Abstract
Prior to ligand activation, the unactivated aryl hydrocarbon receptor (AhR) exists in a heterotetrameric 9S core complex consisting of the AhR ligand-binding subunit, a dimer of hsp90, and an unknown subunit. Here we report the purification of an ∼38-kDa protein (p38) from COS-1 cell cytosol that is a member of this complex by coprecipitation with a FLAG-tagged AhR. Internal amino acid sequence information was obtained, and p38 was identified as the hepatitis B virus X-associated protein 2 (XAP2). The simian ortholog of XAP2 was cloned from a COS-1 cDNA library; it codes for a 330-amino-acid protein containing regions of homology to the immunophilins FKBP12 and FKBP52. A tetratricopeptide repeat (TPR) domain in the carboxy-terminal region of XAP2 was similar to the third and fourth TPR domains of human FKBP52 and the Saccharomyces cerevisiae transcriptional modulator SSN6, respectively. Polyclonal antibodies raised against XAP2 recognized p38 in the unliganded AhR complex in COS-1 and Hepa 1c1c7 cells. It was ubiquitously expressed in murine tissues at the protein and mRNA levels. It was not required for the assembly of an AhR-hsp90 complex in vitro. Additionally, XAP2 did not directly associate with hsp90 upon in vitro translation, but was present in a 9S form when cotranslated in vitro with murine AhR. XAP2 enhanced the ability of endogenous murine and human AhR complexes to activate a dioxin-responsive element–luciferase reporter twofold, following transient expression of XAP2 in Hepa 1c1c7 and HeLa cells.
The aryl hydrocarbon receptor (AhR) is a ligand-inducible transcription factor that is a member of the bHLH-PAS (basic helix-loop-helix Per-Arnt-Sim) regulatory protein superfamily and is central in regulating the response to polycyclic aromatic compounds and halogenated aromatic hydrocarbons such as 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) (for reviews, see references 19, 47, and 49). The AhR is a unique member of this superfamily in that it is activated by ligand and interacts with hsp90. The N-terminal half of the AhR contains the bHLH domain; the HLH region mediates dimerization with its heterodimerization binding partner, the AhR nuclear translocator (Arnt), and the basic regions are involved in the ability of the AhR-Arnt heterodimer to recognize and bind to dioxin-responsive elements (DREs) (12, 13, 16). The PAS region is composed of A and B repeats that have homology to the Drosophila proteins Per and Sim (4). This region has been suggested to be involved in both ligand binding and AhR-Arnt heterodimerization (13, 16). The C-terminal half of these proteins contains a transactivation domain which mediates transcriptional activation (23, 25).
The AhR exists in a heterotetrameric complex in the cytoplasm in a 9S form, consisting of the AhR ligand-binding subunit, a dimer of hsp90, and a protein of ∼43 kDa prior to ligand binding (7). hsp90, which represents 1 to 2% of all cytoplasmic proteins, has been demonstrated to bind to the unliganded AhR as a dimer in chemical cross-linking and immunoabsorption assays (7, 35). hsp90 binds to both the bHLH and PAS domains in the AhR in vitro (16, 37). In the presence of hsp90, the AhR has a higher affinity for ligand, and its absence allows the AhR to bind to DNA (31). Following the binding of agonist, the AhR binds to Arnt in the nucleus of the cell. It has been suggested that during this process the AhR dissociates from the hsp90 dimer before heterodimerizing with Arnt (16, 41, 46). This heterodimer binds to DREs in the enhancer regions of genes such as Cyp1a1 and Cyp1a2, which code for enzymes used in xenobiotic metabolism. The characteristics and properties of the AhR-Arnt heterodimer have been well documented. Less is known, however, about the tetrameric unliganded 9S AhR core complex and the precise role of each protein in the complex.
Like the AhR, several other unliganded cellular receptors, such as the glucocorticoid receptor (GR), have been demonstrated to exist in multiprotein complexes containing an hsp90 dimer and other proteins. Chemical cross-linking studies of the untransformed GR were used to demonstrate that it was composed of hsp90 and a polypeptide of ∼50 to 55 kDa (3, 45). The 50- to 55-kDa protein was later demonstrated to be an FK506-binding protein, now referred to as the immunophilin FKBP52 (also known as hsp56, p59, and FKBP59) (55). Another immunophilin, Cyp-40, has also been isolated in untransformed GR complexes (34). Other proteins have been shown to complex with the GR, including hsp70, which is required for GR-hsp90 assembly (22). Following binding of ligand to the untransformed GR, the GR is then converted to a DNA-binding form which binds to specific GRE sequences in target gene promoters.
The untransformed progesterone receptor (PR) was initially demonstrated by chemical cross-linking analysis to exist in a complex with a dimer of hsp90 and a 59-kDa protein (2, 43, 44). Like the GR complex, this protein was later identified as FKBP52. The immunophilins Cyp-40 and FKBP54 have also been observed in PR complexes (32, 53). Other studies identified a 23-kDa protein associated with the unliganded PR (24). These and other studies suggest that the untransformed PR complex consists of a dimer of hsp90, an immunophilin component, and other associated proteins. In the presence of hormone, the progesterone receptor dissociates from hsp90 and is then capable of dimerization and binding to DNA.
These untransformed steroid receptor complexes are all ligand-inducible transcription factors that bind to specific DNA elements following the binding of ligand and release of hsp90 (for a review, see reference 40). Each of these complexes contains an immunophilin component. Immunophilins are a family of intracellular receptors that are able to bind to the immunosuppressive drugs cyclosporine, FK506, and rapamycin. Cyclosporine binds to cyclophilins, whereas FK506 and rapamycin bind to FK506-binding proteins (FKBPs). Two characteristics of immunophilins are peptidyl-prolyl-cis,trans-isomerase (PPiase, or rotamase) activity and regions containing tetratricopeptide repeat (TPR) motifs. The TPR motif was first identified in the Saccharomyces cerevisiae cell division cycle proteins CDC23, nuc2+, and CDC16, which are required for completion of mitosis, and in the SSN6 and SK13 gene products, which are involved in RNA synthesis (20, 51). Since their discovery, TPR motifs have been found in proteins involved in protein import, transcriptional repression, stress response, protein kinase inhibition, and Drosophila development (18, 28). Characteristics of the TPR motif include the size, hydrophobicity, and spacing of specific amino acid residues located in two domains, A and B, which form helix-turn structures thought to mediate protein-protein interactions (28).
Here we establish that the unliganded AhR exists in a core complex with a dimer of hsp90 and a newly identified subunit, the hepatitis B virus X-associated protein 2 (XAP2), which has strong homology to the human immunophilins FKBP12 and FKBP52. In vitro, XAP2 was not required for the assembly of AhR-hsp90 complexes and sedimented at 0 to 4S. However, cotranslation of murine AhR (mAhR) with XAP2 in vitro resulted in XAP2 shifting to a 9S complex with the AhR and hsp90. XAP2 acted to increase the ability of endogenous human AhR (hAhR) and mAhR to transactivate a DRE-luciferase reporter construct upon ligand treatment.
MATERIALS AND METHODS
Bacterial strains and cell culture.
Escherichia coli DH5α (Gibco BRL) was used for all plasmid preparations, and E. coli DH5αF′IQ (Gibco BRL) was used for expression of the fusion protein pGST-XAP2. COS-1, Hepa 1c1c7 (Hepa 1), and HeLa cells were grown in alpha minimum essential medium supplemented with 10% fetal bovine serum, 100 IU of penicillin per ml, and 0.1 mg of streptomycin per ml at 37°C in 94% air–6% CO2.
Construction and expression of plasmids.
pcDNA3/βmAhR (15) was used for expression of mAhR. The FLAG amino acid epitope (DYKDDDDK) was added to the mAhR by using the T7 forward primer 5′-CCTACAAACCAGAGGTGGACAGTG-3′ and the reverse FLAG primer 5′-CCGCTCGAGTCACTTGTCATCGTCGTCCTTGTAGTCACTCTGCACCTTGCTTAG-3′ in a PCR with pcDNA3/βmAhR as the template. The PCR-generated fragment was digested with HindIII and XhoI and inserted into a HindIII-XhoI site in pcDNA3/β to generate pcDNA3/BmAhR-FLAG. pSVSPORT containing the hAhR was digested with KpnI and SalI and cloned into pCI (Promega) to generate pCI/hAhR. The FLAG amino acid epitope was added to the human AhR by using the T7 forward primer 5′-TAATACGACTCACTATAGGG-3′ and the reverse FLAG primer 5′-CCGCTCGAGTCACTTGTCATCGTCGTCCTTGTAGTCCAGGAATCCACTGGATGT-3′ in a PCR with pCI/hAhR as the template. The PCR-generated product was digested with KpnI and XhoI and inserted into a KpnI-SalI site in pCI to generate pCI/hAhR-FLAG. The presence of the FLAG epitope was confirmed by transcribing and translating these constructs in vitro with a TNT T7 rabbit reticulocyte lysate kit (Promega) followed by immunoblotting with the anti-FLAG M2 monoclonal antibody (IBI Kodak, New Haven, Conn.). The nucleotide sequences of all the PCR-subcloned products were confirmed by DNA sequencing. To generate pCI-XAP2, pCI was digested with KpnI and XhoI and the human XAP2 cDNA containing KpnI and XhoI ends, which was excised from pGEM-XAP2, was inserted. pCI-XAP2 was transcribed and translated in vitro in the presence of [35S]methionine (>1,000 mCi/mmol; Amersham) with the TNT T7 rabbit reticulocyte lysate kit, followed by autoradiography.
Transient-transfection assays.
COS-1 and HeLa cells were transfected at 80% confluency in 6-cm2 tissue culture plates by a lipofectAMINE procedure as specified by the manufacturer (Gibco BRL). Hepa 1 cells were transfected in 24-well tissue culture plates. pGudLuc6.1 was used at 100 ng/well, which contains the DRE-luciferase reporter construct in which the DRE insert was subcloned into the pGL3 vector (29). pcDNA3.1/lacZ/his (Invitrogen) was used at 100 ng/well for transfection efficiency normalization with a β-galactosidase assay kit (Promega). Following transfection, cells were induced with TCDD or dimethyl sulfoxide at a final concentration of 10 nM for 12 h. Following induction, the cells were washed twice in 1× phosphate-buffered saline (PBS), incubated in 400 μl of 1× lysis buffer (Promega), stored at −80°C, thawed, and harvested. Total protein was evaluated by the bicinchoninic acid assay (Pierce).
β-Galactosidase normalization and luciferase assays.
Lysate at 0.75 mg/ml in lysis buffer was incubated with an equal volume of β-galactosidase substrate (Promega) at 37°C for 30 min. The reaction was stopped by the addition of 500 μl of sodium carbonate, and the β-galactosidase activity was measured at 420 nm. Luciferase activity in lysate protein was measured in a TD-20e luminometer (Turner Designs). Luciferase expression was determined relative to β-galactosidase activity.
Transient transfections of COS-1 cells for AhR complex immunoprecipitations.
COS-1 cells were transfected at 80% confluency in 10-cm2 tissue culture plates by the lipofectAMINE procedure as specified by the manufacturer. Transfected COS-1 cells were harvested with trypsin-EDTA and washed once in 1× PBS. The cells were resuspended in MENG buffer (25 mM morpholinepropanesulfonic acid [MOPS], 2 mM EDTA, 0.02% NaN3, 10% glycerol [pH 7.5]) plus 1.25 μg of leupeptin per ml, 1.8 μg of pepstatin A per ml, and 25 μg of aprotonin per ml and homogenized in a Dounce homogenizer. The cell homogenate was centrifuged at 100,000 × g for 60 min at 4°C, and the supernatant was collected to yield the cytosolic fraction.
Immunoprecipitation and gel electrophoresis.
Cytosol isolated from transfected or nontransfected COS-1 cells was immunoprecipitated with the anti-FLAG M2 affinity gel (IBI Kodak). For control reactions, 90 nmol of FLAG peptide (IBI Kodak) was preincubated with M2 affinity gel for 1 h. Cytosol in the presence of MENG buffer–250 mM NaCl–0.5% Nonidet P-40 (NP-40) was incubated for 4 h in 50 μl of packed gel with rocking at 4°C. The M2 affinity gel was subsequently washed with MENG buffer–500 mM NaCl–1% NP-40 four times and with MENG buffer once. To the pellet, 50 μl of 2× Tricine sample buffer (TSB) was added, and the samples were heated at 94°C for 4 min. The immunoabsorbed complexes were resolved by Tricine sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (8% polyacrylamide). The gels were either silver stained or transferred to a polyvinylidene difluoride (PVDF) membrane for immunoblot analysis (58). For immunoabsorption of the unliganded AhR complex from Hepa 1 cells, polyclonal AhR antibody (15 μg) was incubated with 50 μl of protein A-agarose (Pierce) at 4°C for 1 h and washed twice with MENG buffer–500 mM NaCl. Hepa 1 cytosol was incubated with the affinity gel for 3 h, then washed three times with MENG buffer–500 mM NaCl, subjected to sucrose extrusion through a MENG buffer–1 M sucrose–500 mM NaCl–1% NP-40 cushion, and washed once in MENG buffer. The pellet was heated for 4 min in 50 μl of 2× TSB, resolved by Tricine SDS-PAGE, and transferred to a PVDF membrane for immunoblot analysis.
Purification and sequence analysis of p38.
COS-1 cells transfected with pcDNA3/β mAhR-FLAG from a total of 80 10-cm2 tissue culture dishes were collected and homogenized in MENG buffer–1.25 μg of leupeptin per ml–1.8 μg of pepstatin A per ml–25 μg of aprotonin per ml in a Dounce homogenizer, and the cytosolic fraction was isolated as described above. The total cytosolic fraction was incubated with 200 μl (packed-bed volume) of anti-FLAG M2 affinity gel for 4 h in MENG by the immunoprecipitation method described above. Following the first 4 h, the cytosol was again incubated for an additional 4 h in 200 μl (packed-bed volume) of anti-FLAG M2 affinity gel at 4°C and then washed as described above. Both samples were resuspended in 2× TSB, heated for 5 min at 94°C, and resolved by Tricine SDS-PAGE (10% polyacrylamide). The gel was stained with Coomassie brilliant blue and destained, and the p38 band was excised. A gel fragment at the same molecular weight was also excised as a control, and all the samples were stored at −80°C. These samples were then sent to the Howard Hughes Medical Institute W. M. Keck Biopolymer Facility at Yale University for in-gel tryptic digestion and microsequencing of p38. The peptide fragments obtained from in-gel tryptic digestion of p38 were resolved by reverse high-pressure liquid chromatography on a C18 microbore column. Peaks corresponding to individual peptides were selected and subjected to microsequencing.
Cloning and sequencing of simian XAP2 cDNA.
Total RNA was isolated from COS-1 cells using Tri-Reagent (Molecular Research Center, Cincinnati, Ohio). Poly(A)+ mRNA was isolated with the PolyAT Tract kit (Promega). This poly(A)+ mRNA was then used to generate a COS-1 cDNA library with linker adaptors, using the Marathon cDNA amplification kit (Clontech). A 5-μl aliquot of a 1:50 dilution of this library was used in a PCR to amplify the simian XAP2 cDNA with primers 5′-ACGCTGCACAGTGACGACGAG-3′ and 5′-GGGTCCAGCTCCAGCACTTTG-3′, which correspond to the N-terminal peptide (peptide 1) and the C-terminal peptide (peptide 2) of p38, respectively. This resulted in the generation of a 781-bp fragment. After amplification of this partial cDNA, the full-length cDNA was obtained in 5′ and 3′ rapid amplification of cDNA ends (RACE) reactions. The 5′ RACE product was isolated with the AP1 adapter primer, 5′-CCATCCTAATACGACTCACTATAGGGC-3′ (Clontech), and a second reverse primer in the XAP2 human cDNA, 5′-GGGTCCAGCTCCAGCACTTTG-3′. Reamplification of this product with the nested adaptor primer 2 (AP2), 5′-ACTCACTATAGGGCTCGAGCGGC-3 (Clontech), and the reverse primer in the XAP2 human cDNA, 5′-CTTGCCCCGCTTGAAGTAGGC-3′, resulted in a specific 5′ RACE product of 901 bp. The 3′ RACE product was generated by using AP1 and the forward primer 5′-ACGCTGCACAGTGACGACGAG-3′ to generate a 3′ RACE fragment. The final 3′ RACE product was generated with this fragment as a template and with the nested primers AP2 and forward primer 5′-CTCCGCAACATCGCGGCGGGC-3′, which generated an 812-bp fragment. Each of these fragments was cloned into the T-Easy vector (Promega) and sequenced on an ABI Prism 310 genetic analyzer (Applied Biosystems).
Antibody production.
Polyclonal antisera raised against the AhR and hsp90 (hsp86/84) have been described previously (38, 39). Mouse polyclonal ascites was raised against XAP2 by the following method. DH5αF′IQ/pGST-XAP2 was grown to an optical density at 595 nm of 0.3 and induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma) for 3 h. The induced cells were centrifuged at 9,800 × g and washed once in 1× PBS. The pellet was resuspended in sonication buffer (20 mM Tris, 1 mM EDTA [pH 8.0]) and sonicated (48). pGST-XAP2 remained in the insoluble pellet as determined by SDS-PAGE. The pellet was solubilized in lysis buffer (50 mM Tris, 1 mM EDTA, 100 mM NaCl [pH 8.0]) containing 0.1% SDS and 8 M urea (48). The dissolved pellet was incubated for 1 h at room temperature and then centrifuged at 100,000 × g for 1 h at room temperature. The soluble portion was dialyzed against lysis buffer plus 0.1% SDS for 2 h at room temperature with several changes of buffer. The soluble GST-XAP2 fusion protein was then cleaved with factor Xa (New England Biolabs) in 20 mM Tris–100 mM NaCl–2 mM CaCl2–0.1% SDS. Cleaved protein products were then resolved by Tricine SDS-PAGE, transferred to a nitrocellulose membrane (Bio-Rad), and stained with Ponceau S (Sigma). A band at ∼38 kDa that corresponded to XAP2 was excised and used as the antigen. Nitrocellulose fragments of XAP2 were dissolved in dimethyl sulfoxide and injected subcutaneously into several sites on female BALB/c mice (Jackson Laboratories) for polyclonal ascites production essentially as described previously (36). XAP2 polyclonal ascites was affinity purified by incubating ascites with GST-XAP2 fusion protein (2 mg/ml) coupled to CNBr-activated Sepharose overnight. The gel was washed once with 1× PBS, packed into a column, and washed extensively with 1× PBS. The polyclonal antibody was eluted with 0.1 M glycine, and 1.0-ml fractions were collected in tubes containing 200 μl of 1 M Tris (pH 8.0).
Northern blot analysis.
Poly(A)+ mRNA isolated from COS-1 cells was resolved on 0.8% borate–formaldehyde gels with molecular weight standards (Gibco BRL) and transferred to a nitrocellulose membrane by standard procedures (48). The simian XAP2 cDNA was radiolabeled with the random-primed DNA-labeling kit (Boehringer Mannheim) with [α-32P]dATP (>6,000 Ci/mmol; Amersham) and used as a probe. The blot and probe were hybridized at 67°C for 16 h in 7% SDS–0.5 M Na2PO4–1% bovine serum albumin (pH 7.3) essentially as described elsewhere, washed three times at room temperature, incubated for 15 min at 67°C in 40 mM Na2PO4–10% SDS, and washed again at room temperature (8). The blot was then subjected to autoradiography.
QRT-PCR analysis.
For quantitative reverse transcriptase (QRT)-PCR, total RNA was used from various mouse tissues purchased from Ambion (Austin, Tex.). The level of XAP2 mRNA was examined by QRT-PCR. The quantitation of mRNA by QRT-PCR required the use of internal standards to negate tube-to-tube variability in amplification efficiency. A PCR-based method for generating recombinant mRNA templates (rcRNA) for use as internal standards in quantitative PCR was used (56). By this procedure, rcRNA molecules which contain forward and reverse primer sequences for the target XAP2 gene were synthesized. These internal standards produced a PCR product which was easily resolved from that of target mRNA by agarose gel electrophoresis.
Competitive QRT-PCR was performed essentially as described previously with some modifications (17, 57). Reverse transcription of RNA was carried out in a final volume of 20 μl containing 25 mM Tris (pH 8.3), 50 mM (NH4)2SO4, 0.2% β-mercaptoethanol, 0.1 mg of bovine serum albumin per ml, 5 mM MgCl2, 1 mM each deoxynucleoside triphosphate, 1 U of RNase inhibitor, 2.5 U of mouse mammary tumor virus RT, 2.5 mM oligo(dT), 0.1 mg of total RNA, and various amounts of rcRNA internal standard. The samples were incubated at 42°C for 15 min, and RT was inactivated by heating to 99°C for 5 min. To these cDNA samples, a PCR master mix was added to bring the final volume to 50 μl. The final MgCl2 concentration in the PCR mixture was 4 mM, and the mixture contained 2.5 U of Taq polymerase and 10 pmol each of forward and reverse primers. The primer sequences for XAP2 were 5′-ACCCACAGCCTCTCATCTTCC-3′ (forward primer) and 5′-AGGCGCCAGGGCAGGGTCTA-3′ (reverse primer), which recognize nucleotides 640 to 660 and 1109 to 1090 of the AIP (murine XAP2) cDNA sequence (30), respectively. Amplification of cDNA with these primers resulted in a 470-bp product from XAP2 and a 308-bp product from the internal standard.
The reaction mixtures were heated to 94°C for 3 min and immediately cycled 30 times through a 20-s denaturing step at 94°C, a 30-s annealing step at 58°C, and a 40-s elongation step at 72°C. After the final cycle, a 5-min elongation step at 72°C was performed. Aliquots of the PCR mixture were electrophoresed on 3% NuSieve–agarose (3:1 [wt/wt]; FMC BioProducts, Rockland, Maine) gels and the PCR fragments were visualized by ethidium bromide staining and digitized for subsequent densitometry (Eagle Eye II; Stratagene). The amount of target mRNA present was determined essentially as described previously and later modified (17, 56, 59).
Expression and immunoabsorption of XAP2 and mAhR-FLAG in a reticulocyte lysate.
pcDNA3/βmAhR-FLAG and pCI-XAP2 were transcribed and translated in vitro with a TNT coupled transcription-translation rabbit reticulocyte lysate kit (Promega) with T7 polymerase at 30°C for 1 h as specified by the manufacturer. Each mixture was diluted 1/10 in incubation buffer (25 mM MOPS, 1 mM EDTA, 2 mg of bovine serum albumin per ml, 2 mg of ovalbumin per ml, 0.5% Tween, 50 mM NaCl, 10% glycerol, 10 mM sodium molybdate) for 2 h at 4°C and then incubated in 25 μl of M2 affinity gel for 4 h. The complexes were washed four times in incubation buffer and twice in MENG buffer. All the mixtures were resolved by Tricine SDS-PAGE and transferred to a PVDF membrane.
Sucrose density gradient analysis.
Cytosol isolated from COS-1 cells or in vitro translation mixtures was layered on 5.1-ml 10 to 30% sucrose gradients prepared in MENG buffer with 20 mM sodium molybdate. The sealed centrifuge tubes were centrifuged in a Beckman VTi65.2 rotor at 416,000 × gmax for 135 min at 4°C. After centrifugation, 0.2-ml fractions were collected with an Isco model 640 density gradient fractionator. Then 75 μl of each fraction was mixed with an equal volume of 2× TSB and heated at 95°C for 4 min. The samples were subjected to Tricine SDS-PAGE and transferred to a PVDF membrane for immunoblot analysis. Bovine serum albumin (4.4S) and catalase (11.3S) were resolved on separate sucrose gradients and used as sedimentation standards.
RESULTS
The goal of this study was to purify and identify the p43 subunit of the nonactivated AhR complex previously shown to coimmunoprecipitate from Hepa 1 cell extracts (7). Initial studies were performed in Hepa 1 cells to immunopurify large amounts of unliganded AhR complex. These procedures suffered from high backgrounds and relatively poor yields (data not shown). Several other purification schemes were examined, one of which was the use of a transiently expressed FLAG-tagged AhR in COS-1 cells. This cell line was chosen over Hepa 1 cells due to the lower nonspecific binding of COS-1 cytosolic proteins to the anti-FLAG M2 affinity gel and the ability to express larger amounts of AhR.
A transfected mAhR in COS-1 cells is responsive to TCDD.
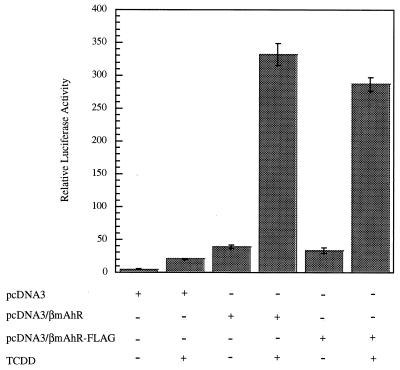
To determine if the AhR signaling pathway could be studied in COS-1 cells which contain very low levels of endogenous AhR, COS-1 cells were cotransfected with plasmids containing the mAhR, a DRE-luciferase reporter construct, and a β-galactosidase gene used for normalization of transfection efficiency. Following transfection, the cells were either induced with the ligand TCDD or treated with carrier solvent. An eightfold induction in luciferase activity above background following TCDD treatment was observed compared to the result for cells treated with carrier solvent (Fig. 1). This established that the AhR was functional in COS-1 cells and was capable of activating a DRE-luciferase reporter following induction by agonist. To characterize the unliganded AhR complex in COS-1 cells, the mAhR gene was epitope tagged at the C terminus with a FLAG sequence (mAhR-FLAG) by PCR. The presence of the FLAG sequence did not significantly interfere with the ability of the AhR to enhance transactivation of the DRE-reporter construct (Fig. 1).
FIG. 1.
A transfected AhR is responsive to TCDD in COS-1 cells. COS-1 cells were transfected with pcDNA3, pcDNA3/βmAhR, or pcDNA3/βmAhR-FLAG in the presence of pGudLuc 6.1 and pcDNA3.1/lacZ/his plasmids. Relative luciferase activity was measured following induction by TCDD or carrier solvent. Each transfection was performed in triplicate.
Transfection of a mAhR-FLAG construct results in the assembly of a heterotetrameric complex in COS-1 cells.
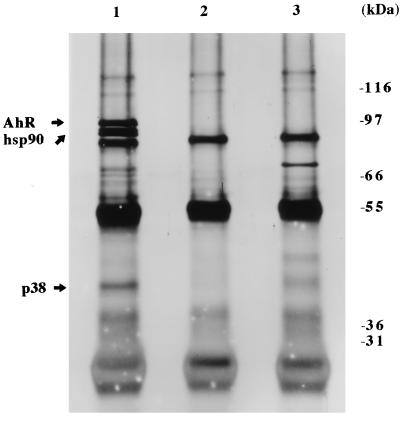
The next question was whether the AhR would assemble into a heterotetrameric complex in COS-1 cells as previously observed in Hepa-1 cells (7). To determine if the same members of this complex were present in COS-1 cells, COS-1 cells were transfected with pcDNA3/βmAhR-FLAG, cytosol was isolated, and AhR-FLAG complexes were immunoabsorbed with the anti-FLAG M2 affinity gel. The affinity gel was then washed in the presence of 500 mM NaCl, and bound complexes were resolved by SDS-PAGE followed by silver staining, which allowed the visualization of all protein species that were stably associated with the AhR. Four prominent bands were observed (Fig. 2, lane 1) compared to the control lanes, which contained the M2 affinity gel preblocked with FLAG peptide (lane 2) or with vector alone (lane 3). The calculated molecular masses of these bands were ∼97, 86, 84, and 38 kDa. This observation was consistent with a previous study in which [35S]methionine-labeled Hepa 1 cytosol was subjected to immunoabsorption with an anti-AhR polyclonal antibody which yielded proteins with molecular masses of ∼97, 86, 84, and 43 kDa (7). The 97-, 86-, and 84-kDa bands from COS-1 cytosol were identified as mAhR-FLAG, hsp86, and hsp84 (hsp90), respectively (data not shown), as demonstrated in Hepa 1 cells (7). In several experiments, a band was detected at ∼70 kDa, although this was variable among several experiments (lane 1). A reassessment of the molecular mass of the 43-kDa protein (p43) from Hepa 1 cytosol compared to the 38-kDa protein in COS-1 cells was performed, and it was determined that these proteins migrate at the same apparent molecular mass of ∼38 kDa. Because the p38 protein was uncharacterized, we wished to isolate it and clone the corresponding gene.
FIG. 2.
The AhR exists in a heterotetrameric complex in COS-1 cells. COS-1 cells were transfected with 9 μg of pcDNA3/βmAhR-FLAG or pcDNA3; cytosolic fractions were isolated, immunoabsorbed with M2 affinity gel, and resolved by SDS-PAGE; and the gel was silver stained. Lanes: 1, pcDNA3/βmAhR-FLAG; 2, pcDNA3/βmAhR-FLAG with M2 affinity gel blocked with FLAG peptide; 3, pcDNA3.
Isolation and purification of p38.
To isolate and identify the 38-kDa protein, COS-1 cells were transfected with pcDNA3/βmAhR-FLAG and mAhR-FLAG complexes were immunoabsorbed with the M2 affinity gel, using a large-scale transfection protocol. The p38 protein was resolved with the other members of the AhR complex immunoabsorbed from ∼100 mg of COS-1 cytosol by Tricine SDS-PAGE and visualized with Coomassie brilliant blue. A total of ∼1.5 μg of p38 and a control fragment at the same molecular mass were excised and subjected to in-gel tryptic digestion. Tryptic peptides were resolved by reverse-phase HPLC on a C18 microbore column. Peaks corresponding to two tryptic peptides of 15 and 14 amino acid residues each were selected and used for microsequencing. The amino acid sequences of peptide 1, TLHSDDEGTVLDDSR, and peptide 2, VLELDPALAPVVSR, were obtained. Alignment of these amino acid sequences with the National Institutes of Health BLAST algorithm revealed 100% homology to human XAP2 (1). A BLAST search with the human XAP2 amino acid sequence yielded strong homology to the immunophilins FKBP52 and FKBP12.
Cloning of simian XAP2.
Peptides 1 and 2 of p38 were aligned with the coding sequence of human XAP2, which mapped to the N-terminal (residues 40 to 54) and C-terminal (residues 291 to 304) ends, respectively. Based on the human nucleotide sequence of XAP2 in the regions corresponding to peptides 1 and 2, respective oligonucleotide primers were designed and used in a PCR with a COS-1 cDNA library to amplify a fragment of simian XAP2. A 781-bp fragment was generated, which corresponded identically to the distance between these primers in the human XAP2 nucleotide sequence. To obtain a full-length cDNA clone, 5′ and 3′ RACE reactions were performed. The full-length simian XAP2 cDNA is composed of 1,242 nucleotides with an open reading frame of 993 nucleotides encoding a predicted 330-amino-acid protein (Fig. 3A). The amino acid sequences of peptides 1 and 2 isolated from the p38 protein were present from residues 40 to 54 and 291 to 304, respectively, confirming that this cDNA contained regions that encoded the two tryptic fragments isolated from p38. This cDNA contained a 5′ untranslated region in which a Kozak consensus sequence for translation initiation upstream of the putative 5′ start site was located, as well as a poly(A) tail consisting of at least 19 nucleotides in its 3′ untranslated region (26).
FIG. 3.
Predicted coding sequence for simian XAP2. (A) Nucleotide and predicted amino acid sequence of simian XAP2. Singly underlined regions indicate homology to FKBP12; the doubly underlined region indicates a conserved TPR domain. (B) Alignment of XAP2 carboxy-terminal TPR, TPR3 of FKBP59, and TPR4 of SSN6. The consensus TPR was generated from CDC16, CDC23, CDC27, SSN6, and SK13 (28).
The N-terminal end of simian XAP2 contains a region of homology to both FKBP12 and FKBP52. Residues 45 to 87 of XAP2 were 34% identical to residues 32 to 74 in human FKBP12, and residues 46 to 90 in the same region were 28% identical to residues 63 to 107 in FKBP52. In addition, a short amino acid sequence of 11 residues in XAP2 (positions 148 to 158) was 63% identical to residues 98 to 108 in human FKBP12. It was also determined that residues 165 to 328 in the carboxy-terminal end of XAP2 were 26% identical to residues 256 to 415 in FKBP52. This region corresponds to domain III of FKBP52, which contains three TPR domains (42). A single TPR domain matching the TPR consensus sequence (-W-LG-Y-A-F-A-P) was identified in the C-terminal portion of simian XAP2 from residues 265-298 (28). This TPR domain was very similar to the third TPR domain of FKBP52 and the fourth TPR in the Saccharomyces cerevisiae transcriptional modulator SSN6. A striking similarity was observed, especially in domain B (Fig. 3B).
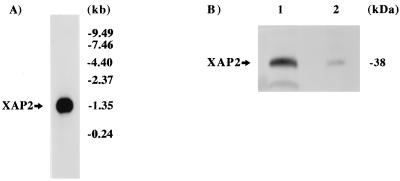
Northern blot analysis was then performed to determine the mRNA transcript size of the simian XAP2 cDNA. A single 1.35-kbp fragment was observed in this analysis with the coding region of the simian XAP2 cDNA as a probe (Fig. 4A). This transcript size was that predicted for a 330-amino-acid protein, including 5′ and 3′ untranslated regions, which corresponded to the full-length XAP2 cDNA.
FIG. 4.
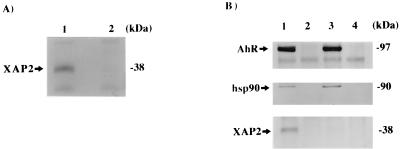
Presence of XAP2 in COS-1 cells. (A) Northern blot analysis of XAP2 mRNA. Poly(A)+ mRNA (4 μg) from COS-1 cells was probed with simian XAP2 cDNA. (B) Immunoblot analysis of XAP2. Lanes: 1, XAP2 translated in vitro; 2, 100 μg of COS-1 cytosol.
To further characterize XAP2, polyclonal antibodies were generated against the intact protein. XAP2 was transcribed and translated in vitro in a rabbit reticulocyte lysate system (Fig. 4B, lane 1). These antibodies were able to detect a single band of ∼38 kDa in an immunoblot analysis. Additionally, these antibodies recognized XAP2 at ∼38 kDa from COS-1 cytosol (lane 2). These results confirmed that XAP2 has an apparent molecular mass of ∼38 kDa, which is in close agreement with the calculated molecular weight of 37,627 encoded by the simian XAP2 cDNA.
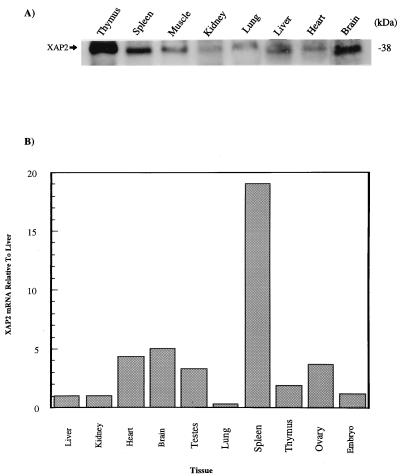
Tissue distribution of XAP2.
Because the AhR is located in a wide variety of tissues (6), we wished to determine whether the tissue distribution of XAP2 would be similar. Immunoblot analysis with the anti-XAP2 polyclonal antibody identified the highest levels of XAP2 protein in the mouse thymus and spleen, with the lowest levels in skeletal muscle and heart (Fig. 5A). To determine the level of XAP2 mRNA, QRT-PCR was performed on total RNA isolated from mouse tissues and XAP2 mRNA was detected in each tissue. It was determined that the spleen, brain, heart, and ovaries had the highest levels of XAP2 mRNA whereas the lungs had the lowest level (Fig. 5B).
FIG. 5.
XAP2 is ubiquitously expressed in murine tissues. (A) Cytosolic extracts (100 μg) from the tissues indicated were resolved on SDS-PAGE, transferred to a PVDF membrane, and immunoblotted with XAP2 polyclonal antibodies. (B) QRT-PCR analysis of XAP2 mRNA in the murine tissues indicated. All values were normalized to XAP2 mRNA expressed in the liver.
XAP2 is a subunit of the unliganded AhR complex.
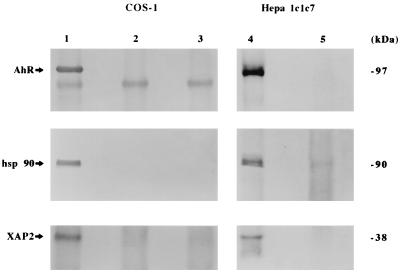
To demonstrate that XAP2 was the p38 protein subunit of the unliganded AhR, cytosol from COS-1 cells transfected with pcDNA3/βmAhR-FLAG were immunoabsorbed with the anti-FLAG M2 affinity gel. To determine if p38 was present in the endogenous unliganded AhR complex, AhR polyclonal antibodies were bound to protein A agarose and used to immunoabsorb endogenous unliganded AhR complexes from Hepa 1 cytosol. Immunoblot analysis with the AhR monoclonal antibody RPT1 identified the AhR in COS-1 and Hepa 1 cells (Fig. 6, lanes 1 and 4) but did not recognize the AhR in control lanes (lanes 2, 3, and 5). Polyclonal antibodies raised against hsp86/84 were used to detect hsp90, which is resolved as a doublet by Tricine SDS-PAGE in COS-1 and Hepa 1 cells (lanes 1 and 4, respectively) (38). hsp90 was not detected in control lanes (lanes 2, 3, and 5). The XAP2 polyclonal antibody recognized XAP2 in the AhR complexes in COS-1 and Hepa 1 cells (lanes 1 and 4), but XAP2 was not detected in control lanes (lanes 2, 3, and 5). This would indicate that XAP2 is a subunit that is stably associated with the endogenous 9S unliganded AhR complex in Hepa 1 cells and in COS-1 cells expressing mAhR-FLAG.
FIG. 6.
XAP2 is a subunit stably associated with the unliganded AhR core complex in COS-1 and Hepa 1 cells. (A) COS-1 cells were transfected with 9 μg of pcDNA3/βmAhR-FLAG or pcDNA3, immunoabsorbed with M2 affinity gel, resolved on SDS-PAGE, blotted, and probed with antibodies raised against the AhR (RPT1), hsp90 (hsp 86/84), and XAP2. Lanes: 1, pcDNA3/βmAhR-FLAG; 2, pcDNA3/βmAhR-FLAG with M2 resin blocked with FLAG peptide; 3, pcDNA3 (cytosol from Hepa 1 cells was immunoabsorbed with polyclonal AhR antisera or rabbit immunoglobulin G, resolved on SDS-PAGE, and used for immunoblot analysis); 4, polyclonal AhR antisera; 5, rabbit IgG.
The unliganded AhR assembles with hsp90 in the absence of XAP2 in vitro.
Because XAP2 was demonstrated to be a stable member of the unliganded AhR complex, we wished to determine if it was required for the assembly of the AhR-hsp90 complex. To assess this question, a rabbit reticulocyte lysate (RL) system was used to study this assembly process in vitro. Previous studies have demonstrated that the AhR associates with hsp90 when transcribed and translated in vitro in RL (16, 31, 37). Once complexed with hsp90 in vitro, and following ligand activation, the AhR is able to transform to the 6S form and bind to a DRE (12, 16). XAP2 polyclonal antibodies were used in an immunoblot analysis to detect XAP2 in RL alone or in an RL mixture containing transcribed and translated XAP2. These antibodies were unable to detect XAP2 in RL alone (Fig. 7A, lane 2) but could recognize XAP2 at ∼38 kDa when it was transcribed and translated in RL (lane 1). To confirm that the AhR assembled with hsp90 in the absence of XAP2, pcDNA3/βmAhR-FLAG alone was transcribed or pcDNA3/βmAhR-FLAG and pCI/XAP2 were cotranscribed and translated in vitro in RL, subjected to immunoabsorption with the M2 affinity gel, washed, resolved on SDS-PAGE, and analyzed for the presence of the AhR, hsp90, and XAP2 by immunoblot analysis. In both cases, the AhR assembled with hsp90 (Fig. 7B, lanes 1 and 3) compared to control lanes (lanes 2 and 4). In the RL reaction lacking the in vitro-transcribed and -translated XAP2, essentially no endogenous XAP2 was detected (lane 3), whereas it was detected in the RL mixture containing in vitro-transcribed and -translated XAP2 (lane 1). Thus, XAP2 is not required for AhR-hsp90 complex formation in vitro.
FIG. 7.
XAP2 is not required for assembly of the AhR with hsp90 in vitro. (A) Immunoblot analysis of XAP2 in RL. Lanes: 1, XAP2 transcribed and translated in vitro in RL; 2, RL alone. Each reaction was resolved by SDS-PAGE and used for immunoblot analysis with XAP2 polyclonal antibodies. (B) Immunoabsorption of AhR-hsp90-XAP2 complex. Lanes: 1, pcDNA3/βmAhR-FLAG cotranscribed and translated in vitro with XAP2 in RL; 2, same mixture as in lane 1 incubated with M2 affinity gel blocked with FLAG peptide; 3, pcDNA3/βmAhR-FLAG transcribed and translated in vitro in reticulocyte lysate; 4, same mixture as in lane 3 incubated with M2 affinity gel blocked with FLAG peptide. Each translation mixture was subjected to immunoabsorption with the M2 affinity gel, washed, resolved by SDS-PAGE, and used in an immunoblot analysis with the antibodies raised against the AhR (RPT1), hsp90 (hsp86/84), and XAP2.
XAP2 is not associated with hsp90 in the absence of the AhR in vitro.
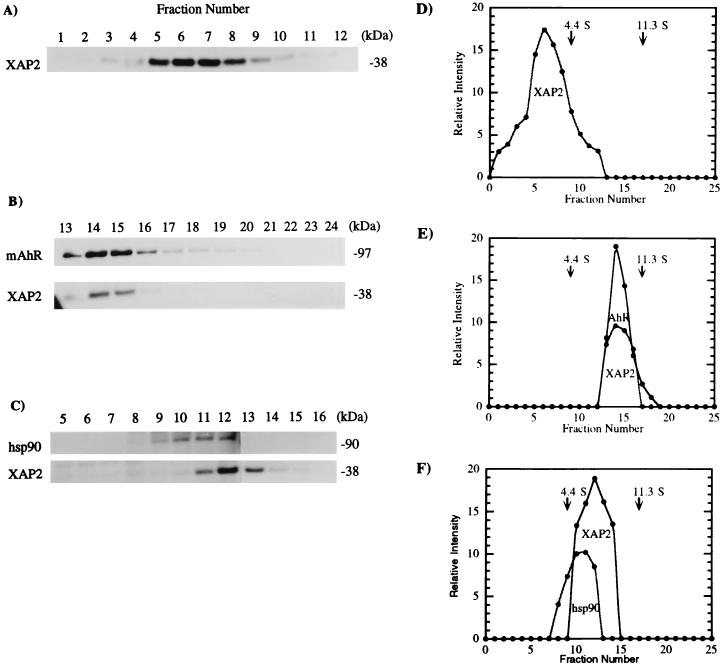
Because XAP2 assembled with the AhR-hsp90 complex in vitro, we wished to determine if XAP2 was able to associate directly with hsp90 in the absence of the AhR. To test this possibility, pCI/XAP2 was transcribed and translated in vitro in RL, which lacks the AhR but contains hsp90, in the presence of [35S]methionine, the reaction was resolved on a sucrose density gradient, and fractions were collected, resolved by SDS-PAGE, and analyzed by autoradiography. XAP2 was detected in the 0S to 4S fraction (Fig. 8A and D) and was not present in the 6S fraction, where hsp90 sediments. Thus, under these conditions, XAP2 did not directly interact with hsp90 in RL. To determine if XAP2 would shift to a 9S form in the presence of the AhR, pCI/XAP2 and pcDNA3/βmAhR-FLAG were cotranscribed and translated in vitro and the translated proteins were analyzed by the same procedure. It was observed that XAP2 was present in the 9S form under these new conditions (Fig. 8B and E). These results suggested that XAP2 becomes associated in a 9S complex only in the presence of the AhR.
FIG. 8.
XAP2 exists in a 0S to 4S complex in RL and shifts to 9S in the presence of mAhR. (A) pCI-XAP2 was transcribed and translated in rabbit RL with [35S]methionine and resolved on a sucrose density gradient, and fractions were collected, resolved by SDS-PAGE, and analyzed by autoradiography. (B) XAP2 was cotranscribed and translated with mAhR in RL by the same procedure. (C) Cytosol isolated from COS-1 cells was resolved on a sucrose density gradient, and fractions were collected, resolved by SDS-PAGE, blotted, and probed with antibodies against hsp90 (3B6p90) or XAP2 followed by incubation with [125I]GAM secondary antibody. (D to F) Quantitative assessments of panels A to C, respectively.
Due to the very low level of endogenous AhR activity and the significant level of XAP2 expression in COS-1 cells, we wished to determine if the interaction of XAP2 with the AhR was dependent upon a preassembled hsp90-XAP2 complex. Cytosol isolated from COS-1 cells was resolved on a sucrose density gradient, and fractions were collected, resolved on SDS-PAGE, and used in an immunoblot analysis to detect XAP2 and hsp90. XAP2 was found to fractionate at ∼6S, where hsp90 sediments (Fig. 8C). This implied that XAP2 may directly interact with hsp90 in vivo. Immunoabsorption assays with COS-1 cell cytosol were then performed with polyclonal anti-hsp90 antibodies and an anti-hsp90 monoclonal antibody, 3G3, to determine if XAP2 would coprecipitate with hsp90. Although hsp90 was immunoabsorbed, no XAP2 was detected with either antibody preparation. This may be due to the inability of these hsp90 antibodies to recognize hsp90-XAP2 complexes.
XAP2 enhances AhR-mediated transcription in Hepa 1 cells.
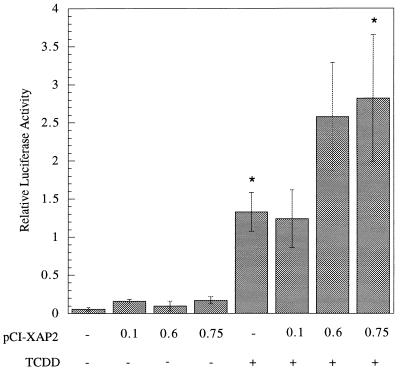
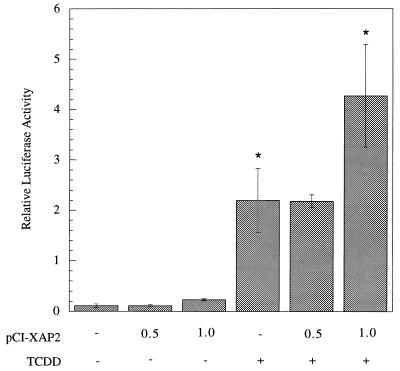
We wished to determine the role of XAP2 in the AhR signaling pathway. Because it was demonstrated that XAP2 associated with the mAhR in COS-1 cells and Hepa 1 cells, we wished to determine the effect that the expression of XAP2 would have on the endogenous mAhR in Hepa 1 cells. We chose Hepa 1 cells as a model system because their endogenous complex contains XAP2 and has been used extensively to characterize the properties of AhR signal transduction. Increasing amounts of pCI/XAP2 were expressed in Hepa 1 cells, followed by induction with TCDD or carrier solvent (Fig. 9). Despite the endogenous level of XAP2 in Hepa 1 cells (30), it was observed that relative luciferase activity increased twofold in a statistically significant manner. This suggested that XAP2 acted to increase the ability of the mAhR to transactivate the DRE-luciferase reporter construct.
FIG. 9.
XAP2 acts a transcriptional enhancer in Hepa 1 cells. Hepa 1 cells were transfected with plasmids pGudLuc 6.1, pcDNA3.1/lacZ/his, and pCI-XAP2. The amount of pCI-XAP2 transfected is given in micrograms, and each transfection was brought to 0.75 μg with pCI vector. Relative luciferase activity was measured following induction by TCDD or carrier solvent. Each transfection was performed in triplicate. Asterisks indicate that a statistically significant difference was obtained (P < 0.05).
XAP2 is capable of binding to hAhR and enhances hAhR-mediated transcription.
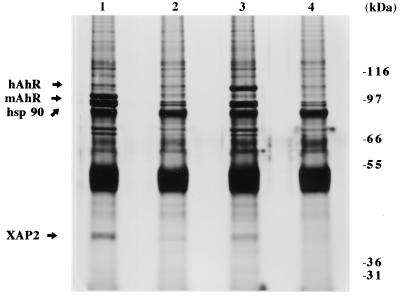
Due to the observation that the hAhR exhibits different physicochemical properties from the mAhR, including higher molecular weight, lower complex stability, and a lower affinity for ligand, we wished to determine if XAP2 was able to associate with the hAhR. To determine if XAP2 was capable of complexing with the hAhR, COS-1 cells were transfected with pCI/hAhR-FLAG, immunoabsorbed with the M2 affinity gel, resolved by Tricine SDS-PAGE, and silver stained. This resulted in the immunoabsorption of four specific bands having molecular masses of ∼106, 86, 84, and 38 kDa (Fig. 10). Immunoblot analysis was used to demonstrate that these proteins were the hAhR ligand-binding subunit, hsp86 and hsp84 (hsp90), and XAP2, respectively (data not shown). Thus, XAP2 can also interact with the hAhR, indicating that the AhR-XAP2 interaction is conserved across mammalian species.
FIG. 10.
XAP2 is a member of the hAhR complex in COS-1 cells. COS-1 cells were transfected with 9 μg of pCI/hAhR-FLAG, pcDNA3/βmAhR-FLAG, or pcDNA3; cytosol was isolated, immunoabsorbed with M2 affinity gel, and resolved by SDS-PAGE; and the gel was silver stained. Lanes: 1, pcDNA3/βmAhR-FLAG; 2, pcDNA/βmAhR-FLAG with M2 affinity gel blocked with FLAG peptide; 3, pCI/hAhR-FLAG; 4, pCI/hAhR-FLAG with M2 affinity gel blocked with FLAG peptide.
Considering that XAP2 is able to associate with the hAhR, we wished to determine if overexpression of XAP2 in a human cell line containing an endogenous hAhR complex could increase the transcriptional activation properties similar to that observed in the murine Hepa 1 cell line. HeLa cells were selected on the basis of previous studies that have characterized the endogenous hAhR complex and demonstrated significant endogenous levels of XAP2 (27, 52). With increasing amounts of pCI/XAP2, activation of the DRE-luciferase construct increased twofold following induction by TCDD in a statistically significant manner. Additionally, the level of activation increased slightly in the absence of exogenous ligand (Fig. 11). This suggests that XAP2 acted to increase the ability of the hAhR to transactivate the DRE-luciferase reporter. Thus, like the mAhR, the ability of the hAhR to transactivate the DRE-luciferase construct was enhanced by XAP2.
FIG. 11.
XAP2 acts as a transcriptional enhancer in HeLa cells. HeLa cells were transfected with plasmids pGudLuc6.1, pcDNA3.1/lacZ/his, and pCI-XAP2. The amount of pCI-XAP2 transfected is given in micrograms and each transfection was brought to 3.0 μg with pCI vector. Relative luciferase activity was measured following induction by TCDD or carrier solvent. Each transfection was performed in triplicate. ∗, a statistically significant difference was obtained (P < 0.05).
DISCUSSION
Several strategies were initially performed to biochemically isolate the ∼38-kDa protein (p38) present in the unliganded AhR complex in Hepa 1 cells, where this protein was first observed (7). However, these efforts resulted in poor yields of purified unliganded AhR complex and high levels of nonspecific protein binding to the M2 affinity gel (data not shown). We sought a different cell line, COS-1, that was able to express high levels of transfected pcDNA3/βmAhR and had a lower level of nonspecific binding to the affinity gel. The AhR was demonstrated to function properly in COS-1 cells, including binding to DREs (Fig. 1). Additionally, a FLAG epitope-tagged AhR was equally able to induce the DRE-luciferase reporter in the presence of TCDD (Fig. 1). Thus, FLAG-tagged AhR was subsequently used to immunoabsorb unliganded AhR complexes assembled in COS-1 cells. This strategy allowed for the large-scale immunoabsorption of unliganded AhR complexes from COS-1 cells and subsequent biochemical purification of p38. To identify the stable subunits of the unliganded AhR complex, immunoabsorbed AhR-FLAG complexes were washed under relatively high-salt conditions, resolved by SDS-PAGE and evaluated by silver staining. This high-resolution technique was used to visualize all protein species that were stably associated with the AhR. This had an advantage over immunoblot analysis with antibody probes, due to the ability to observe all individual proteins stably associated with the complex rather than attempting to identify potential proteins that may coprecipitate with the AhR. Four prominent bands were observed, corresponding to the AhR ligand-binding subunit, a dimer of hsp90, and p38 (Fig. 2). This was consistent with a previous observation in Hepa 1 cells, where the unliganded AhR was demonstrated to be a heterotetramer in immunoabsorption and chemical cross-linking analysis (7). The AhR-hsp90-p38 complexes were purified by a large-scale transfection procedure, and amino acid sequence data was obtained from two tryptic fragments of p38. A BLAST search (1) was performed with these protein sequences, which matched the human hepatitis B virus (HBV) X-associated protein 2 (XAP2) with 100% identity. This protein was termed XAP2 because it was the second protein to be isolated by the yeast two-hybrid screening method to detect cellular factors that interacted with the HBV X protein (27). It is possible that other proteins interact with the unliganded AhR, perhaps through weak interactions. At least two other proteins have been shown to bind to the AhR. It has been reported that pp60c-src is associated with the AhR (14). Additionally, a protein of ∼45 kDa isolated from mouse liver cytosol was shown to interact with the bHLH region of the AhR by using the bHLH region of the AhR on an affinity column (21).
During the preparation of this manuscript, the yeast-two-hybrid system was used by two independent groups as a method to identify potential proteins that interacted with the AhR. A human cDNA, termed ARA9, and a murine cDNA, termed AIP, were shown to interact with the AhR by this method. ARA9 is 99.9% identical (1 residue difference) to human XAP2, and AIP is the murine homolog of XAP2 (5, 30). Alignment of simian XAP2 with human and murine XAP2 revealed 98 and 95% identity, respectively. These studies demonstrated that AIP could interact with the AhR in vivo in yeast in the absence of ligand, whereas ARA9 was shown to interact with the AhR only in the presence of ligand (5, 30). By using a rabbit RL system, it was demonstrated that ARA9/AIP could bind to the AhR in a ligand-dependent or -independent manner (5, 30). Immunoabsorption studies with anti-hsp90 antibodies demonstrated that ARA9 could coprecipitate in the presence of the AhR but not in its absence in rabbit RL (5). Similarly, immunoabsorption of AIP from Hepa 1 cytosol resulted in immunoabsorption of the AhR and hsp90 (30). In both studies, binding of Arnt to ARA9/AIP was not detected (5, 30).
A TPR motif that corresponds to the consensus TPR motif (-W-LG-Y-A-F-A-P) was identified from residues 265 to 298 in simian XAP2. This TPR had a strong similarity to the third TPR domain of FKBP52 and the fourth TPR domain in the S. cerevisiae transcriptional modulator SSN6, especially in domain B (Fig. 3B). SSN6 is a gene functionally related to the SNF1 protein kinase in S. cerevisiae (50). It is a member of the SSN6/TUP1 global regulator complex involved in repression of α-cell-specific gene expression, which is thought to organize repressive regions of chromatin (9). The TPR motifs in SSN6 have been demonstrated to mediate interaction with the S. cerevisiae cell type regulator α2, a homeodomain protein involved in recruiting SSN6/TUP1 complexes, but it is unknown what role, if any, the fourth TPR plays in this interaction (54). The TPR motifs in FKBP52 have been demonstrated to mediate protein-protein interactions with hsp90 (42). This TPR was conserved with 100% identity in the human, simian, and murine XAP2 clones. The functional significance of this well-conserved TPR motif is unknown. The simian XAP2 coding sequence was also analyzed for other regulatory regions. A sequence corresponding to PPiase activity was lacking, although whether XAP2 has this activity remains to be determined.
XAP2 was identified in the AhR complex in COS-1 cells and in the endogenous unliganded AhR complex in Hepa 1 cells (Fig. 6). These results also confirmed that XAP2 is the p38 subunit observed in silver-staining analysis. To determine if XAP2 plays a role in regulating the assembly of the AhR with hsp90, a coupled transcription and translation was performed in vitro in RL. Three groups have independently demonstrated that after the transcription and translation of the AhR in this system, the AhR assembles with hsp90 and is competent to undergo transformation (16, 31, 37). Thus, it was not known if XAP2 played a role in this assembly process. The RL itself was examined for the presence of XAP2 and compared to XAP2 transcribed and translated in RL, by using XAP2 polyclonal antisera in an immunoblot analysis. No detectable levels of XAP2 were observed in RL alone. This suggested that, due to its absence, XAP2 was not required for assembly of the AhR with hsp90. To confirm this observation, AhR-FLAG was expressed in the RL system, in either the absence or the presence of coexpressed XAP2, and immunoabsorbed with the M2 affinity gel in an attempt to coimmunoabsorb any XAP2 in RL. In both cases, the AhR assembled with hsp90 to the same extent whether XAP2 was present or absent. This strengthened the observation that XAP2 was not required for AhR-hsp90 assembly. These results are similar to data obtained with the immunophilin FKBP52 (FKBP59), which is not required for GR-hsp90 assembly (11).
Because XAP2 was demonstrated to be a stable member of the unliganded AhR core complex, we wanted to assess whether XAP2 interacted directly with hsp90 and/or with the AhR. In rabbit RL, XAP2 sedimented at 0S to 4S and was not located where hsp90 sediments at ∼6S. When pCI/XAP2 and pcDNA3/βmAhR-FLAG were coexpressed by using the same procedure and analyzed by sucrose density gradient analysis, the XAP2 band shifted to a ∼9S form, where the AhR-hsp90 complex sediments. Because XAP2 is a member of the AhR-hsp90 complex, it has yet to be determined if protein-protein interactions occur between XAP2-hsp90, XAP2-AhR, or both. XAP2 sedimented at ∼6S when COS-1 cytosol was resolved by sucrose density gradient analysis, suggesting the possibility that XAP2 can associate with hsp90 or other cellular factors in vivo. To determine if there was an interaction between hsp90 and XAP2 directly, polyclonal and monoclonal antibodies raised against hsp90 were used to immunoabsorb hsp90. Immunoblot analysis revealed immunoabsorption of hsp90 but no coimmunoabsorption of XAP2 when using XAP2 polyclonal antibodies (data not shown). However, hsp90 has been demonstrated to coimmunoabsorb with an epitope-tagged murine XAP2 (AIP) transiently expressed in Hepa 1 cells (30). Thus, our results suggest that the inability of XAP2 to coimmunoabsorb with hsp90 may be due to blockage of an epitope-binding site on hsp90 recognized by XAP2.
XAP2 has been demonstrated to interact with two different proteins, the X-protein from HBV (27) and the murine and human AhR (see above). It is unknown what structural similarities, if any, exist between the AhR and the X protein. Alignment of these two proteins failed to find a distinct region or regions of homology between the AhR and the HBV-X protein. The exact role of XAP2 function remains unknown. XAP2 acts to repress the transactivation function of the X protein (27) yet acts to enhance the activity of the AhR. It is suggested by sucrose density gradient analysis that XAP2 exists in the cytosolic fraction as an ∼6S species. Thus, it is possible that XAP2 exists in the cytoplasm bound to hsp90 in a preassembled complex before binding to another protein, such as the AhR or the X protein.
To determine the role of XAP2 in the AhR signaling pathway, XAP2 was expressed in Hepa 1 cells, which contain an endogenous mAhR complex and have been used extensively to study the AhR signaling pathway. A twofold increase in the level of DRE-driven reporter construct was observed following induction by TCDD. This may be considered a substantial increase in activity considering that these cells already have a significant level of XAP2 (30). Expression of AIP (murine XAP2) in Hepa 1 cells with a viral transfection system followed by slot-blot analysis of an endogenous target gene of the AhR, Cyp1a1, also resulted in a 1.5- to 2-fold increase in Cyp1a1 mRNA expression (30). Thus, results presented here, obtained with a DRE-luciferase reporter construct, are consistent with the increased level of mRNA expression observed in an endogenous AhR-inducible gene. Several explanations of the mechanism by which XAP2 increases AhR activity can be considered. First, it may occur by further stabilizing the AhR complex, thereby allowing more AhR to be available for ligand activation and subsequent transactivation via DREs. Second, XAP2 may help target the activated AhR to the nucleus, as has been suggested for chaperone components such as FKBP52 and other immunophilins (10, 33; reviewed in reference 40). This is plausible since XAP2 has been detected in the cytoplasm of HeLa cells by immunocytochemistry (27). Third, XAP2 may enhance the ability of the AhR to heterodimerize with Arnt, creating a larger pool of 6S DNA-binding heterodimer. Fourth, XAP2 may interact with other cellular factors, as yet unidentified but involved in the AhR-mediated signaling pathway. Fifth, XAP2 may alter the affinity of ligand-AhR interactions. Finally a striking theme is emerging with respect to the role of immunophilins or immunophilin-like proteins in unliganded receptor complexes, including those in the steroid receptor superfamily and in the bHLH-PAS domain gene family.
ACKNOWLEDGMENTS
We thank Christopher A. Bradfield for pSVSPORT/hAhR, Michael Denison for the DRE-luciferase reporter construct pGudLuc 6.1, Oliver Hankinson for pcDNA3/βmAhR, Webb Miller for helpful assistance in alignments of XAP2 with human FKBP52 and FKBP12, Richard Pollenz for polyclonal AhR antibodies, Steve Safe for TCDD, and Edward Seto for pGST-XAP2 and pGEM-XAP2 constructs. William P. Long and Sheo S. Singh are acknowledged for providing excellent technical assistance. The HHMI W. M. Keck Biopolymer Facility at Yale University performed in-gel tryptic digestions and subsequent microsequencing of p38.
This work was supported by grants ES04869 and ES07799 from NIEHS.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Aranyi P, Radanyi C, Renoir M, Devin J, Baulieu E-E. Covalent stabilization of the nontransformed chick oviduct cytosol progesterone receptor by chemical cross-linking. Biochemistry. 1988;27:1330–1336. doi: 10.1021/bi00404a036. [DOI] [PubMed] [Google Scholar]
- 3.Bresnick E H, Dalman F C, Pratt W B. Direct stoichiometric evidence that the untransformed Mr 300 000, 9S, glucocorticoid receptor is a core unit derived from a larger heteromeric complex. Biochemistry. 1990;29:520–527. doi: 10.1021/bi00454a028. [DOI] [PubMed] [Google Scholar]
- 4.Burbach K M, Poland A, Bradfield C A. Cloning of the Ah-receptor cDNA reveals a distinctive ligand-activated transcription factor. Proc Natl Acad Sci USA. 1992;89:8185–8189. doi: 10.1073/pnas.89.17.8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carver L A, Bradfield C A. Ligand-dependent interaction of the aryl hydrocarbon receptor with a novel immunophilin homolog in vivo. J Biol Chem. 1997;272:11452–11456. doi: 10.1074/jbc.272.17.11452. [DOI] [PubMed] [Google Scholar]
- 6.Carver L A, Hogenesch J B, Bradfield C A. Tissue specific expression of the rat Ah-receptor and ARNT mRNAs. Nucleic Acids Res. 1994;22:3038–3044. doi: 10.1093/nar/22.15.3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H-S, Perdew G H. Subunit composition of the heteromeric cytosolic aryl hydrocarbon receptor complex. J Biol Chem. 1994;269:27554–27558. [PubMed] [Google Scholar]
- 8.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper J P, Roth S Y, Simpson R T. The global transcriptional regulators, SSN6 and TUP1, play distinct roles in the establishment of a repressive chromatin structure. Genes Dev. 1994;8:1400–1410. doi: 10.1101/gad.8.12.1400. [DOI] [PubMed] [Google Scholar]
- 10.Czar M J, Lyons R H, Welsh M J, Renoir J-M, Pratt W B. Evidence that the FK506-binding immunophilin heat shock protein 56 is required for trafficking of the glucocorticoid receptor from the cytoplasm to the nucleus. Mol Endocrinol. 1995;9:1549–1560. doi: 10.1210/mend.9.11.8584032. [DOI] [PubMed] [Google Scholar]
- 11.Dittmar K D, Hutchison K A, Owens-Grillo J K, Pratt W B. Reconstitution of the steroid receptor-hsp90 heterocomplex assembly system of rabbit reticulocyte lysate. J Biol Chem. 1996;271:12833–12839. doi: 10.1074/jbc.271.22.12833. [DOI] [PubMed] [Google Scholar]
- 12.Dolwick K M, Swanson H I, Bradfield C A. In vitro analysis of Ah receptor domains involved in ligand-activated DNA recognition. Proc Natl Acad Sci USA. 1993;90:8566–8570. doi: 10.1073/pnas.90.18.8566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong L, Ma Q, Whitlock J P., Jr DNA binding by the heterodimeric Ah receptor. J Biol Chem. 1996;271:7942–7948. doi: 10.1074/jbc.271.14.7942. [DOI] [PubMed] [Google Scholar]
- 14.Enan E, Matsumura F. Identification of c-Src as the integral component of the cytosolic Ah receptor complex, transducing the signal of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) through the protein phosphorylation pathway. Biochem Pharmacol. 1996;52:1599–1612. doi: 10.1016/s0006-2952(96)00566-7. [DOI] [PubMed] [Google Scholar]
- 15.Fukunaga B N, Hankinson O. Identification of a novel domain in the aryl hydrocarbon receptor required for DNA binding. J Biol Chem. 1996;271:3743–3749. doi: 10.1074/jbc.271.7.3743. [DOI] [PubMed] [Google Scholar]
- 16.Fukunaga B N, Probst M R, Reisz-Porszasz S, Hankinson O. Identification of functional domains of the aryl hydrocarbon receptor. J Biol Chem. 1995;270:29270–29278. doi: 10.1074/jbc.270.49.29270. [DOI] [PubMed] [Google Scholar]
- 17.Gilliland G S, Perrin K, Bunn H F. Competitive PCR for quantitation of mRNA. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 60–66. [Google Scholar]
- 18.Goebl M, Yanagida M. The TPR snap helix: a novel protein repeat motif from mitosis to transcription. Trends Biochem Sci. 1991;16:173–177. doi: 10.1016/0968-0004(91)90070-c. [DOI] [PubMed] [Google Scholar]
- 19.Hankinson O. The aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- 20.Hirano T, Kinoshita N, Morikawa K, Yanagida M. Snap helix with knob and hole: essential repeats in S. pombe nuclear protein nuc2+ Cell. 1990;60:319–328. doi: 10.1016/0092-8674(90)90746-2. [DOI] [PubMed] [Google Scholar]
- 21.Hossain A, Kikuchi H, Ikawa S, Sagami I, Watanabe M. Identification of cellular protein that can interact specifically with the basic helix-loop-helix domain of the aromatic hydrocarbon receptor. Biochem Biophys Res Commun. 1995;215:405–411. doi: 10.1006/bbrc.1995.2479. [DOI] [PubMed] [Google Scholar]
- 22.Hutchison K A, Dittmar K D, Czar M J, Pratt W B. Proof that hsp70 is required for assembly of the glucocorticoid receptor into a heterocomplex with hsp90. J Biol Chem. 1994;269:5043–5049. [PubMed] [Google Scholar]
- 23.Jain S, Dolwick K M, Schmidt J V, Bradfield C A. Potent transactivation domains of the Ah receptor and the Ah receptor nuclear translocator map to their carboxyl termini. J Biol Chem. 1994;269:31518–31524. [PubMed] [Google Scholar]
- 24.Johnson J L, Beito T G, Krco C J, Toft D O. Characterization of a novel 23-kDa protein of unactive progesterone receptor complexes. Mol Cell Biol. 1994;14:1956–1963. doi: 10.1128/mcb.14.3.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ko H P, Okino S T, Ma Q, Whitlock J P., Jr Dioxin-induced CYP1A1 transcription in vivo: the aromatic hydrocarbon receptor mediates transactivation, enhancer-promoter communication, and changes in chromatin structure. Mol Cell Biol. 1996;16:430–436. doi: 10.1128/mcb.16.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8132. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuzhandaivelu N, Cong Y-S, Inouye C, Yang W-M, Seto E. XAP2, a novel hepatitis B virus X-associated protein that inhibits X transactivation. Nucleic Acids Res. 1996;24:4741–4750. doi: 10.1093/nar/24.23.4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamb J R, Tugendreich S, Hieter P. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem Sci. 1995;20:257–259. doi: 10.1016/s0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- 29.Long, W. P., M. G. Pray-Grant, J. C. Tsai, and G. H. Perdew. Protein kinase C activity is required for aryl hydrocarbon receptor pathway mediated signal transduction. Submitted for publication. [DOI] [PubMed]
- 30.Ma Q, Whitlock J P., Jr A novel cytoplasmic protein that interacts with the Ah receptor, contains tetratricopeptide repeat motifs, and augments the transcriptional response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. J Biol Chem. 1997;272:8878–8884. [PubMed] [Google Scholar]
- 31.McGuire J, Whitelaw M L, Pongratz I, Gustafsson J-A, Poellinger L. A cellular factor stimulates ligand-dependent release of hsp90 from the basic helix-loop-helix dioxin receptor. Mol Cell Biol. 1994;14:2438–2446. doi: 10.1128/mcb.14.4.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Milad M, Sullivan W, Diehl E, Altmann M, Nordeen S, Edwards D P, Toft D O. Interaction of the progesterone receptor with binding proteins for FK506 and cyclosporin A. Mol Endocrinol. 1995;9:838–847. doi: 10.1210/mend.9.7.7476967. [DOI] [PubMed] [Google Scholar]
- 33.Owens-Grillo J K, Czar M J, Hutchison K A, Hoffmann K, Perdew G H, Pratt W B. A model of protein targeting mediated by immunophilins and other proteins that bind to hsp90 via tetratricopeptide repeat domains. J Biol Chem. 1996;271:13468–13475. doi: 10.1074/jbc.271.23.13468. [DOI] [PubMed] [Google Scholar]
- 34.Owens-Grillo J K, Hoffmann K, Hutchison K A, Yem A W, Deibel M R, Jr, Handschumacher R E, Pratt W B. The cyclosporin A-binding immunophilin Cyp-40 and the FK506-binding immunophilin hsp56 bind to a common site on hsp90 and exist in independent cytosolic heterocomplexes with the untransformed glucocorticoid receptor. J Biol Chem. 1995;270:20479–20484. doi: 10.1074/jbc.270.35.20479. [DOI] [PubMed] [Google Scholar]
- 35.Perdew G H. Chemical cross-linking of the cytosolic and nuclear forms of the Ah receptor in hepatoma cell line 1c1c7. Biochem Biophys Res Commun. 1992;182:55–62. doi: 10.1016/s0006-291x(05)80111-1. [DOI] [PubMed] [Google Scholar]
- 36.Perdew G H. Production of murine anti-peptide polyclonal antibodies utilizing a nonantigenic adjuvant. Anal Biochem. 1994;220:214–216. doi: 10.1006/abio.1994.1323. [DOI] [PubMed] [Google Scholar]
- 37.Perdew G H, Bradfield C A. Mapping of the 90 kDa heat shock protein binding region of the Ah receptor. Biochem Mol Biol Int. 1996;39:589–593. doi: 10.1080/15216549600201651. [DOI] [PubMed] [Google Scholar]
- 38.Perdew G H, Hord N, Hollenback C E, Welsh M J. Localization and characterization of the 86- and 84-kDa heat shock proteins in Hepa 1c1c7 cells. Exp Cell Res. 1993;209:350–356. doi: 10.1006/excr.1993.1320. [DOI] [PubMed] [Google Scholar]
- 39.Pollenz R S, Sattler C A, Poland A. The aryl hydrocarbon receptor and aryl hydrocarbon receptor nuclear translocator protein show distinct subcellular localizations in Hepa1c1c7 cells by immunofluorescence microscopy. Mol Pharmacol. 1993;45:428–438. [PubMed] [Google Scholar]
- 40.Pratt W B, Toft D O. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- 41.Probst M R, Reisz-Porszasz S, Agbunag R V, Ong M S, Hankinson O. Role of the aryl hydrocarbon receptor nuclear translocator protein in aryl hydrocarbon (dioxin) receptor action. Mol Pharmacol. 1993;44:511–518. [PubMed] [Google Scholar]
- 42.Radanyi C, Chambraud B, Baulieu E-E. The ability of the immunophilin FKBP59-HBI to interact with the 90-kDa heat shock protein is encoded by its tetratricopeptide repeat domain. Proc Natl Acad Sci USA. 1994;91:11197–11201. doi: 10.1073/pnas.91.23.11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rehberger P, Rexin M, Gehring U. Heterotetrameric structure of the human progesterone receptor. Proc Natl Acad Sci USA. 1992;89:8001–8005. doi: 10.1073/pnas.89.17.8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Renoir J-M, Radanyi C, Jung-Testas I, Faber L E, Baulieu E-M. The nonactivated progesterone receptor is a nuclear heterooligomer. J Biol Chem. 1990;265:14402–14406. [PubMed] [Google Scholar]
- 45.Rexin M, Busch W, Segnitz B, Gehring U. Tetrameric structure of the nonactivated glucocorticoid receptor in cell extracts and intact cells. FEBS Lett. 1988;241:234–238. doi: 10.1016/0014-5793(88)81068-8. [DOI] [PubMed] [Google Scholar]
- 46.Reyes H, Reisz-Porszasz S, Hankinson O. Identification of the Ah receptor nuclear translocator protein (Arnt) as a component of the DNA binding form of the Ah receptor. Science. 1992;256:1193–1195. doi: 10.1126/science.256.5060.1193. [DOI] [PubMed] [Google Scholar]
- 47.Rowlands C J, Gustafsson J-A. Aryl hydrocarbon receptor-mediated signal transduction. Crit Rev Toxicol. 1997;27:109–134. doi: 10.3109/10408449709021615. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 49.Schmidt J V, Bradfield C A. Ah receptor signaling pathways. Annu Rev Cell Dev Biol. 1996;12:55–89. doi: 10.1146/annurev.cellbio.12.1.55. [DOI] [PubMed] [Google Scholar]
- 50.Schultz J, Carlson M. Molecular analysis of SSN6, a gene functionally related to the SNF1 protein kinase of Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:3637–3645. doi: 10.1128/mcb.7.10.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sikorski R S, Boguski M S, Goebl M, Hieter P. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell. 1990;60:307–317. doi: 10.1016/0092-8674(90)90745-z. [DOI] [PubMed] [Google Scholar]
- 52.Singh S S, Hord N G, Perdew G H. Characterization of the activated form of the aryl hydrocarbon receptor in the nucleus of HeLa cells in the absence of exogenous ligand. Arch Biochem Biophys. 1996;329:47–55. doi: 10.1006/abbi.1996.0190. [DOI] [PubMed] [Google Scholar]
- 53.Smith D F, Albers M W, Schrieber S L, Leach K L, Deibel M R., Jr FKBP54, a novel FK506-binding protein in avian progesterone receptor complexes and HeLa extracts. J Biol Chem. 1993;268:24270–24273. [PubMed] [Google Scholar]
- 54.Smith R L, Redd M J, Johnson A D. The tetratricopeptide repeats of Ssn6 interact with the homeo domain of α2. Genes Dev. 1995;9:2903–2910. doi: 10.1101/gad.9.23.2903. [DOI] [PubMed] [Google Scholar]
- 55.Tai, P-K. K., M. W. Albers, H. Chang, L. E. Faber, and S. L. Schreiber. Association of a 59-kilodalton immunophilin with the glucocorticoid receptor complex. Science 256:1315–1318. [DOI] [PubMed]
- 56.Vanden Heuvel J P, Tyson F L, Bell D A. Constructions of recombinant RNA templates for use as internal standards in quantitative RT-PCR. BioTechniques. 1993;14:395–398. [PubMed] [Google Scholar]
- 57.Vanden Heuvel J P, Clark G C, Tritscher A M, Greenlee W F, Lucier G W, Bell D A. Dioxin-responsive genes: examination of dose-response relationships using quantitative reverse transcriptase-polymerase chain reaction. Cancer Res. 1994;54:62–68. [PubMed] [Google Scholar]
- 58.Wray W, Boulikas T, Wray V P, Hancock R. Silver staining of proteins in polyacrylamide gels. Anal Biochem. 1981;118:197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]
- 59.Zachar V, Thomas R A, Goustin A S. Absolute quantification of target DNA: a simple competitive PCR for efficient analysis of multiple samples. Nucleic Acids Res. 1993;21:2017–2018. doi: 10.1093/nar/21.8.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]