Abstract
BACKGROUND:
Endothelial cells are constantly exposed to mechanical forces in the form of fluid shear stress, extracellular stiffness, and cyclic strain. The mechanoresponsive activity of YAP (Yes-associated protein) and its role in vascular development are well described; however, whether changes to transcription or epigenetic regulation of YAP are involved in these processes remains unanswered. Furthermore, how mechanical forces are transduced to the nucleus to drive transcriptional reprogramming in endothelial cells is poorly understood. The YAP target gene, AmotL2 (angiomotin-like 2), is a junctional mechanotransducer that connects cell-cell junctions to the nuclear membrane via the actin cytoskeleton.
METHODS:
We applied mechanical manipulations including shear flow, stretching, and substrate stiffness to endothelial cells to investigate the role of mechanical forces in modulating YAP transcription. Using in vitro and in vivo endothelial depletion of AmotL2, we assess nuclear morphology, chromatin organization (using transposase-accessible chromatin sequencing), and whole-mount immunofluorescent staining of the aorta to determine the regulation and functionality of YAP. Finally, we use genetic and chemical inhibition to uncouple the nuclear-cytoskeletal connection to investigate the role of this pathway on YAP transcription.
RESULTS:
Our results reveal that mechanical forces sensed at cell-cell junctions by the YAP target gene AmotL2 are directly involved in changes in global chromatin accessibility and activity of the histone methyltransferase EZH2, leading to modulation of YAP promotor activity. Functionally, shear stress–induced proliferation of endothelial cells in vivo was reliant on AmotL2 and YAP/TAZ (transcriptional coactivator with PDZ-binding motif) expression. Mechanistically, uncoupling of the nuclear-cytoskeletal connection from junctions and focal adhesions led to altered nuclear morphology, chromatin accessibility, and YAP promotor activity.
CONCLUSIONS:
Our findings reveal a role for AmotL2 and nuclear-cytoskeletal force transmission in modulating the epigenetic and transcriptional regulation of YAP to maintain a mechano-enforced positive feedback loop of vascular homeostasis. These findings may offer an explanation as to the proinflammatory phenotype that leads to aneurysm formation observed in AmotL2 endothelial deletion models.
Keywords: aorta, cell junctions, chromatin, endothelial cells, epigenetic regulation, nuclear membrane, YAP-signaling
Highlights.
AmotL2 (angiomotin-like 2) influences endothelial nuclear morphology and chromatin accessibility.
AmotL2 regulates YAP (Yes-associated protein) expression in the aortic endothelium in vivo.
Loss of AmotL2 modulates H3K27me3 (trimethylation of histone H3 at lysine 27) levels at the YAP promotor to regulate transcription in an EZH2 (enhancer of zeste 2 polycomb repressive complex 2 subunit)-dependent manner.
Endothelial proliferation induced by mechanical forces is dependent on the AmotL2-YAP axis.
YAP transcription is regulated by the nuclear-cytoskeletal connection, including focal adhesions and cell-cell junctions.
The endothelium is constantly exposed and responds to the mechanical force exerted by blood flow. Laminar flow maintains a homeostatic anti-inflammatory and antiproliferative state of vessels, such as the descending aorta, whereas regions of the vasculature exposed to disturbed flow such as bifurcations and the inner arch of the aorta are prone to inflammation, endothelial cell (EC) proliferation, and the development of disease.1,2 The mechanical forces are sensed at the EC junctions by mechanosensory complexes, allowing adaptation of the endothelium to changes in mechanical stimuli via genetic programs. However, how these mechanical forces sensed at the EC junctions are transmitted to the nucleus to influence transcriptional output is poorly understood.
Key examples of vascular junctional mechanosensors include the transmembrane VEGFR2 (vascular endothelial growth factor receptor 2), VE (vascular endothelial)-cadherin, and PECAM1 (platelet endothelial cell adhesion molecule-1)3 complex, along with Plexin D1, which sense fluid shear stress (FSS) in a conformation-dependent manner and shape the EC response.4 Recently, the intracellular mechanosensor AmotL2 (angiomotin-like 2), which localizes to EC junctions, was shown to regulate cell and nuclear morphology of aortic ECs, by forming a complex between the actin cytoskeleton, VE-cadherin, p120, and the nuclear lamina through binding of lamin A.5 Interestingly, mice deleted for endothelial AmotL2 developed abdominal aortic aneurysms (AAAs) due to impaired response of aortic ECs to FSS, highlighting the significance of correct sensing and transducing of forces in vascular homeostasis.5
Mechanical forces are transmitted to the nucleus via the actin cytoskeleton, which is directly tethered to the outer nuclear membrane via the LINC (linker of nucleoskeleton and cytoskeleton) complex, which spans the nuclear membrane and associates with the nuclear lamina of the inner nuclear membrane to directly influence chromatin conformation.6,7 In ECs, AmotL2 also connects the actin cytoskeleton to nuclear lamin A (but not B-type lamins).5 Furthermore, a recent study indicated that SUN1 (sad1 and UNC84 domain containing 1), of the LINC complex, can inversely influence junctional stability of ECs via the microtubule-based network.8 Via these mechanisms, mechanical inputs transduced between cell junctions and the nucleus impact on nuclear morphology, import and export of molecules, nuclear pore conformation, envelope composition, and consequently, chromatin accessibility and transcriptional regulation.9–12 However, how mechanical forces transduced to the nucleus are converted to changes in transcriptional programs in ECs in response to FSS remains poorly understood.
YAP (Yes-associated protein) and its paralogue TAZ (transcriptional coactivator with PDZ-binding motif) are transcriptional coactivators that regulate a multitude of important EC processes, including angiogenesis.13–15 The activity of YAP is regulated via intracellular kinase-mediated pathways and via mechanical forces that affect its nucleocytoplasmic shuttling.16 However, whether changes to transcription or epigenetic control of the YAP gene are involved in these processes remains unanswered. Here, we show that mechanical forces shape the YAP promotor accessibility through modulation of histone activity. Signals transduced via the junctional molecule AmotL2, focal adhesions, and nuclear lamina are indispensable for YAP transcription. Our results reveal that mechanical forces sensed at cell-cell junctions by AmotL2 are directly intervened into changes in global chromatin accessibility and EZH2 (enhancer of zeste 2 polycomb repressive complex 2 subunit) activity, leading to modulation of YAP promotor activity. Moreover, since Yap was required for AmotL2 transcription in ECs in the mouse aorta, our results reveal a mechano-enforced positive feedback loop of transcriptional control of vascular homeostasis.
MATERIALS AND METHODS
Data Availability
All data required to evaluate the conclusions are available in the main text or the Supplemental Material. Sequencing data derived from this study are deposited in the Gene Expression Omnibus under the given accession code (GEO accession: GSE253761). Previously published assay for transposase-accessible chromatin sequencing (ATAC-seq), scRNA-seq (single cell RNA-sequencing), or chromatin immunoprecipitation sequencing (ChIP-seq) data are available at the relevant websites, atlases, and portals outlined in the article along with the relevant accession codes. All other data are available from the corresponding author on reasonable request.
Human AAA Samples RNA-seq
Aortic tissue samples from patients with AAA were obtained from the surgery performed at Karolinska Hospital in Stockholm. Samples of the abdominal aorta of beating-heart, solid organ transplant donors were used as controls. Ethical permit was granted by Regionala etikprövningsnämnden (regional ethical review boards) in Stockholm, and informed consent was obtained from all participants. RNA extraction was performed from the medial layer of the aorta and subsequently sequenced by the Human Transcriptome Array 2.0 (Affymetrix) platform as described previously.17
Animals
For Yap/Taz iΔEC, Wwtr1 flox/flox; Yap flox/flox mice (The Jackson Laboratory) were crossed to Cdh5(BAC)-CreERT218 transgenic mice. Mice were used in a mixed background or after backcrossing to C57BL/6J according to the published protocol (The Jackson Laboratory) and were fed ad libitum with 2016 Teklad Global 16% Protein Rodent Diet with carbohydrate as the main constituent. Yap/Taz wild-type (WT) mice refers to Wwtr1 flox/flox; Yap flox/flox mice only. To induce endothelial-specific Yap/Taz gene inactivation, tamoxifen (Sigma; T5648) in corn oil (Sigma; C8267) was administered by oral gavage for 5 continuous days in 8-week-old mice (2 mg/mouse per day). Cre-positive tamoxifen-treated Wwtr1 flox/flox; Yap flox/flox mice were compared with control mice that were tamoxifen-treated Cre-negative fl/fl. Analysis of aortic samples was performed 7 to 9 weeks after tamoxifen-induced deletion. Data from both males and females were combined to improve power, and sex difference and interaction were not determined in the study. All animal experiments were approved by the National Animal Experiment Board in Finland. Ethical permits: ESAVI/15852/2022 and ESAVI/4975/2019.
For AmotL2 iΔEC, amotl2 flox/flox mice with loxP-flanked amotl2 gene were crossed with Cdh5(PAC)-CreERT219 and ROSA26-EYFP (yellow fluorescent protein) transgenic mice. Mice were maintained on a diet of Special Diets Services standard CRM (P) 801722 pelleted diet (where the main ingredient is wheat) and were fed ad libitum. To induce endothelial-specific amotl2 deletion, tamoxifen was administered by intraperitoneal injection for 5 continuous days. For adult mice over 6 weeks of age, 100 µL of tamoxifen (20 mg/mL) was administered, and analysis of aortic samples was performed 4 weeks following injections. All mice in this study were of the C57BL/6J background and included both female and male mice. To improve power, data from both males and females were combined and sex difference and interaction were not determined in the study. Ethical permits were obtained from the North Stockholm Animal Ethical Committee, and all experiments were performed in accordance with the guidelines of the Swedish Board of Agriculture. Ethical permits: N129/15, 12931-2020, and 22902-2021.
Cell Culture
Human umbilical vein ECs (HUVECs; Promocell, Heidelberg, Germany) were cultured in EC medium (ScienCell) supplemented with 5% fetal calf serum (v/v), 1% EC growth supplement, and 1% penicillin and streptomycin antibiotic cocktail and were cultured on 0.2% gelatin (Sigma-Aldrich; w/v in PBS)-coated plates. Human umbilical arterial ECs (HUAECs; Promocell) were cultured as above in ECBM (EC basal medium) and MV medium (Promocell) supplemented with 2% fetal calf serum (v/v), 0.4% EC growth supplement, 0.1 ng/mL epidermal growth factor (recombinant human), 1 ng/mL basic fibroblast growth factor (recombinant human), 90 μg/mL heparin, and 1 μg/mL hydrocortisone. Both HUVEC and HUAEC were grown to passage 4 or 5. HUVECs were treated with 10 μM FAK (focal adhesion kinase) inhibitor 14 diluted (Sigma) in sterile H2O and 1, 5, and 10 μM of lonafarnib (Sigma) diluted in DMSO.
siRNA Transfection
HUVECs and HUAECs were transfected with SMARTpool siRNA (Dharmacon) as described previously20 targeting YAP, TAZ, or EZH2 in parallel with a scrambled control siRNA. Transfections were performed using Lipofectamine RNAiMax (Invitrogen) transfection agent and Opti-MEM I Reduced-Serum Medium, GlutaMAX Supplement (Gibco) according to manufacturer’s instructions. Cells were transfected with 15 pmol siRNA in a 12-well plate (1×105 cells/well) and scaled accordingly. Briefly, for a single well of a 12-well plate, 15 pmol siRNA duplexes were made in 125 μL of Opti-MEM medium and incubated at room temperature for 5 minutes. For codepletion of multiple targets (YAP and TAZ), 7.5 pmol per target (total 15 pmol) of siRNA duplexes was used. At the same time, 2 μL lipofectamine was made up in 125 μL of OptiMEM and incubated at room temperature for 5 minutes. siRNA and lipofectamine mix were combined and gently mixed before incubating at room temperature for 20 minutes. Following this, culture medium of cells seeded out the previous day, at a density of 1×105 cells, was aspirated and replaced with 500 μL of OptiMEM. Transfection mix was gently added to the cells dropwise. Cells were incubated in this mixture for 4 to 6 hours, before transfection medium was aspirated and replaced with supplemented normal culture medium. siRNA-treated cells were incubated for 24 to 72 hours before being used in downstream assays. The siRNA molecules used (from Dharmacon/Horizon Discovery) are shown below. ON-TARGETplus Human EZH2 (2146) siRNA–SMARTpool (5′-GAGGACGGCUUCCCAAUAA-3′, 5′-GCUGAAGCCUCAAUGUUUA-3′, 5′-UAACGGUGAUCACAGGAUA-3′, 5′-GCAAAUUCUCGGUGUCAAA-3′); ON-TARGETplus Human WWTR1 (25937) siRNA–SMARTpool (5′-CCGCAGGGCUCAUGAGUAU-3′, 5′-GGACAAACACCCAUGAACA-3′, 5′-AGGAACAAACGUUGACUUA-3′, 5′-CCAAAUCUCGUGAUGAAUC-3′); ON-TARGETplus Human YAP1 (10413) siRNA–Individual siRNA (5′-GCACCUAUCACUCUCGAGA-3′).
Lentivirus Generation
Lentiviral supernatants were produced in HEK293T cells following transfection using Lipofectamine 3000 (Invitrogen) transfection agent with third-generation packaging plasmids and either shScr (shRNA scrambled control; SHC332; Sigma), shYAP (Addgene #42541), FUW-tetO-wtYAP (Addgene #84009), and FUW-tetO-HA-YAP5SA generously gifted by Stefano Piccolo, shAmotL2 (MXS19; Sigma), shAmotL2 #1 (GAACAAGATGGACAGTGAAAT) or shAmotL2 #2 (GAGAGATTGGAATCTGCAAAT; VectorBuilder). HUVECs or HUAECs were transfected with the above lentiviral supernatants diluted (2:1) with ECBM supplemented media with polybrene (8 μg/mL; Sigma) overnight. The following day, viral supernatants were removed and replaced with fresh supplemented media and cultured for 72 to 96 hours post-transduction before downstream analysis.
RT-PCR
Total RNA was isolated from HUVECs and HUAECs using RNeasy Plus Kit according to the manufacturer’s instructions (Qiagen). Harvested mRNA was eluted from RNeasy spin columns in RNase free water and stored at −80 °C. mRNA samples were quantified and purity checked using the NanoDrop Lite Spectrophotometer (Thermo Scientific). Using High-Capacity RNA-to-cDNA Kit (Thermo Scientific), cDNA was synthesized according to the manufacturer’s instructions using 500- to 1000-ng RNA. Master mix and mRNA were combined in 9:11, respectively. Samples were placed into C1000 Thermal Cycler (Biorad) to incubate at 42 °C for 60 minutes followed by 70 °C for 5 minutes.
Gene expression of targets listed in Table S1 was analyzed by SYBR green–based qPCR analysis using Applied Biosystems QuantStudio 7 Real-Time PCR Systems (Thermo Scientific), following the standard PCR cycling sequence using GAPDH as an internal control. Results for qPCR target genes were normalized to internal GAPDH controls. Statistical analysis of qPCR data was performed using the ΔΔCt method.
Western Blotting and Subcellular Fractionation
Protein expression was analyzed using precast Bis-Tris-PAGE (Invitrogen). Cells were washed in 1× PBS before being lysed in 1× RIPA lysis buffer (EMD Millipore Corporation). Cell lysates were boiled at 100 °C for 5 minutes and mixed with the NuPAGE LDS Sample Buffer (4×; Invitrogen), the NuPAGE Sample Reducing Agent 10× (Invitrogen), and total protein separated on a NuPAGE 4% to 12%, Bis-Tris, polyacrylamide gradient gel (Invitrogen) before Western blotting on Pure Nitorcellulose 0.2-µm membranes (PerkinElmer). Membranes were probed with primary antibodies, followed by HRP (horseradish peroxidase) conjugated secondary enhanced chemiluminescence anti-mouse (NA931V; 17438421) or enhanced chemiluminescence anti-rabbit antibodies (NA934V; 17434242; Amersham) for detection of probed proteins using enhanced chemiluminescence (Perkin Elmer). Chemiluminescence was detected using an iBright FL1500 (ThermoFisherScientific) imager. Antibodies used for Western blotting: rabbit pAb anti-AmotL2 (affinity purified as described previously21; Innovagen, Lund, Sweden). Anti-YAP (D8H1X; #14074) XP rabbit mAb, rabbit mAb anti-pYAP Ser127 (also cross-reacts with pTAZ Ser89; D9W2I; #13008), and rabbit mAb anti-TAZ (E8E9G; #83669) were from Cell signaling Technologies. Mouse anti-GAPDH (ab181602) and rabbit anti-FAK (phosphor Y397; ab81298) were from Abcam. Mouse anti-lamin A/C (sc-7292; 636) and anti-YAP/TAZ (sc-101199; 63.7) were from Santa Cruz Biotechnology. Subcellular fractionation for the Western blot analysis was performed using the NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermoscientific; WC319307) kit according to the manufacturer’s instructions.
Flow Experiments
Laminar and disturbed flow was generated using orbital flow as described previously5,22 using a Rotamax 120 (Heidolph) rotating platform placed in a cell culture incubator at 37 °C. HUVECs were seeded to 6-well plates with 2 mL of culture medium and orbitally shaken at 150 rpm for the indicated time periods before lysates were harvested as described by Rickman et al,22 collecting from the periphery and center of the wells for laminar and disturbed flow, respectively.
ChIP-Atlas
Publicly available ChIP-seq data submitted to the NCBI SRA were accessed from the ChIP-Atlas.23 Predicted target genes of YAP were also devised using the ChIP-Atlas database. The following data sets were used for YAP1-ChIP-seq: MCF7 (GSM3577938), T47D (GSM3577945), and HEK293 (GSM3732639). TEAD1 (TEA domain transcription factor 1)-ChIP-seq: MCF7 (GSM2859575), T47D (GSM2859586), and HEK293 (GSM3732641). TEAD4-ChIP-seq: MCF7 (GSM3577954), T47D (GSM3577942), and HEK293 (GSM3732642). Data were visualized using the IGV (Integrative Genomics Viewer) browser, version 2.9.4.
ENCODE ChIP-seq data used to view various histone marks around the YAP promotor included HUVEC Pol II (RNA polymerase II) GSM1305213, HUVEC H3K27ac accession: ENCFF656TFQ experiment: ENCSR000ALB, HUVEC H3K4me3 accession: ENCFF925RAZ experiment: ENCSR000AKN, HUVEC EZH2 accession: ENCFF700IMN experiment: ENCSR000ATA, and HUVEC H3K27me3 (trimethylation of histone H3 at lysine 27) accession: ENCFF938EXZ experiment: ENCSR000AKK, and were visualized using the IGV-Web app, version 1.12.5, igv.js version 2.13.5.
Whole-Mount Staining
Whole-mount staining was performed as described previously.5 Briefly, mice were euthanized and aorta harvested. The chest cavity was pinned open, and piercing of left ventricle, whole-body perfusion using 1× PBS (or in 1 cohort 1% PFA) was performed to clear the aorta of blood. Fat and surrounding adventitial tissue were dissected away and whole aortae further fixed in 4% PFA for 1 hour at room temperature. Tissue was then cut length ways and for the aortic arch dissected as shown in Figure 5E. Tissue was then mounted to wax blocks by pinning and tissue covered with a drop of PBS to ensure the tissue does not dry out while other pieces were mounted. Tissue was then fixed for another 10 minutes once mounted and washed with PBS. Tissue was permeabilized for 20 minutes at RT using 0.1% Triton X-100 in PBS followed by 3× PBS washes at intervals of 15 minutes. Tissue was subsequently blocked in 5% fetal calf serum for 1 hour at RT or overnight at 4 °C. Primary antibody incubation was performed in blocking buffer with the following antibodies overnight at 4 °C. Tissue was then washed 3× with 30-minute intervals with 1× PBS followed by secondary antibody incubation for 4 to 6 hours at RT. Tissue was washed before mounting between 2 coverslips using Fluoroshield with DAPI (4′,6-diamidino-2-phenylindole; Sigma; F6057). Tissue was then imaged using either Zeiss 700 or Leica Thunder microscope. Images were taken with the same microscope settings for all samples within the same experiment as outlined under the image analysis methods. The following antibodies were used for immunofluorescent staining: rabbit pAb anti-AmotL2 (affinity purified as described previously21; Innovagen), anti-YAP (D8H1X; #14074) XP rabbit mAb (Cell Signalling Technology), mouse anti-YAP/TAZ (sc-101199; 63.7; Santa Cruz Biotechnology), rabbit pAb anti-ki67 (ab15580), rabbit pAb anti-VE-cadherin (ab33168), EYFP was stained for using chicken pAb anti-GFP (ab13970) or goat pAb anti-GFP (ab6673), and rabbit anti-ERG (ETS-related gene) mAb (ab92513) were from Abcam. TexasRed phalloidin (T7471; Invitrogen) and phalloidin-Atto 647 N (65906; Sigma) were used to label the actin cytoskeleton. Rat anti-Cd31 (MEC 13.3; 553370) and rat anti-Cd144/VE-cadherin (1104.1; 555289) were from BD Biosciences.
Figure 5.
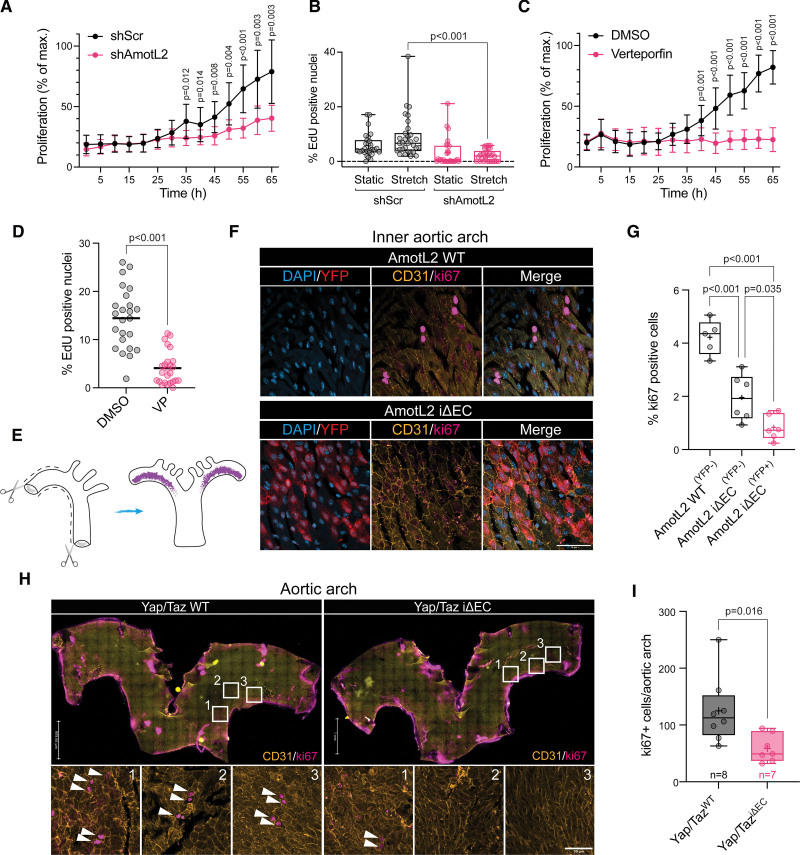
Endothelial proliferation is regulated by AmotL2 (angiomotin-like 2) and YAP (Yes-associated protein). A, Proliferation of human umbilical vein endothelial cell (HUVEC) replated to gelatin-coated plastic following 48-hour postlentiviral transduction with shScr (shRNA scrambled control) or shAmotL2 lentivirus. n=3 independent experiments (mean±SD, 2-way ANOVA with Sidak multiple comparisons). Error bars indicate SD. B, Proliferation of shScr- and shAmotL2-treated HUVECs exposed to uniaxial 12% stretched for 16 hours before 5-ethynyl-2′-deoxyuridine (EdU) incorporation and imaging. Box plots showing quantification of EdU, where the percentage of EdU-positive cells was calculated against the total number of cells stained with Hoechst. Each data point represents one field of view from n=4 independent experiments (mean±SD, Kruskal-Wallis with Dunnet multiple comparisons). C, Proliferation of HUVECs plated to gelatin-coated plastic and treated with 0.2 μg/mL verteporfin (VP) or DMSO vehicle, n=3, error bars indicate SD. D, Quantification of EdU-positive HUVECs treated with 0.2 μg/mL VP or DMSO vehicle (48 h). Cells were counterstained with Hoechst and quantified where the percentage of EdU-positive cells was calculated against the total number of cells stained with Hoechst (each data point represents 1 field of view from n=4 independent experiments, bar indicates mean, Mann-Whitney U test). E, Schematic depicting dissection of whole aorta for en face whole-mount staining. Purple region indicates the inner aortic arch. Schematic drawn using Adobe Illustrator (v25.4.1). F, Representative images of the inner aortic arch of AmotL2 wild-type (WT) and AmotL2 iΔEC, en face whole mount–stained for YFP (yellow fluorescent protein; red), nucleus (blue), CD31 (yellow), ki67 (magenta), using the ChrisLUT BOP palette (https://github.com/cleterrier/ChrisLUTs). Scale bar, 50 µm. Images representative of n=5 mice per group. G, Quantification of total ki67- and CD31-positive cells per aortic inner arch from AmotL2 WT (YFP −neg), n=5; AmotL2 iΔEC (YFP −neg), n=5; and AmotL2 iΔEC (YFP +pos), n=5 (mean±SD, 1-way ANOVA with Tukey multiple comparisons). H, Representative images of the inner aortic arch of Yap/Taz WT and Yap/Taz iΔEC, en face whole mount–stained for CD31 (yellow), nucleus (blue), and ki67 (magenta). White arrows indicate ki67-positive cells. Scale bar, 50 µm. I, Quantification of total ki67- and CD31-positive cells per aortic inner arch from Yap/Taz WT (n=5) and Yap/Taz iΔEC (n=5; mean±SD, Mann-Whitney U test).
For immunofluorescent staining of cultured ECs, cells were treated or transduced as described before washing and fixing using 4% PFA for 5 to 10 minutes at RT. Cells were then washed 2× with PBS and permeabilized for 10 minutes at RT using 0.1% Triton X-100 in PBS followed by 3× PBS washes. Cells were subsequently blocked in 5% fetal calf serum for 1 hour at RT before primary antibody incubation was performed in blocking buffer with the following antibodies overnight at 4 °C. Cells were then washed 3× in PBS followed by secondary antibody incubation for 1 to 2 hours at RT. Cells were washed before mounting to coverslips using Fluoroshield with DAPI (Sigma; F6057). Slides were then imaged as described above. The following antibodies were used for immunofluorescent staining of cultured cells: rabbit pAb anti-VE-cadherin (ab33168), from Abcam. TexasRed phalloidin (T7471; Invitrogen) was used to label the actin cytoskeleton. Mouse anti-lamin A/C (sc-7292; 636) was from Santa Cruz Biotechnology. Mouse anti-prelamin A (PL-1C7) was from Sigma.
Proximity Ligation Assay
Tissue was fixed and processed as described for immunofluorescence whole-mount staining. Tissue was pinned out, permeabilized, and washed as described previously. Tissue was then blocked as per the manufacturer’s conditions before proceeding with the assay using the Duolink in situ proximity ligation assay (PLA) kit (Sigma). Antibodies used for PLA; anti-TEAD1 mouse mAb (610923) BD Biosciences, anti-YAP (D8H1X; #14074) XP rabbit mAb (Cell Signalling Technology).
Chromatin Immunoprecipitation
HUVECs and HUAECs that were subjected to ChIP were transduced with lentiviral vectors for 72 to 96 hours before being processed using the iDeal ChIP-seq kit for transcription factors (Diagenode), according to the manufacturer’s protocol with modifications. Briefly, cells were cross-linked with 1% formaldehyde at RT for 15 minutes with gentle shaking. Glycine stop was added to the cells at 1:10 according to the protocol for 5 minutes before removal of medium and washing cells once with ice-cold 1× PBS keeping cells on ice. Cells were then lysed in 15 mL lysis buffer iL1 provided by the kit and incubated at 4 °C for 20 minutes with rotation. After centrifugation at 500g for 5 minutes at 4 °C, cell pellets were further lysed under rotation for 10 minutes in lysis buffer iL2. Chromatin was sheared using the Covaris M220 focused ultrasonicator for 15 minutes and stored at −80 °C. For immunoprecipitation, ChIP grade antibodies were incubated with prewashed protein A–coated magnetic beads for 2 to 4 hours at 4 °C under rotation. Sheared chromatin was then added to beads and incubated overnight at 4 °C under rotation. Beads were spun and washed according to the protocol. Immunoprecipitated and input DNA were then eluted and purified according to the manufacturer’s protocol. Real-time PCR was performed using 1% of immunoprecipitated or input DNA per reaction using SYBR green reagents according to the manufacturer’s protocol. Fold enrichment (2−DDCT) was calculated using input controls and IgG controls as background. Antibodies used for ChIP experiments were as follows: YAP (D8H1X) XP rabbit mAb antibody (14074; Cell Signaling Technology), RNA Pol II antibody (mAb; 39097), and histone H3K27me3 antibody (pAb; 39155) were from active motif. Mouse IgG (I8765) was from Sigma. Rabbit IgG (C15410206; Diagenode). anti-histone H3 (acetyl K27; ab4729), and anti-histone H3 (tri methyl K4; ab8580) were from Abcam. Primers for ChIP RT-qPCR were as follows: AmotL2: forward, 5′-TGCCAGGAATGTGAGAGTTTC-3′; reverse, 5′-AGGAGGGAGCGGGAGAAG-3′. CTGF: forward, 5′-GAGCTGAATGGAGTCCTACACA-3′; reverse, 5′-GGAGGAATGCTGAGTGTCAAG-3′. YAP (promotor region 1): forward, 5′-GCCGTCATGAACCCCAAGA-3′; reverse, 5′-GAGAGGGGCAACGAGGTTAC-3′. YAP (promotor region 2; Figure S10B and S10C): forward, 5′-GCCTCTCGGTCCACTTCAG-3′; reverse, 5′-GGTTCTACCCCCACCTCTAAT-3′.
Incucyte Growth Curves
Growth curve analyses of HUVECs transfected with shScr or shAmotl2 lentivirus or treated with verteporfin (0.2 μg/mL) or DMSO control were performed using Incucyte Zoom Live-Cell microscope (Essen Bioscience). Twelve-well plates (1×105 cells/well) were seeded to a 12-well plate 48 hours post-transfection. Cell coverage/confluence was calculated using built-in Incucyte image analysis with masking of cell confluence, which was calculated as a percentage of the maximum value obtained from the end point of the assay, which was indicated when 100% confluency for control conditions was reached. Four regions per well were imaged at hourly intervals for 65 hours using the ×20 objective.
5-Ethynyl-2′-Deoxyuridine Proliferation Assay
5-ethynyl-2′-deoxyuridine (EdU) incorporation was performed according to the manufacturer’s protocol using the Click-iT EdU Alexa Fluor 555 Imaging kit (Invitrogen; lot: 2263481). Briefly, HUVECs were plated to Ibidi chamber slides and grown before being subjected to lentiviral transduction for 72 hours before analysis of proliferation using the Click-iT EdU Cell proliferation kit for imaging Alexa fluor 647. Cells were then replated to 0.2% gelatin-coated Ibidi chamber slides and left overnight to adhere and equilibrate. The following day, cells were incubated with EdU component A for 2 hours at 37 °C before fixing in 4% PFA and subsequent permeabilization, blocking, and staining as dictated by the manufacturer’s protocol. Cells were counterstained with Hoechst and imaged using the Leica Thunder system. Three to 5 images per condition were taken using ×20 air objective using the Leica Thunder microscope.
Hydrogels
Petrisoft easy-coat hydrogel 0.2- or 50-kPa 6-well plates were coated with 5 μg/mL human fibronectin (Sigma) and RNA lysates harvested as described previously.
Stretch Assay
Custom-made silicon membranes were sterilized and coated with 40 μg/mL human fibronectin in PBS for 3 to 4 hours at RT before seeding cells 1×105 HUVECs. Cells were cultured overnight before transducing with lentiviral supernatants as described previously. Infected HUVECs were cultured with daily medium changes for 72 hours before stretching of 12% of the original length of the chamber at oscillations of 1 cycle of stretch per second for 16 hours (overnight). Cells were then assessed for proliferation using the EdU incorporation assay as described above. Silicon membranes were cut from the chambers and stained and whole mounted to coverslips with DAPI duromount before analysis by Thunder Leica microscopy at ×20 objective.
DNA Methylation Sequencing
HUAECs were subjected to lentiviral knockdown of shScr or shAmotL2 before DNA isolation using the DNeasy Blood & Tissue Kit (Qiagen). DNA was bisulphite converted according to the protocol of the EZ DNA Methylation-Gold Kit (Zymoresearch) and subsequently sent for DNA MethylationEPIC Infinium bead array (Illumina) sequencing. Bioinformatic analysis was performed using the R package RnBeads 2.0.24
ATAC-seq
ATAC-seq was performed as reported previously.25–27 Using the ATAC-seq library preparation, ATAC-seq library preparation kit (Active Motif) HUVECs and HUAECs were transduced with shScr and shAmotL2 for 72 hours before cells were trypsinized and counted using Trypan blue. One hundred thousand cells were centrifuged, and nuclei was extracted using ice-cold ATAC-lysis buffer (according to the kit protocol; Active Motif) and the pellet resuspended in the Tn5 tagmentation master mix. The transposition reaction was incubated at 37 °C for 30 min at 800 rpm using a thermomixer. Tagmented DNA was purified according to the protocol’s column purification step, followed by PCR amplification of tagmented DNA for 10 cycles using Illumina’s Nextera indexed adapters. After the PCR reaction, libraries were purified with the SPRI magnetic beads and library quality, and concentration was assessed with Qubit bioanalyzer. Four independent experiments were prepared for ATAC-seq for both HUVECs and HUAECs, resulting in 16 libraries in total. Libraries were then sequenced with the Illumina Hiseq platform with a 150-bp paired-end repeats protocol.
Nextera adaptor sequences were first trimmed from the reads using skewer (0.2.2). These reads were aligned to a reference genome using BWA, with standard parameters. These reads were then filtered for high quality (MAPQ, ≥13), nonmitochondrial chromosome, and properly paired reads (longer than 18 nt).
All peak calling was performed with macs2 using macs2 callpeak--nomodel--keepdup all--call-summits. For simulations of peaks called per input read, aligned and deduplicated BAM files were used without any additional filtering. KEGG enrichment analysis was performed using KOBAS (v3), with a P value cut off <0.05. Distribution of peak over various functional areas is analyzed by the ChIPseeker software. Bigwig files displaying sequenced tracks were visualized using the browser Integrative Genomics Viewer (v.1.12.5). Data were overlaid with existing ChIP-seq data of RNAP II, H3K27ac, H3K4me3, EZH2, and H3K27me3, to give contextual chromatin features of the YAP promotor.
Image Analysis
Confocal images were acquired using a scanning laser confocal Zeiss LSM700 microscope. Settings used were as follows: standard laser configuration, objectives-c-ApoChromat 40×/1.2 W Korr M27 water immersion lens, line averaging 2, Z stack 5 slices at 0.5-μM step, resolution (1024×1024) pinhole diameter: 1 airy unit. For whole-mount aortic arches imaged using tiled scan, Leica Thunder LED–based microscopy was used using a ×20 air objective; higher resolution regions of the aortic arch where then imaged using scanning laser confocal Zeiss LSM700 microscopy using the settings outlined above. Microscope settings were kept constant across conditions of experiments and between different independent tissue samples within an experiment.
For YAP nuclear quantification, fluorescent intensity across the major of axis of the cell was measured using DAPI as a marker of the nuclear region and intensity profiles exported and averaged using GraphPad Prism. Fluorescent intensity of 60 cells in total was analyzed from the aorta and inferior vena cava (IVC) of 3 individual mice. To analyze the nuclear YAP:TEAD1 PLA signal, the ImageJ plugin BioVoxxel>Speckle Inspector was used. Images were subject to maximum intensity projection before thresholding and then analysis of dots/nuclei. Junctional staining and quantification were analyzed in ImageJ by drawing a line across the image (3 lines per image) and quantifying the staining intensity profile using the plot profile function. The x and y coordinates were exported and average staining intensity calculated using the area under the curve in Graphpad Prism. For YAP intensity profiles of YAP/TAZ iΔEC quantification, whole image YAP intensity was analyzed.
For analysis of nuclear circularity in YAP/TAZ iΔEC mice, confocal images were analyzed using the DAPI staining of the nucleus using ImageJ. Images were analyzed using the MorphoLibJ plugin: MorphoLibJ>Segmentation>Morphological segmentation. Images were processed as follows: Z stacks were stacked with maximum projection; background was subtracted using subtract background with sliding paraboloid and rolling ball radius of 50 pixels. A threshold was applied to the image to obtain a masked image before the application of gaussian blur (Sigma: 2). The plugin MorphoLibJ was then used to segment images based on morphology using the following settings: Run MorphoLibJ>Segmentation>Morphological segmentation (settings: [object image], gradient—morphological, radius 1, watershed segmentation—tolerance 15, overlaid basins>create image). The segmented image was then used to analyze nuclear parameters using MorphoLibJ>label map followed by MorphoLibJ>Analyze tools. Data were then imported to GraphPad Prism for statistical analysis.
Statistical Analysis
All statistical analyses were performed using GraphPad Prism (v9), with the exception of ATAC-seq data sets in which P values were generated with the software outlined in Table S2 and human AAA samples where the correlation between AmotL2:YAP and AmotL2:TAZ was analyzed using Pearson correlation and Pearson correlation coefficient generated using R (https://www.r-project.org/index.html). Statistical tests used to analyze differences between groups are outlined in each figure legend and consist of unpaired Student t tests, Mann-Whitney U test, and 1- or 2-way ANOVA with correction for multiple comparisons (Tukey, Sidak, or Dunnet) as specified in the figure legends. Normal distribution was tested using the D’Agostino-Pearson and Shapiro-Wilk tests and nonparametric tests applied when appropriate. All tests were 2 tailed and unpaired unless specified otherwise. The number of independent biological replicates is indicated in the figure legends and is referred to as n. P values are displayed in the figures. Data are displayed as means±SD unless otherwise specified. Sample sizes were not predetermined using any tests. Experiments were not randomized or blinded in any way.
For image analysis, a minimum of 3 replicate images (from the same sample) were taken from at least 3 independent experiments. Representative images shown are of a minimum of 3 mice or 3 independent experiments for in vitro cell cultures.
RESULTS
AmotL2 and YAP Are Highly Expressed in the Aorta Compared With the Vena Cava
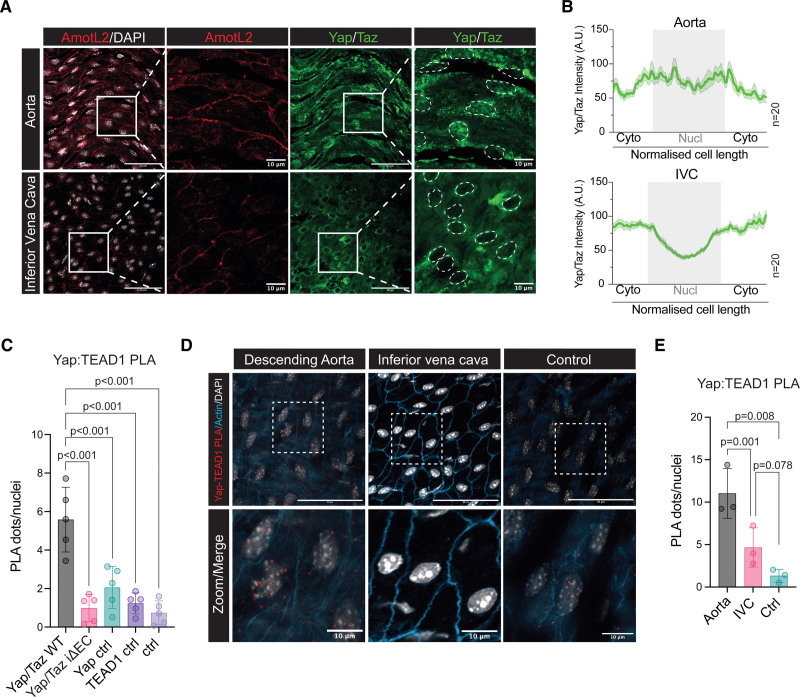
Previous work has shown that YAP regulates AmotL2 expression in epithelial cells,28,29 whereas work from our laboratory has shown that AmotL2 is highly expressed in the aorta compared with the IVC.5 Therefore, we hypothesized that increased expression of AmotL2 may be due to increased YAP activity in the aorta.13,30,31 In silico screening using RNA Annotation and Mapping of Promotors for Analysis of Gene Expression32,33 indicated that the YAP promotor was most active in the thoracic aorta in comparison to other tissues (Figure S1A). Immunofluorescence indicated increased AmotL2 levels and greater nuclear YAP in the aorta versus IVC (Figure 1A and 1B; Figure S1B).
Figure 1.
The murine aorta exhibits high AmotL2 (angiomotin-like 2) expression and YAP (Yes-associated protein) activity compared with the vena cava. A, Confocal imaging of en face whole-mount staining of the descending aorta (top) and inferior vena cava (bottom). AmotL2 is shown in red and YAP/TAZ (transcriptional coactivator with PDZ-binding motif) in green. Nuclei are outlined by white dotted lines for clarity. Scale bar, 50 µm. In zoomed panels, scale bar, 10 µm. B, Quantification of nuclear (Nucl) and cytoplasmic (Cyto) staining of YAP/TAZ. Histogram graph depicts the average intensity (mean±SEM) of n=20 cells from 1 representative animal. A total of 60 cells per aorta or inferior vena cava (IVC) were measured from n=3 mice. See Figure S1B for full quantification. C, Quantification ex vivo en face proximity ligation assay (PLA) of YAP-TEAD1 (TEA domain transcription factor 1) PLA of wild-type (WT) and YAP/TAZ iΔEC and indicated controls (Ctrls; images shown in full in Figure S1C), where each data point represents an individual animal (n=5 mice per group, mean±SD, 1-way ANOVA with Sidak multiple comparisons). D, Representative images of ex vivo en face PLA of YAP:TEAD1 interactions in the endothelium of the descending aorta and inferior vena cava. Positive interactions are indicated by red dots, nuclei in gray, and actin in blue. E, Quantification of d, from n=3 independent WT mice (mean±SD, 1-way ANOVA with Tukey multiple comparisons). Scale bar, 50 and 10 µm, for ×40 and indicated zoomed regions, respectively.
To explore the nuclear YAP localization of the aorta in more detail, we used the PLA, to probe for YAP and TEAD1 interactions. The proximity of YAP-TEAD1 is indicative of YAP activity, which is able to indirectly bind to DNA via TEAD1 and drive transcription.28,34–36 To validate the specificity of YAP-TEAD1 PLA on ex vivo aortic tissue, we utilized inducible endothelial-specific Cdh5(BAC)CreERT2 transgenic mice crossed with Wwtr1 flox/flox; Yap flox/flox (creating the Yap/Taz iΔEC mice after tamoxifen administration), which were compared with Cdh5(BAC)CreERT2 negative Wwtr1 flox/flox; Yap flox/flox mice (control). The genetic deletion of one of the targets of the PLA reaction (here, YAP) resulted in the ablation of PLA signal in Yap/Taz iΔEC mice (Figure S1C). Quantification of YAP-TEAD1 PLA signal in Yap/Taz iΔEC mice, versus WT (Yap/Taz WT), indicated a significant reduction in PLA signal, comparable to single antibody, and double-negative controls (Figure 1C), suggesting high specificity of the technique. We next compared the aorta and IVC using YAP-TEAD1 PLA as a readout of YAP activity. Results indicated increased PLA signal in the aorta, compared with the IVC (Figure 1D and 1E), confirming the immunofluorescent stainings showing increased nuclear YAP. These results suggest that the aorta exhibits increased YAP activity and consequently increased AmotL2 expression, compared with the IVC.
Endothelial YAP Is Required for AmotL2 Expression in Mouse Aorta
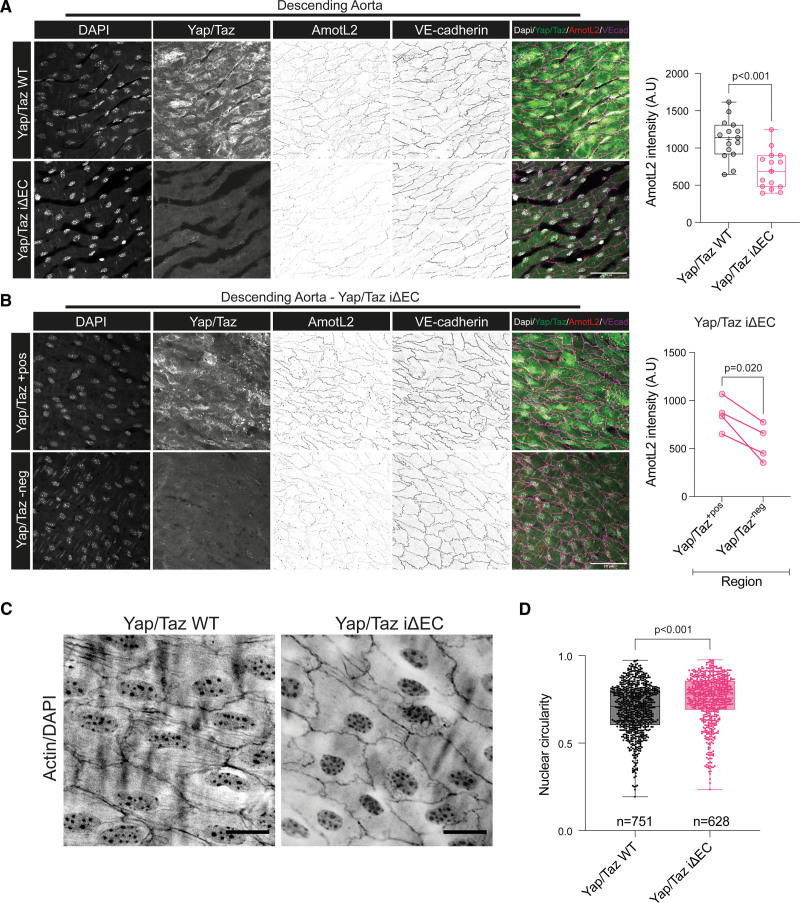
To test whether YAP controls AmotL2 in vivo, we performed immunofluorescent staining of control and Yap/Taz iΔEC aortae. Interestingly, AmotL2, along with Yap/Taz expression, was decreased in Yap/Taz iΔEC mice (Figure S2A; Figure 2A). Assessment of YAP/TAZ by immunofluorescence shows 83.7% knockout efficiency (Figure S2B), therefore, giving rise to regions of the Yap/Taz iΔEC aorta where we observed Yap/Taz positive staining, accompanied by higher expression levels of AmotL2, serving as an internal control within the Yap/Taz iΔEC aorta (Figure 2B). These results confirm that Yap/Taz regulates AmotL2 expression within the aortic endothelium.
Figure 2.
Deletion of endothelial Yap/Taz (transcriptional coactivator with PDZ-binding motif) decreases AmotL2 (angiomotin-like 2) expression and disrupts nuclear shape in aortic endothelial cells in vivo. A, Representative images of en face staining of AmotL2 in the descending aorta of both Yap/Taz wild-type (WT) and Yap/Taz iΔEC mice, n=5 per group. Nucleus (gray), Yap/Taz (green), VE (vascular endothelial)-cadherin (magenta), AmotL2 (red). Scale bar, 50 µm. Bar graph indicates quantification of AmotL2 fluorescent intensity, where each data point represents the intensity profile from 1 image; 3 images per aorta (n=5 mice per group) were analyzed, mean±SD, Mann-Whitney U test. B, Representative images of regions of positive (+pos) and negative (−neg) Yap/Taz staining in Yap/Taz iΔEC mice and corresponding nucleus (gray), Yap/Taz (green), Ve-cadherin (magenta), AmotL2 (red) staining. Scale bar, 50 µm. Graph indicates quantification of AmotL2 fluorescent intensity, where each data point represents average intensity profile from one mouse (n=4 mice per group; paired t test). C, Representative images of en face staining of actin and the nucleus in endothelium of the descending aorta of both Yap/Taz iΔEC and Yap/Taz WT mice (n=5). D, Quantification shown in the box and whisker plot indicates nuclear circularity, measured using ImageJ as described in the methods (n=751 and n=628 nuclei from 5 mice per group, mean±SD, Mann-Whitney U test).
Deletion of endothelial AmotL2 results in reduced actin fibers across the nucleus, perturbed nuclear morphology, and abnormal endothelial sensing of FSS.5 Therefore, we hypothesized that the Yap/Taz deletion would phenocopy the morphological dysfunction observed in AmotL2 iΔEC. Staining of the descending aorta indicated a loss of actin fibers in Yap/Taz iΔEC (Figure 2C) and more rounded nuclei, compared with the elongated morphology of Yap/Taz WT aorta (Figure 2D). These results mirror in vivo endothelial morphology of AmotL2 iΔEC and suggest that Yap is required for AmotL2-dependent response to FSS in the aorta.
We validated our in vivo findings by knockdown of YAP using lentiviral shRNA in HUAECs and HUVECs, respectively, which led to a reduction in AmotL2 and other well-defined YAP target genes such as CTGF and ANKRD1 (Figure S3A and S3B).
Individual and codepletion of YAP and TAZ significantly reduced AmotL2 expression (Figure S3C and S3D); however, no additive effect was observed in codepletion of YAP and TAZ (Figure S3C and S3D). Analysis of protein expression showed that YAP shRNA led to a reduction of AmotL2 in both HUAECs (Figure S3E) and HUVECs (Figure S3F). Conversely, overexpression of a constitutively active YAP mutant (5SA) led to increased AmotL2 (Figure S3G and S3H). To analyze whether mechanical stimulation of YAP modulated Amotl2 levels, HUVECs were plated to soft and stiff hydrogels or exposed to laminar and disturbed flow where resulting qPCR analysis indeed showed upregulated AmotL2 (Figure S3I and S3J).
To verify that YAP binds to the AmotL2 promotor, we analyzed existing ChIP-seq data (https://chip-atlas.org/). Screening of the AmotL2 transcriptional start site for both YAP and TEAD1 binding indicated enrichment across multiple cell lines (Figure S4A), with overlapping enrichment of YAP, TEAD1, and TEAD4 (Figure S4B). Importantly, YAP, TEAD1, and TEAD4 were also enriched at the promotors of CTGF and CYR61 (Figure S4C). Predicted target genes of YAP indicated AmotL2 as the top hit in the ChIP-Atlas database at 1, 5, and 10 kb from the transcriptional start site (Figure S4D). Taken together, these results show that YAP transcriptionally regulates AmotL2 in ECs.
AmotL2 Regulates YAP/TAZ Transcription
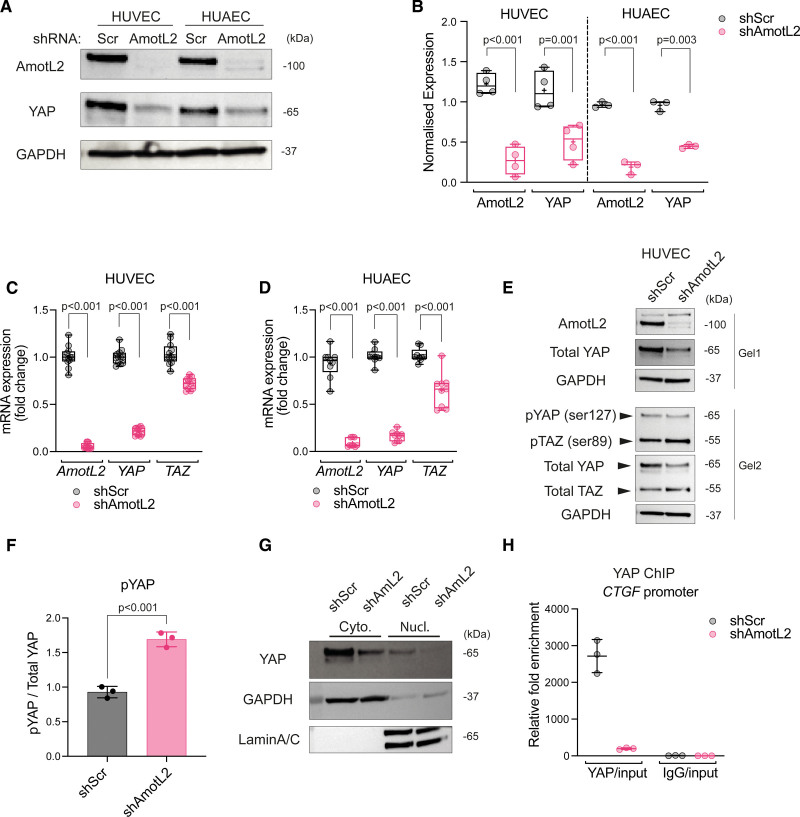
Previous work indicates the existence of a continuous connection linking cell-cell junctions and the nuclear envelope via the actin cytoskeleton, which regulates sensing of FSS.5 We hypothesized that junctional force sensed by this VE-cadherin-p120-AmotL2 complex may regulate YAP activity in the aorta under conditions of mechanical stimulation. We therefore investigated AmotL2-mediated regulation of YAP in ECs using lentiviral shRNA. Intriguingly, the depletion of AmotL2 in HUVECs and HUAECs resulted in a significant reduction in YAP protein (Figure 3A and 3B). Depletion of AmotL2 with 2 additional shRNA constructs showed similar results in HUVECs, with reduced YAP expression (Figure S5A and S5B). In HUAECs, downregulation of YAP was only noted with one of the additional constructs (Figure S5A and S5B). To test the specificity of downregulated YAP, we performed Western blotting using an antibody that detects both YAP and TAZ. Here, AmotL2 depletion led to a marked reduction of YAP, with no effect in TAZ expression (Figure S5C, quantification in Figure S5D). To investigate whether YAP protein regulation was attributable to transcription, RT-qPCR was performed in AmotL2-depleted HUVECs and HUAECs. Results showed a significant reduction in YAP mRNA in both cell lines, suggesting that YAP transcription is AmotL2 dependent (Figure 3C and 3D). This effect appeared irreversible as reexpression of AmotL2 could not rescue YAP expression levels comparable to control conditions (Figure S5E).
Figure 3.
AmotL2 (angiomotin-like 2) regulates YAP (Yes-associated protein)/TAZ (transcriptional coactivator with PDZ-binding motif) transcription. A, Western blot analysis of AmotL2 and YAP in human umbilical vein endothelial cells (HUVECs) and human umbilical arterial endothelial cells (HUAECs) 96 hours post-treatment with shScr (shRNA scrambled control) or shAmotL2 lentivirus. GAPDH was used as a loading control. B, Quantification of AmotL2 and YAP protein levels, relative to GAPDH loading control. HUVECs, n=4; HUAECs, n=3 independent experiments, mean±SD, 2-way ANOVA with Dunnett multiple comparisons. SYBR green RT-qPCR of AmotL2, YAP, and TAZ relative to the housekeeping gene GAPDH, in AmotL2 knockdown HUVECs (C) and HUAECs (D; n=4 independent experiments for HUVECs and n=3 for HUAECs, mean±SD, 2-way ANOVA with Dunnett multiple comparisons). E, Western blot analysis of indicated proteins in HUVECs 96 hours post-treatment with shScr or shAmotL2 lentivirus. Samples were run on 2 gels in parallel and probed with indicated antibodies. GAPDH was used as a loading control. Blots shown are representative of n=3 independent experiments (shown in the supplemental publication material of uncropped Westerns). F, Quantification of pYAP ser127 levels, relative to total YAP (n=3 independent experiments, mean±SD, Mann-Whitney U test). G, Representative Western blot of nuclear:cytoplasmic fractionation detection of indicated proteins in HUVECs 96 hours post-treatment with shScr or shAmotL2 lentivirus. GAPDH and LaminA/C were used as positive and negative controls. H, ChIP showing YAP binding to AmotL2 and CTGF promotor of shScr- or shAmotL2-treated HUVECs. ChIP-qPCR was performed using SYBR green reagents, and quantification was normalized to an IgG control antibody. Plot shown is a representative experiment from n=3 independent experiments (Figure S5J shows 2 further independent experiments). Each data point represents a technical repeat within 1 independent experiment (performed in triplicate; mean±SD).
Given the previous work showing AmotL2 as a negative regulator of YAP phosphorylation,37–41 we were intrigued whether the reduced YAP expression we observed also exhibited changes in phosphorylation. We, therefore, investigated YAP activity by Western blotting for phosphorylated YAP and TAZ in shAmotL2 HUVECs. As before, a reduction in total YAP was observed (Figure 3E), whereas ser127 phosphorylation was found to increase significantly in AmotL2-depleted HUVECs (Figure 3F). No difference was noted in TAZ ser89 phosphorylation upon AmotL2 knockdown (Figure S5G). Subcellular fractionation and analysis of YAP localization did not indicate any difference in the nuclear:cytoplasmic YAP ratio (Figure 3G, quantified in Figure S5H). To further validate AmotL2-dependent YAP activity, we performed YAP-ChIP of the CTGF and AmotL2 promotors. Results indicate a reduction in YAP binding to both promotors, suggesting that endothelial YAP activity is reduced in the absence of AmotL2 (Figure 3H; Figure S5I and S5J). Collectively, these results suggest that transcription of YAP is AmotL2 dependent and that this impacts YAP’s activity as a transcriptional coactivator.
AmotL2 Regulates Yap Expression of the Aortic Endothelium
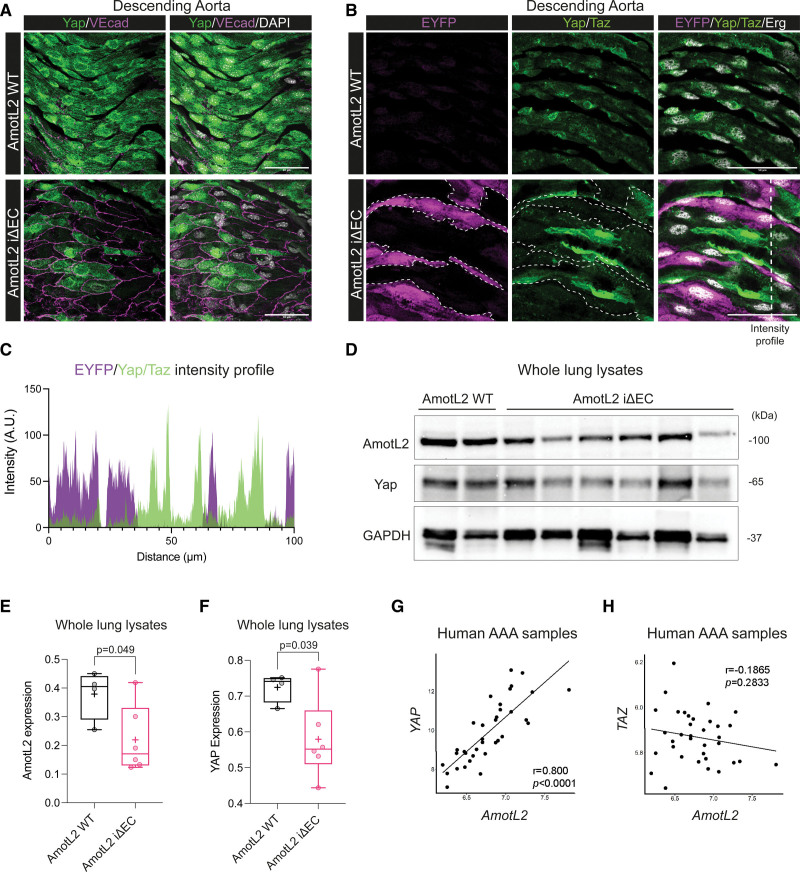
To test whether the Yap expression was also AmotL2 dependent in vivo, we utilized Cdh5(PAC)CreERT2 transgenics42 crossed with amotl2flox/flox and ROSA26-EYFP reporter mice to delete endothelial AmotL2 in adult mice (referred to as AmotL2 iΔEC). As AmotL2 and Yap were both found to be expressed in the aorta (Figure 1), we tested whether AmotL2 deletion resulted in the downregulation of Yap in the aortic endothelium. Using VE-cadherin to confirm the EC identity, immunofluorescence indicated complete ablation of Yap expression in a number of cells of the AmotL2 iΔEC aorta (Figure 4A) accompanied by changes in EC morphology as has been previously documented by our group.5 Using the EYFP reporter for AmotL2 deletion, stainings indicated EYFP+ cells with reduced Yap/Taz, versus neighboring EYFP− cells, exhibiting WT Yap/Taz expression (Figure 4B, quantification in Figure 4C). Importantly, Yap/Taz staining of the descending aorta of tamoxifen-injected CreERT2-EYFP mice indicated homogenous levels of Yap/Taz expression across both EYFP+ and EYFP− cells (Figure S6C). The IVC of AmotL2 iΔEC animals did not indicate a reduction in Yap/Taz staining (Figure S6A) and nuclear:cytoplasmic Yap/Taz of AmotL2 iΔEC, indicated no change in localization or expression levels in the IVC (Figure S6B). Further investigation by Western blotting of lung lysates from AmotL2 WT and iΔEC mice also indicated a reduction in AmotL2 and Yap (Figure 4D, quantification in Figure 4E and 4F), despite the heterogenous nature and the ubiquitous expression of AmotL2 highlighted by scRNA-seq of lungs43,44 (Figure S6D and S6E).
Figure 4.
AmotL2 (angiomotin-like 2) regulates aortic endothelial YAP (Yes-associated protein) in vivo. A, Representative images of en face staining of Yap (green) and VE (vascular endothelial)-cadherin (VEcad; magenta) in the descending aorta of both AmotL2 wild-type (WT) and AmotL2 iΔEC mice. Nucleus (gray), scale bar, 50 µm. Images are representative of n=3 mice per group. B, Representative images of en face staining of EYFP (yellow fluorescent protein; magenta), Yap/Taz (transcriptional coactivator with PDZ-binding motif; green), and Erg (ETS-related gene; gray) in the descending aorta of both AmotL2 WT and AmotL2 iΔEC mice. EYFP-positive cells are outlined for clarity. Scale bar, 50 µm. Images are representative of n=3 mice per group. C, Line profile (indicated by dashed white line in the bottom right-hand panel of B) of immunofluorescent intensity of EYFP and Yap/Taz of AmotL2 iΔEC aorta. D, Western blot analysis of AmotL2, Yap protein expression from AmotL2 WT (n=2), and AmotL2 iΔEC (n=6) whole lung lysates. GAPDH was used as a loading control. E, Box plot indicates quantification derived from WB (shown in the supplemental publication material of uncropped Westerns) of AmotL2 expression relative to GAPDH loading control (AmotL2 WT [n=4] and AmotL2 iΔEC [n=6], mean±SD, unpaired t test). F, As in E for Yap protein expression. AMOTL2 correlation with YAP (G) and TAZ (H) of mRNA expression is presented. The mRNA expression level of the gene was calculated with the mean value of every exon expression detected. For each correlation analysis, samples were obtained from the medial layer of dilated aortae (35 patients with abdominal aortic aneurysm [AAA], 25 male and 10 female patients were enrolled). The correlation between 2 genes was analyzed using the Pearson correlation, and Pearson correlation coefficient is referred to as r. P value and r (microarray analysis in patients with AAA) were calculated using R, version 4.1.1. r and P values are labeled in each individual figure.
AmotL2 is specifically expressed in the endothelium of human aortic sections5 and so to explore the relationship between AmotL2 and YAP expression in the human vasculature, we analyzed transcripts in aortic samples surgically resected from patients undergoing reconstructive surgery of the abdominal aorta. Transcriptomic array data of the medial layer showed a correlation between YAP and AmotL2 (Figure 4G). No correlation between TAZ and AmotL2 was found (Figure 4H), indicating the YAP-specific coregulation with AmotL2. Overall, these data indicate that endothelial YAP expression in vivo is AmotL2 dependent.
AmotL2 Regulates YAP-Dependent Proliferation
Among other functions, YAP is a key regulator of proliferation.15,45–47 To examine the functional implication of AmotL2-dependent YAP regulation, we tested the impact of AmotL2 depletion on EC proliferation. Growth curves of shAmotL2 HUVECs indicated a reduction in confluency over 68 hours (Figure 5A), in line with previous findings.48 Proliferation of untransformed cells is limited by contact inhibition, which, in part, is controlled by YAP.49 As such, analysis of EdU incorporation of subconfluent HUVECs indicated a greater percentage of cells in the S phase over confluent conditions (Figure S7A and S7B), an effect that was significantly reduced in shAmotL2 HUVECs (Figure S7A and S7B). These results were also mirrored on 50-kpa hydrogels (Figure S7C). Furthermore, stretch-induced proliferation is dependent on the YAP/TAZ activity.13 We, therefore, performed uniaxial cyclic stretch on AmotL2-depleted HUVECs and assessed EdU incorporation. Results showed that shAmotL2 subjected to cyclic stretch displayed reduced proliferative capacity compared with shScr controls. Importantly, treatment of HUVECs with the YAP-TEAD competitive inhibitor verteporfin mimicked results of AmotL2 depletion, both in growth kinetics (Figure 5C) and EdU incorporation (Figure S7D, quantified in Figure 5D), indicating the reliance of EC proliferation on YAP-TEAD interactions.
To investigate whether AmotL2 regulates proliferation in vivo, we focused on a subpopulation of proliferating ECs present within the inner arch of the adult aorta50,51 using en face whole-mount staining (Figure 5E). Staining of the inner aortic arch of AmotL2 WT mice with ki67, CD31, and EYFP indicated the presence of proliferating ki67-positive ECs (Figure 5F, bottom). The deletion of AmotL2, indicated by EYFP expression, was accompanied by a reduction in ki67-positive ECs in the inner aortic arch (Figure 5F). Quantification indicated a significant reduction in ki67+/EYFP+ ECs in AmotL2 iΔEC compared with AmotL2 WT, EYFP− littermates (Figure 5G).
To confirm that the proliferation of the ECs of the aortic arch is indeed YAP dependent, we stained the arches of WT Yap/Taz and Yap/Taz iΔEC mice with ki67 and CD31. Staining indicated a reduction in proliferating ECs of Yap/Taz iΔEC versus Yap/Taz WT aortae (Figure 5H, quantification of ki67+/CD31+ cells in Figure 5I). Collectively, these results indicate that mechanically induced endothelial proliferation is regulated by an AmotL2-YAP axis, the absence of which leads to suppressed proliferation.
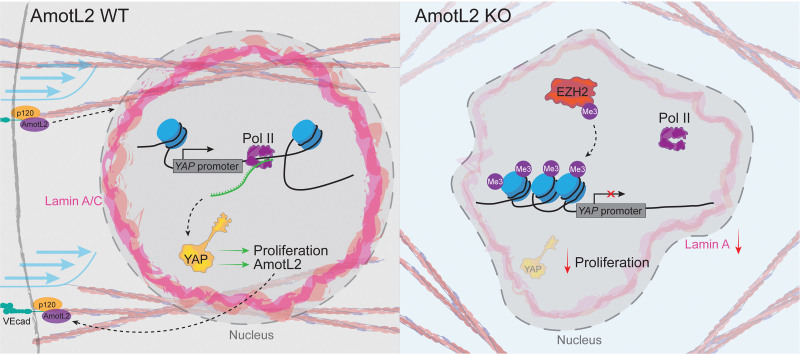
AmotL2 Regulates YAP Transcription by Histone Methylation
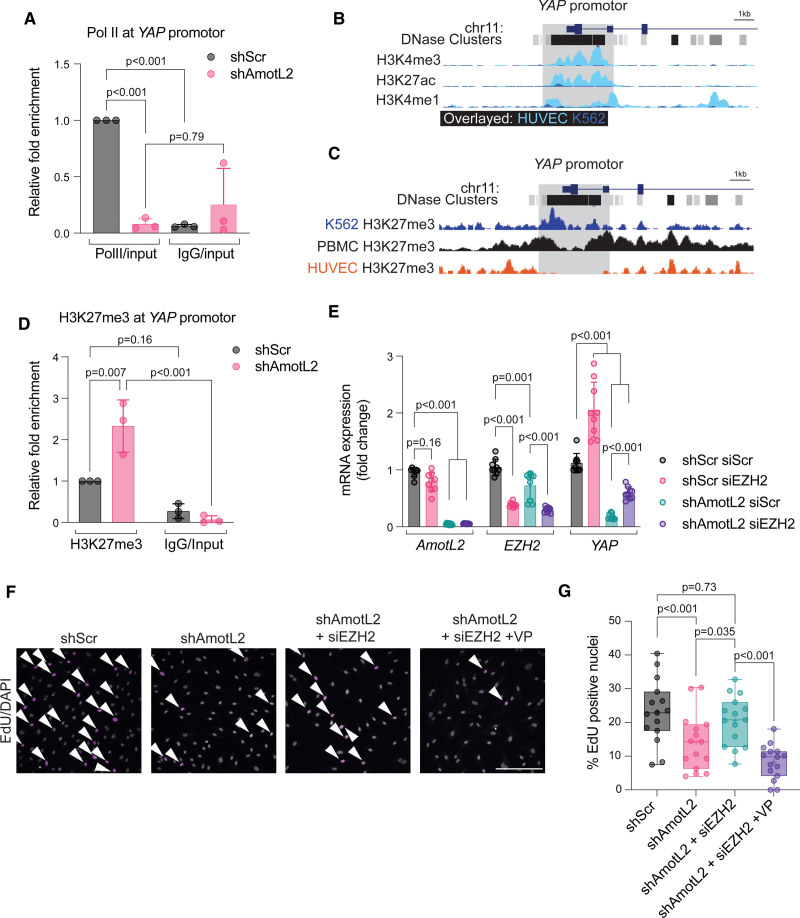
To further understand how AmotL2 regulates YAP transcription, we focused on investigating epigenetic markers associated with transcriptional repression. Initial reporting of the cloning of YAP showed the absence of expression in peripheral blood mononuclear cells52 and the murine spleen.53 Using scRNA-seq of breast tissue from (https://www.proteinatlas.org/), we observed reduced expression of YAP in blood and immune cells, confirming previous findings (Figure S8A). Using a single-cell atlas of chromatin accessibility (http://catlas.org/catlas_hub/), we observed reduced chromatin accessibility around the YAP promotor of CD4+, CD8+, naive T, and NKT (Natural killer T) cells, compared with ECs of several organs (Figure S8B). Open chromatin allows binding of Pol II to the accessible DNA template. We, therefore, tested whether reduced YAP transcription was due to differential YAP promotor-bound Pol II. ChIP-qPCR of shAmotL2 HUVECs indicated a significant reduction in Pol II binding to the YAP promotor, compared with shScr and IgG controls (Figure 6A).
Figure 6.
AmotL2 (angiomotin-like 2) regulates H3K27me3 (trimethylation of histone H3 at lysine 27) to modulate YAP promotor activity. A, ChIP showing Pol II (RNA polymerase II) binding to the YAP promotor of shScr (shRNA scrambled control)- or shAmotL2-treated human umbilical vein endothelial cells (HUVECs). ChIP-qPCR was performed using SYBR green reagents, and quantification was normalized to an IgG control antibody (n=3 independent experiments, mean±SD, 2-way ANOVA with Sidak multiple comparisons). B, Screen grab of the UCSC browser displaying genomic tracks showing published chromatin immunoprecipitation sequencing (ChIP-seq) data of H3K4me3, H3K27ac, and H3K4me1 of ChIP-seq data across HUVEC and K562 (overlayed) within the YAP promotor (data sources are referenced in Methods). C, Screen grab of the UCSD browser displaying genomic tracks showing ChIP-seq data of H3K27me3 of ChIP-seq data across K562, PBMC, and HUVEC within the YAP promotor (data sources are referenced in Methods). D, ChIP-qPCR analysis of H3K27me3 binding to the YAP promotor of shScr- or shAmotL2-treated HUVEC. ChIP-qPCR was performed using SYBR green reagents, and quantification was normalized to an IgG control antibody. Plot shown is representative of n=3 independent experiments, mean±SD, 2-way ANOVA with Sidak multiple comparisons. Each data point represents an independent experiment. E, Codepletion of EZH2 (enhancer of zeste 2 polycomb repressive complex 2 subunit) and AmotL2 rescues YAP mRNA expression. RT-qPCR of indicated targets in HUVECs where knockdown of EZH2 using siRNA was followed by shRNA knockdown using shScr or shAmotL2 (n=3 independent experiments, mean±SD, 2-way ANOVA with Tukey multiple comparisons). F, Representative images of shScr, shAmotL2, shAmotL2+siEZH2, and shAmotL2+siEZH2+verteporfin (VP) at 0.2 μg/mL treated HUVECs 72 hours post-infection with lentiviral vectors, 72 hours post-siRNA transfection, replated to gelatin-coated plastic in subconfluent conditions. Incorporated 5-ethynyl-2′-deoxyuridine (EdU) was detected with secondary antibodies and counterstained with Hoechst. Arrows indicate ki67-positive cells. Scale bar, 250 µm. G, Quantification of F, where each data point represents 1 field of view (5 taken per condition) from n=3 independent experiments, mean±SD, 2-way ANOVA with Tukey multiple comparisons.
DNA methylation is widely accepted as a marker of transcriptional repression. Analysis of published data indicated YAP promotor methylation in immune cells compared with other YAP-expressing cells (Depmap.org; Figure S8C). However, analysis of methylation of AmotL2-depleted HUAECs indicated no change in global differential methylation, and so we turned our attention to histone modifications (Figure S8D).
A plethora of histone modifications are responsible for promotor regulation, many of which are temporal and cell-type specific. We, therefore, used existing ChIP-seq data to compare known YAP-expressing and nonexpressing cell types, to ascertain the main contributors of epigenetic regulation of YAP. Analysis of the YAP promotor revealed the enrichment of markers of active euchromatin; H3K4me3, H3K27ac, and H3K4me1 in HUVECs, compared with low YAP-expressing leukemic cell line K562 (Figure 6B). Further investigation indicated that repressive H3K27me3 was enriched within the YAP promotor of peripheral blood mononuclear cells and K562 but was absent in HUVECs (Figure 6C). We validated these data sets by exploring these modifications within the promotors of genes expressed by ECs (VEGFR2; Figure S8E), immune cells (CD45; Figure S8F), or epithelial cells (CDH1; Figure S8G), which indicated expected patterns of enrichment for the active (H3K4me3, H3K27ac, and H3K4me1) and repressive (H3K27me3) modifications.
We, therefore, hypothesized that AmotL2 regulates YAP transcription through modulation to H3K27 methylation. Depletion of AmotL2 and subsequent H3K27me3 ChIP-qPCR revealed enrichment within the YAP promotor, indicating that repression of YAP may be due to increased repressive histone modifications in HUVECs (Figure 6D) and HUAECs (Figure S9). The subunit EZH2 forms part of the PRC (polycomb repressive complex) and catalyzes the methylation of H3K27.54–57 We performed codepletion of EZH2 and AmotL2, the results of which showed a partial rescue in YAP mRNA (Figure 6E), compared with lenti-shScr and scrambled siRNA conditions. Importantly, the depletion of EZH2 was also able to rescue the proliferative capacity of shAmotL2 HUVECs, an effect that was abrogated by verteporfin treatment, suggesting that the rescue in proliferation was indeed due to increased YAP activity (Figure 6F, quantification in Figure 6G). Collectively, these results indicate that AmotL2 regulates EZH2-mediated H3K27 modifications to regulate YAP transcription.
Chromatin Accessibility Is Regulated by AmotL2
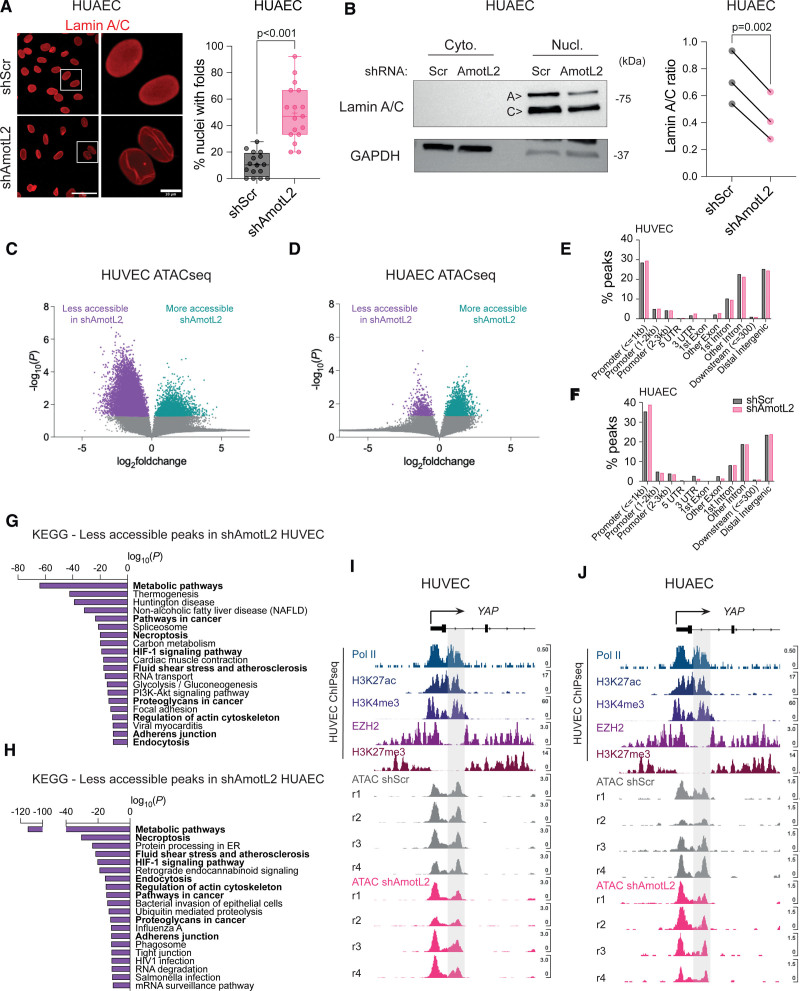
The notion of nuclear shape as a determinant of chromatin landscape and transcriptional activity is becoming increasingly evident.9 AmotL2 links cell-cell junctions to the nuclear envelope via the actin cytoskeleton where deletion of AmotL2 leads to misshapen nuclei of aortic ECs both in vitro and in vivo.5 In agreement with these data, we observed increased presence of nuclear wrinkles in shAmotL2 HUAECs stained for the nuclear membrane protein lamin A/C (Figure 7A). We, therefore, investigated whether AmotL2 regulates lamins to maintain nuclear morphology using subcellular fractionation, which indicated reduced lamin A expression (Figure 7B). We tested whether expression of LMNA and an upstream regulatory metallopeptidase ZMPSTE24, which is responsible for cleavage of farnesylated prelamin A to produce mature lamin A, were perturbed by AmotL2 depletion. Results showed a reduction in LMNA but no change in ZMPSTE24 (Figure S10A), suggesting a direct effect on LMNA transcription rather than altered processing of the protein. Additional results also ruled out changes in LMNB1 expression (Figure S10B).
Figure 7.
Chromatin accessibility is regulated by AmotL2 (angiomotin-like 2). A, Immunofluorescent images showing lamin A/C in human umbilical arterial endothelial cells (HUAECs) with knockdown of AmotL2 or shScr (shRNA scrambled control) lentiviral vectors. Images are representative of n=3 independent experiments. Quantification indicates the number of nuclei with folds or wrinkles, calculated as a percentage of the total number of nuclei within a field of view. Each data point represents 1 field of view (5–7 taken per condition) from n=3 independent experiments (mean±SD, Mann-Whitney U test). B, Representative Western blot of nuclear:cytoplasmic fractionation and detection of lamin A/C in HUAECs 96 hours post-treatment with shScr or shAmotL2 lentivirus. GAPDH were used as positive control for the cytoplasmic fraction. Quantification of lamin A/C ratio between matched shScr and shAmotL2 HUAEC samples. n=3 independent experiments, paired t test. Volcano plot indicating fold change in accessibility of HUVEC (C) or HUAEC (D) peaks of shScr control compared with shAmotL2. Peaks in purple show those that are less accessible in shAmotL2, and more accessible in shScr, while peaks shown in green are more accessible in shAmotL2 and less accessible in shScr. Gray points in the volcano plots represent peaks that are not significantly (P>0.05) changed between each group. Results of peak accessibility regardless of significance can be found in Data S1 (GEO accession: GSE253761). E and F, Distribution of assay for transposase-accessible chromatin sequencing (ATAC-seq) peaks in different regions of the genome of both HUVECs and HUAECs. G and H, KEGG pathway analysis of peaks that are less accessible in HUVEC (G) and HUAEC (H) where genes that are involved in fluid shear stress and atherosclerosis and actin regulation are less accessible in both cell lines and so are highlighted in bold. I and J, IGV (Integrative Genomics Viewer) browser view of the YAP promotor showing publicly available chromatin immunoprecipitation sequencing (ChIP-seq) data of Pol II (RNA polymerase II), H3K27ac, H3K4me3, EZH2 (enhancer of zeste 2 polycomb repressive complex 2 subunit), and H3K27me3 (trimethylation of histone H3 at lysine 27), alongside ATAC-seq data of HUVECs (I) and HUAECs (J) treated with shScr or shAmotL2. r indicates independent biological replicates, of which there are n=4 per condition.
We then asked whether regulation of nuclear morphology by AmotL2 impacts on chromatin accessibility. To do this, we performed genome-wide transposase-accessible chromatin sequencing (ATAC-seq) on shScr and shAmotL2 HUVECs and HUAECs. Figure 7C and 7D indicates volcano plots of peak accessibility in HUVECs and HUAECs, highlighting the widespread changes to chromatin accessibility when knocking down AmotL2. As expected, the distribution of peaks detected indicated enrichment in promotors and distal intergenic regions (Figure 7E and 7F). Despite overall differences in the pattern of AmotL2-regulated chromatin conformation between HUVECs and HUAECs, the KEGG pathway analysis of transcriptional start sites in which peak accessibility was reduced showed several overlapping pathways (Figure 7G and 7H, overlap between groups in bold). Notably, pathways in FSS and atherosclerosis, adherens junctions, and regulation of actin appeared in the 20 most significant less-accessible pathways, in line with the proposed role of AmotL2 as a junctional sensor of FSS. To investigate whether transcriptional regulation of YAP was due to changes in chromatin accessibility of the YAP promotor, we examined shScr and shAmotL2 ATAC-seq, overlayed with published ChIP-seq data in HUVEC of Pol II, H3K27ac, H3K4me3, EZH2, and H3K27me3. ATAC-seq data in both HUVECs and HUAECs showed a bimodal peak of accessibility, in which the downstream peak (highlighted in gray, Figure 7I and 7J) aligned with Pol II binding, active histones (H3K4me3 and H3K27ac), and reduced EZH2 and H3K27me3 repressive histone enrichment. While little difference was noted in the first peak of this bimodal chromatin accessibility, a noticeable reduction in accessibility was observed in the second peak position in shAmotL2 compared with shScr samples (Figure 7I and 7J). Analysis of ranked less-accessible peaks indicated YAP as a transcriptional start site with reduced accessibility in HUVECs (log2[fold change], −1.1596; P=0.0174; Data S1 and S2 [GEO accession: GSE253761]). Additionally, differential KEGG pathways associated with changes in peak accessibility in shScr versus shAmotL2 showed the Hippo signaling pathway in HUVECs (Figure S10C). To investigate whether the changes in chromatin of this specific region of interest reduced Pol II binding, we performed ChIP-qPCR (Figure S10D), which showed that knockdown of AmotL2 indeed reduced Pol II binding to this specific region of the YAP promotor (Figure S10E), suggesting that AmotL2-dependent epigenetic regulation of the YAP promotor may be due to genome-wide changes to chromatin accessibility. Additionally, we found that chromatin around the promotor of LMNA also exhibited condensed conformation upon AmotL2 depletion (Figure S10F), in line with decreased LMNA mRNA of shAmotL2 HUVECs, suggesting that AmotL2 regulates chromatin accessibility of multiple genes including LMNA and YAP.
Mechanical Regulation of Endothelial YAP Transcription
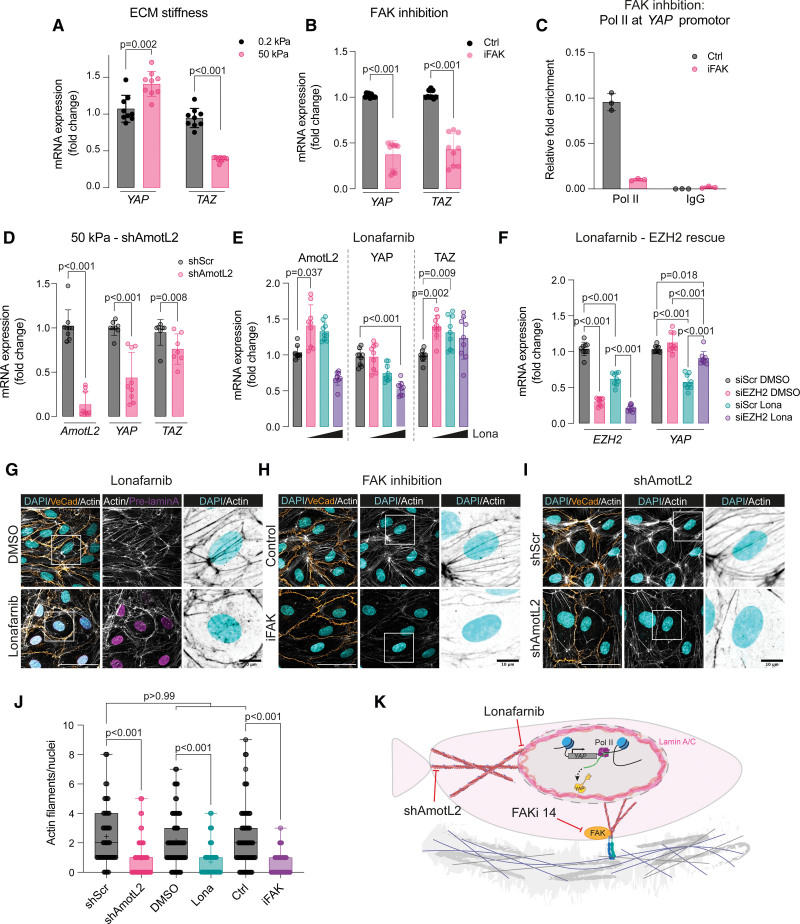
We hypothesized that mechanical forces other than those mediated by the AmotL2 junctional complex may regulate YAP transcription in ECs. We performed shear flow, stretching, and substrate stiffness experiments to delineate the key mechanical determinants that influence YAP transcription. Analysis of the flow-responsive transcription factors KLF2 and KFL4 of HUVECs exposed to laminar flow conditions showed an increase in expression, with reduced expression of KLF4 under conditions of disturbed flow as reported previously58–60 (Figure S11A). However, no change in YAP or TAZ expression was noted under conditions of shear flow at 48 hours (Figure S11A). To investigate the kinetics of the transcriptional response to flow in more detail, we performed flow experiments over a range of time points (with the caveat that consistent confluency cannot be maintained across the time points). As expected, KLF2 showed a maintained increase in expression upon flow stimulation (Figure S11B), whereas AmotL2 exhibited decreased expression over time (Figure S11C), in keeping with the known inactivation of YAP activity reported by Nakajima et al,61 under constant flow conditions. YAP expression exhibited a small but significant increase in expression (Figure S11D), and although this increase could not be replicated in lentiviral-infected cells exposed to the same conditions, AmotL2-dependent YAP transcription was still evident under conditions of shear flow (Figure S11E). To test other forms of mechanical stimulation, we exposed confluent HUVEC monolayers to uniaxial stretching, which did not alter YAP expression compared with static controls (Figure S11F). Here, the reliance of YAP transcription on AmotL2 expression was still apparent under conditions of cellular stretching, as the knockdown of AmotL2 led to a concomitant decrease in YAP expression, irrespective of uniaxial stretching (Figure S11F). To modulate substrate stiffness, we plated cells to hydrogels of 0.2 and 50 kPa stiffness and performed qPCR analysis, which showed that YAP mRNA was upregulated on the stiffer 50-kPa hydrogels (Figure 8A). Interestingly, TAZ showed the inverse, with lower expression on 50-kPa matrices compared with 0.2 kPa. These findings are in line with previous work showing that stiffer matrices upregulate YAP transcription.62,63 Changes in stiffness are sensed by conformational changes to the integrin receptors, leading to downstream activation.64 We, therefore, used an inhibitor of FAK to repress downstream signaling of integrin activation on cells plated to plastic, which reduced YAP transcription, mirroring findings of Jang et al62 (Figure 8B). We performed ChIP analysis to assess whether the downregulation of YAP was due to changes in activity at the YAP promotor and found reduced Pol II binding in FAKi-treated HUVECs (Figure 8C; Figure S11G). We tested whether depletion of AmotL2 in HUVEC cells plated to 50-kPa hydrogels also exhibited downregulated YAP mRNA, which mirrored findings shown on plastic (Figure 8D). To explore whether the FAK activity was regulated by AmotL2, we performed Western blotting of shAmotL2 HUAECs, which showed no difference in pFAK, suggesting that FAK activity and AmotL2 regulate stiffness-induced YAP transcription through distinct pathways (Figure S11H).
Figure 8.
Matrix stiffness and actin-nuclear connection regulate YAP (Yes-associated protein) transcription. A, RT-qPCR of YAP and TAZ (transcriptional coactivator with PDZ-binding motif), relative to the housekeeping gene GAPDH, of HUVECs plated to 0.2- or 50-kPa hydrogels for 18 hours (n=3 independent experiments, mean±SD, multiple Mann-Whitney U tests). B, SYBR green RT-qPCR of YAP in HUVECs treated with 10 µmol/L focal adhesion kinase inhibition (FAKi) for 24 hours before lysing and analysis (n=3 independent experiments, mean±SD, Mann-Whitney U test). C, ChIP showing Pol II (RNA polymerase II) binding to the YAP promotor of control or FAKi-treated HUVECs. ChIP-qPCR was performed using SYBR green reagents, and quantification was normalized to an IgG control antibody. Plot shown is a representative experiment from n=3 independent experiments (Figure S11C shows 2 further independent experiments). Each data point represents a technical repeat within 1 independent experiment (performed in triplicate; mean±SD). D, RT-qPCR of AMOTL2, YAP, and TAZ, relative to the housekeeping gene GAPDH, of AmotL2 knockdown HUVECs plated to 50-kPa hydrogels for 18 hours (n=3 independent experiments, mean±SD, Mann-Whitney U test). E, RT-qPCR of AMOTL2, YAP, and TAZ of HUVECs treated with lonafarnib at 1-, 5-, and 10-µmol/L concentrations compared with DMSO-treated control (n=3 independent experiments, mean±SD, Kruskal-Wallis with Dunn multiple comparisons). F, Depletion of EZH2 (enhancer of zeste 2 polycomb repressive complex 2 subunit) rescues lonafarnib-depleted YAP mRNA expression. RT-qPCR of indicated targets in HUVECs where knockdown of EZH2 using siRNA (72 h) preceded 48 hours of 10 µmol/L lonafarnib treatment (n=3 independent experiments, mean±SD, 2-way ANOVA with Tukey multiple comparisons). G, Immunofluorescent images showing VE (vascular endothelial)-cadherin (orange), actin (gray and black in inverted zoomed panels), prelamin A (magenta), and DAPI (4′,6-diamidino-2-phenylindole; blue) in HUVECs treated with 10 µmol/L lonafarnib for 48 hours. Images are representative of n=3 independent experiments. H, Immunofluorescent images showing VE-cadherin (orange), actin (gray and black in inverted zoomed panels), and DAPI (blue) in HUVECs treated with 10 µmol/L FAKi 14 for 24 hours. Images are representative of n=3 independent experiments. I, Immunofluorescent images as in H of shScr (shRNA scrambled control)- or shAmotL2-treated HUVECs, 72 hours post-infection. Images are representative of n=3 independent experiments. Scale bar, 50 and 10 µm for selected regions of interest. J, Quantification of the total number of actin fibers per nuclei from experiments displayed in G through I. Each data point represents a single nuclei, measured from 3 regions per condition from n=3 independent experiments (mean±SD, Kruskal-Wallis with Dunn multiple comparison). K, Schematic model of proposed action of lonafarnib, FAKi, and shAmotL2 on the actin-nuclear connection and the impact on YAP transcription. Ctrl indicates control; ECM, extracellular matrix; and FAK, focal adhesion kinase.
Cytoskeletal-Nuclear Lamina Interaction Regulates YAP Transcription
Focal adhesions link integrins to the nuclear membrane via the actin cytoskeleton.11 Similarly, the nuclear lamina component lamin A (which associates with the cytoskeleton via the LINC complex) is linked to junctional AmotL2 via the actin cytoskeleton, as has been previously shown by our laboratory.5 We, therefore, suspected that components of the nuclear membrane, which tether actin to the nucleus, may regulate YAP transcription. Lamin A expression was downregulated upon AmotL2 knockdown, and to mimic these changes, we treated HUVECs with the farnesyltransferase inhibitor lonafarnib, which inhibits the processing of prelamin A. Titration of lonafarnib showed a dose-dependent reduction in YAP mRNA (Figure 8E), and Western blotting confirmed that lonafarnib-treated cells reduced mature lamin A, albeit with a modest reduction in YAP protein levels (Figure S11I). To investigate whether AmotL2 depletion and inhibition of lamin A maturation regulate YAP via a common epigenetic mechanism, we performed EZH2 depletion in lonafarnib-treated HUVECs. Results indicated a partial rescue of YAP mRNA in EZH2 knockdown and lonafarnib-treated HUVECs (Figure 8F), similar to that of EZH2-AmotL2 codepletion shown in Figure 6E, suggesting that both inhibition of lamin A maturation and AmotL2 depletion suppress YAP transcription through a similar epigenetic mechanism.
Results of immunofluorescent staining showed that treatment of HUVECs with lonafarnib led to the accumulation of prelamin A compared with vehicle-treated controls (Figure 8G). We also observed a change in the morphology of the actin cytoskeleton, with fewer actin filaments across the nucleus. Similarly, FAKi-treated HUVECs (Figure 8H) and shAmotL2 (Figure 8I) also displayed reduced actin fibers across the nucleus (quantified in Figure 8J), suggesting that inhibition of FAK activity, lamin A processing, and depletion of AmotL2 impairs the mechanosensory connection between the cytoskeleton and to the nucleus, leading to changes in YAP transcription (Figure 8K). Collectively, these results suggest that multiple pathways converge on the nucleus via the cytoskeleton but that overall, they are all required for transcriptional regulation of YAP and appropriate nuclear mechanotransduction.
DISCUSSION
Collectively, our results demonstrate that junctional mechanosensing via AmotL2-cytoskeleton-nuclear connection is integral to YAP transcription and promotor activity, and in turn, AmotL2 expression itself, suggesting a mechano-enforced feedback loop of control of vascular homeostasis. Loss of either side of this feedback loop leads to destabilization of aortic homeostasis; in AmotL2 deletion, this manifests in abdominal aneurysm formation,5 and in YAP/TAZ deletion, impaired mechanotransduction, proliferation, and loss of nuclear shape, which may have detrimental effects on chromatin.
Emerging evidence indicates the important contribution of epigenetic modifications in regulating YAP transcription. A role for DNA methylation of the YAP promotor in gastric cancer cells cultured on soft versus stiff matrices has already been demonstrated,62 and while we could not detect changes to DNA methylation, alterations to histone modifications in the YAP promotor that were AmotL2 dependent were observed. Kaukonen et al described a role for JMJD1A,63 a histone demethylase, in a model of stiffness-dependent regulation of YAP/TAZ transcription through modulation to H3K27ac. More recently, changes in viscosity were associated with transcriptional changes to YAP/TAZ signaling65; however, the precise mechanism of how this transcriptional regulation is elicited was not investigated. Much focus has been given to the localization and activity of YAP; however, these lines of evidence suggest that mechanical stimuli play a direct role in regulating YAP transcription. Our own results show that substrate stiffness plays a predominant role in regulating YAP transcription over other mechanical forces such as stretching and FSS, which are more likely to influence YAP activity.13,61 Indeed, as a target gene of YAP, AmotL2 was found to increase in ECs exposed to disturbed flow, mechanical stretch, and increasing stiffness. Regardless of substrate stiffness acting as the main mechanical force driving YAP transcription, YAP transcription remains AmotL2 dependent, through regulation of nuclear shape, chromatin landscape, and H3K27me3 of the YAP promotor with functional consequence for EC proliferation.
Impaired mechanosensensing of the aortic endothelium results in detrimental EC function, and in the case of AmotL2, leads to the formation of AAAs.5 Work from our laboratory previously outlined that the descending and the ascending aortas exhibit differential ECM (extracellular matrix) composition in both transcriptional and proteomic analysis and that the localized formation of AAA in endothelial AmotL2 deletion is due to impaired flow sensing and increased immune infiltration, leading to degradation of the ECM and spontaneous AAA formation in aged male mice. Changes in ECM composition have previously been shown to modulate AAA incidence.66 Our results suggest that substrate stiffness and FAK activity regulate YAP transcription. Increased degradation of ECM in AmotL2 knock-out may contribute to the loss of YAP expression in the descending aorta as cross talk between junctional mechanosensory complexes and focal adhesion activity under shear stress are well documented.3,4,67–69 While we did not detect any changes in pFAK in AmotL2-depleted cells, we cannot rule out changes to activity of other FA proteins, which may be contributing to changes in YAP transcription. Mutations in the FBN1 gene, which perturbs elastic fiber generation in Marfan syndrome, are at a greater risk of aneurysm formation,70,71 and recent work indicated that reexpression of active YAP in vascular smooth muscle cells of Fbn1C1039G/WT mutant mice was able to prevent degeneration of the aortic tissue.72 Conversely, YAP depletion has been shown to significantly attenuate expression of endothelial proinflammatory factors,73–75 which may explain why we did not observe AAA in the YAP/TAZ-inducible endothelial-specific knockout in our study. Further work is required to determine whether YAP/TAZ-inducible endothelial-specific knockout is more susceptible to AAA formation using either aged mice or models of aneurysm such as angiotensin II infusion.
Regions of the aorta that experience disturbed flow such as bifurcations and the inner aortic arch are prone to inflammation and the development of atherosclerotic lesions. Our results indicate that DF in vitro triggers YAP activity and the upregulation of AmotL2 expression. Furthermore, we showed that proliferation of ECs in the inner aortic arch are reliant of both AmotL2 and YAP/TAZ expression in vivo. Proliferation can be both protective and pathological, the dichotomy of which has been linked to the heterogenous nature of the aortic endothelium, suggesting a balance between populations of protective and disease-promoting ECs. The importance of proliferation in this region to maintain vascular homeostasis is highlighted in JAG1 (jagged canonical notch ligand 1) EC-knock-out mice where increased proliferation protects from the onset of atherosclerosis.51 Whether AmotL2 is protective against atherosclerosis remains to be tested, although given the activation of an inflammatory profile in Amotl2-depleted ECs, we speculate that the latter may be the case.
Junctional remodeling involving VE-cadherin turnover is regulated by YAP/TAZ localization during angiogenesis.13 The importance of junctional force sensing in regulating proliferation and cell density through the regulation of YAP localization is also evident in other cellular contexts such as the skin epidermis76,77 and esophageal epithelia.78 Here, junctional molecules such as α-catenin and plexin-B1 and B2 negatively regulate YAP activity and proliferation in both development and adult homeostasis. Previous work has also shown that AmotL2 is a negative regulator of YAP activity, through several mechanisms including direct binding and modulation of upstream kinases of the Hippo pathway.37–41 Our results indicate that AmotL2 depletion led to increased YAP phosphorylation, albeit with drastically reduced total YAP protein levels. While we focused on elucidating the transcriptional regulation of YAP, it remains possible that AmotL2 may contribute to regulating YAP turnover through phosphorylation and subsequent degradation. AmotL2 depletion led to decreased RhoA GTPase activity,5 a well-characterized regulator of the actin cytoskeleton contractility required for mechanically induced YAP/TAZ nuclear translocation.16 AmotL2 regulation of the RhoA activity, therefore, may explain the decreased YAP phosphorylation and binding to target gene promotors observed in our study.
YAP activity has been shown to be inactivated by steady sustained laminar flow conditions,61 and in agreement with this, AmotL2 (as a target gene of YAP) exhibited decreased expression under sustained laminar flow and conversely increased AmotL2 expression under disturbed flow, concomitant with increased YAP activity. Additionally, our data showed that sustained laminar flow leads to a modest increase in YAP transcription. Whether decreased activity of YAP under sustained laminar flow leads to increased phosphorylation and degradation and triggers modulation to YAP transcription remains unknown. Additionally, uncoupling confluency and cell density from flow responses over prolonged periods in vitro adds difficulty to interpretation as changes in cell density are well known to regulate YAP activity.49 Further work is required to delineate the relationship between YAP protein degradation and turnover and transcriptional regulation in maintaining homeostatic levels of YAP under flow conditions.
Intriguingly, our findings show that AmotL2 regulates LMNA expression, nuclear architecture, and chromatin accessibility. Previous work from our laboratory shows that AmotL2 interacts with components of the nuclear membrane, namely lamin A, in ECs exposed to flow, suggesting an actin cytoskeleton-derived link between cell-cell junctions and the nucleus.5 Additionally, recent work has shown that disruption of the nuclear envelope component SUN1 leads to dysregulation of EC junctions via the microtubule network, highlighting the existence of such a junctional-nuclear connection.8 A direct mechanical link between the nucleus and integrins is well established, allowing the transduction of extracellular forces that subsequently shape chromatin.9 Indeed, inhibition of FAK activity, which has been shown to reduce YAP transcripts,62 phenocopied AmotL2 depletion with reduced actin filaments associated with the nucleus. Additionally, inhibition of lamin A farnesylation leading to reduced incorporation of mature lamin A into the nuclear lamina phenocopied AmotL2 and FAK inhibition, with reduced nuclear actin fibers and EZH2-mediated repression of YAP mRNA, arguing for an intact nuclear-cytoskeletal connection as a pivotal requirement for YAP transcription. YAP itself was also recently shown to maintain nuclear integrity through the actin cap by transcriptional regulation of ACTR2 and the structural nuclear membrane protein LMNB1 with wide implications for aortic mechanosensing and aging.72 In agreement with this phenotype, our findings indicated perturbed nuclei and a reduction in actin fibers in the aortic endothelium of Yap/Taz iΔEC, phenocopying that of AmotL2 iΔEC and may offer an explanation as to the proinflammatory phenotype that leads to aneurysm formation observed in AmotL2 endothelial deletion. While we speculate that changes to nuclear morphology and chromatin are AmotL2 specific, we cannot rule out that the downstream effects of chromatin regulation are partly a consequence of dysregulation of YAP’s ability to regulate chromatin accessibility through SWI/SNF (chromatin remodelling complex) and BRD4 (bromodomain-containing protein 4) interactions.79,80 A further limitation of our study is the assessment of the junctional regulation of YAP transcription, and future work will focus on known interactors of AmotL2, to delineate the specificity of junctional complexes in relaying mechanical force to the nuclear lamina and the impact this has on YAP transcription.
Overall, our current model indicates that mechanical stimulation drives YAP-dependent AmotL2 expression, which is critical for sensing FSS via a junctional-cytoskeleton-nuclear pathway. This in turn is critical for nuclear morphology, chromatin accessibility, and histone modifications within the YAP promotor (Figure 9). To our knowledge, this represents the first example of a junctional molecule that influences epigenetic regulation of YAP and opens the exciting prospect that this signaling axis may play a role in maintaining tissue homeostasis in other cellular contexts. Furthermore, we show that mechanosensing of substrate stiffness by FAK activity, along with proper nuclear lamina composition, plays a direct role in modulating YAP transcription via a cytoskeletal-nuclear–derived connection, highlighting a previously underappreciated layer of regulation to the mechanosensor YAP.
Figure 9.
Model of positive feedback loop that enforces fluid shear stress (FSS)–induced transcription via YAP (Yes-associated protein)-AmotL2 (angiomotin-like 2) coregulation. In AmotL2 wild-type (WT) endothelial cells, a flow sensing junctional-cytoskeletal-nuclear connection maintains nuclear morphology and lamin A expression levels, conferring chromatin accessibility and Pol II (RNA polymerase II) binding to drive YAP transcription and concomitant proliferation. Conversely, deletion of endothelial AmotL2 leads to impaired junctional connections, reduced nuclear lamin A levels, perturbed nuclear shape, changes to global chromatin conformation, and increased H3K27me3 (trimethylation of histone H3 at lysine 27) within the YAP promotor. This leads to decreased Pol II binding to the YAP promotor, reduced YAP transcription, and consequently inhibits proliferative capacity. YAP also transcriptionally regulates AmotL2, overall suggesting a mechano-enforced positive feedback loop involving YAP and AmotL2 to maintain vascular homeostasis.
ARTICLE INFORMATION
Acknowledgments
This study was conceived and designed by A.J. Mannion and L. Holmgren. A.J. Mannion, H. Zhao, and Y. Zhang performed the in vitro experiments. A.J. Mannion, Y. Zhang, and Y. von Wright performed the in vivo experiments. A.J. Mannion and O. Bergman performed the in silico and bioinformatic analyses. J. Roy was involved in the biobank and clinical surgery required for human abdominal aortic aneurysm transcriptomics studies. P. Saharinen provided samples from Yap (Yes-associated protein)/Taz (transcriptional coactivator with PDZ-binding motif) transgenics. A.J. Mannion wrote the manuscript, and all authors contributed to the drafting and editing of the manuscript.
Sources of Funding
This study was supported by grants from the Swedish Heart and Lung Foundation (L. Holmgren and J. Roy), Novo Nordisk Foundation (NNF15CC0018346; L. Holmgren), Swedish Cancer Society (L. Holmgren), Swedish Childhood Cancer Foundation (L. Holmgren), Cancer Society of Stockholm (L. Holmgren), Swedish Research Council (L. Holmgren), Knut and Alice Wallenberg Foundation (L. Holmgren), European Research Council under the EU Horizon 2020 research and innovation programme (grant agreement 773076; P. Saharinen), Sigrid Jusélius Foundation (P. Saharinen), Cancer Foundation Finland (P. Saharinen), Academy of Finland (P. Saharinen).
Disclosures
None.
Supplemental Material
Figures S1–S11
Tables S1 and S2
Data S1 and S2 (GEO accession: GSE253761)
Major Resources Table
Supplementary Material
Nonstandard Abbreviations and Acronyms
- AAA
- abdominal aortic aneurysm
- AmotL2
- angiomotin-like 2
- ATAC-seq
- assay for transposase-accessible chromatin sequencing
- ChIP-seq
- chromatin immunoprecipitation sequencing
- EC
- endothelial cell
- ECM
- extracellular matrix
- EdU
- 5-ethynyl-2′-deoxyuridine
- FAK
- focal adhesion kinase
- FSS
- fluid shear stress
- HUAEC
- human umbilical arterial endothelial cell
- HUVEC
- human umbilical vein endothelial cell
- HRP
- horseradish peroxidase
- IVC
- inferior vena cava
- LINC
- linker of nucleoskeleton and cytoskeleton
- PRC
- polycomb repressive complex
- PLA
- proximity ligation assay
- Pol II
- RNA polymerase II
- WT
- wild type
- YAP
- Yes-associated protein
For Sources of Funding and Disclosures, see page 687.
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/ATVBAHA.123.320300.
REFERENCES
- 1.Davies PF. Flow-mediated endothelial mechanotransduction. Physiol Rev. 1995;75:519–560. doi: 10.1152/physrev.1995.75.3.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Souilhol C, Serbanovic-Canic J, Fragiadaki M, Chico TJ, Ridger V, Roddie H, Evans PC. Endothelial responses to shear stress in atherosclerosis: a novel role for developmental genes. Nat Rev Cardiol. 2020;17:52–63. doi: 10.1038/s41569-019-0239-5 [DOI] [PubMed] [Google Scholar]
- 3.Tzima E, Irani-Tehrani M, Kiosses WB, Dejana E, Schultz DA, Engelhardt B, Cao G, DeLisser H, Schwartz MA. A mechanosensory complex that mediates the endothelial cell response to fluid shear stress. Nature. 2005;437:426–431. doi: 10.1038/nature03952 [DOI] [PubMed] [Google Scholar]
- 4.Mehta V, Pang KL, Rozbesky D, Nather K, Keen A, Lachowski D, Kong Y, Karia D, Ameismeier M, Huang J, et al. The guidance receptor plexin D1 is a mechanosensor in endothelial cells. Nature. 2020;578:290–295. doi: 10.1038/s41586-020-1979-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, Zhang Y, Hutterer E, Hultin S, Bergman O, Kolbeinsdottir S, Jin H, Forteza MJ, Ketelhuth DFJ, Roy J, et al. The VE-cadherin/AmotL2 mechanosensory pathway suppresses aortic inflammation and the formation of abdominal aortic aneurysms. Nat Cardiovasc Res. 2023;2:629–644. doi: 10.1038/s44161-023-00298-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2005;172:41–53. doi: 10.1083/jcb.200509124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martins RP, Finan JD, Farshid G, Lee DA. Mechanical regulation of nuclear structure and function. Annu Rev Biomed Eng. 2012;14:431–455. doi: 10.1146/annurev-bioeng-071910-124638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buglak DB, Bougaran P, Kulikauskas MR, Liu Z, Monaghan-Benson E, Gold AL, Marvin AP, Burciu A, Tanke NT, Oatley M, et al. Nuclear SUN1 stabilizes endothelial cell junctions via microtubules to regulate blood vessel formation. Koh GY, Akhmanova A, eds. Elife. 2023;12:e83652. doi: 10.7554/eLife.83652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dupont S, Wickström SA. Mechanical regulation of chromatin and transcription. Nat Rev Genet. 2022;23:624–643. doi: 10.1038/s41576-022-00493-6 [DOI] [PubMed] [Google Scholar]
- 10.Kalukula Y, Stephens AD, Lammerding J, Gabriele S. Mechanics and functional consequences of nuclear deformations. Nat Rev Mol Cell Biol. 2022;23:583–602. doi: 10.1038/s41580-022-00480-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carley E, King MC, Guo S. Integrating mechanical signals into cellular identity. Trends Cell Biol. 2022;32:669–680. doi: 10.1016/j.tcb.2022.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirby TJ, Lammerding J. Emerging views of the nucleus as a cellular mechanosensor. Nat Cell Biol. 2018;20:373–381. doi: 10.1038/s41556-018-0038-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neto F, Klaus-Bergmann A, Ong YT, Alt S, Vion A-C, Szymborska A, Carvalho JR, Hollfinger I, Bartels-Klein E, Franco CA, et al. YAP and TAZ regulate adherens junction dynamics and endothelial cell distribution during vascular development. Elife. 2018;7:e31037. doi: 10.7554/eLife.31037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sivaraj KK, Dharmalingam B, Mohanakrishnan V, Jeong H-W, Kato K, Schröder S, Adams S, Koh GY, Adams RH. YAP1 and TAZ negatively control bone angiogenesis by limiting hypoxia-inducible factor signaling in endothelial cells. Zaidi M, Rosen CJ, Carmeliet G, eds. Elife. 2020;9:e50770. doi: 10.7554/eLife.50770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J, Kim YH, Kim J, Park DY, Bae H, Lee D-H, Kim KH, Hong SP, Jang SP, Kubota Y, et al. YAP/TAZ regulates sprouting angiogenesis and vascular barrier maturation. J Clin Invest. 2017;127:3441–3461. doi: 10.1172/JCI93825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137 [DOI] [PubMed] [Google Scholar]
- 17.Lindquist Liljeqvist M, Hultgren R, Bergman O, Villard C, Kronqvist M, Eriksson P, Roy J. Tunica-specific transcriptome of abdominal aortic aneurysm and the effect of intraluminal thrombus, smoking, and diameter growth rate. Arterioscler Thromb Vasc Biol. 2020;40:2700–2713. doi: 10.1161/ATVBAHA.120.314264 [DOI] [PubMed] [Google Scholar]
- 18.Okabe K, Kobayashi S, Yamada T, Kurihara T, Tai-Nagara I, Miyamoto T, Mukouyama YS, Sato TN, Suda T, Ema M, et al. Neurons limit angiogenesis by titrating VEGF in retina. Cell. 2014;159:584–596. doi: 10.1016/j.cell.2014.09.025 [DOI] [PubMed] [Google Scholar]
- 19.Sörensen I, Adams RH, Gossler A. DLL1-mediated Notch activation regulates endothelial identity in mouse fetal arteries. Blood. 2009;113:5680–5688. doi: 10.1182/blood-2008-08-174508 [DOI] [PubMed] [Google Scholar]
- 20.Odell AF, Mannion AJ. In vitro co-culture of fibroblast and endothelial cells to assess angiogenesis. Methods Mol Biol. 2022;2441:277–286. doi: 10.1007/978-1-0716-2059-5_21 [DOI] [PubMed] [Google Scholar]
- 21.Hultin S, Zheng Y, Mojallal M, Vertuani S, Gentili C, Balland M, Milloud R, Belting H-G, Affolter M, Helker CSM, et al. AmotL2 links VE-cadherin to contractile actin fibres necessary for aortic lumen expansion. Nat Commun. 2014;5:3743. doi: 10.1038/ncomms4743 [DOI] [PubMed] [Google Scholar]
- 22.Rickman M, Ghim M, Pang K, von Huelsen Rocha AC, Drudi EM, Sureda-Vives M, Ayoub N, Tajadura-Ortega V, George SJ, Weinberg PD, et al. Disturbed flow increases endothelial inflammation and permeability via a frizzled-4-β-catenin-dependent pathway. J Cell Sci. 2023;136:jcs260449. doi: 10.1242/jcs.260449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zou Z, Ohta T, Miura F, Oki S. ChIP-Atlas 2021 update: a data-mining suite for exploring epigenomic landscapes by fully integrating ChIP-seq, ATAC-seq and bisulfite-seq data. Nucleic Acids Res. 2022;50:W175–W182. doi: 10.1093/nar/gkac199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müller F, Scherer M, Assenov Y, Lutsik P, Walter J, Lengauer T, Bock C. RnBeads 2.0: comprehensive analysis of DNA methylation data. Genome Biol. 2019;20:55. doi: 10.1186/s13059-019-1664-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, Satpathy AT, Rubin AJ, Montine KS, Wu B, et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods. 2017;14:959–962. doi: 10.1038/nmeth.4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bajic M, Maher KA, Deal RB. Identification of open chromatin regions in plant genomes using ATAC-seq. Methods Mol Biol. 2018;1675:183–201. doi: 10.1007/978-1-4939-7318-7_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dai X, Liu H, Shen S, Guo X, Yan H, Ji X, Li L, Huang J, Feng XH, Zhao B. YAP activates the Hippo pathway in a negative feedback loop. Cell Res. 2015;25:1175–1178. doi: 10.1038/cr.2015.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grijalva JL, Huizenga M, Mueller K, Rodriguez S, Brazzo J, Camargo F, Sadri-Vakili G, Vakili K. Dynamic alterations in Hippo signaling pathway and YAP activation during liver regeneration. Am J Physiol Gastrointest Liver Physiol. 2014;307:G196–G204. doi: 10.1152/ajpgi.00077.2014 [DOI] [PubMed] [Google Scholar]
- 30.Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Freire Valls A, Schermann G, Shen Y, Moya IM, Castro L, Urban S, Solecki GM, Winkler F, Riedemann L, et al. YAP/TAZ orchestrate VEGF signaling during developmental angiogenesis. Dev Cell. 2017;42:462–478.e7. doi: 10.1016/j.devcel.2017.08.002 [DOI] [PubMed] [Google Scholar]
- 32.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo Y, Hitz BC, Gabdank I, Hilton JA, Kagda MS, Lam B, Myers Z, Sud P, Jou J, Lin K, et al. New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res. 2020;48:D882–D889. doi: 10.1093/nar/gkz1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goulev Y, Fauny JD, Gonzalez-Marti B, Flagiello D, Silber J, Zider A. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr Biol. 2008;18:435–441. doi: 10.1016/j.cub.2008.02.034 [DOI] [PubMed] [Google Scholar]
- 36.Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell. 2008;14:388–398. doi: 10.1016/j.devcel.2008.01.007 [DOI] [PubMed] [Google Scholar]
- 37.Mana-Capelli S, Paramasivam M, Dutta S, McCollum D. Angiomotins link F-actin architecture to Hippo pathway signaling. Mol Biol Cell. 2014;25:1676–1685. doi: 10.1091/mbc.E13-11-0701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paramasivam M, Sarkeshik A, Yates JR, Fernandes MJG, McCollum D. Angiomotin family proteins are novel activators of the LATS2 kinase tumor suppressor. Mol Biol Cell. 2011;22:3725–3733. doi: 10.1091/mbc.E11-04-0300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan SW, Lim CJ, Guo F, Tan I, Leung T, Hong W. Actin-binding and cell proliferation activities of angiomotin family members are regulated by Hippo pathway-mediated phosphorylation. J Biol Chem. 2013;288:37296–37307. doi: 10.1074/jbc.M113.527598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao B, Li L, Lu Q, Wang LH, Liu CY, Lei Q, Guan KL. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 2011;25:51–63. doi: 10.1101/gad.2000111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Artinian N, Cloninger C, Holmes B, Benavides-Serrato A, Bashir T, Gera J. Phosphorylation of the Hippo pathway component AMOTL2 by the mTORC2 kinase promotes YAP signaling, resulting in enhanced glioblastoma growth and invasiveness. J Biol Chem. 2015;290:19387–19401. doi: 10.1074/jbc.M115.656587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pitulescu ME, Schmidt I, Benedito R, Adams RH. Inducible gene targeting in the neonatal vasculature and analysis of retinal angiogenesis in mice. Nat Protoc. 2010;5:1518–1534. doi: 10.1038/nprot.2010.113 [DOI] [PubMed] [Google Scholar]
- 43.He L, Vanlandewijck M, Mäe MA, Andrae J, Ando K, Del Gaudio F, Nahar K, Lebouvier T, Laviña B, Gouveia L, et al. Single-cell RNA sequencing of mouse brain and lung vascular and vessel-associated cell types. Sci Data. 2018;5:180160. doi: 10.1038/sdata.2018.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vanlandewijck M, He L, Mäe MA, Andrae J, Ando K, Del Gaudio F, Nahar K, Lebouvier T, Laviña B, Gouveia L, et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature. 2018;554:475–480. doi: 10.1038/nature25739 [DOI] [PubMed] [Google Scholar]
- 45.Meignin C, Alvarez-Garcia I, Davis I, Palacios IM. The Salvador-Warts-Hippo pathway is required for epithelial proliferation and axis specification in Drosophila. Curr Biol. 2007;17:1871–1878. doi: 10.1016/j.cub.2007.09.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007 [DOI] [PubMed] [Google Scholar]
- 47.Piccolo S, Dupont S, Cordenonsi M. The biology of YAP/TAZ: Hippo signaling and beyond. Physiol Rev. 2014;94:1287–1312. doi: 10.1152/physrev.00005.2014 [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, Li Z, Xu P, Huang L, Tong J, Huang H, Meng A. Angiomotin-like2 gene (amotl2) is required for migration and proliferation of endothelial cells during angiogenesis. J Biol Chem. 2011;286:41095–41104. doi: 10.1074/jbc.M111.296806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Engelbrecht E, Levesque MV, He L, Vanlandewijck M, Nitzsche A, Niazi H, Kuo A, Singh SA, Aikawa M, Holton K, et al. Sphingosine 1-phosphate-regulated transcriptomes in heterogenous arterial and lymphatic endothelium of the aorta. Bautch VL, Stainier DYR, Iruela-Arispe L, eds. Elife. 2020;9:e52690. doi: 10.7554/eLife.52690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Souilhol C, Tardajos Ayllon B, Li X, Diagbouga MR, Zhou Z, Canham L, Roddie H, Pirri D, Chambers EV, Dunning MJ, et al. JAG1-NOTCH4 mechanosensing drives atherosclerosis. Sci Adv. 2022;8:eabo7958. doi: 10.1126/sciadv.abo7958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sudol M, Bork P, Einbond A, Kastury K, Druck T, Negrini M, Huebner K, Lehman D. Characterization of the mammalian YAP (Yes-associated protein) gene and its role in defining a novel protein module, the WW domain *. J Biol Chem. 1995;270:14733–14741. doi: 10.1074/jbc.270.24.14733 [DOI] [PubMed] [Google Scholar]
- 53.Komuro A, Nagai M, Navin NE, Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J Biol Chem. 2003;278:33334–33341. doi: 10.1074/jbc.M305597200 [DOI] [PubMed] [Google Scholar]
- 54.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997 [DOI] [PubMed] [Google Scholar]
- 55.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3 [DOI] [PubMed] [Google Scholar]
- 56.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5 [DOI] [PubMed] [Google Scholar]
- 58.Hamik A, Lin Z, Kumar A, Balcells M, Sinha S, Katz J, Feinberg MW, Gerzsten RE, Edelman ER, Jain MK. Kruppel-like factor 4 regulates endothelial inflammation *. J Biol Chem. 2007;282:13769–13779. doi: 10.1074/jbc.M700078200 [DOI] [PubMed] [Google Scholar]
- 59.Dekker RJ, van Soest S, Fontijn RD, Salamanca S, de Groot PG, VanBavel E, Pannekoek H, Horrevoets AJG. Prolonged fluid shear stress induces a distinct set of endothelial cell genes, most specifically lung Krüppel-like factor (KLF2). Blood. 2002;100:1689–1698. doi: 10.1182/blood-2002-01-0046 [DOI] [PubMed] [Google Scholar]
- 60.Andueza A, Kumar S, Kim J, Kang DW, Mumme HL, Perez JI, Villa-Roel N, Jo H. Endothelial reprogramming by disturbed flow revealed by single-cell RNA and chromatin accessibility study. Cell Rep. 2020;33:108491. doi: 10.1016/j.celrep.2020.108491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nakajima H, Yamamoto K, Agarwala S, Terai K, Fukui H, Fukuhara S, Ando K, Miyazaki T, Yokota Y, Schmelzer E, et al. Flow-dependent endothelial YAP regulation contributes to vessel maintenance. Dev Cell. 2017;40:523–536.e6. doi: 10.1016/j.devcel.2017.02.019 [DOI] [PubMed] [Google Scholar]
- 62.Jang M, An J, Oh SW, Lim JY, Kim J, Choi JK, Cheong JH, Kim P. Matrix stiffness epigenetically regulates the oncogenic activation of the Yes-associated protein in gastric cancer. Nat Biomed Eng. 2021;5:114–123. doi: 10.1038/s41551-020-00657-x [DOI] [PubMed] [Google Scholar]
- 63.Kaukonen R, Mai A, Georgiadou M, Saari M, De Franceschi N, Betz T, Sihto H, Ventelä S, Elo L, Jokitalo E, et al. Normal stroma suppresses cancer cell proliferation via mechanosensitive regulation of JMJD1a-mediated transcription. Nat Commun. 2016;7:12237. doi: 10.1038/ncomms12237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kechagia JZ, Ivaska J, Roca-Cusachs P. Integrins as biomechanical sensors of the microenvironment. Nat Rev Mol Cell Biol. 2019;20:457–473. doi: 10.1038/s41580-019-0134-2 [DOI] [PubMed] [Google Scholar]
- 65.Bera K, Kiepas A, Godet I, Li Y, Mehta P, Ifemembi B, Paul CD, Sen A, Serra SA, Stoletov K, et al. Extracellular fluid viscosity enhances cell migration and cancer dissemination. Nature. 2022;611:365–373. doi: 10.1038/s41586-022-05394-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Steffensen LB, Stubbe J, Lindholt JS, Beck HC, Overgaard M, Bloksgaard M, Genovese F, Holm Nielsen S, Tha MLT, Bang-Moeller SK, et al. Basement membrane collagen IV deficiency promotes abdominal aortic aneurysm formation. Sci Rep. 2021;11:12903. doi: 10.1038/s41598-021-92303-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu Y, Sweet DT, Irani-Tehrani M, Maeda N, Tzima E. Shc coordinates signals from intercellular junctions and integrins to regulate flow-induced inflammation. J Cell Biol. 2008;182:185–196. doi: 10.1083/jcb.200709176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mehta V, Pang KL, Givens CS, Chen Z, Huang J, Sweet DT, Jo H, Reader JS, Tzima E. Mechanical forces regulate endothelial-to-mesenchymal transition and atherosclerosis via an Alk5-Shc mechanotransduction pathway. Sci Adv. 2023;7:eabg5060. doi: 10.1126/sciadv.abg5060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Collins C, Guilluy C, Welch C, O’Brien ET, Hahn K, Superfine R, Burridge K, Tzima E. Localized tensional forces on PECAM-1 elicit a global mechanotransduction response via the integrin-RhoA pathway. Curr Biol. 2012;22:2087–2094. doi: 10.1016/j.cub.2012.08.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pereira L, Andrikopoulos K, Tian J, Lee SY, Keene DR, Ono R, Reinhardt DP, Sakai LY, Biery NJ, Bunton T, et al. Targetting of the gene encoding fibrillin–1 recapitulates the vascular aspect of Marfan syndrome. Nat Genet. 1997;17:218–222. doi: 10.1038/ng1097-218 [DOI] [PubMed] [Google Scholar]
- 71.Dietz HC, Pyeritz RE. Mutations in the human gene for fibrillin-1 (FBN1) in the Marfan syndrome and related disorders. Hum Mol Genet. 1995;4 Spec No:1799–1809. doi: 10.1093/hmg/4.suppl_1.1799 [DOI] [PubMed] [Google Scholar]
- 72.Sladitschek-Martens HL, Guarnieri A, Brumana G, Zanconato F, Battilana G, Xiccato RL, Panciera T, Forcato M, Bicciato S, Guzzardo V, et al. YAP/TAZ activity in stromal cells prevents ageing by controlling cGAS–STING. Nature. 2022;607:790–798. doi: 10.1038/s41586-022-04924-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang KC, Yeh YT, Nguyen P, Limqueco E, Lopez J, Thorossian S, Guan KL, Li YJ, Chien S. Flow-dependent YAP/TAZ activities regulate endothelial phenotypes and atherosclerosis. Proc Natl Acad Sci USA. 2016;113:11525–11530. doi: 10.1073/pnas.1613121113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang L, Luo JY, Li B, Tian XY, Chen LJ, Huang Y, Liu J, Deng D, Lau CW, Wan S, et al. Integrin-YAP/TAZ-JNK cascade mediates atheroprotective effect of unidirectional shear flow. Nature. 2016;540:579–582. doi: 10.1038/nature20602 [DOI] [PubMed] [Google Scholar]
- 75.Li B, He J, Lv H, Liu Y, Lv X, Zhang C, Zhu Y, Ai D. c-Abl regulates YAPY357 phosphorylation to activate endothelial atherogenic responses to disturbed flow. J Clin Invest. 2019;129:1167–1179. doi: 10.1172/JCI122440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, Kreger BT, Vasioukhin V, Avruch J, Brummelkamp TR, et al. Yap1 acts downstream of α-catenin to control epidermal proliferation. Cell. 2011;144:782–795. doi: 10.1016/j.cell.2011.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jiang C, Javed A, Kaiser L, Nava MM, Xu R, Brandt DT, Zhao D, Mayer B, Fernández-Baldovinos J, Zhou L, et al. Mechanochemical control of epidermal stem cell divisions by B-plexins. Nat Commun. 2021;12:1308. doi: 10.1038/s41467-021-21513-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McGinn J, Hallou A, Han S, Krizic K, Ulyanchenko S, Iglesias-Bartolome R, England FJ, Verstreken C, Chalut KJ, Jensen KB, et al. A biomechanical switch regulates the transition towards homeostasis in oesophageal epithelium. Nat Cell Biol. 2021;23:511–525. doi: 10.1038/s41556-021-00679-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chang L, Azzolin L, Di Biagio D, Zanconato F, Battilana G, Lucon Xiccato R, Aragona M, Giulitti S, Panciera T, Gandin A, et al. The SWI/SNF complex is a mechanoregulated inhibitor of YAP and TAZ. Nature. 2018;563:265–269. doi: 10.1038/s41586-018-0658-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zanconato F, Battilana G, Forcato M, Filippi L, Azzolin L, Manfrin A, Quaranta E, Di Biagio D, Sigismondo G, Guzzardo V, et al. Transcriptional addiction in cancer cells is mediated by YAP/TAZ through BRD4. Nat Med. 2018;24:1599–1610. doi: 10.1038/s41591-018-0158-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data required to evaluate the conclusions are available in the main text or the Supplemental Material. Sequencing data derived from this study are deposited in the Gene Expression Omnibus under the given accession code (GEO accession: GSE253761). Previously published assay for transposase-accessible chromatin sequencing (ATAC-seq), scRNA-seq (single cell RNA-sequencing), or chromatin immunoprecipitation sequencing (ChIP-seq) data are available at the relevant websites, atlases, and portals outlined in the article along with the relevant accession codes. All other data are available from the corresponding author on reasonable request.