was transformed with the various plasmids carrying wild-type lacI and its derivatives, respectively. Subsequently, β-galactosidase activities were determined as in Figure 3. Bars 1: R2171 (untransformed strain); bars 2: R2171/pFDX500 (wild-type lacI); bars 3: R2171/pFDX4151 (wild-type lacI with ATG-start codon and NdeI site); bars 4: R2171/pFDX4154 [φ(galK-lacI)]; bars 5: R2171/pFDX4152 [φ(gene8-lacI)]; bars 6: R2171/pFDX4155 [φ(galK-λcI′-lacI)]; and bars 7: R2171/pFDX4156 [φ(gene8-λcI′-lacI)].
Membrane-attached and soluble LacI proteins exhibit similar kinetics of binding to chromosomally located operator lacO1
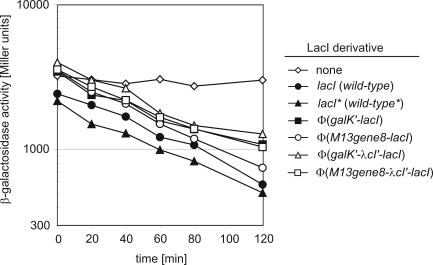
The experiments above demonstrated that the membrane-attached φ(Gene8–LacI) fusion protein is capable of efficiently repressing expression of the lacZ reporter gene and thus of binding to a chromosomally located operator, lacO1. The addition of IPTG led to release from this repression. Moreover, repression as well as inducibility were quite similar with its membrane-anchored fusion derivatives as compared with the wild-type repressor, suggesting that even the equilibrium constants for binding of the operator sequence are similar. However, in the experiments described above, cultures were pregrown without IPTG, split and subsequently grown in the presence or absence of IPTG before enzyme activities were determined. Thus, these data allow no conclusion about the initial rate of operator occupancy. To address this question, cultures of the transformants shown in Figure 4 were pregrown in the presence of IPTG. Subsequently, they were washed free of IPTG to allow establishment of repression, and the kinetics of decline of β-galactosidase as a function of time was analyzed (Figure 5). In the absence of repressor, specific activity remained constant as expected (Figure 5, diamonds). In all other cases, enzyme activities declined with time as a result of re-establishment of repression. In these cases, the remaining activities represent the sum of dilution of enzyme activities by cell growth and of the basal enzyme synthesis under repressing conditions. As can be seen, the slopes of decline of enzyme activities are similar in all cases. The differences that can be seen reflect the differences in repression as revealed by the data in Figure 4 and support the conclusion that the membrane-anchored φ(Gene8–LacI) repressor protein represses the tac promoter at least as efficient as the wild-type repressor. Thus, not only the equilibrium constants but also the initial rate of finding the operator target appears to be similar within the time frame of these kinetics for the membrane-localized and soluble repressor proteins. Taken together, it can be concluded that a membrane-attached Lac repressor protein can find and bind its operator sequence present in the chromosome as efficient as the soluble wild-type LacI protein.
Figure 5.
Kinetics of establishment of repression by the membrane-anchored LacI fusion proteins at the chromosomal tacOP-lacZ cassette. Strain R2171 and its various transformants (Figure 4) were pregrown in the presence of 1 mM IPTG. Subsequently, the cells were washed free of IPTG and growth was continued in IPTG-free medium. Samples were taken after the removal of IPTG at the times indicated and β-galactosidase activities were determined.
DISCUSSION
In this study, we have demonstrated that the E.coli Lac repressor retains its activity when it is artificially fixed with its N-terminus to the inner face of the cytoplasmic membrane. From this location it represses the expression of a tacOP-lacZ reporter cassette present on a plasmid and even in the chromosome with the same efficiency as the wild-type repressor. Moreover, addition of IPTG as an inducer resulted in the same de-repression exhibited by the wild-type repressor (Figures 3 and 4). The data indicate that the equilibrium constants for the occupation of chromosomal lac operator O1 must be similar for the membrane-localized φ(Gene8–LacI) protein and the wild-type Lac repressor (Figures 4 and 5). Pulse-chase experiments verified that the chimeric φ(Gene8–LacI) protein is stable (Figure 1). Subcellular fractionations demonstrated that the chimeric repressor protein is stably anchored to the membrane (Figure 2).
However, we did not demonstrate that the membrane-anchored fusion protein is capable of regulating the authentic lac operon. Indeed, when we replaced the wild-type lacI gene by the gene8-lacI fusion gene within the context of the authentic lac operon in the chromosome, expression of the operon was only weakly repressed. However, very similar results were obtained when the lacI gene was replaced by one of the galK-lacI fusion genes (data not shown). Thus, a fusion to the N-terminus of the Lac repressor encompassing its DNA binding domain generally leads to a defect in repressor function in its authentic context, the lac operon. In this context, full repression requires cooperative binding of LacI to one of two auxiliary operators in addition to operator O1 (20). This cooperative binding requires the tetramerization of LacI (21). It may be concluded from our data that a fusion of a foreign peptide to the N-terminus of LacI in general may negatively interfere with its binding capability to the main operator, O1, when auxiliary binding sites are present.
Nevertheless, our data clearly demonstrate that a membrane-anchored Lac repressor does have unrestricted access to the chromosomally located lacO1 operator. Thus, not only transcriptional antiterminator proteins like the E.coli BglG protein can be engineered to work from the membrane (9) but also transcriptional repressor proteins that have to occupy their binding sites for most of the time and be able to bind back fast in order to achieve efficient repression. Thus, a physical gap between membrane and chromosome does not exist. It appears that either the chromosome already resides in the vicinity of the membrane, making a physical movement of the regulator unnecessary, or that the chromosome dynamically rearranges very fast allowing free access to its sequences from the membrane. This view is in agreement with early studies that reported the chromosome to be associated with the cytoplasmic membrane (22,23).
A major consequence of our results is that membrane sequestration of gene regulators in terms of their physical separation from the chromosome alone cannot account for their inactivation. A well-studied case of a gene activator that shuttles between the chromosome and the membrane is MalT of E.coli. Upon maltose transport MalT is released from the membrane-bound subunit MalK of the maltose transporter which is a prerequisite for MalT activity. It has been known for a long time that even a soluble form of MalK inhibits mal gene expression in vivo, at least when overproduced (24). Recently, it has been demonstrated that soluble MalK is able to inhibit MalT activity in a purified transcription system by blocking the binding of the inducer maltotriose to MalT (25). Thus, membrane sequestration of MalT appears to be just a secondary effect as a consequence of its interaction with the membrane-localized MalK subunit rather than to be directly responsible for MalT inactivity. The quorum-sensing activator protein TraR of A.tumefaciens in the presence of the homoserine lactone signal exists as an active dimer in the cytoplasm, whereas in its absence TraR is sequestered to the membrane in an inactive, monomeric form (6). It, thus, appears that the subcellular localization of TraR modulates its oligomerization state and thereby indirectly regulates the activity of TraR.
In a recent study, it was found that different sites in the chromosome significantly differ in their accessibility to phage λ integrase catalyzed site-specific recombination. In fact, several loci were found that were even designated ‘black holes’ (26). On the other hand, the E.coli chromosome is generally accessible in vivo along its entire length to restriction enzymes suggesting that there are no extensive stably bound structural components that prevent their access (27). Our tests were performed with lac operator O1 integrated into the λ attB site of the E.coli chromosome. We are currently investigating with the aid of a transposon borne tacOP-lacZ cassette whether differences in the accessibility to repression by the membrane-bound Lac repressor exist within the chromosome. Our finding that membrane-bound Lac repressor can tightly bind to its operator should in principal allow to tether almost any DNA sequences containing lac operator to the cytoplasmic membrane and to study the consequences. For example, DNA topology or chromatin structure could be captured between two operator sites and released upon the addition of IPTG, and the consequences for recombination, gene expression or DNA replication could be studied.
Acknowledgments
We thank G. Igloi for DNA sequencing. This work was supported by grants from the Landesschwerpunkt Baden-Württemberg, the Graduiertenkolleg ‘Biochemie der Enzyme’ (DFG) and the Fonds der Chemischen Industrie to B.R. Funding to pay the Open Access publication charges for this article was provided by the Land Baden-Württemberg.
Conflict of interest statement. None declared.
REFERENCES
- 1.Böhm A., Boos W. Gene regulation in prokaryotes by subcellular relocalization of transcription factors. Curr. Opin. Microbiol. 2004;7:151–156. doi: 10.1016/j.mib.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Ostrovsky de Spicer P., Maloy S. PutA protein, a membrane-associated flavin dehydrogenase, acts as a redox-dependent transcriptional regulator. Proc. Natl Acad. Sci. USA. 1993;90:4295–4298. doi: 10.1073/pnas.90.9.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plumbridge J. Regulation of gene expression in the PTS in Escherichia coli: the role and interactions of Mlc. Curr. Opin. Microbiol. 2002;5:187–193. doi: 10.1016/s1369-5274(02)00296-5. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka Y., Itoh F., Kimata K., Aiba H. Membrane localization itself but not binding to IICB is directly responsible for the inactivation of the global repressor Mlc in Escherichia coli. Mol. Microbiol. 2004;53:941–951. doi: 10.1111/j.1365-2958.2004.04179.x. [DOI] [PubMed] [Google Scholar]
- 5.Schlegel A., Bohm A., Lee S.J., Peist R., Decker K., Boos W. Network regulation of the Escherichia coli maltose system. J. Mol. Microbiol. Biotechnol. 2002;4:301–307. [PubMed] [Google Scholar]
- 6.Qin Y., Luo Z.Q., Smyth A.J., Gao P., Beck von Bodman S., Farrand S.K. Quorum-sensing signal binding results in dimerization of TraR and its release from membranes into the cytoplasm. EMBO J. 2000;19:5212–5221. doi: 10.1093/emboj/19.19.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klopprogge K., Grabbe R., Hoppert M., Schmitz R.A. Membrane association of Klebsiella pneumoniae NifL is affected by molecular oxygen and combined nitrogen. Arch. Microbiol. 2002;177:223–234. doi: 10.1007/s00203-001-0379-x. [DOI] [PubMed] [Google Scholar]
- 8.Miller V.L., Taylor R.K., Mekalanos J.J. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 9.Görke B., Rak B. Efficient transcriptional antitermination from the Escherichia coli cytoplasmic membrane. J. Mol. Biol. 2001;308:131–145. doi: 10.1006/jmbi.2001.4590. [DOI] [PubMed] [Google Scholar]
- 10.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 11.Schnetz K., Sutrina S.L., Saier M.H., Jr, Rak B. Identification of catalytic residues in the β-glucoside permease of Escherichia coli by site-specific mutagenesis and demonstration of interdomain cross-reactivity between the β-glucoside and glucose systems. J. Biol. Chem. 1990;265:13464–13471. [PubMed] [Google Scholar]
- 12.Görke B., Rak B. Catabolite control of Escherichia coli regulatory protein BglG activity by antagonistically acting phosphorylations. EMBO J. 1999;18:3370–3379. doi: 10.1093/emboj/18.12.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rak B. A control system which causes alternating turn-off/turn-on of transcription on insertion element IS1. Mol. Gen. Genet. 1983;192:61–65. doi: 10.1007/BF00327647. [DOI] [PubMed] [Google Scholar]
- 14.Diederich L., Rasmussen L.J., Messer W. New cloning vectors for integration into the lambda attachment site attB of the Escherichia coli chromosome. Plasmid. 1992;28:14–24. doi: 10.1016/0147-619x(92)90032-6. [DOI] [PubMed] [Google Scholar]
- 15.Miller J.H. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 16.Schnetz K., Stülke J., Gertz S., Krüger S., Krieg M., Hecker M., Rak B. LicT, a Bacillus subtilis transcriptional antiterminator protein of the BglG family. J. Bacteriol. 1996;178:1971–1979. doi: 10.1128/jb.178.7.1971-1979.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seitz S., Lee S.J., Pennetier C., Boos W., Plumbridge J. Analysis of the interaction between the global regulator Mlc and EIIBGlc of the glucose-specific phosphotransferase system in Escherichia coli. J. Biol. Chem. 2003;278:10744–10751. doi: 10.1074/jbc.M212066200. [DOI] [PubMed] [Google Scholar]
- 18.Leeds J.A., Beckwith J. Lambda repressor N-terminal DNA-binding domain as an assay for protein transmembrane segment interactions in vivo. J. Mol. Biol. 1998;280:799–810. doi: 10.1006/jmbi.1998.1893. [DOI] [PubMed] [Google Scholar]
- 19.Pogliano J., Ho T.Q., Zhong Z., Helinski D.R. Multicopy plasmids are clustered and localized in Escherichia coli. Proc. Natl Acad. Sci. USA. 2001;98:4468–4491. doi: 10.1073/pnas.081075798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oehler S., Eismann E.R., Krämer H., Müller-Hill B. The three operators of the lac operon cooperate in repression. EMBO J. 1990;9:973–979. doi: 10.1002/j.1460-2075.1990.tb08199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthews K.S., Nichols J.C. Lactose repressor protein: functional properties and structure. Prog. Nucleic Acid Res. Mol. Biol. 1998;58:127–164. doi: 10.1016/s0079-6603(08)60035-5. [DOI] [PubMed] [Google Scholar]
- 22.Leibowitz P.J., Schaechter M. The attachment of the bacterial chromosome to the cell membrane. Int. Rev. Cytol. 1975;41:1–28. doi: 10.1016/s0074-7696(08)60964-x. [DOI] [PubMed] [Google Scholar]
- 23.Portalier R., Worcel A. Association of the folded chromosome with the cell envelope of E.coli: characterization of the proteins at the DNA-membrane attachment site. Cell. 1976;8:245–255. doi: 10.1016/0092-8674(76)90008-8. [DOI] [PubMed] [Google Scholar]
- 24.Reyes M., Shuman H.A. Overproduction of MalK protein prevents expression of the Escherichia coli mal regulon. J. Bacteriol. 1988;170:4598–4602. doi: 10.1128/jb.170.10.4598-4602.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joly N., Böhm A., Boos W., Richet E. MalK, the ATP-binding cassette component of the Escherichia coli maltodextrin transporter, inhibits the transcriptional activator MalT by antagonizing inducer binding. J. Biol. Chem. 2004;279:33123–33130. doi: 10.1074/jbc.M403615200. [DOI] [PubMed] [Google Scholar]
- 26.Garcia-Russell N., Harmon T.G., Le T.Q., Amaladas N.H., Mathewson R.D., Segall A.M. Unequal access of chromosomal regions to each other in Salmonella: probing chromosome structure with phage lambda integrase-mediated long-range rearrangements. Mol. Microbiol. 2004;52:329–344. doi: 10.1111/j.1365-2958.2004.03976.x. [DOI] [PubMed] [Google Scholar]
- 27.Postow L., Hardy C.D., Arsuaga J., Cozzarelli N.R. Topological domain structure of the Escherichia coli chromosome. Genes Dev. 2004;18:1766–1779. doi: 10.1101/gad.1207504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]