Abstract
The high efficiency and specificity of enzymes make them play an important role in life activities, but the high cost, low stability and high sensitivity of natural enzymes severely restrict their application. In recent years, nanozymes have become convincing alternatives to natural enzymes, finding utility across diverse domains, including biosensing, antibacterial interventions, cancer treatment, and environmental preservation. Nanozymes are characterized by their remarkable attributes, encompassing high stability, cost-effectiveness and robust catalytic activity. Within the contemporary scientific landscape, metal–organic frameworks (MOFs) have garnered considerable attention, primarily due to their versatile applications, spanning catalysis. Notably, MOFs serve as scaffolds for the development of nanozymes, particularly in the context of bacterial detection and treatment. This paper presents a comprehensive review of recent literature pertaining to MOFs and their pivotal role in bacterial detection and treatment. We explored the limitations and prospects for the development of MOF-based nanozymes as a platform for bacterial detection and therapy, and anticipate their great potential and broader clinical applications in addressing medical challenges.
In this review, we focus on the application of MOF-based nanozymes with natural enzyme activity in bacterial detection and therapy, and we believe that MOF-based nanozymes will have a broader development space in the biomedical field in the future.
1. Introduction
Enzymes, whether proteins or ribonucleic acids (RNAs), exhibit remarkable specificity and catalytic efficiency, enabling them to mediate a wide array of biochemical reactions with precision and vigor, thus constituting essential players in the orchestration of life activities.1,2 Although widely used, natural enzymes suffer from intrinsic drawbacks, including poor stability, elevated costs, intricate production processes, susceptibility to environmental factors, and challenges associated with retrieval and recycling, thereby imposing significant constraints on their practical application.3 With advancements in research technologies, nanomaterials have been discovered and the concept of “nanozymes” was first introduced in 2004 in a study on new catalysts based on gold nanoparticles (AuNPs).4 Nanozymes, being artificial nanomaterials, possess intrinsic enzymatic attributes and mimic the structure and function of their natural counterparts.5,6 Compared with natural enzymes, nanozymes can be synthesized using simpler methods at reduced costs while concurrently displaying heightened catalytic activity and stability.7–9 Nanozymes have demonstrated immense potential across diverse domains, such as diagnostic detection,10,11 antibacterial agents,12–14 biosensing,5,15,16 and tumor therapy.17–19 However, despite the extensive body of research surrounding nanozymes, the design of nanozymes with ideal performance remains a formidable challenge.20 Nanozymes often exhibit uneven surface structures and enzymatic activity levels that fall short of those achieved by natural enzymes.21 Moreover, due to their structural complexity, nanozymes often struggle to faithfully recreate the intricate three-dimensional nature of natural enzymes, frequently resulting in a deficiency of equivalent active sites.22 These impediments impede the broader application of nanozymes.
In order to solve the above problems, it is necessary to design and synthesize nanozymes with catalytic properties. Nanozymes constructed based on metal–organic frameworks (MOFs) offer a partial solution to these challenges. MOFs represent a relatively new class of crystalline porous nanomaterials comprising a blend of metal ions or ion clusters interconnected by organic ligands.23,24 Distinguishing themselves from conventional nanomaterials, MOFs feature a flexible structure and composition, high specific surface area, adjustable porosity, and an abundance of active sites.25 Their porous architecture facilitates the ingress of small molecule substrates, enabling extensive interaction with active sites and promoting product diffusion. Owing to their distinctive structures and excellent catalytic properties, MOFs are regarded as promising materials for the synthesis of new nanozymes. Notably, MOF-based nanozymes exhibit superior activity and enhanced stability in comparison to conventional counterparts.26
To the best of our knowledge there are many excellent studies reporting the application of MOFs and nanozymes, but there are no overviews that provide a comprehensive and specialized review of their advances in the diagnosis and treatment of bacteria.9,18,27–30 This review offered a concise exploration of the applications of MOF-based nanozymes in bacterial detection and therapy. First, we expounded upon the application of MOF-based nanozymes in bacterial detection, including colorimetric assay, fluorescence assay, ELISA, and their incorporation into electrochemical and microfluidic sensors. Subsequently, we delved into their role in inhibiting bacterial proliferation, examining antibacterial, antibiofilm, and antifungal aspects. We contended that MOF-based nanozymes exhibit potent antibacterial effects, harboring the potential to replace antibiotics, thus presenting promising avenues within the domain of bacterial diagnosis and treatment.
2. Application of MOF-based nanozymes in bacterial detection
Bacterial infections stand as formidable threats to global public health, frequently leading to elevated morbidity and mortality rates.31–33 In view of the pathogenicity of bacterial infections, the timely and accurate identification of infection sources, disease prevention, and targeted treatment are of paramount importance. At present, the gold standard for clinical bacterial detection is still bacterial culture.34,35 However, this conventional approach entails significant resource consumption, demands substantial manpower, and is marred by prolonged incubation periods, often exceeding 72 hours.34,36 Given these limitations, there is an imperative to develop new and efficient pathogen detection methods that can either complement or supplant bacterial culture. In recent years, various novel detection techniques have emerged, such as enzyme-linked immunosorbent assay (ELISA),37,38 immunochromatography (ICA),39 polymerase chain reaction (PCR),38 loop-mediated isothermal amplification (LAMP)40,41 and biosensors.42 Biosensors, in particular, represent a category of transduction devices highly sensitive to biological substances, converting them into electrical signals. Various biosensors are often reported for pathogen detection.42–44
MOF-based nanozymes are considered ideal biosensors for catalytic recognition of target analytes and mediate signal amplification or conversion due to their biocatalytic characteristics and species diversity, and have great potential for development in the field of biosensing. MOF-based nanozymes have developed into a new type of nanozyme, which is relatively simple to prepare, more flexible in design and more abundant in active site than carbon group, metal, and transition metal compound nanozymes. The catalytic capacity of MOF-based nanozymes mainly comes from the following two aspects: on the one hand, the redox reaction of metal ions such as Fe, Cu, Co, Ni and Ce has the catalytic activity; on the other hand, it is the natural enzyme catalytic reaction process simulated by special organic ligands (organic ligands as the electronic medium to transfer the electrons accepted by one substrate to another substrate). Depending on the enzyme activities, researchers have developed MOF-based nanozymes with different enzyme activities, mainly including peroxidase (POD),45,46 glucose oxidase (GOx),16,47 oxidase,48–50 superoxide dismutase (SOD),51–53 and catalase (CAT).54,55 We summarize the basic features of MOF-based nanozymes in Table 1. MOF-based nanozymes have better stability than natural enzymes, but the former is more cytotoxic and is difficult to use in vivo analysis, while the latter is easily affected by PH. In addition, many MOF-based nanozymes can have two or even multiple enzyme activities. Simultaneously, and can be used in the analysis of multiple enzyme cooperative reactions, such as the catalytic cascade. In Table 2, we organize the applications of MOF-based nanozymes in bacterial detection.
Basic features of MOF-based nanozymes.
| Enzyme-like activity | Catalytic mechanism | Categorization | Example of MOFs | Ref. |
|---|---|---|---|---|
| Peroxidase | H2O2 catalyzes substrate oxidation | Oxidoreductase | Cu-MOF, Zn-MOF, Fe-MOF, Zr-MOF, Ce-MOF | 63, 92, 97, 145, 160 |
| Oxidase | Oxidizes the substrate while reducing oxygen to H2O2 | Oxidoreductase | Zn-MOF, Zr-MOF, Fe/Mn-MOF | 66, 67, 162 |
| Catalase | Catalyzes the decomposition of H2O2 into oxygen and water | Oxidoreductase | Mn-MOF, Zn-MOF, Ce-MOF | 106, 164, 175 |
| Superoxide dismutase | Catalyzes the disproportionation of superoxide anion radicals to generate oxygen and H2O2 | Oxidoreductase | Mn-MOF, Ce-MOF | 107, 175 |
Summarized applications of MOF-based nanozymes of bacterial detection.
| MOF materials | Enzyme-like characteristics | Microorganisms | Detection method | Linearity range | Limit of detection | Time | Ref. |
|---|---|---|---|---|---|---|---|
| Ab2/AuNPs/MOFs | Peroxidase | S. aureus | Electrochemical biosensors | 10–7.5 × 107 CFU mL−1 | 6 CFU mL−1 | 45 min | 61 |
| ssDNA-Au/CuMOF | Peroxidase | S. aureus | Electrochemical biosensors and colorimetric assay | 10–108 CFU mL−1 | 5 CFU mL−1 | — | 63 |
| NMOF@AMP–Fc | Oxidase | Vibrio parahaemolyticus | Electrochemical biosensors | 10–107 CFU mL−1 | 4 CFU mL−1 | 30 min | 64 |
| Fe3O4@NMOF-Apt and Au@Fc-PBA | Oxidase | Vibrio parahaemolyticus | Electrochemical biosensors | 10–109 CFU mL−1 | 3 CFU mL−1 | 20 min | 65 |
| Zn-based MOF/CMC/AuNPs | Oxidase | Haemophilus | Electrochemical biosensors | 0.1 pM–10 nM | 1.48 fM | — | 66 |
| Fe/Mn bimetallic MOFs | Oxidase | Salmonella typhimurium | Electrochemical biosensors | — | 0.07 pM | — | 67 |
| Cu-MOF | Peroxidase | S. aureus | Colorimetric assay | 50–10 000 CFU mL−1 | 20 CFU mL−1 | — | 70 |
| Zn/Co-ICP@GOx | Glucose oxidase, peroxidase | Gram-positive, Gram-negative bacteria | Colorimetric assay | 0.01–1.0 mM | 0.005 mM | 40 min | 71 |
| GOx@GA–Fe(ii) | Glucose oxidase, peroxidase | E coli, S. aureus, Listeria, and Salmonella typhimurium | Colorimetric assay | — | 0.43 μM (glucose) | 12 h | 72 |
| Fe3O4@MIL-100(Fe)–Au | Peroxidase | Salmonella | Colorimetric assay | — | — | — | 73 |
| NH2-MIL-101(Fe) | Peroxidase | Salmonella | Microfluidic biosensor | 1.5 × 101–1.5 × 107 CFU mL−1 | 14 CFU mL−1 | 1 h | 74 |
| Fe-MIL-88NH2 MOF | Peroxidase | Salmonella | Microfluidic biosensor | — | 93 CFU mL−1 | <1 h | 75 |
| Eu/Tb (BTC) MOF | Fluorescent MOF | Bacillus | Fluorescence | — | 1087 nM | — | 78 |
| VAN-PEG-FITC/HCAA@UiO-66 | Fluorescent MOF | S. aureus | Fluorescence | 1.05 × 103–1.05 × 107 CFU mL−1 | 12 CFU mL−1 | — | 79 |
| NH2-MIL-53(Al) | Fluorescent MOF | S. aureus | Fluorescence | 50–10 000 CFU mL−1 | 20 CFU mL−1 | — | 80 |
| Zr-mMOF | Fluorescent MOF | Acinetobacter baumannii | Fluorescence | 101–105 CFU mL−1 | 10 CFU mL−1 | 2.5 h | 81 |
| MIL-88 | Horseradish peroxidase | Aflatoxin B1 | ELISA | 0.01–20 ng mL−1 | 0.009 ng mL−1 | — | 82 |
| Co3Fe-MMOF NPs | Peroxidase | Aeromonas hydrophila | ELISA and colorimetric assay | 62–6.2 × 108 CFU mL−1 | 30 CFU mL−1 | — | 83 |
2.1. Electrochemical biosensors
Electrochemical biosensors consist of a biological recognition element, Electrochemical signal transformation element and a signal amplification element, to provide selective quantitative information. A brief introduction of the working principle: when the analyte to spread to the electrode surface of the biometric unit (protein, enzyme, antibody, nucleic acid, etc.), and a series of biochemical reactions occur on the electrode surface, the electrochemical signal through the electrode output and record, to obtain the concentration of the substance to be measured. Electrochemical sensors have been extensively employed in modern bioanalytical chemistry for a wide variety of applications due to their high portability, technical simplicity, sensitivity, cost-effective, and point-of-care testing.56–60 In the realm of electrochemical biosensors, several innovative approaches have emerged to enhance the detection of various bacteria.
2.1.1. Staphylococcus aureus detection
Hu et al. adopted an in situ reduction method to cultivate AuNPs on two-dimensional (2D) MOFs, paving the way for an electrochemical detection method for Staphylococcus aureus (S. aureus). This method hinged on the bioconjugation of the aureus phage protein antibody with AuNPs.61 The 2D MOF-based nanozyme exhibited POD-like activity, rendering it capable of producing a clear electrochemical signal, thereby enabling the specific identification of S. aureus with a minimum limit of detection (LOD) of 6 CFU mL−1. Akash Deep et al. designed and synthesized NH2-MIL-53(Fe)-bacteriophage biosensor for the highly sensitive detection of S. aureus with a LOD of 31 CFU mL−1.62 In addition, combining the catalytic activity of MOF as biosensors for the detection of pathogenic bacteria is also a new idea. Zou et al. designed a ssDNA-Au/CuMOF dual-responsive detection system based on an electrochemical sensor and colorimetry for the detection of S. aureus. They utilized the POD-like activity of ssDNA-Au/CuMOF to detect S. aureus. ssDNA-Au/CuMOF has a detection range of the MOF was in the range of 10 to 108 CFU mL−1. LOD was as low as 5 CFU mL−1.63
2.1.2. Vibrio parahaemolyticus detection
Cao et al. introduced a novel electrochemical aptasensor with oxidase activity.64 They synthesized NMOF@AMP–Fc by incorporating a novel antimicrobial peptide (AMP) and ferrocene (Fc) with nanoscale MOFs (NMOFs). NMOF@AMP–Fc served as a signal label, which, when conjugated with captured Vibrio parahaemolyticus (VP), formed a sandwich complex, generating an electrical signal from the Fc moiety to enable rapid detection. Remarkably, the entire process was completed within 30 min, with a LOD of 4 CFU mL−1, underlining its high sensitivity and specificity. Similarly, Li et al. also used aptamer-labeled magnetic nanoscale MOF (Fe3O4@NMOF) as capture probes, coupled with AuNPs functionalized with phenylboronic acid and ferrocene as nanolabels. Such MOFs with oxidase-like activity are able to detect VP by electrochemical methods.65
2.1.3. Haemophilus influenzae and Salmonella typhimurium detection
Sohrabi et al. contributed to the field by devising electrochemical gene sensors. In the case of Haemophilus influenzae, a zinc-based MOF/CMC/AuNP sensor with oxidase activity was developed, exhibiting a LOD and limit of quantitation (LOQ) of 1.48 fM and 3.23 fM, respectively, under optimal conditions.66 Similarly, Sohrabi et al. designed an electrochemical biosensor with oxidase activity for the detection of Salmonella typhimurium.67 This entailed synthesizing Fe/Mn bimetallic MOFs, combining them with methyl-β-cyclodextrin (MβCD), and incorporating AuNPs on multiwalled carbon nanotubes (MWCNTs), ultimately yielding nanocomposites immobilized onto an Au electrode surface. Under optimal conditions, this sensor demonstrated a LOD of 0.07 pM and LOQ of 0.21 pM.
The preparation of the above electrochemical sensors is very simple, low cost, short detection time, and can be used for point-of-care testing of bacteria. Those electrochemical sensors mainly through using the oxidase or POD activity of MOFs to realize the amplification strategy of the original signal, and play a role in improving the detection sensitivity. Considering such recognition, discovery of new classes of MOFs to modify the electrode's surface with high sensitivity, large surface area, and reproducibility, and electronic, unique catalytic, and biocompatibility properties is the direction of research in this class of electrochemical sensing. Electrochemical biosensors have gained wide popularity in bacterial detection techniques, including but not limited to S. aureus, Hemophilus, VP, and Salmonella typhimurium.
2.2. Colorimetric assay
Colorimetric assay is also composed of biological recognition elements, signal sensing amplification elements and signal readout system, which immediately detects the presence of sample through the color change or simple instruments such as ultraviolet-visible spectrophotometer. The process is very fast.68,69 Signal amplification is also an important strategy to improve its detection sensitivity.
Tan et al. synthesized Cu-MOF particles with POD-like activity using by a mixed solvothermal method, and this new MOF nanozyme combines a catalyzed chromogenic reaction with an aptamer, providing a new colorimetric assay for the detection of S. aureus.70 It has a high sensitivity with a linear range of 50 to 10 000 CFU mL−1 and a LOD of 20 CFU mL−1 for S. aureus.
Qiu et al. synthesized Zn/Co-ICP@GOx, a composite material formed by embedding GOx in Zn–Co infinite coordination polymers. This multifunctional polymer exhibits GOx and POD activities and has been used in the colorimetric detection of glucose. Since glucose metabolism is a ubiquitous phenomenon among bacteria, residual glucose levels indirectly provide insights into the viability of Gram-negative and Gram-positive bacteria. This method exhibits a linear range of 0.01 to 1.0 mM and a LOD of 0.005 mM.71 Similarly, Zhang et al. engineered GOx@GA–Fe(ii) nanozymes by combining gallic acid and GOx.72 These GOx@GA–Fe(ii) nanozymes possess GOx and POD activities with remarkable specificity and sensitivity. They were shown to be colorimetric probes for quantifying glucose content and facilitating microbial detection in food systems.
Meteku et al. contributed to the field by devising Fe3O4@MIL-100(Fe)–Au nanostructures. These nanostructures possess POD-like activity, achieved by the incorporation of magnetic Fe3O4 nanorods, MIL-100(Fe) and Au.73 The resulting structures were conjugated with Salmonella antibodies, thereby enabling the capture of Salmonella pathogens, facilitated by magnetic targeting within a magnetic field. In addition, the researchers leveraged these nanostructures for the colorimetric detection of hydrogen peroxide and the catalytic reduction of selected organic pollutants. The results of their efforts showed substantial enhancements in bacterial capture rates, reaction speed, and catalytic efficiency.
The above colorimetric method designs different types of MOF-based nanozymes, especially by designing different POD and GOx activities to achieve the enzyme cascade reaction, to finally realize the signal amplification. Due to the biocatalytic properties of MOF-based nanozymes, the use of MOF-based nanozymes to detect bacteria by colorimetric assay is highly attractive and provides a new idea for bacterial detection.
2.3. Microfluidic biosensors
With the development and progress of microprocessing technology, microfluidics plays an increasing role in pathogen examination. Due to its significant advantages, such as fast reaction, high automatic operation degree and small volume. At the same time, the combination of microfluidic and sensor technology can improve the integration degree of instruments, so it is easy to carry around for on-site screening.
Qi et al. introduced a microfluidic biosensor that leveraged NH2-MIL-101(Fe) possessing pseudo-POD activity and immunomagnetic nanorods (MNB). This combination facilitated the selective separation and concentration of Salmonella, culminating in the formation of MNB-Salmonella MOF complexes. These complexes catalyzed the conversion of colorless o-phenylenediamine and H2O2 into yellow 2,3-diaminophenazine. Impressively, the sensor exhibited the capability to detect Salmonella typhimurium over a wide concentration range, spanning from 1.5 × 101 to 1.5 × 107 CFU mL−1 of within 1 h, boasting a LOD of 14 CFU mL−1.74
Guo et al. introduced an innovative microfluidic immunosensor optimized for the rapid detection of Salmonella. This approach entailed the utilization of Fe-MIL-88NH2 MOF nanocubes (NCs), stimulating POD activity, augmented by platinum NPs for signal amplification. Notably, the incorporation of smartphone thermal imaging allowed for the real-time monitoring of temperature changes. The sensor exhibited remarkable sensitivity, capable of detecting Salmonella at concentrations as low as 93 CFU mL−1 within 1 h.75
In addition to the use of various traditional biosensor tests. Combined with microfluidic technology, instrument integration and automation can be achieved. MOFs with POD-like activity as a signal amplifier enables rapid detection of Salmonella in less than 1 h. This is an innovative breakthrough, which also provides new ideas for the detection of other foodborne pathogens.
2.4. Fluorescence assay
Fluorescence detection has attracted widespread attention due to its advantages of high sensitivity, fast response, and low cost. Fluorescent MOFs show large specific surface area, high porosity, great possibility of structural modification and functionalization. In particular, emission centers in fluorescent MOFs, including metal ions or ligands, are extremely sensitive to changes in the environment, which allows high sensitivity detection.
Dipicolinic acid (DPA) serves as a prominent marker for spore-forming pathogens,76 making it a crucial indicator in the detection of infectious spores in samples.77 In this context, Zhuang et al. synthesized a novel nanoscale dual-emission lanthanide Eu/Tb (BTC) MOF.78 This newly synthesized MOF exhibited remarkable sensitivity, selectivity, and precision, rendering it particularly suited for the selective detection of DPA as a fluorescent probe with a LOD of 1087 nM.
For the specific detection of S. aureus, Li et al. investigated a highly efficient fluorescent nanoprobe, VAN-PEG-FITC/HCAA@UiO-66, which exhibited high selectivity even in real samples and had a LOD of 12 CFU mL−1.79 The MOF material, NH2-MIL-53(Al), prepared by Fu et al. can release a large number of fluorescent signals after alkaline hydrolysis, which can be used to quantitatively identify S. aureus using different sites.80 This method is highly sensitive with a linear range of 50 to 10 000 CFU mL−1 and a LOD of 20 CFU mL−1. Li et al. adopted a distinct approach, employing Zr-MOF as a functional coating for magnetic Fe3O4 NPs, thereby creating a modified surface (Zr-mMOF). This innovation paved the way for the generation of a fluorescent signal (F@UIO-66–NH2), facilitating the detection of Acinetobacter baumannii in blood samples.81 This method enabled the enrichment and detection of Acinetobacter baumannii within 2.5 h, featuring a LOD of 10 CFU mL−1 and a linear range of 101 to 105 CFU mL−1.
In summary, fluorescent MOFs are one of the most promising materials for high-sensitivity fluorescence detection. MOFs with fluorescence emission properties are sensitive to changes in the environment, have high sensitivity, and have obtained excellent detection results in bacterial detection, making them promising materials for fluorescence detection.
2.5. ELISA
Owing to its advantages such as simplicity and ease of operation, enzyme-linked immunosorbent assay (ELISA) has been a general tool in biological assays. However, the relatively low sensitivity and accuracy are the primary limitations of traditional ELISA. Fortunately, by combining MOFs and ELISA, the performance of ELISA has been improved a lot. In a remarkable integration of the ELISA, Xu et al. harnessed ELISA's potential by merging it with MIL-88, a material endowed with POD activity. This strategic pairing enabled the sensitive detection of aflatoxin B1.82 The pivotal innovation involved coupling MIL-88 with an antibody specifically engineered to replace horseradish peroxidase (HRP) within the catalytic color development system. This ingenious adaptation successfully mitigated the issue of false positives that could arise during the detection process, leading to a substantial enhancement in detection accuracy. Notably, this method achieved a LOD of 0.009 ng mL−1, featuring a linear working range of 0.01 to 20 ng mL−1, which provided a 20-fold increase in sensitivity compared with the conventional ELISA. In addition, ELISA can be combined with colorimetric assay to detect bacteria. Researchers synthesized Co3Fe-MMOF NPs featuring prominent POD-like activity through a solvothermal method. These NPs effectively catalyzed the yellow color reaction of TMB in the presence of H2O2. By combining the chromogenic reaction of Co3Fe-MMOF with antibody-based recognition and magnetic separation, the colorimetric detection method demonstrated a LOD of 30 CFU mL−1 for Aeromonas hydrophila. This encompassed a wide detection range spanning 62 to 6.2 × 108 CFU mL−1.83
ELISA is a common method for bacterial detection, but this method is poorly stabilized and prone to false positives.84,85 The combination of MOFs with POD activity and ELISA improves the stability and sensitivity of the material and is an innovative method for detecting bacteria.
3. Application of MOF-based nanozymes in antibacterial therapy
We summarize the different kinds of MOF-based nanozymes in antimicrobial therapy in Table 3.
Summarized applications of MOF-based nanozymes of bacterial therapy.
| MOF | Loaded compound | Enzyme-like characteristics | Microorganisms | Antibacterial activity in vitro (doses) | Antimicrobial principle | Ref. |
|---|---|---|---|---|---|---|
| N-CNTs@Co | — | Oxidase | S. aureus, E. coli | 30 μg mL−1 | Catalyzes oxygen to produce free radicals and generate ROS | 91 |
| PEG@Zn/Pt–CN | Pt nanozymes | Peroxidase | S. aureus (99.63%), E. coli (98.74%) | 150 μg mL−1 | — | 92 |
| Fe-MOF–Ag | AgNPs | Peroxidase | S. aureus (86–91%), E. coli (98–99%) | 100 μg mL−1 | Catalyzes the decomposition of low concentrations of H2O2 to ˙OH | 97 |
| PDA@AgNPs@bilayer hydrogel | AgNPs, PDA, PTT | Peroxidase | S. aureus, E. coli | — | Enhancement of enzyme activity by PTT therapy | 99 |
| UsAuNPs/MOFs hybrid | AuNPs | Peroxidase | S. aureus, E. coli | 1 mg mL−1 | Catalyzes H2O2 to ˙OH | 103 |
| Mn-ZIF-8 | Mn2+/Mn4+ | Superoxide dismutase, catalase | S. aureus, E. coli | 20 μg mL−1 | Scavenges ROS, regulating the shift of macrophage polarization from M1 phenotype to M2 phenotype | 106 |
| ZIF8/Au–GOx NPs | AuNPs, GOx | Catalase | S. aureus (100%), E. coli (100%) | 4 μg mL−1, 8 μg mL−1 | Enhanced H2O2 production and zinc ion release in acidic environments | 112 |
| GOx/Ag@ZIF-HA | AgNPs, GOx | Glucose oxidase | S. aureus (>99%), E. coli (>99%) | 10 μg mL−1, 5 μg mL−1 | Glucose oxidase catalyzes the production of hydrogen peroxide and AgNPs release silver ions | 116 |
| NH2-MIL-125–GO–Pt | PTT | — | S. aureus (99.94%), E. coli (99.12%) | 2.5 mg mL−1 | Improvement of ROS generation by photocatalysis and synergistic photothermal effect antimicrobials | 130 |
| PEG@Zr-Fc MOF hydrogel | PTT | Peroxidase | S. aureus (100%), E. coli (99.2%) | 50 μg mL−1 | Synergistic damage to bacteria through photothermal effects and peroxidase reactions | 131 |
| Pd@Pt-T790 | SDT | Catalase | MRSA | — | Control 1O2 productivity and thus SDT effects | 13 |
| UiO-66–NH–CO–MoS2 | PDT, PTT | Peroxidase | MRSA (96.7%), AREC (99.7%) | 20 μg mL−1 | Generates 1O2 and ˙OH to disrupt the cell integrity of bacteria | 145 |
| CoS2/MoS2NSs | — | Oxidase, peroxidase | S. aureus | 10 μg mL−1, 1 mg mL−1 | Massively production of ˙O2− damaged toward the lipid of the cell membrane and cause death | 146 |
| E. coli | ||||||
| PCN-222Pt | — | Oxidase, peroxidase | S. aureus (98.69%), E. coli (99.91%) | 200 μg mL−1 | Catalyzes O2 and H2O2 to produce ROS in the dark | 162 |
| Cu-MOF (Cu ions and 2-methylimidazole) | — | Peroxidase | S. aureus (99.9%) | 4 μg mL−1 | Oxidizes proteins and lipids on the surface of bacteria, inducing bacterial death | 163 |
| Pd-MOF@PAzo@SNP | Nitric oxide | Oxidase, peroxidase | S. aureus (85%), E. coli (83%) | 64 μg mL−1 | Catalyzes O2 and H2O2 to produce ROS | 165 |
| pGNP-Fe2 | — | Oxidase, peroxidase | S. aureus (85%), E. coli (83%) | — | Adsorption to cell membranes leading to physical strains and then using ROS to kill bacteria | 166 |
| Ce-MOF | — | Catalase, superoxide dismutase, peroxidase | A. flavus (93.3%), A. niger (96.3%), A. terreus (99.3%), C. albicans (93.3%), R. glutinis (96%) | 40 μg mL−1 | — | 175 |
3.1. Antibacterial properties
The persistent threat of bacterial infections looms large, endangering not only human and animal health but also the delicate ecological equilibrium.32,86,87 While antibiotics have played a pivotal role in curbing bacterial infections to a certain extent, the widespread misuse of these antibiotics has led to the emergence of multidrug-resistant bacterial strains, casting a formidable shadow over the progress of antibiotic-based treatments.88 Consequently, bacterial infections, often evolving into untreatable infectious diseases, have claimed a devastating toll on human lives.87,89 To tackle this formidable challenge, there arises an urgent imperative to explore and develop novel therapeutic strategies capable of supplanting antibiotics, ultimately paving the way for the elimination of drug-resistant bacterial strains. Within this evolving landscape, MOF-based nanozymes, characterized as a new type of material, have surfaced as promising contenders, wielding substantial therapeutic potential in the battle against bacterial infections.
3.1.1. Synergy between MOFs and metal ions/NPs in antibacterial applications
Introducing different metal ions or metal nanoparticles into MOF-based nanozymes can modulate their enzymatic activities and properties. There are a variety of methods for modifying metal nanoparticles onto MOFs materials, such as in situ growth method and co-precipitation method, which are synthesized in a way that allows control of the size and dispersion of the nanoparticles.90 Among the metal nanoparticles, AuNPs have enzyme-like activity on their own, and their addition to MOFs can turn the latter into MOF-based nanozymes. In contrast, AgNPs have a broad and generalized antimicrobial effect, which can have a synergistic effect when combined with MOF-based nanozymes. The effective combination of these metal nanoparticles with MOF-based nanozymes can produce enhanced antimicrobial effects under certain conditions, which is an antimicrobial idea worth investigating.
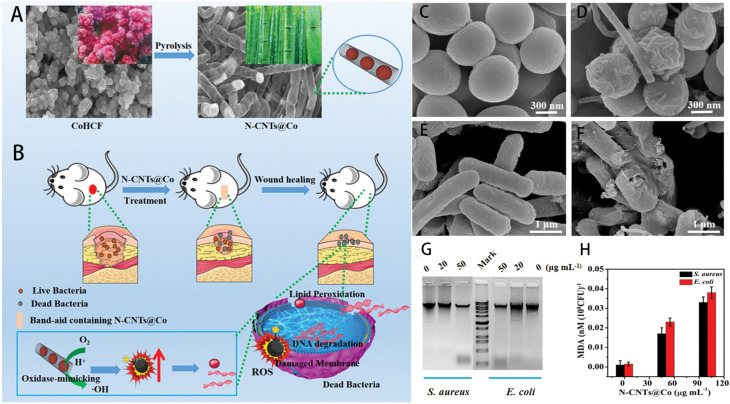
He et al. used cobalt cyanide as a raw material to fabricate carbon nanotubes (N-CNTs@Co) coated with cobalt NPs by high-temperature pyrolysis (Fig. 1A and B).91 N-CNTs@Co is an artificial nanozyme, characterized by highly efficient mimic oxidase activity, which are biocompatible and. Can catalyze oxygen to generate copious reactive oxygen species (ROS) under acidic conditions. As shown in Fig. 1C and E, the cell walls of S. aureus and E. coli after treatment with HAc–NaAc buffer were intact. However, the cell walls of S. aureus and Escherichia coli (E. coli) after treatment with N-CNTs@Co (Fig. 1D and F) were damaged. As shown in Fig. 1G and H, the level of DNA degradation and malondialdehyde content increased with the increase of N-CNTs@Co concentration, and the content of malondialdehyde in the cells was positively proportional to the degree of cell damage. All of the above indicated the excellent antimicrobial effect of N-CNTs@Co against S. aureus and E. coli. Wang et al. decorated platinum nanozymes onto zinc-based photosensitizers to produce a POD-active of hybrid MOF-derived nanozymes, opening new avenues for combating infections.92
Fig. 1. (A) Schematic diagram of N-CNTs@Co preparation and (B) antibacterial application of N-CNTs@Co; (C) S. aureus incubation in HAc–NaAc buffer and (D) N-CNTs@Co; (E) E. coli incubation in HAc–NaAc buffer and (F) N-CNTs@Co; (G) Gel electrophoresis of DNA from S. aureus and E. coli treated with different concentrations of N-CNTs@Co; (H) analysis of MDA content after treatment of S. aureus and E. coli with different concentrations of N-CNTs@Co. Error bars represent the standard deviation of three measurements.91.
Silver NPs (AgNPs), celebrated for their exceptional antibacterial properties encompassing Gram-positive, Gram-negative, and multidrug-resistant bacterial strains,93–95 play a central role in recent developments. Pham et al. synthesized Fe-based MOF crystal Fe-MIL-88B–NH2 using 2-aminoterephthalic acid and ferric chloride as precursor materials.96 On this basis, Hu et al. and Zhang et al. introduced Ag into this MOF, thereby creating Fe-MOF–Ag, each manifesting antibiotic capabilities with excellent POD-like activity.97,98 These innovative materials catalyze the decomposition of H2O2 to generate highly toxic hydroxyl radicals, culminating in outstanding antibacterial properties against E. coli and S. aureus, all while preserving biocompatibility. Li et al. introduced an Ag nanozyme-based bilayer hydrogel by incorporating polydopamine (PDA) to reduce Ag to AgNPs. This hydrogel represents a formidable bulwark against bacterial infections and expedites wound healing, offering the added benefit of near-infrared light (NIR) absorption at 808 nm. This feature results in hyperthermal and light-enhanced POD activity, reinforcing its antibacterial efficacy.99
AuNPs, renowned for their oxidase and POD properties, have the intrinsic capability to combat bacteria by generating ROS. Furthermore, they hold promise as candidates in the quest to address bacterial resistance.100–102 Hu et al. synthesized UsAuNPs/MOF by in situ reduction of ultrathin AuNPs (UsAuNPs) on 2D MOFs, which exhibited remarkable POD-like activity.103 Similarly, Liao et al. fused AuNPs with copper MOFs (Cu-MOFNs), crafting a plasmonic nanozyme.104 Cu-MOFNs demonstrate POD-like activity, while AuNPs have unique localized surface plasmon properties. This composite material exhibits the capacity for in vitro bacterial inhibition and in vivo healing of infected wounds while sidestepping significant biotoxicity.
Expanding beyond the scope of metals, the combination of MOF-based nanozymes and manganese metals also yields antibacterial effects. Aryanejad et al. spearheaded the synthesis of a stable heterogeneous catalyst and Mn-MOF nanomaterials, which exhibit good antibacterial activity against Bacillus cereus and E. coli, namely UoB-4.105 Wan et al. introduced Mn2+/Mn4+ into zeolite imidazolate framework-8 (ZIF-8), engendering a MOF nanozyme with multifaceted antibacterial and inflammatory regulatory functions.106 Zn2+ in ZIF-8 has an antibacterial effect on S. aureus and E. coli, while the addition of Mn2+/Mn4+ offer inflammation regulation capabilities, ameliorating excessive inflammatory responses. Mn-ZIF-8 showcases excellent CAT and SOD activities, further orchestrating macrophage polarization by scavenging ROS. This culminates in the suppression of excessive inflammation, a reshaping of inflammatory immunity, and an enhanced defense against bacterial infections. Impressively, these attributes translated into favorable outcomes for wound healing.
In summary, we found that in recent studies, there are many types of nanoparticles that can bind with MOF-based nanozymes, mainly including AgNPs, AuNPs, cobalt NPs, manganese NPs and so on. Some of these NPs have their own antibacterial or enzymatic activity, while others synergize well with the MOF-based nanozymes to enhance the antibacterial effect.
3.1.2. Synergy between MOFs and natural enzymes in antibacterial applications
The combination of MOFs and natural enzymes can also exert a favorable antimicrobial therapeutic effect. Some MOFs possess quasi-POD abilities, enabling the generation of toxic ROS when exposed to H2O2 for local sterilization.107 This approach circumvents potential harm to normal cells and tissues associated with the direct use of high-concentration hydrogen peroxide,108 thus offering promising prospects for wide-range antibacterial advantages. However, nanozymes with POD-mimetic activity exhibit optimal reactivity under strongly acidic conditions,109 while bacterial infections typically manifest in local environments with neutral to mildly alkaline pH values,110 necessitating innovative solutions for in vivo antimicrobial applications.
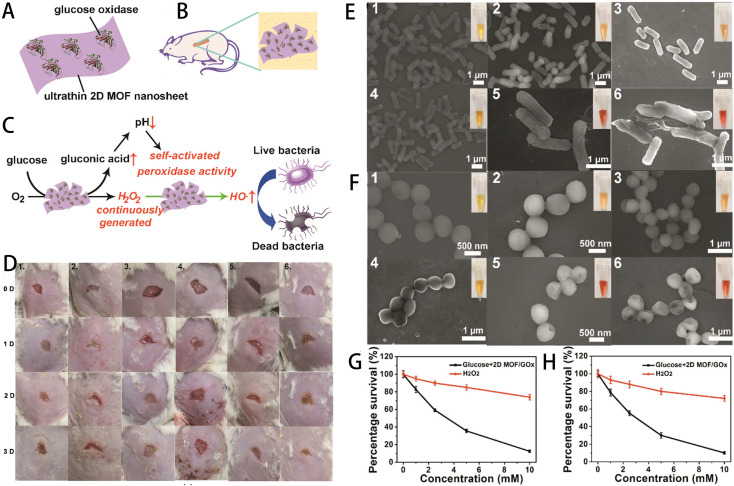
To address this challenge, researchers have embarked on extensive experimentation. Liu et al. pioneered the synthesis of hybrid 2D MOF/GOx nanocatalysts through the physical adsorption of GOx onto 2D MOF nanosheets, specifically 2D Cu-TCPP(Fe) nanosheets (Fig. 2A).111 Within this hybrid system, GOx orchestrates the continuous conversion of nontoxic glucose into abundant gluconic acid and H2O2. The resulting gluconic acid lowers the pH of the environment to 3–4, significantly activating the POD-like activity of 2D Cu-TCPP(Fe) nanosheets. This activation catalyzes the release of highly toxic hydroxyl radicals from H2O2, ultimately leading to bacterial death. Compared to the control group, the experimental mice did not produce any erythema during wound healing (Fig. 2D). As shown in Fig. 2E and F, untreated E. coli and S. aureus had smooth surfaces with intact cell walls, which were wrinkled and damaged after treatment with glucose + 2D MOF/GOx. As shown in Fig. 2G and H, at the same concentration of glucose and H2O2, the bacterial viability was significantly lower after glucose + 2D MOF/GOx treatment, which also proved the antimicrobial effect of the nanomaterial. Similarly, Wang et al. devised ZIF-8/Au–GOx (ZAG) NPs, leveraging a triple synergistic antibacterial strategy.112 In an acidic milieu facilitated by gluconic acid, ZAG NPs engage in cascade catalytic ROS production, further enhanced by Zn2+ release from ZIF-8. This synergistic approach results in an overall improved antibacterial performance, enabling ZAG NPs to eradicate 100% (106 CFU mL−1) of E. coli and S. aureus at concentrations of 8 μg mL−1 and 4 μg mL−1, respectively. In a similar vein, Zhou et al. developed a MOF-based nanocatalytic material termed MnFe2O4@MIL/Au&GOx (MMAG), which exhibited exceptional antibacterial activity. In addition to the role of GOx, MnFe2O4NPs contribute to the process by deleting GSH, effectively weakening the bacterial intracellular defense systems.113
Fig. 2. (A) Composition of the hybrid 2D MOF/GOx nanocatalyst. (B) Application of the 2D MOF/GOx as a band-aid for wound healing in mice. (C) Illustration of the benign self-activating cascade mechanism of the hybrid 2D MOF/GOx nanocatalyst. (D) Sequential photographs of S. aureus-infected mouse wounds at various time intervals. Six groups were subjected to different treatments: (1) blank band-aid, (2) glucose + blank band-aid, (3) glucose + 2D MOF band-aid, (4) 2D MOF/GOx band-aid, (5) glucose + GOx band-aid, (6) glucose + 2D MOF/GOx band-aid. Statistically significant differences in the obtained data (P < 0.05) are indicated. (E) Scanning electron microscope (SEM) images of E. coli and (F) S. aureus. The inset shows the corresponding photograph after the addition of methyl red. Bacterial samples from groups 1 to 6 are incubated with (1) PBS, (2) glucose, (3) glucose + 2D MOF nanosheets, (4) 2D MOF/GOx, (5) glucose + GOx, (6) glucose + 2D MOF/GOx. (G) Comparison of E. coli survival and (H) S. aureus survival treated with H2O2 and glucose + 2D MOF/GOX. Errors are represented as standard deviations from the mean (n = 3).111.
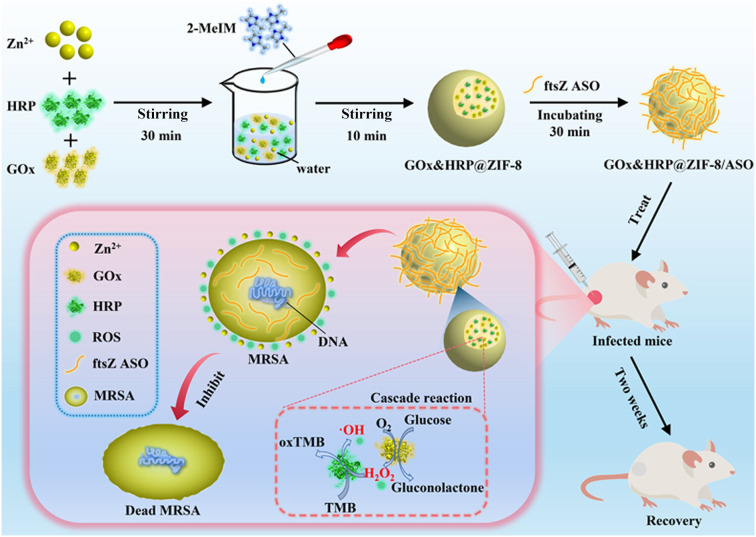
Zhang et al. pioneered a novel nanomaterial designed to combat drug-resistant bacterial infections (Fig. 3).114 This innovative approach involved encapsulating GOx and HRP within the ZIF-8, followed by complexation with antisense oligonucleotides (ASOs) to generate GOx&HRP@ZIF-8/ASO NPs. Extensive in vitro testing using the checkerboard method demonstrated the efficacy of these nanomaterials against E. coli, S. aureus, and methicillin-resistant S. aureus (MRSA) under the action of glucose. Notably, the minimum inhibitory concentration for MRSA is only 16 μg mL−1. In addition, in the presence of glucose, GOx&HRP@ZIF-8/ASO exhibits the potential to promote wound healing effectively and demonstrate excellent in vivo biocompatibility. Likewise, Cheng et al. employed a collaborative approach by con-encapsulating GOx and l-arginine (l-Arg) within Cu-MOF (CuBDC) to synergistically achieve potent antibacterial effects.115l-Arg/GOx@CuBDC exhibits remarkable antibacterial efficiency, with inactivation rates of 98% against E. coli (at 38 μg mL−1) and 97% against S. aureus (at 3.8 μg mL−1). In vivo experiments in mice confirmed the biocompatibility of l-Arg/GOx@CuBDC nanomaterials. Furthermore, Li et al. designed a new nanozyme by loading individual Ag NCs (50 nm) and GOx in a ZIF, subsequently coating them with hyaluronic acid (HA).116 This composite material displays excellent biocompatibility and selectivity, effectively inhibiting the growth of two model bacterial strains at low concentrations (5 μg mL−1 for E. coli and 10 μg mL−1 for S. aureus).
Fig. 3. Schematic diagram of the preparation route and synergistic sterilization of GOx&HRP@ZIF-8/ASO NPs in the treatment of MRSA wound infection.114.
In summary, natural enzymes are poorly stabilized and easily inactivated, and combining natural enzymes with MOFs can improve the stability of the materials and expand their application scenarios. Combining MOFs materials with POD-like enzyme activity with GOx can produce a cascade reaction that utilizes the generated ROS to achieve bacterial damage and elimination. This also provides new types of strategy for designing and constructing new MOF-based nanozymes.
3.1.3. Synergistic antibacterial strategies: combining composite MOF-based nanozymes with PTT, PDT and SDT
In the realm of antibacterial therapies, the potential of photothermal therapy (PTT), photodynamic therapy (PDT) and sonodynamic therapy (SDT) has garnered significant attention alongside well-established nanozyme-based approaches.117–119 These innovative treatments offer non-invasive alternatives that have shown promise in combating bacterial infections. PTT exploits the properties of photothermal agents to induce localized hyperthermia effectively. Under NIR light irradiation, PTT can combat drug-resistant bacteria and dismantle bacterial biofilms. What sets PTT apart is its rapid action and reduced susceptibility to antibiotic resistance.120–122 However, PTT's efficacy hinges on achieving elevated temperatures,123 which necessitates high-intensity light and substantial doses of photothermal agents. While effective against pathogens, these conditions may inadvertently inflict harm to healthy tissues.124,125 Notably, MOF-based nanozymes have emerged as promising adjuncts to PTT. When integrated with PTT, MOF-based nanozymes synergistically enhance bacterial eradication and wound healing, presenting a novel antibacterial therapy.126 PDT and SDT produce large amounts of reactive ROS, such as singlet oxygen or hydroxyl radicals, in the presence of photosensitizers and acoustic sensitizers. These ROS molecules, in turn, oxidize and destroy surrounding biomolecules, ultimately leading to the elimination of pathogenic microorganisms.127–129
3.1.3.1. Advancing antibacterial potential with MOF-based nanozyme composites in photothermodynamic applications
In the realm of combating bacterial infections, researchers have explored innovative approaches that harness the combined power of MOFs and photothermodynamic methods. These endeavors aim to bolster antibacterial efficacy while minimizing the risk of resistance development, offering a promising frontier in the battle against drug-resistant bacteria. One notable achievement comes from Wu et al., who employed a straightforward hydrothermal method to synthesize NH2-MIL-125–GO–Pt.130 This MOF composite, enriched with graphene oxide (GO) and platinum NPs, exhibits exceptional photocatalytic efficiency and photothermal effect. Intriguingly, when subjected to white light irradiation for just 20 min, NH2-MIL-125–GO–Pt showcased an impressive antibacterial performance, achieving reductions of 99.94% for S. aureus and 99.12% for E. coli.
In another pioneering study, Wang et al. crafted PEG@Zr-Fc MOF hydrogels by modifying zirconium-ferrocene MOF nanosheets with polyethylene glycol dicarboxylic acid. These hydrogels harnessed the intrinsic capability of ROS to capture E. coli and S. aureus while facilitating the decomposition of H2O2. This synergistic interplay resulted in a substantial enhancement of photothermal performance and catalytic activity, leading to the effective eradication of bacterial pathogens.131
Moreover, Han et al. introduced a novel composite consisting of UIO-66-coated gold nanorods (AuNRs) enveloped within a silica shell. Engineering for the controlled release of iodine—an antimicrobial agent free from susceptibility to drug resistance—this composite demonstrates exceptional photothermal efficiency.132 Upon exposure to NIR light, the photothermal effect induced by AuNRs under irradiation triggered the controlled release of iodine, exerting light-triggered antibacterial activity against S. aureus and E. coli.
3.1.3.2. Advancing PDT with MOF-based nanozyme composites
Furthermore, we delved into the innovative approaches that harness the potential of MOF-based nanozyme composites to bolster PDT in antibacterial applications. Raf et al. embarked on the synthesis of nanofibers of Cu-based coordination polymer [Cu(HBTC)(H2O)3] by a microwave-assisted hydrothermal method and prepared large particles and bulk crystals of [Cu(HBTC)(H2O)3].133 These nanomaterials harnessed a dual-action antibacterial mechanism rooted in the generation of ROS and the controlled release of Cu2+ ions. Impressively, when the concentration of these composite nanomaterials reached 250 μg mL−1, the nanofibers exhibit remarkable antibacterial rates of 99.9% against E. coli and 99.1% S. aureus. Equally noteworthy, the larger particles exhibit significant antibacterial efficacy, with rates of 96.7% for E. coli and 96.2% for S. aureus against these respective bacterial strains.
In another innovative approach, Wang et al. crafted an environmentally friendly nanohybrid material, denoted as ZnDMZ, by ingeniously combiningZIF-8 and Zn-doped MoS2 (Zn–MoS2) nanosheets. ZnDMZ exhibited enhanced photocatalytic prowess under 660 nm illumination, leading to the generation of an amplified quantity of ROS capable of targeting bacteria.134 After 20 min of light irradiation, ZnDMZ achieved an impressive bacteriostatic rate of 99.9% against S. aureus, underscoring its robust bactericidal potential and its ability to expedite wound healing in cases of bacterial infections.
Zhang et al. introduced an antibacterial nanomaterial, designated PCN-224-Ag–HA, by coating HA onto Ag+-loaded photosensitive MOF.135 HA, featuring a negative charge, functions as a reservoir for Ag ions, effectively controlling their release. When encountering target bacteria that secret hyaluronidase, the HA coating on PCN-224-Ag–HA underwent degradation. This triggered the release of Ag ions, which, when combined with the ROS produced by the nanomaterials under visible light irradiation, exert potent antibacterial actions. This pioneering approach exhibits promising results in the treatment of wounds infected by multi-drug resistant bacteria in mouse models.
3.1.3.3. Advancing photothermal photodynamic therapy with MOF-based nanozyme composites
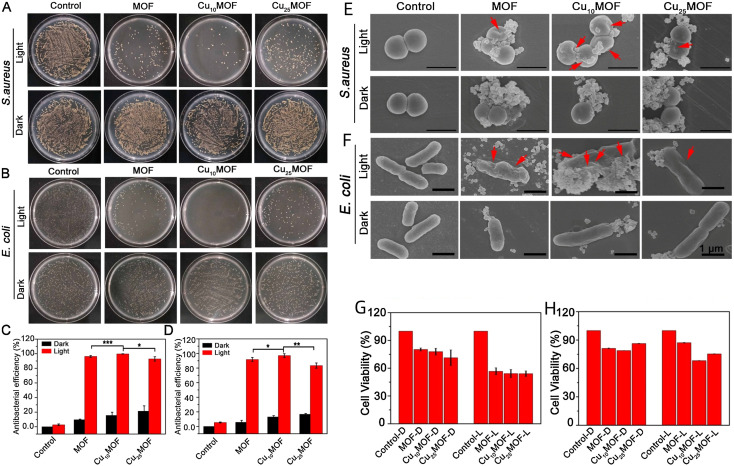
Based on the therapeutic modalities and characteristics of PDT and PTT, combining them simultaneously in MOF-based nanozymes has the potential to achieve an amplified sterilizing effect. Han et al. took a unique route by introducing Cu2+ into the porphyrin ring of PCN224, bolstering heat generation and catalytic performance of MOF and increasing the production of ROS (Fig. 4).136 They observed significant antimicrobial effects of MOF, Cu10MOF and Cu25MOF by plate coating (Fig. 4A and B) and SEM (Fig. 4E and F) with light at 660 nm. Cu10MOF had the best antimicrobial effect after using 20 min of light, with antimicrobial efficacy against S. aureus and E. coli of 99.71% and 97.14%, respectively (Fig. 4C and D). While ensuring the antibacterial effect, the three materials had no obvious cytotoxicity (Fig. 4G and H) and even promoted wound healing in rats.
Fig. 4. In vitro antibacterial activity of MOFs against S. aureus and E. coli. (A) S. aureus and (B) E. coli mixed with MOF (500 ppm) and subjected to either 660 nm (0.4 W cm−2) light exposure or kept in darkness for 20 min. The bacterial mixtures are spread on LB agar plates and incubated at 37 °C for 24 h. (C) Statistical analysis results of antibacterial activity against S. aureus and (D) E. coli. (*P < 0.1, **P < 0.05, ***P < 0.001, t-test; all experiments conducted at least thrice). The bacteria are mixed with 500 ppm MOF and subjected to 660 nm (0.4 W cm−2) light exposure or kept in the dark for 20 min before and after treatment. (E) Morphology and structure of S. aureus and (F) E. coli. Observed through scanning electron microscopy after 20 min of exposure to 660 nm (0.4 W cm−2) light or darkness (n ≥ 3). Viability assessed using the MTT method after (G) 1 d and (H) 3 days of treatment.136.
Xiong et al. pursued a different path, synthesizing NCs by subjecting ZIF-8 to pyrolysis at 800 °C, followed by surface modification with Ag2S, leading to the creation of Ag2S/NCs.137 Ag2S/NCs exhibit commendable photothermal conversion efficiency and potent photodynamic function. Irradiation with 808 nm NIR light rapidly induced both heat generation and ROS production, resulting in a notable 97.3% increase in resistance to S. aureus within 20 min. To further amplify antibacterial potency, Yu et al. employed an in situ sulfidation technique, embedding CuS NPs within a Cu-based MOF structure, known as HKUST-1.138 This MOF, with its remarkable photodynamic and photothermal attributes, achieved a striking bactericidal efficiency rate of 99.70% against S. aureus and 99.80% against E. coli under 20 min of NIR light exposure.
Enhancing antibacterial activity through the incorporation of PDA into MOF NPs proved to be another successful strategy. Han et al. synthesized CuS within the Cu-based MOF-HKUST NP structure and subsequently coated them with PDA, resulting in CuS@HKUST-PDA.139 This combination synergy significantly enhanced photothermal and photocatalytic performance, amplifying the material's capacity for free radical generation. As a result, it exhibited robust antibacterial efficiency, killed 99.77% of S. aureus and E. coli under light exposure. Furthermore, Han et al. innovatively synthesized a novel MOF material using benzoic acid, meso-tetra(4-carboxyphenyl)porphine (H4TCPP) and zirconium oxychloride octahydrate, and then modified it with PDA through a simple self-polymerization reaction, yielding MOF-PDA.140 This unique combination unlocked enhanced photothermal and photocatalytic performance, leading to increased ROS production. Under 660 nm light irradiation for 20 min, MOF-PDA exhibited remarkable antibacterial effectiveness, achieving rates of 99.62% and 99.97% against S. aureus and E. coli, respectively.
3.1.3.4. Pioneering sonodynamic therapy using MOF-based nanozyme composites
SDT is a cutting-edge approach that utilizes ultrasound to trigger sonosensitizers, stimulating the production of ROS for therapeutic purposes. Notably, SDT boasts the advantage of deeper tissue penetration, setting it apart from PDT, which has been primarily employed in cancer diagnosis and treatment.141–143 Recent strides in nanotechnology have extended the utility of SDT to address bacterial infections.
Sun et al. achieved a groundbreaking advancement with their development of Pd@Pt-T790 MOF-based nanozyme constructs, exhibiting formidable antibacterial efficacy.13 These nanozymes autonomously trigger CAT-like activity in response to ultrasonic radiation, facilitating an ample oxygen supply. This innovation empowers SDT to induce ROS generation, unlocking its therapeutic potential for the treatment of deep-seated MRSA infections.
In a pioneering study, Pan et al. used ZIF-8-derived carbon@TiO2 NPs (ZTNs) as inhalable sonosensitizers to combat bacterial pneumonia.144 Under ultrasound irradiation, ZTNs generate ROS in vitro, effectively eliminating multidrug-resistant Gram-negative bacteria. Innovative intratracheal nebulization techniques enabled precise delivery of ZTNs to the site of lung infections, where SDT efficiently cleared multidrug-resistant Gram-negative bacteria in immunocompetent and immunocompromised mouse models. Remarkably, ultrasound-irradiated ZTNs achieved a 100% survival rate in severely immunodeficient mice with life-threatening bacterial pneumonia, with no obvious toxicity observed at the cellular and animal levels.
3.1.3.5. Advancements in antibacterial therapy using MOF-based nanozyme composites
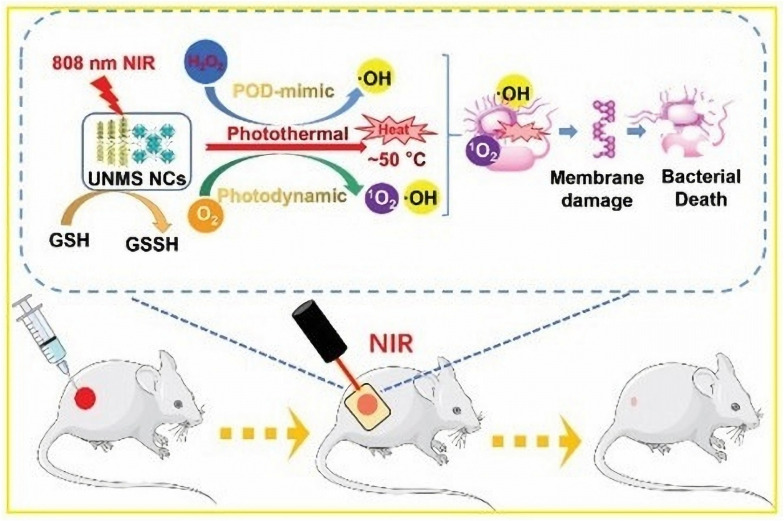
In a seminal advancement, Zhang et al. constructed Zr-MOF-based UiO-66–NH–CO–MoS2 nanocomposites (UNMS NCs).145 These innovative NCs, bearing a cationic charge, proficiently entrapped and immobilized bacteria through electrostatic interactions. Notably, UNMS NCs harnessed the synergistic potential of photothermal, photodynamic, and POD-like activities collectively culminating in the effective annihilation of bacterial pathogens under irradiation with 808 nm NIR light (Fig. 5). The application of NIR light induced a remarkable bactericidal rate, reaching 99.7% against MRSA and 96.7% against ampicillin-resistant E. coli (AREC). In addition, the high temperature induced by 808 nm radiation in UNMS NCs expedited the oxidation process of GSH, effectively incapacitating bacterial intercellular defense mechanisms and significantly augmenting the antibacterial efficiency. In a parallel endeavor, Wang et al. designed and synthesized new MOF nanosheets (CoS2/MoS2 NSs).146 These nanosheets demonstrated exceptional photocatalytic properties and concurrently exhibited both oxidase and POD-like activities when exposed to ultrapure water. CoS2/MoS2 NSs exhibited robust antibacterial potential, effectively targeting E. coli and S. aureus. Moreover, Hatamie et al. embarked on a solvothermal synthesis to fabricate graphene oxide/cobalt metal–organic framework (GO/Co-MOF) composites.147 This composite material, born from the combination of cobalt salt and terephthalic acid, effectively curbed the proliferation of E. coli and S. aureus.
Fig. 5. Schematic diagram of the bactericidal mechanism of UNMS NCs and their application in wound healing.145.
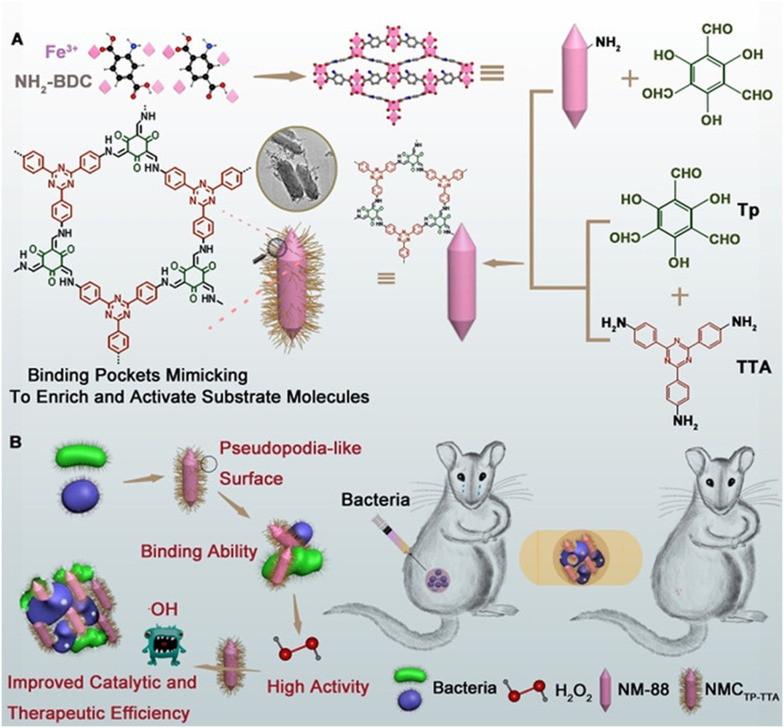
In an intriguing innovation, Zhang et al. introduced a MOF@COF nanozyme, which exhibited a pseudo-POD effect (Fig. 6).148 Covalent organic frameworks (COFs), recognized for their morphology-controlled organic porous polymers, find application in catalytic reactions.149,150 By imitating the functions of amino acid residues and providing a hydrophobic spatial pore structure, COFs tailored the pore microenvironment around the active center. MOF@COF, replete with active sites in this customized microenvironment and a pseudopod-like surface, amplified the catalytic potential of MOF-based nanozymes. Consequently, it substantially elevated therapeutic efficiency, thereby enhancing bacterial inhibition.
Fig. 6. (A) Synthesis of hybrid MOF@COF nanozymes. (B) Application of hybrid MOF@COF nanozymes to inhibit bacteria.148.
In summary, the increase of drug-resistant bacterial infections arrives a great challenge for clinical treatment, and it is difficult to achieve the ideal therapeutic effect with the current single antimicrobial modality. Combining MOF-based nanozymes with PTT, PDT and SDT approaches can enhance bacterial membrane permeability and promote ROS production, and this multimodal synergistic therapy has great potential for clinical applications in bacterial infections.
3.2. Antibiotic membrane targeting bacterial biofilms with MOF-based nanozymes
Biofilm formation stands as the predominant manifestation of microbial growth, and biofilms can colonize not only living organisms but also various environmental substrates.151 This intricate relationship bacterial infections and biofilms plays a pivotal role in the emergence of infectious diseases in various systems and organs.152,153 Additionally, biofilm generation significantly contributes to bacterial resistance to antibiotics.154 A bacterial biofilm constitutes a complex and robust aggregation wherein bacteria adhere to a surface, engendering copious extracellular matrix secretion to encase themselves within.155 This matrix, comprising an intricate amalgamation of proteins, lipids, nucleic acids (eDNA and eRNA), polysaccharides, and other biomolecules, imparts formidable protection to bacterial colonies. This multifaceted extracellular matrix serves as a bulwark against environmental adversities, evades host immune responses, and withstands antibiotic attacks.156,157 The resultant resistance mechanisms, distinct from conventional antimicrobial resistance, render biofilms recalcitrant and arduous to eliminate.154,158,159 Therefore, investigating and formulating antibacterial agents with a specific focus on biofilm eradication holds substantial promise. Notably, MOF-based nanozymes have emerged as potent candidates for disrupting and eliminating bacterial biofilms.
Liu et al. designed a series of MOF/Ce nanozymes with DNase and POD mimetic activities.160 Cerium(iv) complexes, functioning as deoxyribonucleases, effectively catalyze the hydrolysis of eDNA, consequently destabilizing mature biofilms. In parallel, MOFs with POD-like activity efficiently eliminated bacteria exposed within dispersed biofilms in the presence of H2O2, precluding bacterial reactivation, colonization, and recurrent biofilm formation. The synergistic interplay between these two nanozymes improves the anti-biofilm efficacy, substantially impeding the formation of bacterial biofilms. In a separate innovative approach, Qiu et al. designed CeO2-modified PCN-224@CeO2.157 CeO2 disrupts bacterial initial adhesion by inhibiting the function of extracellular ATP (eATP), while cytotoxic ROS produced by MOF materials resist bacterial populations. This dual-action mechanism has demonstrated remarkable efficacy in preventing the formation of biofilms.
Zheng et al. pioneered an innovative antimicrobial agent, effective against Gram-positive and Gram-negative bacteria, by coupling gold nanoclusters (AuNCs) within titanium carbide (MXene).161 MXene disrupts bacterial membranes, while AuNC, once inside bacterial cells, generates a high concentration of ROS, leading to membrane rupture. Furthermore, the unique wrinkled structure of MXene–AuNCs effectively inhibited the formation of biofilms. Yu et al. designed a PCN-222-Pt nanozyme composite characterized by superior biocompatibility.162 PCN-222-Pt, boasting robust oxidase and POD-like activities, spontaneously generates ROS, exhibiting compelling anti-biofilm properties in vitro. Experimental data underscored the ability of PCN-222-Pt to resist biofilm formation, achieving reductions of bacterial biofilm populations, with rates of 98.69% against S. aureus and 99.91% against E. coli within 1 h. Wang et al. harnessed a straightforward one-step methodology to fabricate two-dimensional Cu-MOF nanosheets (Cu-MOF NSs) composed of Cu ions and 2-methylimidazole.163 The two-dimensional morphology of Cu-MOF NSs with POD-like activity provides a high density of Cu2+ surface active sites, facilitating the efficient oxidation of proteins and lipids on the bacterial surface, ultimately inducing bacterial death. Experimental results underscored the capacity of prepared 2D Cu-MOF NSs to effectively eradicate the biofilm of S. aureus, achieving a remarkable reduction of up to 99.9% of bacteria at a concentration of 4 μg mL−1.
Yu et al. introduced a pioneering acid-responsive ROS composite nanomaterial for biofilm introduced a pioneering one-pot synthesis. This material amalgamated folic acid (FA) and lysine carbon dots (Lys-CD) to form ZIF-8@Lys-CD@FA.164 ZIF-8@Lys-CD@FA exhibited CAT-like activity, engendering superoxide radicals and hydroxyl radicals, thus inflicting more potent oxidative stress and effectively dismantling mature biofilms.
As functional nanomaterials, MOF-based nanozymes have great potential to replace antibiotics for effective treatment of biofilm infections caused by bacteria. Different nanozymes can exhibit different enzymatic activities, such as oxidase-like, POD-like and CAT-like activities, they can all produce chemical reactions that effectively attack the bacterial biofilm, thus realizing the complete elimination of bacteria. The rational construction and design of MOF-based nanozymes to eliminate biofilms provide a new antimicrobial idea on the way to treat bacterial infections.
3.3. Antibacterial and anti-biofilm properties of MOF-based nanozymes
MOF-based nanozymes exhibit multifaceted capabilities, wielding their influence against bacteria through direct antibacterial mechanisms and the disruption of biofilm formation. These versatile nanozymes not only curtail bacterial proliferation but also dismantle and eradicate biofilms, offering a holistic approach to antibacterial strategies.
Huang et al. in situ encapsulated palladium (Pd) nanocrystals, endowed with oxidase and POD activity, within the intricate structure of MOF UiO-66. Subsequent surface modification with polyazobenzene (PAzo) yielded the Pd-MOF@PAzo@SNP nanoplatform.165 This nanozyme acts synergistically complemented NO therapy, manifesting exceptional antibacterial and antibiofilm characteristics, along with an accelerated wound healing capability.
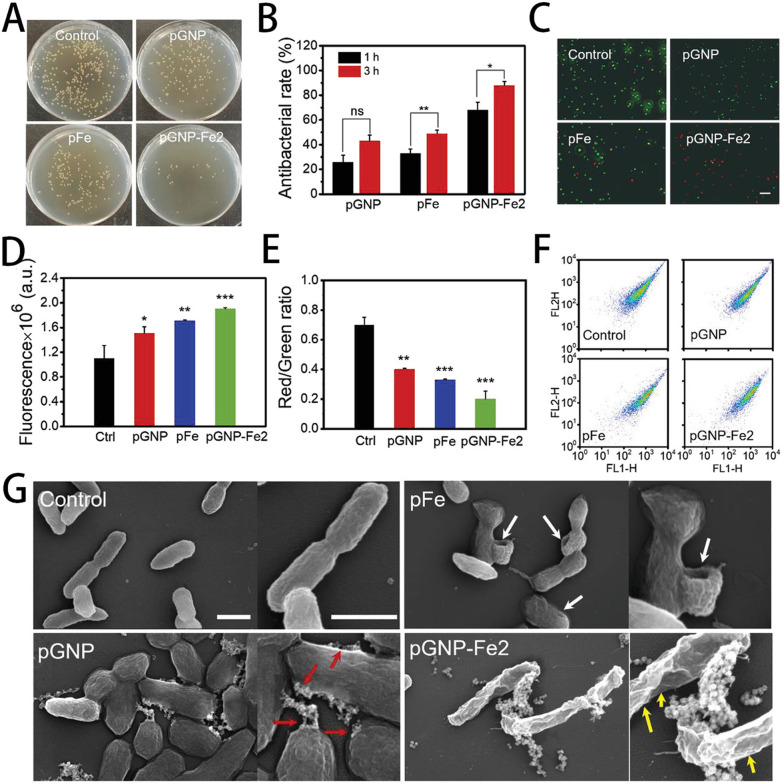
Liu et al. prepared an innovative method to synthesize AuNPs (pGNP-Fe) using phenolic compounds extracted from botanical sources as a dual-purpose reducing and blocking agent, accompanied by Fe3+ as a complexing agent. pGNP-Fe efficiently adheres to bacterial membranes, exerting mechanical stresses that cause cellular deformation and membrane impairment.166 This cascade process, augmented by its oxidase and POD properties, engendered the production of ROS while collaborating with the Fenton reaction, ultimately heightening its antibacterial efficiency. The antimicrobial effect of pGNP-Fe2 was demonstrated to be higher than that of pGNP and pFe groups, and the antibacterial effect increased with time (Fig. 7A–C). As shown in Fig. 7D–F, the pGNP-Fe2 group had the highest penetration rate, which was more favorable for the interaction between bacteria and samples. As shown in Fig. 7G, the bacteria in the control group had a typical rod shape. Bacteria in the other three groups were damaged to varying degrees, with the pGNP-Fe2-treated bacteria being the most severely damaged of the three groups. Significantly, this material achieves high-efficiency and long-lasting antibacterial effects, independent of external antimicrobial agents and antibiotics (Fig. 7). Huang et al. reported a MOF-derived nanocarbon composite, composed of Zn and a graphitic carbon framework, that incorporated AgNPs through a displacement reaction between Zn and Ag+.167 Upon NIR light irradiation, this composite generated substantial heat, which disrupted bacterial membranes, while concurrently releasing Zn+ and Ag+, causing chemical damage to bacterial intracellular components. Antibacterial experiments underscored its nearly 100% bactericidal efficacy against high bacterial concentrations at exceedingly low doses (0.16 mg mL−1).
Fig. 7. Antibacterial efficiency and bacterial membrane interactions: (A) optical images of colony-forming units of E. coli after 3 h of treatment. (B) Antibacterial rates after 1 h and 3 h of treatment. (C) Live/dead fluorescence images of treated bacteria (scale bar = 20 μm). (D) Effects of different treatments on the permeability of the outer membrane of E. coli. (E) The red/green ratio representing the membrane potential. (F) Flow cytometry arrays of different treatments. (G) SEM images of treated bacteria (scale bar = 1 μm).166.
Nong et al. took NH2-MIL-88B(Fe) a step further by incorporating Fe3O4, resulting in the synthesis of the Fe3O4@PVP@MIL-88B(Fe)–NH–lysozyme/carvacrol (FPMLC) nanozyme.168 FPMLC hydrolyzed the peptidoglycan layer of the bacterial cell wall through lysozymes, releasing carvacrol upon NIR light irradiation and causing bacterial cell membrane disruption. Even at a low dose (100 μg mL−1) and brief NIR light exposure (10 min), FPMLC exhibits 100% inactivation of E. coli and S. aureus (106 CFU mL−1) without obvious cytotoxicity.
Wang et al. designed a phototherapeutic antibacterial material by embedding peptides within a hydrogel network, with copper sulfide nanodots (CuS NDS) distributed throughout the hydrogel.169 Antimicrobial peptides executed their function, while CuS NDS responded to NIR light exposure with ROS generation and photothermal effects. The resultant heat and ROS, acting as non-contact antimicrobial agents, coupled with direction action by antimicrobial peptides, culminated in irreversible membrane disruption, cell content disruption, and bacterial thermal ablation.
Li et al. loaded GOx and bovine hemoglobin (BHb) on ZIF-8, culminating in a MOF-based cascade nanoreactor (ZIF-8@GOx@BHb).170 GOx orchestrated the production of H2O2 while consuming glucose, thus depriving bacteria of nutrients, while BHb continuously delivered O2 to GOx. Capitalizing on the remarkable POD-like activity of ZIF-8@GOx@BHb, this nanomaterial effectively impeded the growth of MRSA and E. coli and obliterated MRSA biofilms, offering a robust defense against bacterial infections. Notably, this constructed nanoreactor underwent degradation and excretion via feces with negligible biological toxicity.
Li et al. prepared hybrid MOF/enzyme nanoreactors (MIL@GOx-MIL NRs) based on MIL NPs and GOx by a two-step synthesis method.171 GOx catalyzed the conversion of glucose to gluconic acid, lowering the pH from 7.4 to around 4. At pH 4, MIL@GOx-MIL NRs produced more hydroxyl radicals, suppressing bacterial growth. Experimental results illustrated that even at a concentration of 5 μg mL−1, MIL@GOx-MIL NRs curtailed MRSA proliferation, while 80 μg mL−1 of MIL@GOx-MIL NRs eradicated MRSA biofilm formation, as confirmed via plate counting methods.
In summary, ROS leads to irreversible membrane damage to scavenge bacteria and can also cause oxidative damage directly to the intracellular, and this dual-mode pathway to enhance antimicrobial efficacy makes MOF-based nanozymes even more advantageous in the antimicrobial field.
3.4. Antifungal properties of MOF-based nanozymes
Fungus are eukaryotic organisms that can release their spores into the air and cause fungal infections. Common fungal strains include Aspergillus and Candida albicans, which pose a significant threat to human health.172 But the current drug resistance of the fungus poses a serious obstacle to its treatment.173 We need to explore new antifungal methods to treat fungal infections. In addition to their well-established antibacterial and anti-biofilm attributes, MOF-based nanozymes exhibit notable antifungal potential.
Bouson et al. employed a facile hydrothermal synthesis method to fabricate a Cu-based benzene tricarboxylate MOF (Cu–BTC MOF).174 Cu–BTC MOF displayed significant inhibitory effects against a spectrum of fungal strains, encompassing Candida albicans, Aspergillus niger, Aspergillus oryzae and Fusarium oxysporum. Intriguingly, the most effective inhibition of Candida albicans was achieved after a 60 min incubation at 500 ppm. It is postulated that the antifungal mechanism of Cu–BTC involves the reduction of O2 to generate ROS. In a parallel vein, Abdelhamid et al. undertook the synthesis of a Ce-MOF-based nanozyme (AU-1) using a solvothermal method.175 Au-1 demonstrated commendable SOD, CAT, and POD activities, rendering it proficient in fungi, such as Aspergillus flavus, Aspergillus niger, Aspergillus terreus, Candida albicans, and Rhodotorula glutinis. Impressively, AU-1 exhibited bactericidal efficiencies ranging from 93.3% to 99.3%.
The above studies demonstrated that MOF-based nanozymes were able to inhibit the growth of Aspergillus and Candida albicans to a certain extent and had excellent antifungal activity. Research of MOF-based nanozymes in antifungal field is still in the initial exploratory stage, which requires more attention and efforts to explore the application of MOF-based nanozymes in antifungal field. We expect that MOF-based nanozymes will take a new step forward in the antifungal field in the near future, thus promoting the application of MOF-based nanozymes in nanomaterials and biomedical fields.
4. Conclusion and outlook
In conclusion, MOFs represent a versatile class of materials with tunable physical, chemical, and structural properties, rendering them highly amenable to applications as nanozymes in the biomedical field. Our review has explored the burgeoning field of MOF-based nanozymes with a particular focus on their excellent antibacterial activity. As evidenced by numerous experimental studies, MOF-based nanozymes have emerged as promising candidates for bacterial diagnosis and treatment. We have surveyed a selection of recent investigations into the antibacterial capabilities of MOF-based nanozyme, revealing their potential to revolutionize the field of antibacterial therapy. While substantial progress has been made in harnessing MOF-based nanozymes for antibacterial purposes, several significant challenges and opportunities remain. Researchers must continue to explore strategies to enhance material stability and refine synthesis techniques. When acting MOF-based nanozyme in vivo, it is necessary to fully evaluate the degradability, biocompatibility, and cytotoxicity of the material to ensure that the material works without damaging the organism.
As a relatively new research direction, the unique properties of MOF-based nanozymes give them great potential for bacterial diagnosis and therapy. Accurate and effective diagnosis is as important as antimicrobial therapy in the prevention and treatment of diseases caused by bacterial infections. And as bacterial resistance increases, we need to develop new antimicrobial agents to deal with bacterial infections. The translation of MOF-based nanozyme antibacterial activity into clinically viable diagnostic and therapeutic modalities is a promising frontier that necessitates continued investigation. The emergence of MOF-based nanozymes has given us a new way of thinking when it comes to fighting bacterial infections. They can accelerate infected wound healing, treat orthopedic implant-associated infections and even cancer therapy. Furthermore, in addition to its application in living organisms, it can also expand its scope of application in food safety inspection and packaging, drinking water testing and disinfection, packaging and storage of pharmaceuticals, air sterilization and filtration, and surface disinfection in public areas, thereby mitigating the risk of pathogens to humans. Given the multifaceted functionality and catalytic prowess inherent to MOF-based nanozymes, it is reasonable to anticipate their pivotal role in shaping the landscape of bacterial infection diagnosis and treatment and even in the biomedical field in the foreseeable future. We hope that this review will help researchers to fully understand the current situation and further advance the development of MOF-based nanozymes in bacterial diagnostics and therapy.
Author contributions
Yiwei An: conceptualization, date curation, investigation, visualization, writing original draft, writing review & editing. Jie Cheng: software, supervision, visualization. Xuankun Fang: visualization, software, validation. Shuiyuan Yang: formal analysis. Zuanguang Chen: resources, formal analysis. Yanli Tong: conceptualization, resources, supervision, writing review & editing, funding acquisition.
Conflicts of interest
The authors have no conflicts to declare.
Supplementary Material
Acknowledgments
This study is reinforced via funding from National Natural Science Foundation of China (No. 82304435) and Guangzhou Municipal (Academy) Enterprise Joint Key Projects (No. 2023A03J0882).
References
- Garcia-Viloca M. Gao J. Karplus M. Truhlar D. G. Science. 2004;303:186–195. doi: 10.1126/science.1088172. [DOI] [PubMed] [Google Scholar]
- Kuah E. Toh S. Yee J. Ma Q. Gao Z. Q. Chem. – Eur. J. 2016;22:8404–8430. doi: 10.1002/chem.201504394. [DOI] [PubMed] [Google Scholar]
- Lin Y. H. Ren J. S. Qu X. G. Acc. Chem. Res. 2014;47:1097–1105. doi: 10.1021/ar400250z. [DOI] [PubMed] [Google Scholar]
- Manea F. Houillon F. B. Pasquato L. Scrimin P. Angew. Chem., Int. Ed. 2004;43:6165–6169. doi: 10.1002/anie.200460649. [DOI] [PubMed] [Google Scholar]
- Wu J. J. X. Wang X. Y. Wang Q. Lou Z. P. Li S. R. Zhu Y. Y. Qin L. Wei H. Chem. Soc. Rev. 2019;48:1004–1076. doi: 10.1039/C8CS00457A. [DOI] [PubMed] [Google Scholar]
- Wei H. Wang E. K. Chem. Soc. Rev. 2013;42:6060–6093. doi: 10.1039/C3CS35486E. [DOI] [PubMed] [Google Scholar]
- Kotov N. A. Science. 2010;330:188–189. doi: 10.1126/science.1190094. [DOI] [PubMed] [Google Scholar]
- Huang Y. Y. Ren J. S. Qu X. G. Chem. Rev. 2019;119:4357–4412. doi: 10.1021/acs.chemrev.8b00672. [DOI] [PubMed] [Google Scholar]
- Huang X. Zhang S. T. Tang Y. J. Zhang X. Y. Bai Y. Pang H. Coord. Chem. Rev. 2021;449:214216. doi: 10.1016/j.ccr.2021.214216. [DOI] [Google Scholar]
- Chen Q. M. Li S. Q. Liu Y. Zhang X. D. Tang Y. Chai H. X. Huang Y. M. Sens. Actuators, B. 2020;305:127511. doi: 10.1016/j.snb.2019.127511. [DOI] [Google Scholar]
- Liu W. D. Chu L. Zhang C. H. Ni P. J. Jiang Y. Y. Wang B. Lu Y. Z. Chen C. X. Chem. Eng. J. 2021;415:128876. doi: 10.1016/j.cej.2021.128876. [DOI] [Google Scholar]
- Cao F. F. Zhang L. Wang H. You Y. W. Wang Y. Gao N. Ren J. S. Qu X. G. Angew. Chem., Int. Ed. 2019;58:16236–16242. doi: 10.1002/anie.201908289. [DOI] [PubMed] [Google Scholar]
- Sun D. Pang X. Cheng Y. Ming J. Xiang S. J. Zhang C. Lv P. Chu C. C. Chen X. L. Liu G. Zheng N. F. ACS Nano. 2020;14:2063–2076. doi: 10.1021/acsnano.9b08667. [DOI] [PubMed] [Google Scholar]
- Wei F. Cui X. Y. Wang Z. Dong C. C. Li J. D. Han X. J. Chem. Eng. J. 2021;408:127240. doi: 10.1016/j.cej.2020.127240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng R. Chang R. Zou Q. L. Shen G. Z. Jiao T. F. Yan X. H. Small. 2021;17:2008114. doi: 10.1002/smll.202008114. [DOI] [PubMed] [Google Scholar]
- Xu W. Q. Jiao L. Yan H. Y. Wu Y. Chen L. J. Gu W. L. Du D. Lin Y. H. Zhu C. Z. ACS Appl. Mater. Interfaces. 2019;11:22096–22101. doi: 10.1021/acsami.9b03004. [DOI] [PubMed] [Google Scholar]
- Dong S. M. Dong Y. S. Jia T. Liu S. K. Liu J. Yang D. He F. Gai S. L. Yang P. P. Lin J. Adv. Mater. 2020;32:2002439. doi: 10.1002/adma.202002439. [DOI] [PubMed] [Google Scholar]
- Zhang X. L. Li G. L. Wu D. Li X. L. Hu N. Chen J. Chen G. Wu Y. N. Biosens. Bioelectron. 2019;137:178–198. doi: 10.1016/j.bios.2019.04.061. [DOI] [PubMed] [Google Scholar]
- Li S. S. Shang L. Xu B. L. Wang S. H. Gu K. Wu Q. Y. Sun Y. Zhang Q. H. Yang H. L. Zhang F. R. Gu L. Zhang T. R. Liu H. Y. Angew. Chem., Int. Ed. 2019;58:12624–12631. doi: 10.1002/anie.201904751. [DOI] [PubMed] [Google Scholar]
- Fan K. L. Wang H. Xi J. Q. Liu Q. Meng X. Q. Duan D. M. Gao L. Z. Yan X. Y. Chem. Commun. 2017;53:424–427. doi: 10.1039/C6CC08542C. [DOI] [PubMed] [Google Scholar]
- Wang Z. R. Zhang R. F. Yan X. Y. Fan K. L. Mater. Today. 2020;41:81–119. doi: 10.1016/j.mattod.2020.08.020. [DOI] [Google Scholar]
- Wang Q. Q. Wei H. Zhang Z. Q. Wang E. K. Dong S. J. TrAC, Trends Anal. Chem. 2018;105:218–224. doi: 10.1016/j.trac.2018.05.012. [DOI] [Google Scholar]
- Yaghi O. M. Li H. J. Am. Chem. Soc. 1995;117:10401–10402. doi: 10.1021/ja00146a033. [DOI] [Google Scholar]
- Yaghi O. M. O'Keeffe M. Ockwig N. W. Chae H. K. Eddaoudi M. Kim J. Nature. 2003;423:705–714. doi: 10.1038/nature01650. [DOI] [PubMed] [Google Scholar]
- Niu X. H. Li X. Lyu Z. Y. Pan J. M. Ding S. C. Ruan X. F. Zhu W. L. Du D. Lin Y. H. Chem. Commun. 2020;56:11338–11353. doi: 10.1039/D0CC04890A. [DOI] [PubMed] [Google Scholar]
- Ali A. Ovais M. Zhou H. G. Rui Y. K. Chen C. Y. Biomaterials. 2021;275:120951. doi: 10.1016/j.biomaterials.2021.120951. [DOI] [PubMed] [Google Scholar]
- Hou J. J. Xianyu Y. L. Small. 2023:2302640. doi: 10.1002/smll.202302640. [DOI] [PubMed] [Google Scholar]
- Li Y. Y. Zhu W. X. Li J. S. Chu H. T. Colloids Surf., B. 2021;198:111465. doi: 10.1016/j.colsurfb.2020.111465. [DOI] [PubMed] [Google Scholar]
- Han D. L. Liu X. M. Wu S. L. Chem. Soc. Rev. 2022;51:7138–7169. doi: 10.1039/D2CS00460G. [DOI] [PubMed] [Google Scholar]
- Quijia C. R. Alves R. C. Hanck-Silva G. Frem R. C. G. Arroyos G. Chorilli M. Crit. Rev. Microbiol. 2022;48:161–196. doi: 10.1080/1040841X.2021.1950120. [DOI] [PubMed] [Google Scholar]
- Gaynes R. Edwards J. R. Natl Nosocomial I. Clin. Infect. Dis. 2005;41:848–854. doi: 10.1086/432803. [DOI] [PubMed] [Google Scholar]
- Gupta A. Mumtaz S. Li C. H. Hussain I. Rotello V. M. Chem. Soc. Rev. 2019;48:415–427. doi: 10.1039/C7CS00748E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochado A. R. Telzerow A. Bobonis J. Banzhaf M. Mateus A. Selkrig J. Huth E. Bassler S. Beas J. Z. Zietek M. Ng N. Foerster S. Ezraty B. Py B. Barras F. Savitski M. M. Bork P. Göttig S. Typas A. Nature. 2018;559:259–263. doi: 10.1038/s41586-018-0278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerill F. R. Wilson J. W. Vetter E. A. Goodman K. M. Torgerson C. A. Harmsen W. S. Schleck C. D. Ilstrup D. M. Washington J. A. Wilson W. R. Clin. Infect. Dis. 2004;38:1724–1730. doi: 10.1086/421087. [DOI] [PubMed] [Google Scholar]
- Mwaigwisya S. Assiri R. A. M. O'Grady J. Expert Rev. Mol. Diagn. 2015;15:681–692. doi: 10.1586/14737159.2015.1029459. [DOI] [PubMed] [Google Scholar]
- Harbarth S. Hawkey P. M. Tenover F. Stefani S. Pantosti A. Struelens M. J. Int. J. Antimicrob. Agents. 2011;37:110–117. doi: 10.1016/j.ijantimicag.2010.10.022. [DOI] [PubMed] [Google Scholar]
- Plummer M. Franceschi S. Vignat J. Forman D. de Martel C. Int. J. Cancer. 2015;136:487–490. doi: 10.1002/ijc.28999. [DOI] [PubMed] [Google Scholar]
- Tang Y. J. Ali Z. Zou J. Jin G. Zhu J. C. Yang J. Dai J. G. RSC Adv. 2017;7:51789–51800. doi: 10.1039/C7RA09064A. [DOI] [Google Scholar]
- Yu J. L. Su J. Zhang J. Wei X. T. Guo A. L. RSC Adv. 2017;7:17819–17823. doi: 10.1039/C7RA00821J. [DOI] [Google Scholar]
- Zhong L. L. Zhou Q. Tan C. Y. Roberts A. P. Ahmed M. A. E. Chen G. P. Dai M. Yang F. Xia Y. Liao K. Liang Y. J. Yang Y. Q. Feng S. Y. Zheng X. B. Tian G. B. Infect. Drug Resist. 2019;12:1877–1887. doi: 10.2147/IDR.S210226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D. C. Zhu Y. Z. Li N. Lu Y. Cheng J. Xu Y. C. Sens. Actuators, B. 2020;310:127834. doi: 10.1016/j.snb.2020.127834. [DOI] [Google Scholar]
- Zheng L. Y. Cai G. Z. Wang S. Y. Liao M. Li Y. B. Lin J. H. Biosens. Bioelectron. 2019;124:143–149. doi: 10.1016/j.bios.2018.10.006. [DOI] [PubMed] [Google Scholar]
- Ali A. A. Altemimi A. B. Alhelfi N. Ibrahim S. A. Biosensors. 2020;10:58. doi: 10.3390/bios10060058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesewski E. Johnson B. N. Biosens. Bioelectron. 2020;159:112214. doi: 10.1016/j.bios.2020.112214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. J. Sun D. W. Pu H. B. Small. 2022;18:2200178. doi: 10.1002/smll.202200178. [DOI] [PubMed] [Google Scholar]
- Yu K. Li M. J. Chai H. N. Liu Q. Hai X. Tian M. W. Qu L. J. Xu T. L. Zhang G. Y. Zhang X. J. Chem. Eng. J. 2023;451:138321. doi: 10.1016/j.cej.2022.138321. [DOI] [Google Scholar]
- Huang Y. Zhao M. T. Han S. K. Lai Z. C. Yang J. Tan C. L. Ma Q. L. Lu Q. P. Chen J. Z. Zhang X. Zhang Z. C. Li B. Chen B. Zong Y. Zhang H. Adv. Mater. 2017;29:1700102. doi: 10.1002/adma.201700102. [DOI] [PubMed] [Google Scholar]
- Wang Z. Y. Huang Y. C. Xu K. Q. Zhong Y. Y. He C. H. Jiang L. P. Sun J. K. Rao Z. Zhu J. N. Huang J. Xiao F. Liu H. F. Xia B. Y. Nat. Commun. 2023;14:69. doi: 10.1038/s41467-022-35721-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X. X. Li Y. L. Qi S. Y. Chen Y. Yin M. Zhang L. Tian X. X. Gong S. H. Wang F. K. Zhu Y. W. Liu Y. Qiu J. X. Xu D. P. J. Inorg. Organomet. Polym. Mater. 2022;32:3595–3600. doi: 10.1007/s10904-022-02422-w. [DOI] [Google Scholar]
- Yao R. H. Li Z. J. Li J. Tuo K. Zhang D. B. Fan C. B. Liu G. Deng Y. H. Pu S. Z. Microchem. J. 2023;193:109214. doi: 10.1016/j.microc.2023.109214. [DOI] [Google Scholar]
- Yang B. W. Yao H. L. Yang J. C. Chen C. Shi J. L. Nat. Commun. 2022;13:1988. doi: 10.1038/s41467-022-29735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y. H. Han Y. Zhao J. C. Lv Y. F. Fan C. Y. Zheng L. Zhang Z. S. Liu Z. G. Li C. Lin Y. H. Nano-Micro Lett. 2023;15:112. doi: 10.1007/s40820-023-01059-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. Zhang Y. Wang Z. Z. Cao F. F. Sang Y. J. Dong K. Pu F. Ren J. S. Qu X. G. Mater. Horiz. 2019;6:1682–1687. doi: 10.1039/C9MH00339H. [DOI] [Google Scholar]
- Yang Y. Zhu D. M. Liu Y. Jiang B. Jiang W. Yan X. Y. Fan K. L. Nanoscale. 2020;12:13548–13557. doi: 10.1039/D0NR02800B. [DOI] [PubMed] [Google Scholar]
- Yin S. Y. Song G. S. Yang Y. Zhao Y. Wang P. Zhu L. M. Yin X. Zhang X. B. Adv. Funct. Mater. 2019;29:1901417. doi: 10.1002/adfm.201901417. [DOI] [Google Scholar]
- Ivnitski D. Abdel-Hamid I. Atanasov P. Wilkins E. Stricker S. Electroanalysis: An International Journal Devoted to Fundamental and Practical Aspects of Electroanalysis. 2000;12:317–325. doi: 10.1002/(SICI)1521-4109(20000301)12:5<317::AID-ELAN317>3.0.CO;2-A. [DOI] [Google Scholar]
- Zelada-Guillén G. A. Tweed-Kent A. Niemann M. Göringer H. U. Riu J. Rius F. X. Biosens. Bioelectron. 2013;41:366–371. doi: 10.1016/j.bios.2012.08.055. [DOI] [PubMed] [Google Scholar]
- Wang S. D. Liu Y. D. Zhu A. W. Tian Y. Anal. Chem. 2023;95:388–406. doi: 10.1021/acs.analchem.2c04541. [DOI] [PubMed] [Google Scholar]
- Laraib U. Sargazi S. Rahdar A. Khatami M. Pandey S. Int. J. Biol. Macromol. 2022;195:356–383. doi: 10.1016/j.ijbiomac.2021.12.052. [DOI] [PubMed] [Google Scholar]
- Karimi-Maleh H. Orooji Y. Karimi F. Alizadeh M. Baghayeri M. Rouhi J. Tajik S. Beitollahi H. D. Agarwal S. Gupta V. K. Rajendran S. Ayati A. Fu L. Sanati A. L. Tanhaei B. Sen F. Shabani-nooshabadi M. Asrami P. N. Al-Othman A. Biosens. Bioelectron. 2021;184:113252. doi: 10.1016/j.bios.2021.113252. [DOI] [PubMed] [Google Scholar]
- Hu W. C. Pang J. Biswas S. Wang K. Wang C. Xia X. H. Anal. Chem. 2021;93:8544–8552. doi: 10.1021/acs.analchem.1c01261. [DOI] [PubMed] [Google Scholar]
- Bhardwaj N. Bhardwaj S. K. Mehta J. Kim K. H. Deep A. ACS Appl. Mater. Interfaces. 2017;9:33589–33598. doi: 10.1021/acsami.7b07818. [DOI] [PubMed] [Google Scholar]
- Chen Y. Chen Y. Yi H. C. Gu H. W. Yin X. L. Xiang D. L. Zou P. Microchem. J. 2023;190:108681. doi: 10.1016/j.microc.2023.108681. [DOI] [Google Scholar]
- Hu J. H. Wu D. Z. Li T. H. Cao Y. T. Wang X. Gan N. Sens. Actuators, B. 2022;352:130987. doi: 10.1016/j.snb.2021.130987. [DOI] [Google Scholar]
- Wang W. H. Tan L. Wu J. Y. Li T. H. Xie H. Z. Wu D. Gan N. Anal. Chim. Acta. 2020;1133:128–136. doi: 10.1016/j.aca.2020.08.006. [DOI] [PubMed] [Google Scholar]
- Sohrabi H. Majidi M. R. Nami F. Asadpour-Zeynali K. Khataee A. Mokhtarzadeh A. Microchim. Acta. 2021;188:1–16. doi: 10.1007/s00604-021-04757-6. [DOI] [PubMed] [Google Scholar]
- Sohrabi H. Majidi M. R. Asadpour-Zeynali K. Khataee A. Mokhtarzadeh A. Chemosphere. 2022;287:132373. doi: 10.1016/j.chemosphere.2021.132373. [DOI] [PubMed] [Google Scholar]
- Wang S. J. Xu D. P. Ding C. C. Tian Y. C. Ge K. J. Guo L. Li J. Dong Q. L. Huang Y. Liu Q. Microchim. Acta. 2020;187:1–10. doi: 10.1007/s00604-019-3921-8. [DOI] [PubMed] [Google Scholar]
- Ren R. R. Cai G. N. Yu Z. Z. Zeng Y. Y. Tang D. P. Anal. Chem. 2018;90:11099–11105. doi: 10.1021/acs.analchem.8b03538. [DOI] [PubMed] [Google Scholar]
- Wang S. Q. Deng W. F. Yang L. Tan Y. M. Xie Q. J. Yao S. Z. ACS Appl. Mater. Interfaces. 2017;9:24440–24445. doi: 10.1021/acsami.7b07307. [DOI] [PubMed] [Google Scholar]
- Qiu P. P. Yuan P. Deng Z. C. Su Z. Q. Bai Y. He J. C. Microchim. Acta. 2021;188:1–10. doi: 10.1007/s00604-021-04981-0. [DOI] [PubMed] [Google Scholar]
- Zhang Q. P. Wang X. Z. Kang Y. Sun H. Liang Y. M. Liu J. Su Z. H. Dan J. Luo L. P. Yue T. L. Wang J. L. Zhang W. T. Foods. 2021;10:2596. doi: 10.3390/foods10112596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meteku B. E. Huang J. Zeng J. B. Aslam S. Zhang Y. Zhang X. Cui B. W. Wen C. Y. Yan Z. F. Chin. Chem. Lett. 2021;32:3245–3251. doi: 10.1016/j.cclet.2021.03.019. [DOI] [Google Scholar]
- Qi W. Z. Zheng L. Y. Wang S. H. Huang F. C. Liu Y. J. Jiang H. Y. Lin J. H. Biosens. Bioelectron. 2021;178:113020. doi: 10.1016/j.bios.2021.113020. [DOI] [PubMed] [Google Scholar]
- Guo R. Y. Xue L. Cai G. Z. Qi W. Z. Liu Y. J. Lin J. H. ACS Appl. Nano Mater. 2021;4:5115–5122. doi: 10.1021/acsanm.1c00574. [DOI] [Google Scholar]
- Håkansson K. Lindahl M. Svensson G. Albertsson J. Brunvoll J. Spanget-Larsen J. Milanova R. K. Nakata H. Nasiri A. Okada Y. J. A. C. S. Acta Chem. Scand. 1993;47:449–455. doi: 10.3891/acta.chem.scand.47-0449. [DOI] [Google Scholar]
- Evanoff D. D. Heckel J. Caldwell T. P. Christensen K. A. Chumanov G. J. Am. Chem. Soc. 2006;128:12618–12619. doi: 10.1021/ja0642717. [DOI] [PubMed] [Google Scholar]
- Wu M. N. Zhuang Y. X. Liu J. B. Chen W. W. Li X. Y. Xie R. J. Opt. Mater. 2020;106:110006. doi: 10.1016/j.optmat.2020.110006. [DOI] [Google Scholar]
- Qiao J. Chen X. B. Xu X. L. Fan B. Guan Y. S. Yang H. Li Q. J. Mater. Chem. B. 2023;11:8519–8527. doi: 10.1039/D3TB01428B. [DOI] [PubMed] [Google Scholar]
- Wei J. Y. Zhang L. X. Yang H. L. Wang L. Fu Z. F. Nanoscale. 2021;13:12546–12552. doi: 10.1039/D1NR02108G. [DOI] [PubMed] [Google Scholar]
- Yang S. S. Guo Y. C. Fan J. C. Yang Y. J. Zuo C. Bai S. L. Sheng S. C. Li J. J. Xie G. M. Microchim. Acta. 2020;187:1–10. doi: 10.1007/s00604-019-3921-8. [DOI] [PubMed] [Google Scholar]
- Xu Z. Long L. L. Chen Y. Q. Chen M. L. Cheng Y. H. Food Chem. 2021;338:128039. doi: 10.1016/j.foodchem.2020.128039. [DOI] [PubMed] [Google Scholar]
- Xu D. P. Ge K. J. Chen Y. Qi S. Y. Tian Y. C. Wang S. Y. Qiu J. X. Wang X. Dong Q. L. Liu Q. Microchem. J. 2020;154:104591. doi: 10.1016/j.microc.2019.104591. [DOI] [Google Scholar]
- Wu L. Li G. H. Xu X. Zhu L. Huang R. M. Chen X. Q. TrAC, Trends Anal. Chem. 2019;113:140–156. doi: 10.1016/j.trac.2019.02.002. [DOI] [Google Scholar]
- Wu K. F. Ma C. B. Zhao H. Chen M. J. Deng Z. Y. Food Chem. 2019;277:273–278. doi: 10.1016/j.foodchem.2018.10.130. [DOI] [PubMed] [Google Scholar]
- Gao W. W. Gopala L. Bheemanaboina R. R. Y. Zhang G. B. Li S. Zhou C. H. Eur. J. Med. Chem. 2018;146:15–37. doi: 10.1016/j.ejmech.2018.01.038. [DOI] [PubMed] [Google Scholar]
- Uruén C. Chopo-Escuin G. Tommassen J. Mainar-Jaime R. C. Arenas J. Antibiotics. 2021;10:3. doi: 10.3390/antibiotics10010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiple I. B. Zhang Z. Y. Jakubec P. Langlois-Mercier A. Wright P. M. Hog D. T. Yabu K. Allu S. R. Fukuzaki T. Carlsen P. N. Kitamura Y. Zhou X. Condakes M. L. Szczypinski F. T. Green W. D. Myers A. G. Nature. 2016;533:338–345. doi: 10.1038/nature17967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talebi Bezmin Abadi A. Rizvanov A. A. Haertlé T. Blatt N. L. BioNanoScience. 2019;9:778–788. doi: 10.1007/s12668-019-00658-4. [DOI] [Google Scholar]
- Li S. Q. Liu X. D. Chai H. X. Huang Y. M. TrAC, Trends Anal. Chem. 2018;105:391–403. doi: 10.1016/j.trac.2018.06.001. [DOI] [Google Scholar]
- He S. Y. Huang J. Q. Zhang Q. Zhao W. Xu Z. A. Zhang W. Adv. Funct. Mater. 2021;31:2105198. doi: 10.1002/adfm.202105198. [DOI] [Google Scholar]
- Wang X. Sun X. Y. Bu T. Wang Q. Z. Jia P. Dong M. N. Wang L. Composites, Part B. 2022;229:109465. doi: 10.1016/j.compositesb.2021.109465. [DOI] [Google Scholar]
- Bruna T. Maldonado-Bravo F. Jara P. Caro N. Int. J. Mol. Sci. 2021;22:7202. doi: 10.3390/ijms22137202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes T. A. Costa I. F. M. Jorge P. Sousa A. C. André V. Cerca N. Kirillov A. M. ACS Appl. Mater. Interfaces. 2021;13:12836–12844. doi: 10.1021/acsami.0c19446. [DOI] [PubMed] [Google Scholar]
- Guo C. P. Cheng F. Liang G. L. Zhang S. Jia Q. J. He L. H. Duan S. X. Fu Y. K. Zhang Z. H. Du M. Chem. Eng. J. 2022;435:134915. doi: 10.1016/j.cej.2022.134915. [DOI] [Google Scholar]
- Pham M. H. Vuong T. Vu A. T. Do T. O. Langmuir. 2011;27:15261–15267. doi: 10.1021/la203570h. [DOI] [PubMed] [Google Scholar]
- Hu Z. L. Liu X. Jiao L. Wei X. Q. Wang Z. Huang N. Y. Li J. New J. Chem. 2021;45:17772–17776. doi: 10.1039/D1NJ02088A. [DOI] [Google Scholar]
- Zhang W. T. Ren X. Y. Shi S. Li M. Liu L. Z. Han X. M. Zhu W. X. Yue T. L. Sun J. Wang J. L. Nanoscale. 2020;12:16330–16338. doi: 10.1039/D0NR01471K. [DOI] [PubMed] [Google Scholar]
- Li Y. Fu R. Z. Duan Z. G. Zhu C. H. Fan D. D. Colloids Surf., B. 2022;210:112230. doi: 10.1016/j.colsurfb.2021.112230. [DOI] [PubMed] [Google Scholar]
- Hu Y. H. Cheng H. J. Zhao X. Z. Wu J. J. Muhammad F. Lin S. C. He J. Zhou L. Q. Zhang C. P. Deng Y. Wang P. Zhou Z. Y. Nie S. M. Wei H. ACS Nano. 2017;11:5558–5566. doi: 10.1021/acsnano.7b00905. [DOI] [PubMed] [Google Scholar]
- Hu S. Jiang Y. N. Wu Y. P. Guo X. Y. Ying Y. Wen Y. Yang H. F. ACS Appl. Mater. Interfaces. 2020;12:55324–55330. doi: 10.1021/acsami.0c12988. [DOI] [PubMed] [Google Scholar]
- Li K. Lin C. C. Li M. H. Xu K. He Y. Mao Y. L. Lu L. Geng W. B. Li X. M. Luo Z. Cai K. Y. ACS Nano. 2022;16:2381–2398. doi: 10.1021/acsnano.1c08664. [DOI] [PubMed] [Google Scholar]
- Hu W. C. Younis M. R. Zhou Y. Wang C. Xia X. H. Small. 2020;16:2000553. doi: 10.1002/smll.202000553. [DOI] [PubMed] [Google Scholar]
- Liao X. W. Xu Q. Y. Sun H. J. Liu W. Y. Chen Y. M. Xia X. H. Wang C. J. Phys. Chem. Lett. 2022;13:312–323. doi: 10.1021/acs.jpclett.1c03804. [DOI] [PubMed] [Google Scholar]
- Aryanejad S. Bagherzade G. Moudi M. New J. Chem. 2020;44:1508–1516. doi: 10.1039/C9NJ04977K. [DOI] [Google Scholar]
- Wan Y. Fang J. Wang Y. Sun J. Sun Y. Sun X. L. Qi M. L. Li W. Li C. Y. Zhou Y. M. Xu L. Dong B. Wang L. Adv. Healthcare Mater. 2021;10:2101515. doi: 10.1002/adhm.202101515. [DOI] [PubMed] [Google Scholar]
- Yang Y. Wu X. Z. Ma L. He C. Cao S. J. Long Y. P. Huang J. B. Rodriguez R. D. Cheng C. Zhao C. S. Qiu L. Adv. Mater. 2021;33:2005477. doi: 10.1002/adma.202005477. [DOI] [PubMed] [Google Scholar]
- Yin W. Y. Yu J. Lv F. T. Yan L. Zheng L. R. Gu Z. J. Zhao Y. L. ACS Nano. 2016;10:11000–11011. doi: 10.1021/acsnano.6b05810. [DOI] [PubMed] [Google Scholar]
- Ji S. F. Jiang B. Hao H. G. Chen Y. J. Dong J. C. Mao Y. Zhang Z. D. Gao R. Chen W. X. Zhang R. F. Liang Q. Li H. J. Liu S. H. Wang Y. Zhang Q. H. Gu L. Duan D. M. Liang M. M. Wang D. S. Yan X. Y. Li Y. D. Nat. Catal. 2021;4:407–417. doi: 10.1038/s41929-021-00609-x. [DOI] [Google Scholar]
- Schneider L. A. Korber A. Grabbe S. Dissemond J. Arch. Dermatol. Res. 2007;298:413–420. doi: 10.1007/s00403-006-0713-x. [DOI] [PubMed] [Google Scholar]
- Liu X. P. Yan Z. Q. Zhang Y. Liu Z. W. Sun Y. H. Ren J. S. Qu X. G. ACS Nano. 2019;13:5222–5230. doi: 10.1021/acsnano.8b09501. [DOI] [PubMed] [Google Scholar]
- Wang M. X. Zhou X. Li Y. H. Dong Y. Q. Meng J. S. Zhang S. Xia L. B. He Z. Z. Ren L. Chen Z. W. Zhang X. C. Bioact. Mater. 2022;17:289–299. doi: 10.1016/S0261-3069(97)00025-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. Zhang S. Liu Y. Meng J. S. Wang M. X. Sun Y. J. Xia L. B. He Z. Z. Hu W. X. Ren L. Chen Z. W. Zhang X. C. ACS Appl. Mater. Interfaces. 2022;14:11104–11115. doi: 10.1021/acsami.1c24231. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Lai L. G. Liu Y. J. Chen B. N. Yao J. Zheng P. W. Pan Q. S. Zhu W. F. ACS Appl. Mater. Interfaces. 2022;14:6453–6464. doi: 10.1021/acsami.1c23808. [DOI] [PubMed] [Google Scholar]
- Cheng X. Q. Zhang S. Liu H. H. Chen H. M. Zhou J. H. Chen Z. W. Zhou X. Xie Z. X. Kuang Q. Zheng L. S. ACS Appl. Mater. Interfaces. 2020;12:36996–37005. doi: 10.1021/acsami.0c12159. [DOI] [PubMed] [Google Scholar]
- Li Y. J. Gao Z. G. Zhang Y. Chen F. H. An P. J. Wu H. S. You C. Q. Sun B. W. Chem. Eng. J. 2021;416:127610. doi: 10.1016/j.cej.2020.127610. [DOI] [Google Scholar]
- Yougbare S. Chou H. L. Yang C. H. Krisnawati D. I. Jazidie A. Nuh M. Kuo T. R. J. Hazard. Mater. 2021;407:124617. doi: 10.1016/j.jhazmat.2020.124617. [DOI] [PubMed] [Google Scholar]
- Huo J. J. Jia Q. Y. Huang H. Zhang J. Li P. Dong X. C. Huang W. Chem. Soc. Rev. 2021;50:8762–8789. doi: 10.1039/D1CS00074H. [DOI] [PubMed] [Google Scholar]
- Li M. Li L. Q. Su K. Liu X. M. Zhang T. J. Liang Y. Q. Jing D. D. Yang X. J. Zheng D. Cui Z. D. Li Z. Y. Zhu S. L. Yeung K. W. K. Zheng Y. F. Wang X. B. Wu S. L. Adv. Sci. 2019;6:1900599. doi: 10.1002/advs.201900599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. Ma L. Cheng C. Deng Y. Y. Huang J. B. Fan X. Nie C. X. Zhao W. F. Zhao C. D. Adv. Funct. Mater. 2018;28:1705708. doi: 10.1002/adfm.201705708. [DOI] [Google Scholar]
- Chen Y. Gao Y. J. Chen Y. Liu L. Mo A. C. Peng Q. J. Controlled Release. 2020;328:251–262. doi: 10.1016/j.jconrel.2020.08.055. [DOI] [PubMed] [Google Scholar]
- Gao Q. Zhang X. Yin W. Y. Ma D. Q. Xie C. J. Zheng L. R. Dong X. H. Mei L. Q. Yu J. Wang C. Z. Gu Z. J. Zhao Y. L. Small. 2018;14:1802290. doi: 10.1002/smll.201802290. [DOI] [PubMed] [Google Scholar]
- Kennedy J. Blair I. S. McDowell D. A. Bolton D. J. J. Appl. Microbiol. 2005;99:1229–1235. doi: 10.1111/j.1365-2672.2005.02697.x. [DOI] [PubMed] [Google Scholar]
- Wang Y. Q. Jin Y. Y. Chen W. Wang J. J. Chen H. Sun L. Li X. Ji J. Yu Q. Shen L. Y. Wang B. L. Chem. Eng. J. 2019;358:74–90. doi: 10.1016/j.cej.2018.10.002. [DOI] [Google Scholar]
- Guo Z. R. Liu Y. N. Zhang Y. L. Sun X. Y. Li F. Bu T. Wang Q. Z. Wang L. Biomater. Sci. 2020;8:4266–4274. doi: 10.1039/D0BM00706D. [DOI] [PubMed] [Google Scholar]
- Hao L. W. Jiang R. J. Fan Y. Xu J. N. Tian L. M. Zhao J. Ming W. H. Ren L. Q. ACS Sustainable Chem. Eng. 2020;8:15834–15842. doi: 10.1021/acssuschemeng.0c03384. [DOI] [Google Scholar]
- Zhang J. Liu C. L. Liu J. J. Bai X. H. Cao Z. K. Yang J. Yu M. Ramakrishna S. Long Y. Z. Nanoscale. 2021;13:6105–6116. doi: 10.1039/D1NR00179E. [DOI] [PubMed] [Google Scholar]
- Liu W. Z. Zhang Y. X. You W. W. Su J. Q. Yu S. H. Dai T. Huang Y. M. Chen X. Y. Song X. R. Chen Z. Nanoscale. 2020;12:13948–13957. doi: 10.1039/D0NR01073A. [DOI] [PubMed] [Google Scholar]
- Jia Q. Y. Song Q. Li P. Huang W. Adv. Healthcare Mater. 2019;8:1900608. doi: 10.1002/adhm.201900608. [DOI] [PubMed] [Google Scholar]
- Luo Y. Li B. Liu X. M. Zheng Y. F. Wang E. R. Li Z. Y. Cui Z. D. Liang Y. Q. Zhu S. L. Wu S. L. Bioact. Mater. 2022;18:421–432. doi: 10.1016/j.bioactmat.2022.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Sun X. Y. Bu T. Wang Q. Z. Zhang H. Jia P. Li L. W. Wang L. Acta Biomater. 2021;135:342–355. doi: 10.1016/j.actbio.2021.08.022. [DOI] [PubMed] [Google Scholar]
- Han X. Boix G. Balcerzak M. Moriones O. H. Cano-Sarabia M. Cortés P. Bastús N. Puntes V. Llagostera M. Imaz I. Maspoch D. Adv. Funct. Mater. 2022;32:2112902. doi: 10.1002/adfm.202112902. [DOI] [Google Scholar]
- Raf A. Ye J. W. Zhang S. Q. Qi Y. Wang G. Y. Che Y. Ning G. T. Dalton Trans. 2019;48:17810–17817. doi: 10.1039/C9DT03649K. [DOI] [PubMed] [Google Scholar]
- Wang C. F. Luo Y. Liu X. M. Cui Z. D. Zheng Y. F. Liang Y. Q. Li Z. Y. Zhu S. L. Lei J. Feng X. B. Wu S. L. Bioact. Mater. 2022;13:200–211. doi: 10.1016/j.bioactmat.2021.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Sun P. P. Zhang L. Wang Z. Z. Wang F. M. Dong K. Liu Z. Ren J. S. Qu X. G. Adv. Funct. Mater. 2019;29:1808594. doi: 10.1002/adfm.201808594. [DOI] [Google Scholar]
- Han D. L. Han Y. J. Li J. Liu X. M. Yeung K. W. K. Zheng Y. F. Cui Z. D. Yang X. J. Liang Y. Q. Li Z. Y. Zhu S. L. Yuan X. B. Feng X. B. Yang C. Wu S. L. Appl. Catal., B. 2020;261:118248. doi: 10.1016/j.apcatb.2019.118248. [DOI] [Google Scholar]
- Xiong K. Li J. Tan L. Cui Z. D. Li Z. Y. Wu S. L. Liang Y. Q. Zhu S. L. Liu X. M. Colloid Interface Sci. Commun. 2019;33:100201. doi: 10.1016/j.colcom.2019.100201. [DOI] [Google Scholar]
- Yu P. L. Han Y. J. Han D. L. Liu X. M. Liang Y. Q. Li Z. Y. Zhu S. L. Wu S. L. J. Hazard. Mater. 2020;390:122126. doi: 10.1016/j.jhazmat.2020.122126. [DOI] [PubMed] [Google Scholar]
- Han D. L. Yu P. L. Liu X. M. Xu Y. D. Wu S. L. Rare Met. 2022;41:663–672. doi: 10.1007/s12598-021-01786-1. [DOI] [Google Scholar]
- Han D. L. Li Y. Liu X. M. Yeung K. W. K. Zheng Y. F. Cui Z. D. Liang Y. Q. Li Z. Y. Zhu S. L. Wang X. B. Wu S. L. J. Mater. Sci. Technol. 2021;62:83–95. doi: 10.1016/j.jmst.2020.05.055. [DOI] [Google Scholar]
- Zhu P. Chen Y. Shi J. L. ACS Nano. 2018;12:3780–3795. doi: 10.1021/acsnano.8b00999. [DOI] [PubMed] [Google Scholar]
- Xu M. M. Zhou L. Q. Zheng L. Zhou Q. Liu K. Mao Y. H. Song S. S. Cancer Lett. 2021;497:229–242. doi: 10.1016/j.canlet.2020.10.037. [DOI] [PubMed] [Google Scholar]
- Qian X. Q. Zheng Y. Y. Chen Y. Adv. Mater. 2016;28:8097–8129. doi: 10.1002/adma.201602012. [DOI] [PubMed] [Google Scholar]
- Pan X. T. Wu N. Tian S. Y. Guo J. Wang C. H. Sun Y. Huang Z. Z. Chen F. Z. Wu Q. Y. Jing Y. Yin Z. Zhao B. H. Xiong X. L. Liu H. Y. Zhou D. S. Adv. Funct. Mater. 2022;32:2112145. doi: 10.1002/adfm.202112145. [DOI] [Google Scholar]
- Liao Z. Y. Xia Y. M. Zuo J. M. Wang T. Hu D. T. Li M. Z. Shao N. N. Chen D. Song K. X. Yu X. Zhang X. Y. Gao W. W. Adv. Healthcare Mater. 2022;11:2101698. doi: 10.1002/adhm.202101698. [DOI] [PubMed] [Google Scholar]
- Wang J. Wang Y. Zhang D. Chen C. Yang Z. X. ACS Appl. Nano Mater. 2021;4:7698–7711. doi: 10.1021/acsanm.1c00910. [DOI] [Google Scholar]
- Hatamie S. Ahadian M. M. Zomorod M. S. Torabi S. Babaie A. Hosseinzadeh S. Soleimani M. Hatami N. Wei Z. H. Mater. Sci. Eng., C. 2019;104:109862. doi: 10.1016/j.msec.2019.109862. [DOI] [PubMed] [Google Scholar]
- Zhang L. Liu Z. W. Deng Q. Q. Sang Y. J. Dong K. Ren J. S. Qu X. G. Angew. Chem., Int. Ed. 2021;60:3469–3474. doi: 10.1002/anie.202012487. [DOI] [PubMed] [Google Scholar]
- Liu S. N. Hu C. L. Liu Y. Zhao X. Y. Pang M. L. Lin J. Chem. – Eur. J. 2019;25:4315–4319. doi: 10.1002/chem.201806242. [DOI] [PubMed] [Google Scholar]
- Ding S.-Y. Gao J. Wang Q. Zhang Y. Song W.-G. Su C.-Y. Wang W. J. Am. Chem. Soc. 2011;133:19816–19822. doi: 10.1021/ja206846p. [DOI] [PubMed] [Google Scholar]
- Flemming H. C. Wuertz S. Nat. Rev. Microbiol. 2019;17:247–260. doi: 10.1038/s41579-019-0158-9. [DOI] [PubMed] [Google Scholar]
- Bjarnsholt T. APMIS. 2013;121:1–58. doi: 10.1111/apm.12099. [DOI] [PubMed] [Google Scholar]
- Vestby L. K. Gronseth T. Simm R. Nesse L. L. Antibiotics. 2020;9:59. doi: 10.3390/antibiotics9020059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies D. Nat. Rev. Drug Discovery. 2003;2:114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- Karygianni L. Ren Z. Koo H. Thurnheer T. Trends Microbiol. 2020;28:668–681. doi: 10.1016/j.tim.2020.03.016. [DOI] [PubMed] [Google Scholar]
- Flemming H. C. Wingender J. Nat. Rev. Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Qiu H. Pu F. Liu Z. W. Deng Q. Q. Sun P. P. Ren J. S. Qu X. G. Small. 2019;15:1902522. doi: 10.1002/smll.201902522. [DOI] [PubMed] [Google Scholar]
- Koo H. Allan R. N. Howlin R. P. Stoodley P. Hall-Stoodley L. Nat. Rev. Microbiol. 2017;15:740–755. doi: 10.1038/nrmicro.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart P. S. Costerton J. W. Lancet. 2001;358:135–138. doi: 10.1016/S0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- Liu Z. W. Wang F. M. Ren J. S. Qu X. G. Biomaterials. 2019;208:21–31. doi: 10.1016/j.biomaterials.2019.04.007. [DOI] [PubMed] [Google Scholar]
- Zheng K. Y. Li S. Jing L. Chen P. Y. Xie J. P. Adv. Healthcare Mater. 2020;9:2001007. doi: 10.1002/adhm.202001007. [DOI] [PubMed] [Google Scholar]
- Yu Y. Cheng Y. Tan L. Liu X. M. Li Z. Y. Zheng Y. F. Wu T. Liang Y. Q. Cui Z. D. Zhu S. L. Wu S. L. Chem. Eng. J. 2022;431:133279. doi: 10.1016/j.cej.2021.133279. [DOI] [Google Scholar]
- Wang Z. Y. Guo W. Zhang K. Ye Y. M. Wang Y. M. Sui D. D. Zhao N. N. Xu F. J. Sci. China: Technol. Sci. 2022;65:1052–1058. doi: 10.1007/s11431-021-1963-3. [DOI] [Google Scholar]
- Yu M. Z. Zhang G. K. Li P. L. Lu H. J. Tang W. T. Yang X. Huang R. B. Yu F. Wu W. Z. Xiao Y. H. Xing X. D. Mater. Sci. Eng., C. 2021;127:112225. doi: 10.1016/j.msec.2021.112225. [DOI] [PubMed] [Google Scholar]
- Huang T. Yu Z. Yuan B. Jiang L. Liu Y. Sun X. Liu P. Jiang W. Tang J. Mater. Today Chem. 2022;24:100831. doi: 10.1016/j.mtchem.2022.100831. [DOI] [Google Scholar]
- Liu P. Wu Y. Z. Mehrjou B. Tang K. W. Wang G. M. Chu P. K. Adv. Funct. Mater. 2022;32:2110635. doi: 10.1002/adfm.202110635. [DOI] [Google Scholar]
- Yang Y. Wu X. Z. He C. Huang J. B. Yin S. Q. Zhou M. Ma L. Zhao W. F. Qiu L. Cheng C. Zhao C. S. ACS Appl. Mater. Interfaces. 2020;12:13698–13708. doi: 10.1021/acsami.0c01666. [DOI] [PubMed] [Google Scholar]
- Nong W. Q. Chen Y. L. Lv D. Y. Yan Y. T. Zheng X. Shi X. M. Xu Z. Guan W. L. Wu J. Guan Y. G. Chem. Eng. J. 2022;431:134003. doi: 10.1016/j.cej.2021.134003. [DOI] [Google Scholar]
- Wang X. Qiu L. Wang C. Gao Z. H. Zhou S. W. Cui P. F. Jiang P. J. Hu H. Z. Ni X. Y. Du X. C. Wang J. H. Xia J. Biomater. Sci. 2022;10:654–664. doi: 10.1039/D1BM01533H. [DOI] [PubMed] [Google Scholar]
- Li D. X. Chen T. Zhang Y. F. Xu Y. H. Niu H. T. Adv. Healthcare Mater. 2021;10:2100716. doi: 10.1002/adhm.202100716. [DOI] [PubMed] [Google Scholar]
- Li T. Qiu H. Q. Liu N. Li J. W. Bao Y. H. Tong W. J. Colloids Surf., B. 2020;191:111001. doi: 10.1016/j.colsurfb.2020.111001. [DOI] [PubMed] [Google Scholar]
- Perlin D. S. Rautemaa-Richardson R. Alastruey-Izquierdo A. Lancet Infect. Dis. 2017;17:E383–E392. doi: 10.1016/S1473-3099(17)30316-X. [DOI] [PubMed] [Google Scholar]
- Berman J. Krysan D. J. Nat. Rev. Microbiol. 2020;18:319–331. doi: 10.1038/s41579-019-0322-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouson S. Krittayavathananon A. Phattharasupakun N. Siwayaprahm P. Sawangphruk M. R. Soc. Open Sci. 2017;4:170654. doi: 10.1098/rsos.170654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelhamid H. N. Mahmoud G. A. E. Sharmoukh W. J. Mater. Chem. B. 2020;8:7548–7556. doi: 10.1039/D0TB00894J. [DOI] [PubMed] [Google Scholar]